Abstract
We previously reported the isolation of a mutant poliovirus lacking the entire genomic RNA 3′ noncoding region. Infection of HeLa cell monolayers with this deletion mutant revealed only a minor defect in the levels of viral RNA replication. To further analyze the consequences of the genomic 3′ noncoding region deletion, we examined viral RNA replication in a neuroblastoma cell line, SK-N-SH cells. The minor genomic RNA replication defect in HeLa cells was significantly exacerbated in the SK-N-SH cells, resulting in a decreased capacity for mutant virus growth. Analysis of the nature of the RNA replication deficiency revealed that deleting the poliovirus genomic 3′ noncoding region resulted in a positive-strand RNA synthesis defect. The RNA replication deficiency in SK-N-SH cells was not due to a major defect in viral translation or viral protein processing. Neurovirulence of the mutant virus was determined in a transgenic mouse line expressing the human poliovirus receptor. Greater than 1,000 times more mutant virus was required to paralyze 50% of inoculated mice, compared to that with wild-type virus. These data suggest that, together with a cellular factor(s) that is limiting in neuronal cells, the poliovirus 3′ noncoding region is involved in positive-strand synthesis during genome replication.
Paralytic poliomyelitis is the consequence of the destruction of motor neurons during a poliovirus infection. Only 1 to 2% of individuals infected with poliovirus develop poliomyelitis, suggesting that factors affecting virus tropism and systemic distribution play a significant role in the pathogenic outcome of an infection (7). The cell- and tissue-specific tropism of poliovirus during a systemic infection may be restricted by tissue accessibility (i.e., the blood-brain barrier), cellular attachment and entry, or intracellular interactions between the virus and the cell. In the late 1950s, Holland and colleagues demonstrated that purified poliovirus genomic RNA introduced into nonprimate cells by transfection leads to a single replicative cycle (21). Their data showed that cells which are not normally susceptible to poliovirus infections can be productively infected when receptor-mediated entry of the virus is bypassed, suggesting that the poliovirus receptor (PVR) plays a major role in virus tropism. These conclusions were further supported by the finding that transgenic mice expressing the human PVR (hPVR) are susceptible to poliovirus infection (27, 44). Furthermore, the pathology in PVR transgenic mice infected with attenuated or wild-type poliovirus resembles the pathology observed in infected primates, the natural host of poliovirus (22, 43). These observations suggested a strong correlation between PVR-mediated tropism and poliovirus pathology.
Although PVR expression may be a major determinant of poliovirus tropism, it is clear that additional factors can influence virus tropism and disease pathology. Assays for virus binding activity in tissue homogenates revealed that binding of virus can occur in tissues where poliovirus replication does not take place (20, 28). Indeed, whether assayed in an infected primate or PVR transgenic mice, there is no absolute correlation between susceptibility to virus infection and the expression of PVR (15, 25, 35, 37, 40, 42). The inability of virus to spread to tissues such as kidney, heart, and lung may be a consequence of host range restrictions that are independent of receptor binding. Determinants of the attenuation phenotypes for the Sabin strains of poliovirus have been mapped to the 5′-proximal portion of genomic RNAs of all three strains (4, 8, 14, 18, 23, 24, 33, 39, 54). Shiroki et al. induced a host-range phenotype of poliovirus by introducing mutations in the internal ribosomal entry site of the poliovirus genome (47). While the mutations in the Sabin 5′ noncoding regions (NCRs) have been shown to affect translation efficiency of the viral genomes (48-50) with the resulting loss of translation efficiency proposed to contribute to the mechanism of attenuation (55), all elements contributing to cell-specific attenuation have yet to be fully elucidated.
Much of the effort to identify the molecular mechanism(s) of attenuation following infections with the Sabin strains of poliovirus has focused on the 5′ ends of the genomes. However, mutations in the 3′ NCR of poliovirus Sabin 1 may contribute to the attenuation phenotype (2, 11, 16). Previously, Todd et al. had shown that poliovirus genomic RNA lacking the entire genomic 3′ NCR is infectious in HeLa cells (51). The resulting mutant virus (Δ3′ NCR PV1) replicated surprisingly well in HeLa cells, considering the nature of the lesion, although not to wild-type virus levels. There is an apparent discrepancy between the selective pressures that have resulted in the conservation of the 3′ NCR among the three serotypes of poliovirus and the relatively efficient replication properties of the deletion mutant in HeLa cells. The functions provided by the 3′ NCR of poliovirus RNA may, in part, be duplicated by other viral or cellular elements in HeLa cells, therefore rendering the 3′ NCR dispensable for virus replication in these cells.
To further understand the role of the poliovirus 3′ NCR during viral replication and to investigate a possible cell-specific role of this region of the genome, we examined the replication properties of the Δ3′ NCR PV1 in a neuronal cell line and in PVR transgenic mice. While deletion of the complete 3′ NCR impaired virus replication in HeLa cells, the replication defect was much more pronounced in a neuroblastoma cell line (SK-N-SH). The complete genomic 3′ NCR deletion had little or no effect on poliovirus genome translation; however, the slight defect in genomic RNA synthesis previously observed in HeLa cells was more severe in neuroblastoma cells. This cell-specific replication defect resulted in a lower ratio of positive-strand to negative-strand RNA accumulation following infection. Furthermore, the deletion mutation had a profound effect on the ability of the virus to replicate and induce pathology in PVR transgenic mice. These data suggest that removal of the 3′ NCR of the poliovirus genome restricts positive-strand synthesis in cells of neuronal origin.
MATERIALS AND METHODS
Virus stocks and plaque assay.
The generation and characterization of the plaque-isolated Δ3′ NCR PV1 stock was described previously (51) and is referred to here as Δ3′ NCR PV1. Plaque assays were performed as described previously (9).
One-step growth cycle analysis.
One-step growth analysis was performed on HeLa (R19) or SK-N-SH monolayers in 60-mm plates at 37°C as described previously (52). Briefly, HeLa (R19) or SK-N-SH monolayers were infected at a multiplicity of infection (MOI) of 20 or 17. Virus was adsorbed for 30 min at room temperature in 60-mm plates and then washed with phosphate-buffered saline. Following the infection of HeLa (R19) cell monolayers, 3 ml of minimal essential medium plus 8% newborn calf serum with 1% nonessential amino acids was added. Infections of SK-N-SH monolayers were followed by the addition of 3 ml of Dulbecco's modified Eagle medium plus 20% fetal bovine serum. Infected cell monolayers were harvested at the indicated times, virus was released from the cells by five freeze-thaw cycles, cellular debris was pelleted, and the viral titer of the clarified supernatant was determined by plaque assay on HeLa (R19) cells.
Total cellular RNA extraction.
Total cellular RNA was harvested using TriReagent (Molecular Research Center, Inc., Cincinnati, Ohio), developed from methods described previously (10). Briefly, HeLa (R19) or SK-N-SH monolayers were infected at an MOI of 17 or 20 at 37°C. At the indicated times postinfection, 600 μl of TriReagent was added to the cell monolayers and the cell lysates were harvested. Total cellular RNA was extracted from the lysates by chloroform extraction of the aqueous phase. The RNA was precipitated with ethanol, pelleted, dried, and resuspended in diethylpyrocarbonate-treated H2O.
RNA analysis.
Northern blot analysis was performed with 5 or 10 μg of glyoxal-treated total cellular RNAs (34). Hybridization and washing conditions were as described previously (9). 32P-labeled probes were used to detect viral genomic RNA.
RNase protection assays were performed as previously described (38) to measure positive-strand RNA accumulation. A 108-ng aliquot of total RNA from infected or mock-infected cells was hybridized with 25 fmol of 32P-labeled RNA probe, designated the (+) probe, containing nucleotides complementary to positive-sense sequences 5601 to 5809 of poliovirus viral RNA. In addition to virus- specific sequences, the probe contained 49 nucleotides of vector sequence. Hybridization reactions were denatured for 10 min at 85°C in hybridization buffer (1 mM EDTA, 0.4 M NaCl, 80% formamide, 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) [PIPES]; pH 6.4) and then incubated overnight at 60°C. The mixture was then cooled to room temperature, 300 μl of RNase digestion cocktail (0.5 M NaCl, 5 mM EDTA, 4.5 μg of RNase A/ml, 350 U of T1/ml, 10 mM Tris; pH 7.5) was added, and the reaction was incubated for 1 h at 7°C. Twenty microliters of 10% sodium dodecyl sulfate (SDS) and 100 μg of proteinase K were then added, and samples were incubated for 30 min at 37°C. Protected RNAs were then phenol-chloroform extracted, ethanol precipitated in the presence of 20 μg of carrier tRNA, resuspended in 15 μl of formamide loading buffer, and resolved on an 8% polyacrylamide gel containing 7 M urea.
Negative-strand RNA accumulation was detected in a two-cycle RNase protection assay also described by Novak and Kirkegaard (38). A 1.3-μg aliquot of total RNA from infected or mock-infected cells was denatured for 10 min at 85°C in hybridization buffer (1 mM EDTA, 0.4 M NaCl, 80% formamide, 40 mM PIPES; pH 6.4) and then incubated overnight at 60°C in the absence of radiolabeled probe. The samples were then subjected to RNase treatment, proteinase K digestion, phenol-chloroform extraction, and ethanol precipitation as described above. Samples were then hybridized to 75 fmol of 32P-labeled probe complementary to nucleotides 5601 to 5809 of the negative-strand viral RNA and subjected to the same procedure employed to detect plus-strand viral RNA.
Translation and immunoprecipitation of [35S]methionine-labeled viral proteins.
Immunoprecipitation utilizing extracts from [35S]methionine-labeled infected cells was carried out as described previously (46). One hour prior to harvesting of infected SK-N-SH cells, 50 μCi of [35S]methionine was added to cell monolayers in 60-mm plates. Cells were harvested in phosphate-buffered saline and pelleted, and the supernatant was discarded. The cell pellets were resuspended in 55 μl of Laemmli sample buffer (29) and boiled for 3 min. Twenty microliters was removed and subjected to electrophoresis on an SDS-12.5% polyacrylamide gel. The remaining volume of radiolabeled lysate was diluted to 400 μl in TENN buffer (5 mM EDTA, 0.5% Nonidet P-40, 150 mM NaCl, 50 mM Tris; pH 7.4), and 20 μl of protein A-agarose (Roche) was added and incubated on ice for 10 min. Nonspecific protein A complexes were cleared by centrifugation for 1 min. Ten microliters of anti-3C serum (EΔ3C) (45) was added to the supernatant, vortexed, and placed on ice for 1 h. Forty microliters of protein A-agarose was added, vortexed, and placed on ice for 10 min. Following this incubation, the mixture was centrifuged and the protein A-agarose beads were washed three times with 500 μl of SNNTE buffer (5% sucrose, 1% Nonidet P-40, 0.5 mM NaCl, 5 mM EDTA, 50 mM Tris; pH 7.4) and once with 500 μl of NTE buffer (50 mM NaCl, 1 mM EDTA, 10 mM Tris; pH 7.4). The pellet was resuspended in 50 μl of Laemmli sample buffer, boiled for 3 min, and pelleted for 1.5 min, and the supernatant was subjected to electrophoresis on a 12.5% polyacrylamide gel containing SDS.
PD50 determination in hPVR transgenic mice.
The hPVR transgenic mouse line hPVR1-17 has been described previously (44). Mice were given 50-μl intracerebral (IC) inoculations of virus. The mice were observed once a day for signs of paralysis or death. Paralyzed mice were sacrificed immediately, and any remaining mice were sacrificed at 14 days. Genotyping was performed as previously described (44). PD50 values (viral titers that would paralyze 50% of the mice inoculated) were calculated as previously described (8).
RESULTS
Cell-specific virus growth phenotype of a 3′ NCR deletion mutant poliovirus.
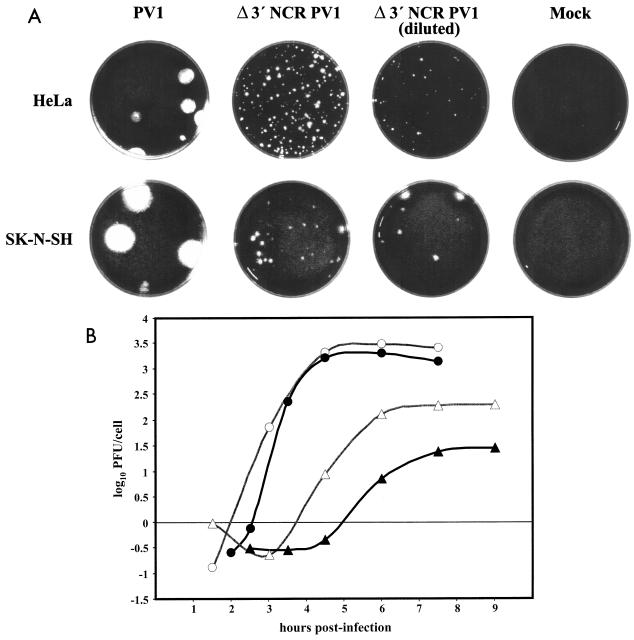
The Δ3′ NCR PV1 harbors a deletion of the complete 3′ NCR maintaining the first stop codon and the U of the second stop codon and is followed by the genome-encoded 3′-terminal poly(A) tract. We have shown that Δ3′ NCR PV1 replicates with slightly slower kinetics and grows to a lower maximum titer than wild-type poliovirus in HeLa cells (51). However, in a human host (or transgenic mouse), cervical epithelial cells (the precursor of HeLa cells) are not infected by poliovirus and are therefore not relevant to poliovirus pathogenesis. To examine the phenotype of the deletion mutant in a cell line that is more relevant to a poliovirus infection, we compared the plaque morphology in infected SK-N-SH and HeLa cell monolayers. In both cell lines, the deletion mutant virus had a small-plaque phenotype compared to that of wild-type poliovirus (Fig. 1A). Plaques produced by wild-type virus were larger on SK-N-SH monolayers than on HeLa monolayers, as were those produced by Δ3′ NCR PV1. Interestingly, equal-titer (assayed on HeLa cell monolayers) infections with Δ3′ NCR PV1 produced approximately 1/10 the number of plaques on SK-N-SH cells than on HeLa cells. These data suggested that there is a difference in the ability of the deletion mutant to infect SK-N-SH cells compared to HeLa cells.
FIG. 1.
Δ3′ NCR mutant virus plaque phenotype and growth. The generation and characterization of the plaque-isolated Δ3′ NCR PV1 stock was described previously (40). Plaque assays were performed as described in Materials and Methods. (A) Plaque assays were performed with virus stocks diluted to yield isolated plaques on monolayers. HeLa cells and SK-N-SH cells were infected with wild-type or Δ3′ NCR poliovirus and incubated at 37°C for 3 days. (B) One-step growth analysis was performed on HeLa (R19) or SK-N-SH monolayers in 60-mm plates at 37°C as described previously (52). Cells were infected at an MOI of ∼20 with either wild-type poliovirus (○ or •) or Δ3′ NCR PV1 (▵ or ▴). Infected SK-N-SH cells (• or ▴) or HeLa cells (○ or ▵) were harvested at the indicated times. Viruses were released from the cells by five freeze-thaw cycles, and the accumulation of virus was determined by plaque assays on HeLa cells. Virus titer is reported as the log10 PFU per cell.
To determine if the defect in the efficiency of plaque formation of the mutant virus was due to a replication deficiency, we performed a single-cycle growth analysis in HeLa cells and SK-N-SH cells. Figure 1B shows the single-cycle growth curves of wild-type or deletion mutant virus in both cell lines. Wild-type poliovirus accumulated at nearly the same rate and had a similar total accumulation profile in both cell lines (Fig. 1B). When compared to wild-type virus, the deletion mutant virus was defective for growth in HeLa cells and in SK-N-SH cells (Fig. 1B). The titer of the deletion mutant was lower than that of wild-type virus in HeLa cells by approximately 1 log10 unit but was reduced by approximately 2 log10 units compared to wild-type virus titers in SK-N-SH cells. These data suggested that the lower titer of the Δ3′ NCR PV1 in infected neuroblastoma cells was likely due to a virus replication defect.
Cell-specific defect in genome replication of the Δ3′ NCR mutant.
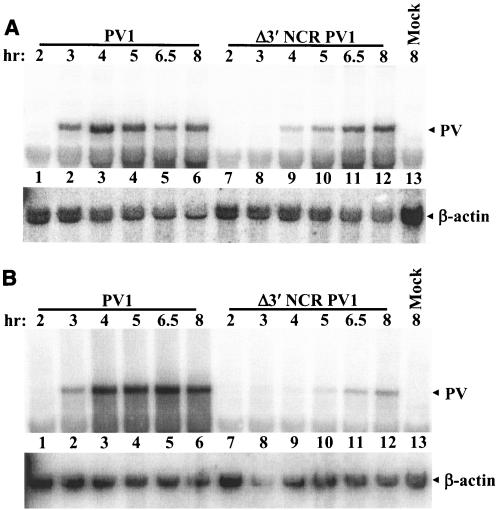
To investigate whether the cell-dependent defect in virus growth was due to a deficiency in mutant virus genome replication and accumulation, HeLa and SK-N-SH cells were infected at a high MOI and total cellular RNA was subjected to Northern blot analysis (Fig. 2). As observed previously (51), Δ3′ NCR virus genome accumulation was slower than that of wild type following infection of HeLa cells (Fig. 2A, compare lane 2 to lane 9). Although slightly reduced, the maximum level of RNA accumulation in the deletion-mutant-infected cells was comparable to that with wild-type virus (Fig. 2A, compare lane 3 to lane 12). The defect in replication kinetics and accumulation of mutant viral RNA was more pronounced in SK-N-SH cells than in HeLa cells. The maximum RNA levels achieved also demonstrated that the replication of the Δ3′ NCR genome in SK-N-SH cells was grossly defective compared to that of the wild type (Fig. 2B, compare lane 5 to lane 12). These data show that the slight defect in mutant genome replication in HeLa cells is significantly exacerbated in SK-N-SH cells (compare Fig. 2A, lanes 7 to 12, with B, lanes 7 to 12). The maximum RNA levels, normalized to the β-actin loading controls, were quantitated by phosphorimager analysis. The maximum deletion mutant RNA level was ∼80% of the maximum wild-type RNA level in infected HeLa cells. In contrast, the maximum deletion mutant RNA level in SK-N-SH cells was only ∼15% of the maximum wild-type RNA level. The differences in the ratios of deletion mutant to wild-type RNA genome accumulation represented a >5-fold reduction in the ability of the deletion mutant genome RNA to replicate in infected SK-N-SH cells compared to infected HeLa cells. Thus, the mutant virus growth impairment in SK-N-SH cells appears to be due to a cell-specific defect in viral RNA replication.
FIG. 2.
Northern blot analysis of viral genome accumulation in infected cells. (A) HeLa cell monolayers were mock infected (lane 13) or infected at an MOI of 20 with either wild-type poliovirus (lanes 1 to 6) or Δ3′ NCR PV1 (lanes 7 to 12). Total cellular RNA was harvested at the indicated times following infection using TriReagent (Molecular Research Center, Inc.), as developed from methods described previously (10). Northern blot analysis was performed with 5 μg of glyoxal-treated total cellular RNA (34). Hybridization and washing conditions were as described previously (9). Two 32P-labeled labeled probes, L391*(-) and PV7350(-) (complementary to the positive-strand RNA genomes at positions 253 to 272 and 7350 to 7369, respectively) were used to detect viral genomic RNA. Cellular β-actin mRNA was probed as an equal RNA loading control using random-primed 32P-labeled DNA probes. (B) Northern blot analysis was performed using total cellular RNA from infected SK-N-SH cells as described for panel A.
Strand-specific replication defect of Δ3′ NCR PV1.
The deletion of the genomic 3′ NCR may result in a defect in either positive- or negative-strand RNA synthesis, or the mutation may be equally deleterious to the synthesis of both strands of RNA. To more clearly define the nature of the RNA replication defect, we determined the ratio of positive- to negative-strand RNA accumulation during an infection. The ratio of positive- to negative-strand RNAs that accumulate during an infection with wild-type poliovirus has been reported to be approximately 35:1 to 65:1 at peak replication times after infection of HeLa cells (5, 17, 31, 38). The large molar excess of positive-strand RNA effectively competes with probes designed to detect negative-strand RNA accumulation in Northern blotting and RNase protection assays. In order to accurately measure the accumulation of negative-strand RNA following an infection, we used a two-cycle RNase protection assay previously described by Novak and Kirkegaard (38). This assay allows the sensitive detection of negative-strand RNAs by removal of excess positive-strand RNAs prior to hybridization with labeled RNA probes designed to detect viral negative strands. Using this assay (and a standard RNase protection assay to detect positive-stand viral RNAs), we determined the levels of positive-strand and negative-strand RNAs at different times after infection of HeLa cells or SK-N-SH cells (Fig. 3A and B). Molar amounts of RNA were determined by phosphorimager analysis, comparing each band intensity to that of a probe of known concentration. The data presented here are the result of two independent experiments, but only one set of data are displayed in Fig. 3. In HeLa cells, the maximum level of positive-strand RNA detected after infection with the deletion mutant virus was ∼25% of the maximum level found after wild-type poliovirus infection (Fig. 3A, compare lane 4 to lane 8). In contrast, the maximum level of negative-strand deletion mutant RNA was nearly identical to that of negative-strand wild-type RNA (Fig. 3B, compare lane 4 to lane 8). In HeLa cells infected with wild-type virus, the molar ratio of positive- to negative-strand RNA was found to be ∼35:1 at 5 h postinfection (compare Fig. 3A, lane 4, to B, lane 4). Following HeLa cell infections with Δ3′ NCR PV1, the ratio of positive-strand to negative-strand RNA was ∼10:1 at the peak of infection (compare Fig. 3A, lane 8, to B, lane 8).
FIG. 3.
Analysis of positive- and negative-strand viral RNA accumulation and positive-strand RNA stability in wild-type- or Δ3′ NCR-infected cells. For the RNase protection analysis shown in panels A and B, RNA samples were derived from the experiment displayed in Fig. 2. (A) Positive-strand viral RNA accumulation was determined in a standard RNase protection assay as described in Materials and Methods. Marker RNAs (with sizes indicated in nucleotides on the left side of the panel) are shown in lane 1. A 5.3-fmol aliquot of undigested probe was used for comparison to quantitate the molar amounts of RNA loaded in each lane by phosphorimager analysis (lane 2). Equal amounts (108 ng) of total cellular RNA were protected by 25 fmol of probe complementary to positive-sense poliovirus RNA sequences 5601 to 5809 (lanes 3 to 16). HeLa cells (lane 3 to lane 9) or SK-N-SH cells (lane 10 to lane 16) were infected with either wild-type poliovirus (lanes 3 to 5 and 10 to 12) or Δ3′ NCR PV1 (lanes 6 to 8 and 13 to 15). The RNA harvest times (hours postinfection) are indicated at the top of each lane. Mock-infected cells were harvested at 8 h postinfection (lanes 9 and 16). (B) Negative-strand RNA levels were determined by subjecting 1.3 μg of total cellular RNA to a two-cycle RNase protection assay as described in Materials and Methods. RNA samples were separated on 7 M urea-8% polyacrylamide gels. (C) RNA stability of positive-strand RNAs derived from infection of monolayers of SK-N-SH cells with wild-type or Δ3′ NCR mutant virus. Cells were infected at an MOI of ∼20 in the presence of 2 mM guanidine-HCl, and total RNA was isolated at the times indicated as described in Materials and Methods. Glyoxal-treated RNA (10 μg) was subjected to Northern blot analysis following electrophoresis on a 1.0% agarose gel in the presence of 10 mM sodium phosphate. Poliovirus and β-actin RNAswere detected by hybridization to 32P-labeled random hexamer-primed DNA probes. Hybridization and washing conditions were as described elsewhere (9).
The profile of viral RNA accumulation in infected SK-N-SH cells was very different from that observed in infected HeLa cells. In infected SK-N-SH cells, the maximum level of deletion mutant positive-strand RNA was only ∼10% of the level of wild-type RNA (Fig. 3A, compare lane 11 to lane 15). However, the maximum level of deletion mutant negative-strand RNA was ∼80% of wild-type negative-strand RNA in infected SK-N-SH cells (Fig. 3B, compare lane 12 to lane 15). In SK-N-SH cells infected with wild-type virus, the ratio of positive- to negative-strand viral RNA was reduced slightly compared to that of infected HeLa cells. We calculated the ratio of positive- to negative-strand viral RNA to be ∼30:1 (compare Fig. 3A, lane 12, to B, lane 12) in infected neuroblastoma cells. Significantly, the ratio of positive- to negative-strand viral RNA produced following infection with the 3′ NCR deletion mutant was only ∼5:1 in infected SK-N-SH cells, suggesting a defect in positive-strand viral RNA synthesis in the absence of a 3′ NCR. However, our RNase protection assay does not account for possible differences in stability between positive-strand RNAs harboring a 3′ NCR and those that have this region deleted. To address this possibility, we infected SK-N-SH cell monolayers with wild-type PV1 or the Δ3′ NCR mutant virus in the presence of guanidine-HCl (an inhibitor of poliovirus RNA synthesis). At the times indicated in Fig. 3C, total RNA was harvested and subjected to Northern blot analysis following electrophoretic separation on an agarose gel. The data displayed in Fig. 3C demonstrate that there were no significant differences in stability between wild-type and Δ3′ NCR RNAs in the absence of viral RNA replication, ruling out differential turnover of positive-strand RNAs as a possible cause for the differences observed in our RNase protection experiments. Overall, these data allow us to conclude that the viral replication defect observed in the absence of the poliovirus genomic 3′ NCR is primarily due to a strand-specific defect in RNA synthesis which is more pronounced in infected SK-N-SH cells than in infected HeLa cells.
Viral translation and protein processing in Δ3′ NCR PV1-infected cells.
We had previously shown that genomes of the 3′ NCR deletion mutant and wild-type virus were translated with equal efficiency in infected HeLa cells and in S-10 extracts derived from HeLa cells (51). To determine if the reduced mutant genome replication in SK-N-SH cells was due to a defect in viral translation (leading to a secondary effect on RNA synthesis), we examined the translation and processing efficiencies of wild-type and Δ3′ NCR mutant genomes. SK-N-SH cells were infected with either wild-type or deletion mutant virus at a high MOI, and an hour before cells were harvested translation products were labeled by incubation with [35S]methionine. During a poliovirus infection, host cell cap-dependent translation is shut down by the activity of a viral proteinase. Total host cell translation shutoff was not observed in Δ3′ NCR PV1-infected SK-N-SH cell cultures, resulting in a high background of cellular translation products (Fig. 4A, lanes 9 to 15). To increase the signal-to-noise ratio for viral translation products, viral proteins were immunoprecipitated with a polyclonal antibody that recognizes the viral protein 3C (45). Figure 4B shows viral translation and processing products immunoprecipitated by the anti-3C serum. These products include the polypeptide precursors P3 and 3CD, the alternative processing product 3C′, and the mature product, 3C. Compared to infection by wild-type virus, there was a delay in achieving maximum levels of translation and protein processing products in Δ3′ NCR PV1-infected cells (Fig. 4B, compare lanes 4 and 5 to lane 11). Although the translation efficiency of the mutant virus did not result in wild-type levels of polypeptides in infected SK-N-SH cells (Fig. 4B, compare lanes 4 to 10 with lanes 11 to 17), the deficiency was minor compared to the RNA synthesis defect (Fig. 2B) or the virus production defect (Fig. 1B) in the same cell line.
FIG. 4.
Viral translation and processing kinetics in cell culture. SK-N-SH monolayers in 60-mm dishes were infected at an MOI of 25 with either wild-type poliovirus (lanes 2 to 8) or Δ3′ NCR virus (lanes 9 to 15). An hour prior to cell harvest, the products of translation were labeled by the addition of 50 μCi of [35S]methionine. At the indicated times, cells were harvested and boiled in Laemmli sample buffer (29) for 3 min. (A) Portions (one-third of the total) of the radiolabeled translation products were analyzed on an SDS-12.5% polyacrylamide gel, which was subsequently subjected to phosphorimager analysis (Personal Molecular Imager FX; Bio-Rad). (B) Immunoprecipitation of poliovirus 3C-containing translation products from the remainder of the cellular lysates was carried out as described previously (46) using 10 μl of anti-3C serum (EΔ3C) (45) and protein A-agarose (Roche). These protein products included precursor polyproteins P3 and 3CD, the mature cleavage product 3C, and the alternatively processed product 3C′. The positions of these proteins on the gel are indicated on the left side of the figure. Labeled proteins were analyzed on an SDS-12.5% polyacrylamide gel, and the gel was subjected to phosphorimager analysis.
Δ3′ NCR virus replication in PVR transgenic mice.
SK-N-SH and HeLa cells are immortalized, both having been derived from human tumors. In addition, these cells have undergone selection pressures to survive in cell culture. Therefore, the cell line-specific replication phenotype of Δ3′ NCR PV1 may not represent a pathologically relevant observation. To examine whether the deletion mutation has an attenuation phenotype in vivo and to measure the degree of any such attenuation, we employed a transgenic mouse model expressing the hPVR. The PVR transgenic mouse has been established as an excellent model for poliovirus infection and poliovirus-induced pathology (1, 22, 26, 27, 42-44). To analyze the ability of the deletion mutant virus to cause disease, we determined the viral titer (in PFU) that would paralyze approximately 50% of the mice inoculated (PD50) (41). Transgenic mice were given IC inoculations with dilutions of Δ3′ NCR PV1 stocks and observed for signs of paralysis. The PD50 of wild-type poliovirus was 6 × 103 PFU, while Δ3′ NCR PV1 had a PD50 of 2.7 × 107 PFU. When nontransgenic mice were inoculated IC with 9 × 106 or 9 × 107 PFU of the mutant virus, none of the mice developed signs of paralysis, nor did they display any signs of infection. These data suggest that the paralysis induced by Δ3′ NCR PV1 in PVR transgenic mice was due to receptor-mediated infections in the central nervous system. Furthermore, the titer of deletion mutant virus per gram of brain tissue from a paralyzed PVR transgenic mouse was approximately 3 log10 units lower than that in a PVR transgenic mouse infected with wild-type poliovirus (data not shown). These data demonstrate a significant attenuation phenotype of the Δ3′ NCR virus in PVR transgenic mice compared to that of wild-type virus.
DISCUSSION
Based upon the assumption that the 3′ NCR of poliovirus acts as a cis-acting signal, Todd et al. had previously attempted to define the minimal element necessary for viral genome replication (51). We deleted the entire genomic 3′ NCR and found that this mutant virus replicated to within 1 log10 unit of wild-type virus levels in HeLa cells (51). This finding was surprising, because the 3′ NCR of the three poliovirus serotypes is well conserved despite the fact that the viral RNA-dependent RNA polymerase has a high error rate (12, 13, 53). To define a biologically relevant role for the 3′ NCR, we examined the growth of the Δ3′ NCR virus in different cell types. It has been demonstrated that a point mutation in the poliovirus 5′ NCR reduces neurovirulence in mice and causes a replication defect in neuroblastoma tissue culture cells (3, 30). In this report, we showed that Δ3′ NCR PV1 accumulates more slowly and to a lower titer in a neuroblastoma cell line (SK-N-SH) than in HeLa cells as a result of an exacerbated RNA replication defect. The phenotype is not likely due to a cell-dependent translation deficiency, because the virus translation efficiency was decreased only moderately. It is likely that the decrease in translation efficiency in SK-N-SH cells is a by-product of the reduced number of RNA genomes caused by the defect in viral genome replication, although we cannot rule out the contribution of a slight defect in translation to reduced levels of nonstructural proteins available for viral RNA replication functions.
Neurovirulence of the deletion mutant virus was markedly attenuated in PVR transgenic mice. Compared to wild-type virus, greater than 1,000 times more mutant virus was required to induce paralysis following IC inoculations. The level of attenuation of the deletion mutant was comparable to that of the Sabin type 1 vaccine strain. The PD50 of Δ3′ NCR PV1 was ∼3 × 107 PFU, and the PD50 of Sabin 1 in the same PVR transgenic mouse line has been reported to be >5 × 107 (8). Although it is possible that the attenuation phenotype seen in the PVR transgenic mice is the result of a species-specific requirement for the 3′ NCR, the PD50 was in good agreement with the growth characteristics of the mutant virus in the human SK-N-SH cell line. None of the nontransgenic mice showed symptoms of infection when inoculated with high-titer Δ3′ NCR PV1 stocks, allowing us to conclude that the symptoms induced in PVR transgenic mice infected with mutant virus were due to the ability of the transgenic mice to support mutant virus replication. It will be interesting to determine whether deletion mutant virus recovered from the transgenic mice has accumulated any additional mutations that allow higher levels of replication compared to those of input Δ3′ NCR virus. In a related study, Merkle and colleagues recently observed that a mutant coxsackievirus B3 harboring a partial deletion within the 3′ NCR of genomic RNA was attenuated for induction of myocarditis in infected mice, suggesting a role for the coxsackievirus 3′ NCR in viral pathogenesis (36).
It has been reported that the ratio of positive- to negative-strand RNA accumulation is lower in leukocytic and nerve cells following infections with wild-type poliovirus (31). We report a similar finding here, where the ratio of positive- to negative-strand RNA was lower in neuronal cells infected with wild-type virus than in infected HeLa cells. These data suggest that there is a cell-specific contribution to positive-strand RNA synthesis and that a cellular factor(s) involved in positive-strand RNA synthesis is limiting in cells of neuronal origin. After initiation of negative-strand RNA synthesis occurs, the mutant virus faces an exacerbated strand-specific defect in positive-strand RNA synthesis. This was evidenced by the lower ratio of positive-strand to negative-strand viral RNA during an infection with the mutant virus compared to the ratio detected during a wild-type poliovirus infection. In order to observe such a phenotype, the mutant virus would either have to synthesize higher levels of negative-strand RNAs or lower levels of positive-strand RNAs during genome replication. The data shown in Fig. 3B do not reveal significant differences in the levels of negative-strand RNAs found in cells infected with wild-type or Δ3′ NCR PV1. In addition, the results shown in Fig. 3C indicate that we did not detect any differences in stability of positive-strand viral RNAs during infection of neuronal cells by wild-type or mutant virus in the absence of viral RNA synthesis. Therefore, it is likely that the lower ratio of positive- to negative-strand RNA is due to reduced levels of positive-strand RNA synthesis.
We found that the strand-specific defect in Δ3′ NCR mutant RNA synthesis was exacerbated in neuronal cells, suggesting that a cellular factor(s) involved in positive-strand RNA synthesis interacts with the poliovirus 3′ NCR during genomic replication. The differences in phenotypic growth patterns may be the result of a lower concentration, posttranslational modification, alternative splicing patterns, or alternative patterns of subcellular sequestering of a host factor(s). Alternatively, the mutant virus growth defect could be due to differences in cellular processes, such as intracellular motility, membranous vesicle trafficking, or differences in cell morphology that could exacerbate a defect caused by a lack of genomic tethering to the replication complexes.
The observation that an RNA structure located at one terminus of the poliovirus genome functions in initiation of RNA synthesis at the opposite terminus is not without precedent. There is evidence that the 5′-terminal cloverleaf structure (also known as stem-loop I) located on the positive strand of poliovirus genomic RNA is involved in initiation of negative-strand RNA synthesis (6, 19, 32). Thus, it is possible that a similar mechanism of 5′-3′ termini interaction or communication is required for positive-strand RNA synthesis and that this putative interaction is provided, in part, by the genomic 3′ NCR. We have not yet determined whether these RNA sequences or secondary structures that are necessary for the initiation of positive-strand synthesis function in the context of the 3′ ends of the positive strands, acting in trans, or in the 5′ ends of negative-strand intermediates, acting in cis. Independent of the precise mechanism of attenuation caused by the strand-specific defect in RNA synthesis, the cell-dependent replication phenotype of the deletion mutant virus reveals a previously unknown property of the poliovirus 3′ NCR.
Acknowledgments
We are indebted to Hung Nguyen and My Phuong Tran for assistance with cell culture. We also thank Bert Flanegan and Raul Andino for helpful discussions about RNA stability.
D.M.B. and C.T.C. were predoctoral trainees of U.S. Public Health Service training grant AI 07319. G.M.J. was a predoctoral trainee of U.S. Public Health Service training grant GM 07311. This work was supported by U.S. Public Health Service grants AI 20017 (to V.R.R.) and AI 22693 (to B.L.S.) from the National Institutes of Health.
REFERENCES
- 1.Abe, S., Y. Ota, S. Koike, T. Kurata, H. Horie, T. Nomura, S. Hashizume, and A. Nomoto. 1995. Neurovirulence test for oral live poliovaccines using poliovirus-sensitive transgenic mice. Virology 206:1075-1083. [DOI] [PubMed] [Google Scholar]
- 2.Agol, V. I., S. G. Drozdov, V. P. Grachev, M. S. Kolesnikova, V. G. Kozlov, N. M. Ralph, L. I. Romanova, E. A. Tolskaya, A. V. Tyufanov, and E. G. Viktorova. 1985. Recombinants between attenuated and virulent strains of poliovirus type 1: derivation and characterization of recombinants with centrally located crossover points. Virology 143:467-477. [DOI] [PubMed] [Google Scholar]
- 3.Agol, V. I., S. G. Drozdov, T. A. Ivannikova, M. S. Kolesnikova, M. B. Korolev, and E. A. Tolskaya. 1989. Restricted growth of attenuated poliovirus strains in cultured cells of a human neuroblastoma. J. Virol. 63:4034-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agol, V. I., V. P. Grachev, S. G. Drozdov, M. S. Kolesnikova, V. G. Kozlov, N. M. Ralph, L. I. Romanova, E. A. Tolskaya, A. V. Tyufanov, and E. G. Viktorova. 1984. Construction and properties of intertypic poliovirus recombinants: first approximation mapping of the major determinants of neurovirulence. Virology 136:41-55. [DOI] [PubMed] [Google Scholar]
- 5.Andino, R., G. E. Rieckhof, and D. Baltimore. 1990. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell 63:369-380. [DOI] [PubMed] [Google Scholar]
- 6.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodian, D., and D. H. Horstmann. 1965. Polioviruses, p. 430-473. In F. L. Horsfall and I. Tamm (ed.), Viral and rickettsial infections of man. Lippincott, Philadelphia, Pa.
- 8.Bouchard, M. J., D. H. Lam, and V. R. Racaniello. 1995. Determinants of attenuation and temperature sensitivity in the type 1 poliovirus Sabin vaccine. J. Virol. 69:4972-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charini, W. A., C. C. Burns, E. Ehrenfeld, and B. L. Semler. 1991. trans rescue of a mutant poliovirus RNA polymerase function. J. Virol. 65:2655-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 11.Christodoulou, C., F. Colbere-Garapin, A. Macadam, L. F. Taffs, S. Marsden, P. Minor, and F. Horaud. 1990. Mapping of mutations associated with neurovirulence in monkeys infected with Sabin 1 poliovirus revertants selected at high temperature. J. Virol. 64:4922-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Torre, J. C., C. Giachetti, B. L. Semler, and J. J. Holland. 1992. High frequency of single-base transitions and extreme frequency of precise multiple-base reversion mutations in poliovirus. Proc. Natl. Acad. Sci. USA 89:2531-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Torre, J. C., E. Wimmer, and J. J. Holland. 1990. Very high frequency of reversion to guanidine resistance in clonal pools of guanidine-dependent type 1 poliovirus. J. Virol. 64:664-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans, D. M., G. Dunn, P. D. Minor, G. C. Schild, A. J. Cann, G. Stanway, J. W. Almond, K. Currey, and J. V. J. Maizel. 1985. Increased neurovirulence associated with a single nucleotide change in a noncoding region of the Sabin type 3 poliovaccine genome. Nature 314:548-550. [DOI] [PubMed] [Google Scholar]
- 15.Freistadt, M. S., G. Kaplan, and V. R. Racaniello. 1990. Heterogeneous expression of poliovirus receptor-related proteins in human cells and tissues. Mol. Cell. Biol. 10:5700-5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgescu, M. M., M. Tardy-Panit, S. Guillot, R. Crainic, and F. Delpeyroux. 1995. Mapping of mutations contributing to the temperature sensitivity of the Sabin 1 vaccine strain of poliovirus. J. Virol. 69:5278-5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giachetti, C., and B. L. Semler. 1991. Role of a viral membrane polypeptide in strand-specific initiation of poliovirus RNA synthesis. J. Virol. 65:2647-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gromeier, M., L. Alexander, and E. Wimmer. 1996. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc. Natl. Acad. Sci. USA 93:2370-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holland, J. J. 1961. Receptor affinities as major determinants of enterovirus tissue tropisms in humans. Virology 15:312-326. [DOI] [PubMed] [Google Scholar]
- 21.Holland, J. J., L. C. McLaren, and J. T. Syverton. 1959. The mammalian cell virus relationship. IV. Infection of naturally insusceptible cells with enterovirus ribonucleic acid. J. Exp. Med. 110:65-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horie, H., S. Koike, T. Kurata, Y. Sato-Yoshida, I. Ise, Y. Ota, S. Abe, K. Hioki, H. Kato, C. Taya, T. Nomura, S. Hashizume, H. Yonekawa, and A. Nomoto. 1994. Transgenic mice carrying the human poliovirus receptor: new animal models for study of poliovirus neurovirulence. J. Virol. 68:681-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawamura, N., M. Kohara, S. Abe, T. Komatsu, K. Tago, M. Arita, and A. Nomoto. 1989. Determinants in the 5′ noncoding region of poliovirus Sabin 1 RNA that influence the attenuation phenotype. J. Virol. 63:1302-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohara, M., S. Abe, T. Komatsu, K. Tago, M. Arita, and A. Nomoto. 1988. A recombinant virus between the Sabin 1 and Sabin 3 vaccine strains of poliovirus as a possible candidate for a new type 3 poliovirus live vaccine strain. J. Virol. 62:2828-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koike, S., H. Horie, I. Ise, A. Okitsu, M. Yoshida, N. Iizuka, K. Takeuchi, T. Takegami, and A. Nomoto. 1990. The poliovirus receptor protein is produced both as membrane-bound and secreted forms. EMBO J. 9:3217-3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koike, S., H. Horie, Y. Sato, I. Ise, C. Taya, T. Nomura, I. Yoshioka, H. Yonekawa, and A. Nomoto. 1993. Poliovirus-sensitive transgenic mice as a new animal model. Dev. Biol. Stand. 78:101-107. [PubMed] [Google Scholar]
- 27.Koike, S., C. Taya, T. Kurata, S. Abe, I. Ise, H. Yonekawa, and A. Nomoto. 1991. Transgenic mice susceptible to poliovirus. Proc. Natl. Acad. Sci. USA 88:951-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunin, C. M., and W. S. Jordan. 1961. In vitro adsorption of poliovirus by noncultured tissues. Effect of species, age and malignancy. Am. J. Hyg. 73:245-257. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.La Monica, N., and V. R. Racaniello. 1989. Differences in replication of attenuated and neurovirulent polioviruses in human neuroblastoma cell line SH-SY5Y. J. Virol. 63:2357-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lopez-Guerrero, J. A., F. Martinez-Abarca, M. Fresno, L. Carrasco, and M. A. Alonso. 1991. Cell type determines the relative proportions of (−) and (+) strand RNA during poliovirus replication. Virus Res. 20:23-29. [DOI] [PubMed] [Google Scholar]
- 32.Lyons, T., K. E. Murray, A. W. Roberts, and D. J. Barton. 2001. Poliovirus 5′-terminal cloverleaf RNA is required in cis for VPg uridylylation and the initiation of negative-strand RNA synthesis. J. Virol. 75:10696-10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macadam, A. J., G. Ferguson, C. Arnold, and P. D. Minor. 1991. An assembly defect as a result of an attenuating mutation in the capsid proteins of the poliovirus type 3 vaccine strain. J. Virol. 65:5225-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMaster, G. K., and G. G. Carmichael. 1977. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc. Natl. Acad. Sci. USA 74:4835-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendelsohn, C. L., E. Wimmer, and V. R. Racaniello. 1989. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 56:855-865. [DOI] [PubMed] [Google Scholar]
- 36.Merkle, I., M. J. van Ooij, F. J. van Kuppeveld, D. H. Glaudemans, J. M. Galama, A. Henke, R. Zell, and W. J. Melchers. 2002. Biological significance of a human enterovirus B-specific RNA element in the 3′ nontranslated region. J. Virol. 76:9900-9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nomoto, A., S. Koike, and J. Aoki. 1994. Tissue tropism and species specificity of poliovirus infection. Trends Microbiol. 2:47-51. [DOI] [PubMed] [Google Scholar]
- 38.Novak, J. E., and K. Kirkegaard. 1991. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J. Virol. 65:3384-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Omata, T., M. Kohara, S. Kuge, T. Komatsu, S. Abe, B. L. Semler, A. Kameda, H. Itoh, M. Arita, and E. Wimmer. 1986. Genetic analysis of the attenuation phenotype of poliovirus type 1. J. Virol. 58:348-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Racaniello, V. R., and R. Ren. 1994. Transgenic mice and the pathogenesis of poliomyelitis. Arch. Virol. Suppl. 9:79-86. [DOI] [PubMed] [Google Scholar]
- 41.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 42.Ren, R., and V. R. Racaniello. 1992. Human poliovirus receptor gene expression and poliovirus tissue tropism in transgenic mice. J. Virol. 66:296-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ren, R., and V. R. Racaniello. 1992. Poliovirus spreads from muscle to the central nervous system by neural pathways. J. Infect. Dis. 166:747-752. [DOI] [PubMed] [Google Scholar]
- 44.Ren, R. B., F. Costantini, E. J. Gorgacz, J. J. Lee, and V. R. Racaniello. 1990. Transgenic mice expressing a human poliovirus receptor: a new model for poliomyelitis. Cell 63:353-362. [DOI] [PubMed] [Google Scholar]
- 45.Roehl, H. H., T. B. Parsley, T. V. Ho, and B. L. Semler. 1997. Processing of a cellular polypeptide by 3CD proteinase is required for poliovirus ribonucleoprotein complex formation. J. Virol. 71:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Semler, B. L., C. W. Anderson, R. Hanecak, L. F. Dorner, and E. Wimmer. 1982. A membrane-associated precursor to poliovirus VPg identified by immunoprecipitation with antibodies directed against a synthetic heptapeptide. Cell 28:405-412. [DOI] [PubMed] [Google Scholar]
- 47.Shiroki, K., T. Ishii, T. Aoki, Y. Ota, W. X. Yang, T. Komatsu, Y. Ami, M. Arita, S. Abe, S. Hashizume, and A. Nomoto. 1997. Host range phenotype induced by mutations in the internal ribosomal entry site of poliovirus RNA. J. Virol. 71:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Svitkin, Y. V., N. Cammack, P. D. Minor, and J. W. Almond. 1990. Translation deficiency of the Sabin type 3 poliovirus genome: association with an attenuating mutation C472→U. Virology 175:103-109. [DOI] [PubMed] [Google Scholar]
- 49.Svitkin, Y. V., S. V. Maslova, and V. I. Agol. 1985. The genomes of attenuated and virulent poliovirus strains differ in their in vitro translation efficiencies. Virology 147:243-252. [DOI] [PubMed] [Google Scholar]
- 50.Svitkin, Y. V., T. V. Pestova, S. V. Maslova, and V. I. Agol. 1988. Point mutations modify the response of poliovirus RNA to a translation initiation factor: a comparison of neurovirulent and attenuated strains. Virology 166:394-404. [DOI] [PubMed] [Google Scholar]
- 51.Todd, S., J. S. Towner, D. M. Brown, and B. L. Semler. 1997. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J. Virol. 71:8868-8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Todd, S., J. S. Towner, and B. L. Semler. 1997. Translation and replication properties of the human rhinovirus genome in vivo and in vitro. Virology 229:90-97. [DOI] [PubMed] [Google Scholar]
- 53.Ward, C. D., M. A. Stokes, and J. B. Flanegan. 1988. Direct measurement of the poliovirus RNA polymerase error frequency in vitro. J. Virol. 62:558-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westrop, G. D., K. A. Wareham, D. M. Evans, G. Dunn, P. D. Minor, D. I. Magrath, F. Taffs, S. Marsden, M. A. Skinner, G. C. Schild, and J. W. Almond. 1989. Genetic basis of attenuation of the Sabin type 3 oral poliovirus vaccine. J. Virol. 63:1338-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wimmer, E., C. U. T. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]