Abstract
Human cytomegalovirus (CMV) establishes latent infections in hematopoietic cells such as granulocyte-macrophage progenitors (GM-Ps). During latency the virus is sequestered in a nonreplicating state, although limited transcriptional activity has been previously reported. In this study we sought to further examine viral gene expression during the latent phase of infection. Using an experimental model of latency, primary human GM-Ps were latently infected with CMV strain Toledo and extracted RNA subjected to reverse transcription-PCR by using CMV gene-specific primers. Using this approach, we detected transcription from the UL111.5A region of the viral genome. This transcription was also detected in GM-Ps latently infected with AD169 and Towne strains, indicating that expression was CMV strain independent. Significantly, we detected UL111.5A-region transcripts in mononuclear cells from healthy bone marrow and mobilized peripheral blood allograft donors, demonstrating expression during natural latent infection. Mapping experiments with RNA extracted from latently infected GM-Ps revealed the expression of a novel UL111.5A region transcript with a splicing pattern that differed from that reported during productive infection of permissive cells. This UL111.5A region transcript expressed during latent infection is predicted to encode a 139-amino-acid protein with homology to the potent immunosuppressor interleukin-10 (IL-10) and to the viral IL-10 homolog that is expressed during productive CMV infection. Expression of a latency-associated cmvIL-10 may confer upon the virus an ability to avoid immune recognition and clearance during the latent phase of infection.
Human cytomegalovirus (CMV) is a species-specific betaherpesvirus that causes severe disease in neonates and in immunosuppressed individuals such as allograft transplant recipients and patients with AIDS (20). In common with all herpesviruses, the life cycle of CMV is characterized by productive, latent, and reactivation phases. During productive infection, viral genes are expressed in a temporarily regulated cascade, resulting in the synthesis of new, infectious virus (17). At some point after initial infection, the virus establishes a lifelong latent infection in myeloid lineage cells, during which time viral gene expression is restricted and infectious virus is not produced (16, 31, 32). Periodically, virus can reactivate from latency, a process that results in the generation of infectious virus and which is the major cause of serious CMV-associated diseases common in recipients of solid organ and bone marrow (BM) allografts. The ability of CMV to persist in a latent state for the life of the host ensures a reservoir of virus for subsequent reactivation and highlights the importance of latency to the success of this virus as a human pathogen. Maintenance of latency in the face of the immune response argues for a dynamic interplay between virus and host where CMV encodes functions during latency that ensure its survival during this phase of infection.
Attempts to understand the molecular basis of CMV latency have included studies to identify and characterize the function(s) of viral genes expressed during latent infection of myeloid progenitor cells. Studies utilizing cultured granulocyte-macrophage progenitors (GM-Ps) in an experimental model of latency identified two classes of CMV latency-associated transcripts (CLTs), sense and antisense CLTs, which originate from the major immediate-early (MIE) region of the viral genome (5, 9-11, 26). CLTs are also expressed during natural latent infection in peripheral blood and BM mononuclear cells from healthy donors (5, 11) and antibodies to open reading frames (ORFs) encoded by these transcripts have been detected in the serum from long-term healthy blood and BM donors (11, 13). However, the functions of the MIE region CLTs have not yet been defined (34).
Few other studies have sought to assess viral gene expression during latency, although transcriptional activity from the US28 region has been reported after experimental nonproductive infection of the monocytic cell line (THP-1) (1). In addition, a study utilizing BM-derived CD34+ cells in a model of CMV latency indicated that viral gene expression was more widespread, with transcriptional activity from no fewer than 66 genes reported at some point between day 1 and day 8 postinfection (4). To date, however, MIE region CLTs are the only transcripts expressed during latent infection that have been cloned and structurally characterized and shown to be expressed in naturally infected individuals.
We recently reported that CMV latent infection of GM-Ps resulted in a reduced expression of cell surface major histocompatibility complex (MHC) class II, suggesting that latent CMV encodes an immune evasion strategy (27). The viral gene(s) responsible for this phenotype was not identified, but the downregulation during latency was independent of the CMV US2 and US3 genes that have been shown to modulate MHC class II expression during productive infection (3, 6, 33). Interestingly, the CMV UL111.5A gene encodes a functional homolog of the potent immunomodulatory cytokine interleukin-10 (denoted cmvIL-10) which is expressed during productive infection of permissive cells (12, 15, 30). Thus, we sought to determine whether the UL111.5A region was expressed during latent CMV infection. UL111.5A region transcription was detected by reverse transcription PCR (RT-PCR) during both experimental and natural CMV latency. RACE (rapid amplification of cDNA ends) mapping and RT-PCR primer-walking experiments revealed the expression of a novel UL111.5A region transcript that differed from that reported to be expressed during productive infection. This transcript is predicted to encode a 139-amino-acid protein with homology to human IL-10, suggesting that CMV may encode a functional viral IL-10 homologue during latency.
MATERIALS AND METHODS
Cells and virus culture.
Human fetal liver hematopoietic cells were prepared and cultured as GM-Ps in suspension as described previously (9). On day 3, cells were mock or latently infected with either the sequenced strain AD169 (2), the high-passage strain TowneVarRIT3 (8, 21), or the low-passage strain Toledo (22) at a multiplicity of infection (MOI) of 3. GM-Ps were collected and transferred two to three times a week for 2 weeks. Human foreskin fibroblasts (HFFs) were used for virus propagation and titer determination by plaque assay.
Samples of BM and mobilized peripheral blood (MPB) were collected from clinically healthy allograft donors at Westmead Hospital. Mononuclear cells were isolated on Ficoll gradients and counted prior to RNA extraction.
RNA extraction and RT-PCR analysis.
Total RNA was extracted by using a RNAqueous kit (Ambion, Inc.). RNA samples (0.2 to 5 μg) were reverse transcribed in a 20-μl volume in the presence of 1× first-strand buffer (50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2), 0.5 mM concentrations of deoxynucleoside triphosphates, 20 mM dithiothreitol, 40 U of RNaseOUT RNase inhibitor, 90 ng of random hexanucleotide primers, and 200 U of SuperScript II reverse transcriptase (Invitrogen) for 10 min at 25°C, followed by 1.5 h at 42°C. The reaction was stopped by incubation for 15 min at 70°C.
A 3-μl aliquot of the resulting cDNAs was subjected to PCR amplification in a 50-μl reaction volume containing 50 mM KCl, 20 mM Tris-HCl (pH 8.4), 1.5 mM MgCl2, 0.1 mM concentrations of deoxynucleoside triphosphates, 0.2 μM concentrations of forward and reverse primers, and 2.5 U of Platinum Taq DNA polymerase (Invitrogen). Briefly, the cycle parameters used in the present study were 94°C for 45 s, 60°C for 1 min, and 72°C for 2 min (cycle A parameters); 94°C for 30 s, 68°C for 30 s, and 72°C for 2 min (cycle B parameters); 94°C for 30 s, 56°C for 30 s, and 72°C for 2 min (cycle C parameters); 94°C for 30 s, 60°C for 30 s, and 72°C for 2 min (cycle D parameters). UL111.5A region primer sequences and predicted PCR product sizes are summarized in Table 1. The primers used for the detection of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were as follows: forward, 5′-CGAGATCCCTCCAAAATCAA-3′; and reverse, 5′-TGTGGTCATGAGTCCTTCCA-3′. PCR products were separated by electrophoresis on 1.5% agarose gels and visualized with ethidium bromide. Where indicated, products were Southern blotted and hybridized with a digoxigenin-ddUTP end-labeled oligonucleotide probe (JAS-R6) according to manufacturer's protocols (Roche). Probe binding was visualized by using the CDP-Star chemiluminescence detection system (Roche).
TABLE 1.
Primers and predicted products for PCR analyses
| Primer | Sequence | Pair | Predicted product size (bp)a
|
||
|---|---|---|---|---|---|
| Unspliced | Spliced intron 1 | Spliced introns 1 and 2 | |||
| JAS-53 | 5′-ACTATTCTAACCGCGGAAG-3′ | JAS-R4 | 712 | 636 | 553 |
| JAS-R5 | 803 | 727 | 644 | ||
| JAS-52 | 5′-CATAAAGGACCACCTACCTGGGA-3′ | JAS-R4 | 655 | 579 | 496 |
| JAS-R5 | 746 | 670 | 587 | ||
| JAS-F1 | 5′-TACAAAGCCGCAGTGTCGTCCAGAGGATTACG-3′ | JAS-R1 | 247 | 171 | N/A |
| JAS-B1 | 346 | 270 | N/A | ||
| JAS-R5 | 588 | 512 | 429 | ||
| JAS-P3 | 5′-CAGATTGCAAGATCTCCGCGTCACCTT-3′ | JAS-R4 | 461 | 385 | 302 |
| JAS-R1 | 5′-CAACAACCAGTCCATGACGCTGCATC-3′ | JAS-F1 | 247 | 171 | N/A |
| JAS-B1 | 5′-GTAGATGGATTCTAGCGTCGAGCGCAT-3′ | JAS-F1 | 346 | 270 | N/A |
| JAS-R4 | 5′-TCCTGAGACAGCCGACTAATCACGGAC-3′ | JAS-53 | 712 | 636 | 553 |
| JAS-52 | 655 | 579 | 496 | ||
| JAS-P3 | 461 | 385 | 302 | ||
| JAS-R5 | 5′-TCTCGAGTGCAGATACTCTTCGAGACGG-3′ | JAS-53 | 803 | 727 | 644 |
| JAS-52 | 746 | 670 | 587 | ||
| JAS-F1 | 588 | 512 | 429 | ||
| JAS-R6 | 5′-GACCACCGTACCGTCGAGCCACACGGAG-3′ | Probe | N/A | N/A | N/A |
NA, not applicable.
3′ RACE.
The 3′ ends of UL111.5A region transcripts were mapped by using the SMART RACE cDNA amplification kit (BD Biosciences). Aliquots (1 μg) of DNase-treated total RNA from CMV-infected GM-Ps were treated according to the manufacturer's recommendations. 3′ RACE products were generated by amplification of poly(dT)-generated cDNAs for 40 cycles (cycle B parameters) with the primers JAS-F1 and UPM (BD Biosciences), followed by a second round of 40 cycles (cycle B parameters) with the primers JAS-P3 and NUP (BD Biosciences). 3′ RACE products were UA cloned into the pDrive cloning vector (Qiagen), and plasmid DNA was extracted by using the QIAprep spin miniprep kit (Qiagen).
RESULTS
CMV UL111.5A region transcripts are expressed during experimental latent infection of GM-Ps.
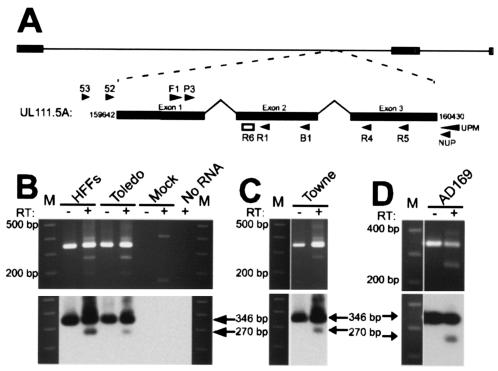
RT-PCR was used to determine whether UL111.5A region transcripts were expressed during experimental latent infection. Human fetal liver-derived GM-Ps were either mock infected or infected with CMV strain Toledo at an MOI of 3. On day 14 postinfection, the total RNA was extracted and reverse transcribed before being amplified for 45 cycles by using the cycle A parameters with the primers JAS-F1 and JAS-B1, which span across UL111.5A intron 1 (Fig. 1A). Amplification of infected GM-P RNA yielded a RT-dependent 270-bp PCR product which was confirmed by Southern blot hybridization to be UL111.5A specific and which corresponded in size to spliced intron 1 transcripts made in productively infected HFFs (Fig. 1B). A 346-bp genomic-sized PCR product was also detected in infected GM-P and HFF samples. Amplification in both the presence or absence of RT indicated that the template for the 346-bp product was either contaminating viral DNA, an unspliced transcript, or a combination of the two. Amplification was not detected in mock-infected GM-Ps or when RNA was omitted from the reaction mixture.
FIG. 1.
(A) Schematic representation of the CMV genome with the UL111.5A transcript expanded to show the position of primers (arrow heads) and probe (open box) used in the present study. (B) RT-PCR analysis of UL111.5A region gene expression in latently infected GM-Ps. Detection of UL111.5A region transcripts in CMV stain Toledo productively infected HFFs (day 4 postinfection) and latently infected GM-Ps (day 14 postinfection) (B) and CMV strain Towne (C) and AD169 (D) latently infected GM-Ps (day 14 postinfection). Panels B to D show ethidium bromide-stained agarose gels (upper panel) and the corresponding Southern blots (lower panel) of products generated from RT-PCR analysis for UL111.5A region transcripts. The sizes of the RT-PCR products are indicated with arrows and the numbers to the left of the gels indicate the size of the adjacent molecular weight size markers (M). The presence (+) or absence (−) of reverse transcriptase (RT) in the reaction mixture is indicated.
Assessment of UL111.5A region transcription was extended to GM-P cultures latently infected with either CMV strain AD169 or strain TowneVarRIT3. Like strain Toledo, RT-PCR and Southern blot hybridization demonstrated that both strain AD169 and strain TowneVarRIT3 expressed spliced UL111.5A region transcripts (Fig. 1C and D). It was concluded that the UL111.5A region is expressed during experimental latent infection of GM-Ps and that expression is a feature common to multiple strains of CMV.
Although the GM-P model of latency used in the present study has been shown to support a nonproductive infection and spontaneous reactivation has never been observed (5, 9, 11, 26, 34), we confirmed that GM-Ps remained free of infectious virus. Supernatants and cell lysates of 1.5 × 105 GM-Ps from latently infected cultures were tested for the presence of infectious virus by plaque assay on permissive HFFs. No evidence of plaque formation was detected in either sample. We also assessed infected GM-P cultures by RT-PCR for evidence of productive gene expression. In contrast to RNA from productively infected HFFs, RNA samples from infected GM-P cultures were consistently negative for the expression of UL120, a spliced γ gene which encodes a putative structural glycoprotein expressed during the productive phase (23; data not shown). These data are consistent with previous reports that failed to detect productive infection in CMV-infected GM-P cultures (9, 26, 27).
Enumeration of latently infected cells expressing UL111.5A region transcripts.
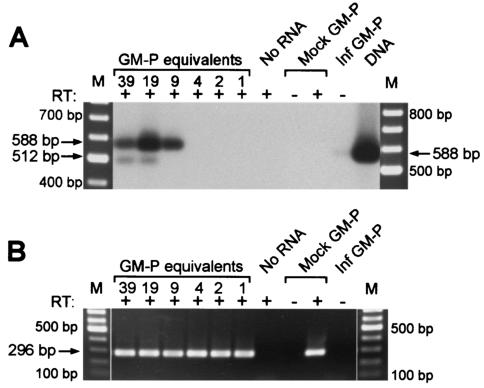
To determine the proportion of GM-Ps expressing UL111.5A region transcripts, RT-PCR was performed on dilutions of total RNA extracted from GM-Ps 14 days after infection with CMV strain Toledo at an MOI of 3. Infected GM-P RNA was treated with DNase before being diluted such that each reaction contained either 39, 19, 9, 4, 2, or 1 infected GM-P equivalents. Mock-infected GM-P RNA was added to each dilution so that the total amount of RNA per reaction remained constant at 0.5 μg. Random-primed cDNA was subjected to 40 cycles of PCR amplification by using the cycle B parameters with the primers JAS-F1 and JAS-R5. An RT-dependent 512-bp product corresponding to spliced UL111.5A transcripts was detected at 39 and 19 infected GM-P equivalents but became undetectable at 9 infected GM-P equivalents (Fig. 2A). A 588-bp genomic sized product was also detected down to nine infected GM-P equivalents. The 588-bp product may represent an unspliced UL111.5A region transcript, although it is possible that it resulted from contaminating viral DNA since a genome-sized product was detected (albeit very weakly) when RT was omitted from a control reaction containing RNA from 20,000 infected GM-Ps. A second round of 30 cycles of PCR by using the cycle B parameters with the primers JAS-P3 and JAS-R4 did not result in detection of either product at a higher dilution (data not shown). Amplification was not detected in mock-infected GM-P samples or when RNA was omitted from the reaction mixture (Fig. 2A). These data suggest that spliced UL111.5A region transcripts are expressed in at least 1 of 19 (5.2%) but in fewer than 1 of 9 (11.1%) experimentally infected GM-Ps. Comparable results were obtained in a further two cell dilution RT-PCR experiments performed with cells from two separate GM-P cultures (at least 2.3% but less than 12.5%, at least 1.3% but less than 6.3%). To determine whether amplifiable RNA was present at each cell dilution, we performed RT-PCR for 40 cycles for cellular GAPDH transcripts by using the cycle D parameters. A 296-bp product representing GAPDH transcripts was readily detected at each GM-P dilution (Fig. 2B). No amplification was detected when either RNA or RT were omitted from the reaction. This analysis confirmed that transcripts could be successfully amplified from each RNA sample.
FIG. 2.
(A) Enumeration of GM-Ps expressing UL111.5A region transcripts. Southern blot of RT-PCR products showing detection of UL111.5A region transcripts. Latently infected GM-Ps were counted and serially diluted to give 39, 19, 9, 4, 2, and 1 cell per reaction as indicated above lanes. Arrows adjacent to the lanes indicate the position of predicted spliced (512 bp) and unspliced (588 bp) RT-PCR products amplified with the primers JAS-F1 and JAS-R5. GM-Ps from a mock-infected (Mock) or latently infected (Inf GM-P) culture, a sample without RNA (No RNA), and the products of a PCR for UL111.5A region DNA were included as controls. The presence (+) or absence (−) of RT in each reaction is indicated. An ethidium bromide-stained 100-bp DNA ladder (M) is shown on both sides of the panel. (B) Ethidium bromide-stained gel showing RT-PCR detection of GAPDH transcripts (296-bp product) in the same RNA samples as panel A.
To assess the number of cells containing the viral genome, cell lysates prepared from serially diluted GM-Ps were subjected to previously described PCR amplification with the primers JAS-F1 and JAS-R5. This analysis revealed detection of the viral genome down to a dilution of two cells (data not shown). This was consistent with previous, more extensive analyses that characterized the levels and distribution of viral DNA in latently infected GM-Ps and showed that the GM-P model of latency used in the present study reproducibly results in the vast majority of the cells harboring viral genomes (26).
Structural analysis of UL111.5A region transcripts.
During productive infection of permissive HFFs, the UL111.5A transcript is comprised of two introns and three exons whose 5′ and 3′ ends have previously been defined (12, 15). This transcript codes for a functional homolog of IL-10 (cmvIL-10) (12, 30). To evaluate the structure of UL111.5A region transcripts expressed during latency, we applied 3′ RACE procedures and 5′ primer-walking RT-PCR to DNase-treated total RNA extracted from GM-Ps 14 days after infection with CMV strain Toledo (MOI = 3).
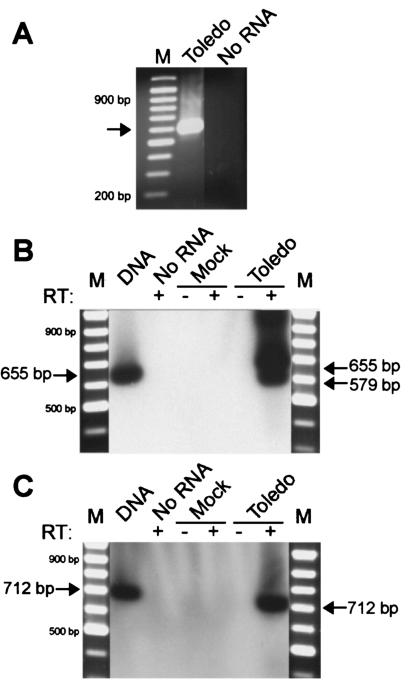
For 3′ RACE analysis, poly(dT)-primed cDNA was subjected to two rounds of PCR amplification. In addition to amplifying the 3′ end of the latency expressed transcript, PCR primers were designed to determine whether this transcript was spliced like that reported during productive infection. Thus, primers spanned across both intron 1 and intron 2. A single PCR product of approximately 600 bp was readily detected, but no product was detected when template was omitted from the PCR (Fig. 3A). The PCR product was UA cloned, and the sequence was determined from four individual clones. These clones all exhibited an identical sequence consistent with a single polyadenylation site located 14 bp downstream of a consensus polyadenylation signal (AATAAA), indicating the same termination site as that used during productive infection (AD169 nucleotide position 160430, accession number X17403). Sequencing of the four clones also revealed a processed transcript with a single spliced region of 76 bp that was identical to intron 1 of the transcript expressed during productive infection, but no other splicing was observed. The same 3′ end and splicing pattern was observed in a further two replicate experiments with RNA from GM-P cultures latently infected with either strain Toledo or strain AD169 (data not shown). These experiments demonstrated that latent UL111.5A region transcripts terminated at the same site reported for productive UL111.5A transcripts but differed with respect to intron 2, which remained unspliced during latency.
FIG. 3.
(A) Determination of 3′ terminus of UL111.5A region transcripts expressed during latent CMV infection of GM-Ps. Ethidium bromide-stained agarose gel of 3′ RACE PCR products derived from RNA extracted from GM-Ps latently infected with CMV strain Toledo. The arrow indicates a single 3′ RACE PCR product of approximately 600 bp after nested amplification with the primers JAS-F1 and UPM for the first round and the primers JAS-P3 and NUP for the second round. A negative control containing no RNA template and a 100-bp DNA ladder (M) are indicated. (B and C) Determination of the 5′ terminus of UL111.5A region transcripts expressed during latent CMV infection of GM-Ps. Southern blots of RT-PCR products were derived from RNA extracted from mock- or CMV strain Toledo-infected GM-Ps. (B) RT-PCR products amplified with the forward primer JAS-52. Arrows indicate the position of 579-bp spliced and 655-bp unspliced products. (C) RT-PCR products amplified with the forward primer JAS-53. Arrows indicate the position of the predicted 712-bp unspliced product, but no spliced product was detected. DNA extracted from productively infected HFFs was included as a positive control for the PCRs. A negative control containing no RNA template and a 100-bp DNA ladder (M) are indicated. The presence (+) or absence (−) of RT in each reaction mixture is indicated.
There are two previously identified transcriptional start sites within the region under examination. These are the start of the UL111.5A transcript at position 159642 (15) and the start of the UL111A transcript 27 bp upstream at position 159615 (2). Both transcripts encode ORFs that originate from the same methionine at position 159678. We determined whether the UL111.5A region transcripts expressed during latency were likely to utilize either of these transcription start sites by performing a series of primer-walking RT-PCRs with the forward primers JAS-52 or JAS-53. Primer JAS-52 lies between the two start sites, with its 5′ end at the UL111A start site and its 3′ end 4 bp upstream of the UL111.5A start site. Primer JAS-53 lies upstream of both start sites, with its 3′ end being 38 bp upstream of the UL111A start site. To determine whether latent transcripts initiated upstream of the UL111.5A start site, GM-P RNA was reverse transcribed and subjected to 40 cycles of PCR by using cycle C parameters with primers JAS-52 and JAS-R5, followed by a second round of 25 cycles of PCR by using cycle C parameters with primers JAS-52 and JAS-R4. Southern blot hybridization revealed the presence an RT-dependent fragment that was smaller than a viral DNA template-sized fragment and was consistent with a predicted 579-bp product derived from a single (intron 1) spliced transcript, as detected during the 3′ RACE mapping (Fig. 3B). No amplification was detected with mock-infected GM-P RNA or when RNA was omitted from the reaction mixture. In addition to the 579-bp spliced transcript fragment, a genomic-template-sized fragment of 655 bp was also amplified from latently infected RNA. The DNase treatment of this sample, together with the dependence on RT, suggests that this fragment was derived from an unspliced transcript. The amplification with forward primer JAS-52 of UL111.5A region transcripts expressed during latent infection suggests that these transcripts initiate upstream of the start of the UL111.5A transcript expressed during productive infection.
The RT-PCR was repeated with 40 cycles of amplification using the cycle C parameters with primers JAS-53 and JAS-R5, followed by a second round of 25 cycles using the cycle C parameters with primers JAS-53 and JAS-R4. A single, RT-dependent 712-bp product consistent with the amplification of an unspliced transcript was detected in infected GM-Ps (Fig. 3C). A spliced UL111.5A region transcript was not detected, and no products were amplified from mock-infected GM-Ps or when RNA was omitted from the reaction. Comparable results were obtained when primer-walking RT-PCRs with the forward primers JAS-52 and JAS-53 were applied to DNase-treated RNA extracted from GM-Ps latently infected with strain TowneVarRIT3 (data not shown). If we assume that binding of the full primer sequence is required for successful amplification, these data indicate that the spliced, latent UL111.5A region transcript initiates within a 38-bp region (between nucleotide positions 159577 and 159615) upstream of the productive UL111.5A start site which contains the UL111A start site (Fig. 4A). These data also suggest that an unspliced latent transcript utilizes a different start site that is upstream of nucleotide position 159558.
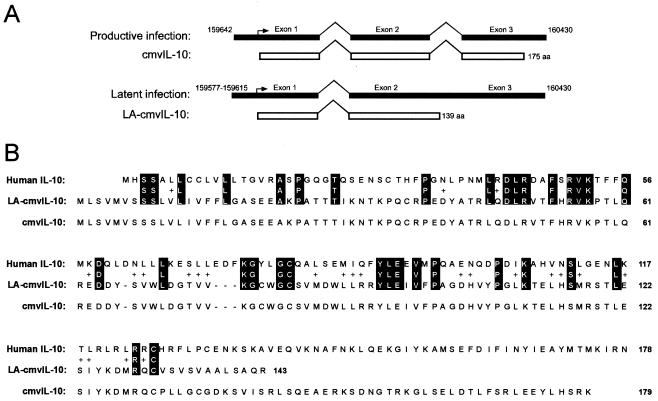
FIG. 4.
(A) Summary of the structure of UL111.5A region transcripts expressed during productive and latent infection. Upper lines show the double-spliced UL111.5A transcript expressed during productive infection and the corresponding 175-amino-acid ORF that encodes cmvIL-10. Lower lines show the single spliced UL111.5A region transcript expressed during latent infection and its corresponding ORF, which encodes a putative 139-amino-acid protein termed LA-cmvIL-10. Black boxes and open boxes depict transcripts and ORFs, respectively. Nucleotide position numbers (AD169 genome) of the start and stop sites of transcription are indicated, and the region encompassing the start site of the spliced transcript expressed during latency is shown as a gray box. The first methionine residue is indicated by a right-angled arrow. (B) Alignment of the amino acid sequences of human IL-10 and LA-cmvIL-10. Identical amino acids are shown in white letters on a black background and conserved amino acids are shown as “+” signs. Amino acid residues are numbered from the first methionine. The amino acid sequence of the IL-10 homolog expressed by CMV during productive infection (cmvIL-10) is also shown.
In summary, structural analyses of UL111.5A region transcripts expressed during latency revealed the presence of a novel transcript with a single intron and two exons that initiates within a small region upstream of UL111.5A start site and is coterminal with UL111.5A transcripts expressed during productive infection. An unspliced transcript with as-yet-undefined ends was also identified. Sequence analysis of the spliced transcript by using the National Center for Biotechnology Information ORF Finder program revealed the presence of a single, 139-amino-acid ORF. A BLAST search of the 139-amino-acid sequence against the human genome demonstrated 27% identity and 46% similarity to human IL-10 over a 124-amino-acid region (Fig. 4B). It was concluded that a novel, spliced UL111.5A region transcript expressed during latent infection encodes a putative cmvIL-10-like protein that we have termed latency associated (LA)-cmvIL-10. Interestingly, in parallel experiments with RNA extracted from productively infected HFFs (72 h postinfection), we found evidence of expression of the LA-cmvIL-10 transcript (data not shown), suggesting that LA-cmvIL-10 may be expressed during both latent and productive phases of infection.
Detection of UL111.5A region transcripts in naturally infected individuals.
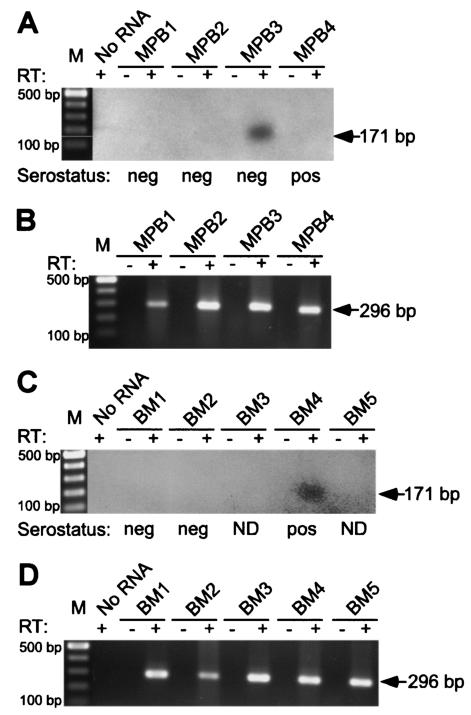
Mononuclear cells were isolated from BM and granulocyte-colony stimulating factor MPB samples collected from healthy allograft donors. Total RNA was extracted from aliquots of 3.5 × 105 to 1.0 × 107 cells and treated with RQ1 DNase to remove contaminating DNA before being subjected to RT-PCR. UL111.5A region transcripts were amplified for 40 cycles from random-primed cDNA using cycle A parameters with primers JAS-F1 and JAS-B1, followed by a second round of amplification for 35 cycles using cycle A parameters with primers JAS-F1 and JAS-R1. Products were resolved by gel electrophoresis before being transferred to nylon membranes and hybridized to an internal end-labeled oligonucleotide probe (JAS-R6). A 171-bp RT-dependent PCR fragment corresponding in size to spliced UL111.5A region transcripts was detected in two of nine donors (Fig. 5A and C). No amplification was observed when RT or RNA was omitted from the reaction mixtures. The CMV serostatus of the donors was provided by Westmead Hospital. Of the two UL111.5A transcript-positive donors, one was CMV seropositive and one was seronegative. It was concluded that the UL111.5A region was transcribed during natural latent CMV infection but that expression did not always correlate with donor serostatus. To evaluate the quality of the RNA extracted from each donor cell sample, we performed RT-PCR with cycle D parameters for transcripts from the cellular gene GAPDH. An RT-dependent, 296-bp product representing GAPDH transcripts was readily amplified from all nine donor RNA samples, confirming that transcripts could be successfully amplified from each RNA sample (Fig. 5B and D).
FIG. 5.
(A and C) Spliced UL111.5A-region transcripts are expressed during natural latent infection. A Southern blot of MPB (MPB1 to MPB4) and BM (BM1 to BM5) samples after heminested PCR amplification with the primers JAS-F1 and JAS-B1 (first round) and with the primers JAS-F1 and JAS-R1 (second round) was carried out. The arrow indicates the position of a predicted 171-bp spliced UL111.5A region transcript product. A negative control containing no RNA template and a 100-bp DNA ladder (M) are indicated. The presence (+) or absence (−) of RT in each reaction mixture is indicated. The CMV serostatus of each donor is indicated as positive (pos), negative (neg), or not determined (ND). (B and D) Detection of GAPDH by RT-PCR in the same RNA samples as in panels A and C. The arrow indicates the position of a predicted 296-bp GAPDH transcript product.
DISCUSSION
This study reports the detection of transcription from the UL111.5A region of the human CMV genome during experimental latent infection of hematopoietic progenitor cells and in BM and MPB cells from naturally infected individuals. Although CMV encodes more than 200 distinct genes, viral gene expression during the latent phase of infection remains poorly defined. To date, UL111.5A region CLTs and the previously identified MIE region CLTs represent the only two classes of viral transcripts that have been detected during natural latent CMV infection and have been subjected to structural analyses. The detection of UL111.5A region CLTs in GM-Ps latently infected with either CMV strain AD169, Towne, or Toledo demonstrates that expression is strain independent and is a conserved feature of CMV. This is further supported by detection in naturally infected cell samples from healthy allograft donors, a finding consistent with a bone fide role for these transcripts during latency.
The function of UL111.5A region CLTs is under investigation, but our current analyses revealed a spliced transcript encoding a single, 139-amino-acid ORF with homology to human IL-10. Human IL-10 is a multifunctional immunomodulatory cytokine that has potent immunosuppressive effects on hematopoietic cells (18). Its primary role is to suppress immune function by inhibiting the synthesis of proinflammatory cytokines such as IL-2, gamma interferon, and tumor necrosis factor alpha. IL-10 can also inhibit MHC class II and costimulatory adhesion molecule expression, resulting in the suppression of antigen specific T-cell proliferation. Given these properties, it is not surprising that IL-10 has been shown to be exploited by a large range of intracellular pathogens as a means of suppressing the immune response (24). Some pathogens induce IL-10 production during infection, while others encode their own IL-10 homologs. The genomes of several herpesviruses, including Epstein-Barr virus (7, 19), equine herpesvirus 2 (25), human CMV (12, 15), and the rhesus macaque, baboon, and African green monkey CMVs (15) have regions with sequence homology to human IL-10. Several of these herpesviruses have been further shown to express a viral IL-10 homolog, although these studies have been restricted to the productive phase of infection in permissive cells. This includes the viral IL-10 homolog encoded by human CMV, designated cmvIL-10, which is secreted by fibroblasts during productive infection (12) and has been subsequently demonstrated to function in a manner similar to human IL-10 (30).
In our present assessment of viral gene expression during latency, the 139-amino-acid ORF was the only ORF identified on UL111.5A region CLTs. The putative protein product, designated LA-cmvIL-10, is predicted to share the same initiation methionine as the cmvIL-10 expressed during productive CMV infection. In addition, like the processed UL111.5A transcript expressed during productive infection, splicing of the UL111.5A region CLT (intron 1) maintains an ORF prior to the identified stop codon of UL111A. However, unlike the UL111.5A transcript, the UL111.5A region CLT that we mapped does not contain a second intron, resulting in an in-frame stop codon at nucleotide position 160171. Thus, both LA-cmvIL-10 and cmvIL-10 are predicted to be colinear for the first 127 amino acids, with divergent sequences for the remaining C-terminal portions. The UL111.5A region latency-associated transcript that we cloned, sequenced, and mapped contains a single intron, but an unspliced transcript with as-yet-undefined ends was also identified, and it remains possible that other variants of this transcript may also be expressed during CMV latency. In this respect, preliminary data suggest that alternately spliced variants may be present in experimentally infected GM-Ps. Further characterization of transcription across the UL111.5A region is therefore a major goal of our future studies.
We have recently demonstrated that latent CMV infection of myeloid progenitor cells results in a specific reduction of cell surface expression of MHC class II proteins (27). The ability of CMV to modulate MHC class II expression during the latent phase of infection represents a strategy that would afford the virus escape from immunosurveillance and increase the chances for lifelong survival, but the viral gene product responsible for this phenotype has not yet been defined. Although speculative at this stage, our identification of a latency-associated viral transcript encoding a putative IL-10 homolog raises the intriguing possibility that LA-cmvIL-10 may function to downregulate MHC class II on latently infected cells. Additional studies will be required to define any role of LA-cmvIL-10 in immune modulation during CMV latency.
GAPDH transcripts were readily detected by RT-PCR in samples extracted from all BM and MPB donors, indicating the presence of amplifiable RNA. The inability to detect UL111.5A region CLTs in cells from all seropositive donors probably reflects a frequency of transcript expression below the limits of sensitivity of our RT-PCR assay. We were unable to detect previously identified sense MIE CLTs in any donor sample (data not shown), a finding also probably due to the relative sensitivity of our RT-PCR assay. The detection of UL111.5A region transcripts in a seronegative donor is consistent with previous reports that have found that seronegative donors are frequently CMV DNA positive by PCR (14, 26, 28, 29).
The GM-P model of latency consistently results in >90% of cells harboring viral genomes (26). Our demonstration that between 1 and 12% of GM-Ps from latently infected cultures expressed detectable UL111.5A region CLTs indicates a dissociation between transcription from this region and the presence of the viral genome, suggesting that latency may proceed in some cells that fail to express these transcripts. This finding is consistent with previous analyses which showed that MIE CLTs are expressed in approximately 2% of latently infected GM-Ps (9, 26). It remains to be determined whether MIE CLTs and UL111.5A region CLTs are coexpressed in the same cells or whether they represent different populations of latently infected GM-Ps with potentially different biological functions.
In summary, we report the first detection of gene expression from the UL111.5A region of the CMV genome during both experimental and natural latent infection. During latency, the virus expresses a novel, singly spliced transcript which is predicted to encode a protein (LA-cmvIL-10) with homology to human IL-10. Although it remains to be determined, expression of LA-cmvIL-10 during the latent phase of infection may contribute to the success of CMV as a human pathogen by suppressing the host's ability to successfully mount an immune response against the virus in its latent form. The development of therapies to inhibit the ability of CMV to subvert the immune response during latency may ultimately lessen its ability to cause disease in allograft recipients.
Acknowledgments
This study was supported by NHMRC grant 153873 (to B.S.). C.J. was the holder of an Australian Postgraduate Award and a Westmead Millennium Foundation Stipend Enhancement Award.
REFERENCES
- 1.Beisser, P. S., L. Laurent, J. L. Virelizier, and S. Michelson. 2001. Human cytomegalovirus chemokine receptor gene US28 is transcribed in latently infected THP-1 monocytes. J. Virol. 75:5949-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, and J. A. Martignetti. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 3.Chevalier, M. S., G. M. Daniels, and D. C. Johnson. 2002. Binding of human cytomegalovirus US2 to major histocompatibility complex class I and II proteins is not sufficient for their degradation. J. Virol. 76:8265-8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodrum, F. D., C. T. Jordan, K. High, and T. Shenk. 2002. Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: a model for latency. Proc. Natl. Acad. Sci. USA 99:16255-16260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hahn, G., R. Jores, and E. S. Mocarski. 1998. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl. Acad. Sci. USA 95:3937-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hegde, N. R., R. A. Tomazin, T. W. Wisner, C. Dunn, J. M. Boname, D. M. Lewinsohn, and D. C. Johnson. 2002. Inhibition of HLA-DR assembly, transport, and loading by human cytomegalovirus glycoprotein US3: a novel mechanism for evading major histocompatibility complex class II antigen presentation. J. Virol. 76:10929-10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu, D. H., R. de Waal Malefyt, D. F. Fiorentino, M. N. Dang, P. Vieira, J. de Vries, H. Spits, T. R. Mosmann, and K. W. Moore. 1990. Expression of interleukin-10 activity by Epstein-Barr virus protein BCRF1. Science 250:830-832. [DOI] [PubMed] [Google Scholar]
- 8.Just, M., A. Buergin-Wolff, G. Emoedi, and R. Hernandez. 1975. Immunization trials with live attenuated cytomegalovirus TOWNE 125. Infection 3:111-114. [DOI] [PubMed] [Google Scholar]
- 9.Kondo, K., H. Kaneshima, and E. S. Mocarski. 1994. Human cytomegalovirus latent infection of granulocyte-macrophage progenitors. Proc. Natl. Acad. Sci. USA 91:11879-11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kondo, K., and E. S. Mocarski. 1995. Cytomegalovirus latency and latency-specific transcription in hematopoietic progenitors. Scand. J. Infect. Dis. Suppl. 99:63-67. [PubMed] [Google Scholar]
- 11.Kondo, K., J. Xu, and E. S. Mocarski. 1996. Human cytomegalovirus latent gene expression in granulocyte-macrophage progenitors in culture and in seropositive individuals. Proc. Natl. Acad. Sci. USA 93:11137-11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotenko, S. V., S. Saccani, L. S. Izotova, O. V. Mirochnitchenko, and S. Pestka. 2000. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc. Natl. Acad. Sci. USA 97:1695-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Landini, M. P., T. Lazzarotto, J. Xu, A. P. Geballe, and E. S. Mocarski. 2000. Humoral immune response to proteins of human cytomegalovirus latency-associated transcripts. Biol. Blood Marrow Transplant. 6:100-108. [DOI] [PubMed] [Google Scholar]
- 14.Larsson, S., C. Soderberg-Naucler, F. Z. Wang, and E. Moller. 1998. Cytomegalovirus DNA can be detected in peripheral blood mononuclear cells from all seropositive and most seronegative healthy blood donors over time. Transfusion 38:271-278. [DOI] [PubMed] [Google Scholar]
- 15.Lockridge, K. M., S. S. Zhou, R. H. Kravitz, J. L. Johnson, E. T. Sawai, E. L. Blewett, and P. A. Barry. 2000. Primate cytomegaloviruses encode and express an IL-10-like protein. Virology 268:272-280. [DOI] [PubMed] [Google Scholar]
- 16.Mendelson, M., S. Monard, P. Sissons, and J. Sinclair. 1996. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J. Gen. Virol. 77:3099-3102. [DOI] [PubMed] [Google Scholar]
- 17.Mocarski, E., C. T. Courcelle. 2001. Cytomegaloviruses and their replication, p. 2629-2673. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 18.Moore, K. W., R. de Waal Malefyt, R. L. Coffman, and A. O'Garra. 2001. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 19:683-765. [DOI] [PubMed] [Google Scholar]
- 19.Moore, K. W., P. Vieira, D. F. Fiorentino, M. L. Trounstine, T. A. Khan, and T. R. Mosmann. 1990. Homology of cytokine synthesis inhibitory factor (IL-10) to the Epstein-Barr virus gene BCRFI. Science 248:1230-1234. (Erratum, 250:494.) [DOI] [PubMed] [Google Scholar]
- 20.Pass, R. 2001. Cytomegalovirus, p. 2675-2705. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 21.Plotkin, S. A., T. Furukawa, N. Zygraich, and C. Huygelen. 1975. Candidate cytomegalovirus strain for human vaccination. Infect. Immun. 12:521-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quinnan, G. V., Jr., M. Delery, A. H. Rook, W. R. Frederick, J. S. Epstein, J. F. Manischewitz, L. Jackson, K. M. Ramsey, K. Mittal, and S. A. Plotkin. 1984. Comparative virulence and immunogenicity of the Towne strain and a nonattenuated strain of cytomegalovirus. Ann. Intern. Med. 101:478-483. [DOI] [PubMed] [Google Scholar]
- 23.Rawlinson, W. D., and B. G. Barrell. 1993. Spliced transcripts of human cytomegalovirus. J. Virol. 67:5502-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redpath, S., P. Ghazal, and N. R. Gascoigne. 2001. Hijacking and exploitation of IL-10 by intracellular pathogens. Trends Microbiol. 9:86-92. [DOI] [PubMed] [Google Scholar]
- 25.Rode, H. J., W. Janssen, A. Rosen-Wolff, J. J. Bugert, P. Thein, Y. Becker, and G. Darai. 1993. The genome of equine herpesvirus type 2 harbors an interleukin 10 (IL10)-like gene. Virus Genes 7:111-116. [DOI] [PubMed] [Google Scholar]
- 26.Slobedman, B., and E. S. Mocarski. 1999. Quantitative analysis of latent human cytomegalovirus. J. Virol. 73:4806-4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slobedman, B., E. S. Mocarski, A. M. Arvin, E. D. Mellins, and A. Abendroth. 2002. Latent cytomegalovirus down-regulates major histocompatibility complex class II expression on myeloid progenitors. Blood 100:2867-2873. [DOI] [PubMed] [Google Scholar]
- 28.Smith, K. L., J. K. Kulski, T. Cobain, and R. A. Dunstan. 1993. Detection of cytomegalovirus in blood donors by the polymerase chain reaction. Transfusion 33:497-503. [DOI] [PubMed] [Google Scholar]
- 29.Soderberg-Naucler, C., K. N. Fish, and J. A. Nelson. 1997. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell 91:119-126. [DOI] [PubMed] [Google Scholar]
- 30.Spencer, J. V., K. M. Lockridge, P. A. Barry, G. Lin, M. Tsang, M. E. Penfold, and T. J. Schall. 2002. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J. Virol. 76:1285-1292. (Erratum, 76:3585.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanier, P., D. L. Taylor, A. D. Kitchen, N. Wales, Y. Tryhorn, and A. S. Tyms. 1989. Persistence of cytomegalovirus in mononuclear cells in peripheral blood from blood donors. BMJ 299:897-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor-Wiedeman, J., J. G. Sissons, L. K. Borysiewicz, and J. H. Sinclair. 1991. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J. Gen. Virol. 72:2059-2064. [DOI] [PubMed] [Google Scholar]
- 33.Tomazin, R., J. Boname, N. R. Hegde, D. M. Lewinsohn, Y. Altschuler, T. R. Jones, P. Cresswell, J. A. Nelson, S. R. Riddell, and D. C. Johnson. 1999. Cytomegalovirus US2 destroys two components of the MHC class II pathway, preventing recognition by CD4+ T cells. Nat. Med. 5:1039-1043. [DOI] [PubMed] [Google Scholar]
- 34.White, K. L., B. Slobedman, and E. S. Mocarski. 2000. Human cytomegalovirus latency-associated protein pORF94 is dispensable for productive and latent infection. J. Virol. 74:9333-9337. [DOI] [PMC free article] [PubMed] [Google Scholar]