Abstract
In the last 20 years, it has become clear that developmental genes and their regulators, noncoding RNAs including microRNAs and long-noncoding RNAs, within signaling pathways play a critical role in the pathogenesis of cancer. Many of these pathways were first identified in genetic screens in Drosophila and other lower organisms. Mammalian orthologs were subsequently identified and genes within the pathways cloned and found to regulate cell growth. The GENES and pathways expressed during embryonic development, including the Notch, Wnt/β-Catenin, TGF-β/BMP, Shh/Patched, and Hippo pathways are mutated, lost, or aberrantly regulated in a wide variety of human cancers, including skin, breast, blood, and brain cancers, including medulloblastoma. These biochemical pathways affect cell fate determination, axis formation, and patterning during development and regulate tissue homeostasis and regeneration in adults.
Medulloblastoma, the most common malignant nervous system tumor in childhood, are thought to arise from disruptions in cerebellar development [reviewed by Marino, S. (2005). Medulloblastoma: Developmental mechanisms out of control. Trends Mol. Med. 11, 17–22]. Defining the extracellular cues and intracellular signaling pathways that control cerebellar neurogenesis, especially granule cell progenitor (GCP) proliferation and differentiation has been useful for developing models to unravel the mechanisms underlying medulloblastoma formation and growth. In this chapter, we will review the development of the cerebellar cortex, highlighting signaling pathways of potential relevance to tumorigenesis.
1. Introduction
In their classical treatise on brain tumors, Bailey and Cushing wrote, “the histogenesis of the brain furnishes the indispensable background for an understanding of its tumors” (Bailey and Cushing, 1926). The idea that tumors form from specific populations of immature neurons suggests that common mechanisms underlie development and tumor formation. In the developing cerebellum, precursors of the granule cell are thought to give rise to medulloblastomas (Bailey and Cushing, 1925) the most common childhood primary CNS tumor (Packer et al.., 1999). Medulloblastomas arise in the cerebellar vermis and spread rapidly through the cerebrospinal pathways, where they form tumors of variable size along the ventricles (Packer et al., 1999). Medulloblastomas are a diverse set of tumors as evidenced by several criteria including differing histopathologies (Louis et al., 2007). The most aggressive forms of the disease occur in infants and young children and long-term survivors are at significant risk of cognitive and psychological deficits (Levisohn et al., 2000) due to the effects of current treatment protocols that include resection, irradiation, and chemotherapy.
2. Cerebellar Development
The cerebellar cortex is a remarkably simple laminar structure, with two principal neurons, the granule cell and the Purkinje cell, and a diverse set of interneurons, which modulate the output of the Purkinje cell to the cerebellar nuclei (Palay and Chan-Palay, 1974). The most prevalent neuronal subclass within the cerebellum, indeed within the entire mammalian central nervous system, is the cerebellar granule neuron. Granule neurons serve an essential role in coordinating afferent input to and motor output from the cerebellum through their excitatory connections with Purkinje neurons (Ito, 2006; Fig. 8.1). While the role of the cerebellum in sensorimotor functions, balance control, and the vestibular ocular reflex have long been appreciated (Ito, 2006), recent studies have revealed a role for the cerebellum in a wide range of cognitive functions, including feed-forward sensory-motor learning, speech, and spatial memory (Boyden et al., 2004; De Zeeuw and Yeo, 2005; du Lac et al., 1995; Fiez, 1996; Fiez and Petersen, 1998; Schmahmann and Caplan, 2006; Timmann et al., 1999). Notably, a loss of spatial memory and other cognitive functions have been reported in children after successful tumor resection (Levisohn et al., 2000). Although the function of the cerebellum in learning and memory is complex, the remarkably simple architectonics of the cerebellum make it an attractive model system for studying CNS tumors, especially developmental tumors such as medulloblastoma.
Figure 8.1.
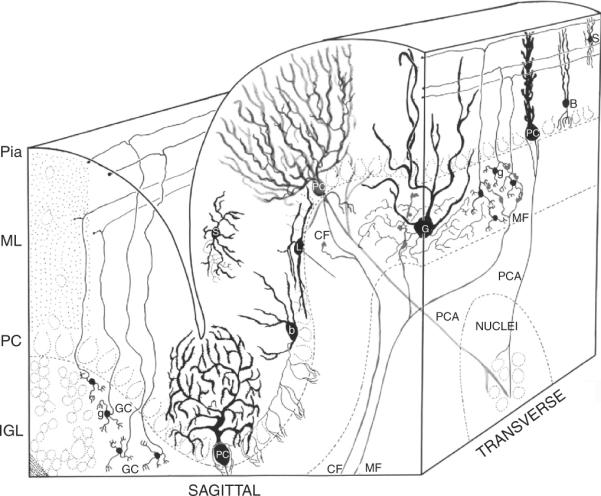
Neurons of the cerebellar cortex. Four granule cells (GC) are shown at the lower left of the sagittal view of the cerebellar cortex. Short, claw-like dendrites extend from the granule cell soma, which projects an ascending axon that bifurcates in a T. The resultant branches extend through the molecular layer (ML) parallel to the long axis of the folium in the transverse plane and form synaptic connections with the Purkinje cell (PC). The dendritic arbors of the PC extend in the sagittal plane. Mossy fiber (MF) afferents from the pontine nucleus and other sources synapse with the dendrites of the granule cells (in the transverse plane). The axons of the Golgi (G) interneuron, the mossy fiber axons, and granule cell dendrites form the synaptic glomeruli of the internal granule cell layer (IGL). PCs align with the cell soma of Bergmann glia (B) in the Purkinje cell layer (PC), as well as several classes of cerebellar interneurons, including basket (b), Lugaro (L), and candelabra (not shown) neurons. Stellate interneurons (S) are located among the parallel fiber axons and Purkinje cell dendrites in the molecular layer (ML). Afferent axons from the olivary nucleus form single climbing fiber projections (CF) onto the Purkinje neurons. Purkinje neurons, the sole output neuron of the cerebellar cortex, project axons (PCA) to the neurons of the cerebellar nuclei (Nuclei). Modified from Palay and Chan-Palay (1974).
2.1. Embryonic cerebellar development: Neurogenesis and patterning in the cerebellar anlagen
Fate mapping and transplantation studies indicate that the cerebellar territory arises from rhombomere 1, delineated by the expression of Hoxa2 (posterior) and Otx2 (anterior) (Wingate and Hatten, 1999). Secreted proteins of the Wnt (McMahon and Bradley, 1990) and fibroblast growth factor (FGF) families (Chi et al., 2003; Crossley et al., 1996; Martinez et al., 1999), including Wnt1, Fgf8, and Fgf15, control the expression of transcription factors that delineate rhombomere 1 (Otx2 and Gbx1; Joyner et al., 2000) and of genes required to establish the cerebellar territory, including the homeobox genes En1 and Pax2/5/8 (Joyner, 1996; Joyner et al., 1989)
The cerebellum, like the cerebrum, consists of an outer cortical structure and set of subcortical nuclei, the cerebellar nuclei, which project to cerebellar targets (Fig. 8.2). During cerebellar histogenesis, a complex pattern of neurogenesis and cell movements generates the cerebellar cortex and cerebellar nuclei (Hatten and Heintz, 1995; Morales and Hatten, 2006). Classical studies indicate that the dorsomedial ventricular zone (VZ) along the fourth ventricle gives rise to the principal output neuron of the cerebellar cortex, the Purkinje cell, neurons of the cerebellar nuclei, and more than half a dozen types of cerebellar interneurons, including Golgi, basket, and stellate cells (Dino et al., 2000; Laine and Axelrad, 2002; Palay and Chan-Palay, 1974). A secondary germinal zone forms along the anterior aspect of the rhombic lip, which generates the cerebellar granule neuron as well as a subpopulation of neurons of the cerebellar nuclei (Fink et al., 2006; Wang et al., 2005) and neurons of several precerebellar nuclei of the “cerebellar system” (Dymecki and Tomasiewicz, 1998; Machold and Fishell, 2005; Wang et al., 2005; Wingate, 2001; Wingate and Hatten, 1999).
Figure 8.2.
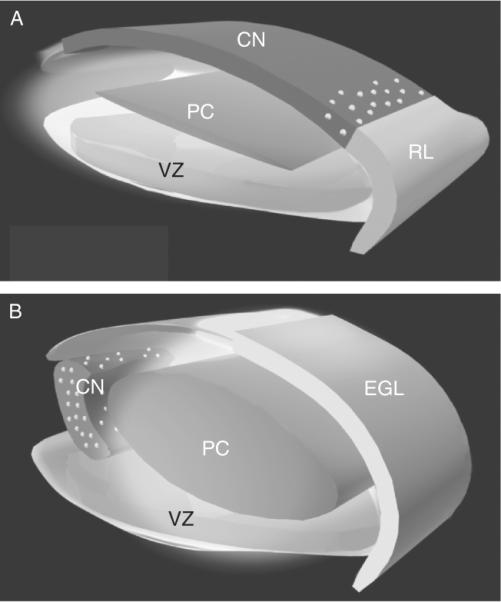
A model for the generation and migratory pathways of progenitors of the cerebellar cortex and the cerebellar nuclei. In the mouse, progenitors of the cerebellar nuclei (CNP) are generated in the VZ of the emerging cerebellar anlagen. Between E10 and E12.5, this progenitor population migrates radially along the glial fiber system (data not shown) to establish a superficial layer (dark gray) Subsequently, progenitors of the Purkinje cell (PCP) are generated in the VZ (E11–E14.5) and migrate radially to form a zone (PC, medium gray) beneath the CNPs. A second germinal zone, the anterior rhombic lip (RL, light gray), generates a subpopulation of precursors of neurons of the cerebellar nuclei, precerebellar nuclei (data not shown), and the granule neurons of the cerebellar cortex. At E12.5, a layer of CNPs (dark gray) generated in the VZ occupy the surface of the embryonic cerebellum. Between E12.5 and E14.5, several populations of RL progenitors move up onto the surface of the anlagen (RL, light gray). These include the bulk of the progenitor population, the precursors of the granule neuron (light gray), and a subpopulation of cells that intercalate among the VZ-derived CNPs (gray zone with white dots). By E14.5, the CNPs (containing both VZ and RL derived progenitors) have migrated off of the surface and formed the nascent cerebellar nuclei. By E16, the cerebellar nuclei (gray zone with white dots) settle beneath the emerging cerebellar cortex (containing the precursors of the granule neurons (EGL, light gray)), the Purkinje neuron (PC, medium gray zone), and the interneurons. Thus, a complex pattern of histogenesis and migration set forth the architecture of the murine cerebellum. From Morales and Hatten (2006).
In the mouse, the earliest cerebellar progenitors exit the cell cycle in the VZ at approximately embryonic (E) day E10.25 and generate neurons of the cerebellar nuclei (Fig. 8.3). Studies on transcription factor expression during cerebellar histogenesis show that postmitotic precursors of neurons of the cerebellar nuclei express the transcription factors Lhx2/9, Meis 1/2, and Irx3 (Morales and Hatten, 2006). By E11.2, this pool of progenitors migrates radially along the emerging glial fiber system to form a superficial zone across the dorsal surface of the cerebellar anlagen. Between E11 and E14, postmitotic precursors of the Purkinje neuron, identified by expression of the LIM transcription factors LHX1 and LHX5, migrate away from the VZ along radial glial fibers and assemble into a broad zone in the core of the anlagen (Morales and Hatten, 2006). Recent genetic loss of function studies and fate mapping analyses demonstrate that Ptf1a expression is required to generate the progenitors of cerebellar GABAergic neurons (Purkinje cells and interneurons) in the cerebellar ventricular zone (Hoshino et al., 2005; Pascual et al., 2007). In humans, PTF1A mutations are associated with cerebellar agenesis (Aldinger and Elsen, 2008).
Figure 8.3.
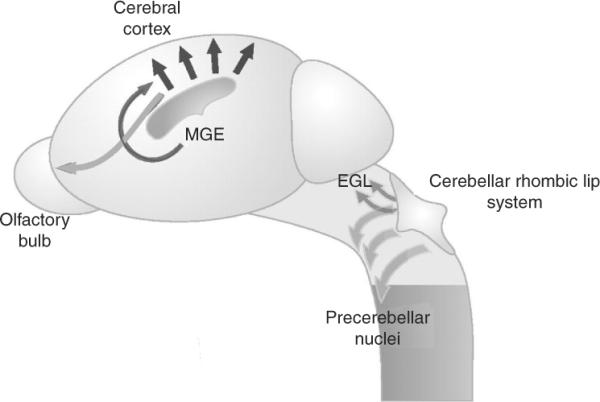
Transverse migratory pathways from the rhombic lip of the cerbellar territory establish the precerebellar nuclei in the brainstem. Progenitors of the cerebellum arise along the rhombic lip at the midbrain/hindbrain junction and migrate dorsally, (dark gray arrows). Progenitors of the precerebellar nuclei in the brainstem arise from more caudal portions of the rhombic lip and migrate ventrally along a netrin1 pathway (light gray arrows). In the forebrain, transverse neuronal migrations include dorsal migration of progenitors from the medial ganglionic eminence (MGE) into the cortex (dark gray arrow), and the posterior to anterior migration of progenitors in the rostral migratory stream (light gray arrow). Radial migrations in the cerebral cortex are also illustrated (black arrows). Adapted from Hatten (2002).
2.2. The rhombic lip, a secondary germinal zone
By E12.5, a secondary neurogenic zone appears along the anterior aspect of the rhombic lip. Cells in this zone express the basic helix-loop-helix (bHLH) transcription factor Atoh1/Math1, (Alder et al., 1996; Machold and Fishell, 2005; Wang et al., 2005) the zinc finger protein Zic1 (Aruga, 2004), and Meis1 as well as the markers Pde1c, and Pcsk9 (Morales and Hatten, 2006). At this stage, proliferating pools of progenitors migrate out of the rhombic lip and spread across the surface of the cerebellar anlagen to form the external granule layer (EGL). As this morphogenetic out of the rhombic lip begins, postmitotic precursors of the cerebellar nuclei along the surface of the anlagen migrate toward the rostral aspect of the anlage (E12.5–E15.5) and inward to a position beneath the emerging zone of Purkinje cell precursors to form the cerebellar nuclei. Fate mapping experiments confirm that a subpopulation of Math1-positive progenitors in the rhombic lip migrate with the pool of cerebellar nuclei progenitors into the deeper aspect of the anlagen as a subpopulation of the neurons of the cerebellar nuclei (Fink et al., 2006; Morales and Hatten, 2006; Wang et al., 2005). The anterior rhombic lip also generates neuronal precursors that migrate ventrally where they form the lateral pontine nucleus and cochlear nucleus, hindbrain nuclei of the “cerebellar system” (Dymecki and Tomasiewicz, 1998; Machold and Fishell, 2005; Morales and Hatten, 2006; Wang et al., 2005; Wingate, 2001; Wingate and Hatten, 1999).
The vast majority of rhombic lip derivatives migrate onto the dorsal surface where they form the EGL (Miale and Sidman, 1961; Palay and Chan-Palay, 1974; Ramon y Cajal, 1995), a zone of proliferating GCPs that generates the cerebellar granule cell (Alder et al., 1996; Alder et al., 1999), the most abundant neuron in the brain (Fig. 8.4). In man, some 45 billion granule neurons are produced during cerebellar development, out of approximately 110 billion neurons in the human brain. Molecular genetic studies demonstrate that Math1 (Ben-Arie et al., 1997) and MycN (Knoepfler et al., 2002) are required for granule cell specification and the expansion of the pool of granule cerebellar progenitors (GCPs) in the early postnatal period of development. Studies on the transcriptional regulation of Math1, by Johnson and colleagues show that ZIC1 binds a conserved site within the sequence of the Math1 enhancer region, and represses Math1 transcription by blocking the autoregulatory activity of Math1 (Ebert et al., 2003; Fig. 8.4).
Figure 8.4.
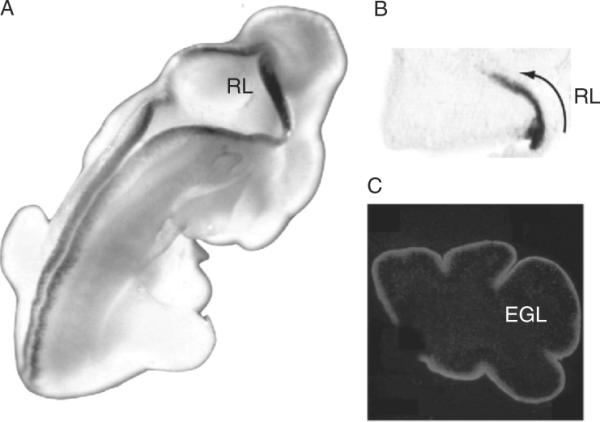
Math1 Expression in the developing cerebellum. Math1 is expressed in the rhombic lip (RL) of the cerebellar territory and the dorsal ridge of the developing CNS in the E12.5 mouse embryo (A). At E14.5 Math1 is expressed in RL progenitors forming the EGL of the developing anlagen (B). In the postnatal cerebellar cortex, Math1 is expressed in proliferating GCPs located in the superficial aspect of the EGL. Photos in panels (A) and (C) were kindly provided by Dr. Jane Johnson.
The specification of granule cell identity also depends on extracellular signals that dorsalize the neural tube, including the BMP family members GDF, Bmp6, and Bmp7 (Hogan, 1996). To analyze the role of BMPs in granule cell specification, we examined the pattern of expression of genes encoding BMP family members in the anterior rhombic lip and adjacent tissues (Alder et al., 1999; Fig. 8.5). These experiments showed that Bmp7 was expressed in the anterior rhombic lip and that Bmp6 and Gdf7 were expressed in cells of the dorsal neuroepithelium adjacent to the rhombic lip (Alder et al., 1999). Treatment of progenitors from the ventral region of the mes/met region where the cerebellar territory forms with Bmp7 induced the expression of both En1 and Math1. In addition, transplantation studies showed the generation of GCPs from neural tube explants treated with BMP7. These studies support the hypothesis that in the developing cerebellum, BMP signaling pathways play a critical role in GCP differentiation (Alder et al., 1999). This view is consistent with recent genetic analyses showing that mice with a targeted deletion of the BMP type 1 receptor genes Bmpr1a and Bmpr1b have a severe loss of granule neurons, resulting in a smaller cerebellar cortex without foliation (Qin et al., 2006).
Figure 8.5.
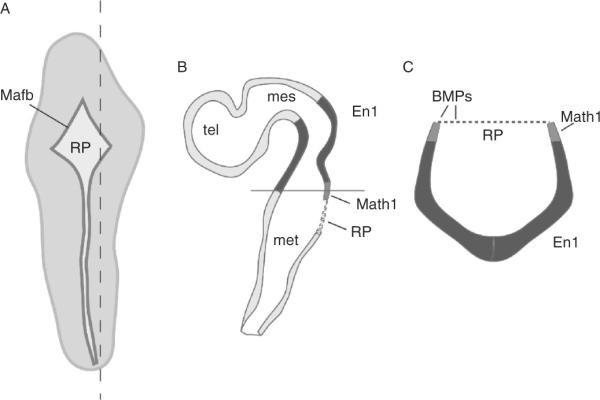
Expression of Bmps in the embryonic cerebellar anlagen. Schematic diagram of an E11 mouse embryo (A), sagittal section (B) through level indicated by dashed line in (A), and a transverse section (C) at the level indicated by the black line in (B). In (A), the roof plate (RP), shown in dark gray, is revealed by expression of the roof plate marker Mafb (Millonig et al., 2000), across the rautenbreite and the dorsal ridge of the embryo. In (B) and (C) En1/2 expression, which marks the cerebellar territory, is shown in black, and the RP is represented as a dotted line. Gdf7, Bmp6, and Bmp7 are expressed along the dorsal edges of the neural tube and in the overlying RP. The only region of overlapping expression between En1/2 (black) and Math1 (gray) is in the rhombic lip, which is adjacent to the RP (for details see Alder et al., 1999). mes, mesencephalon; met, metencephalon; tel, telencephalon.
Other studies, employing both gain of function and loss of function genetic approaches in mice, provide evidence that Notch1 activation increases Math1 expression by antagonizing BMP receptor signaling (Machold et al., 2007). These studies further suggest that the level of Notch1 activity in cerebellar rhombic lip progenitors regulates their fate. These and other studies that have identified changing patterns of gene expression during GCP development (Hatten and Heintz, 1995; Kuhar et al., 1993) that include cell cycle regulators (Cyclins D1,D2/Ccnd1,2), genes required for glial-guided migration of GCPs (Zheng et al., 1996), the GABAβ6 receptor, and the Lynx family of proteins involved in modulating the activity of a subset of parallel fiber-Purkinje cell circuits (Ibanez-Tallon et al., 2002; Miwa et al., 1999, 2006).
2.3. Cerebellar radial glia
Although radial glia are required for the migration of several populations of neuronal progenitors, including Purkinje cell progenitors, in embryonic cerebellar histogenesis (Morales and Hatten, 2006) and Bergmann glia guide the migration of postmitotic GCPs in postnatal cerebellar development (Edmondson and Hatten, 1987; Hatten, 2002; Rakic, 1971), relatively little attention has been given to the role of neuron–glial interactions in cerebellar neurogenesis (Fig. 8.6). Early studies on neuron–glia interactions in cultured GCPs and in cultured glioblastoma cells showed that neurons regulate the proliferation of cerebellar glial cells and of human glioblastoma cell lines (Hatten et al., 1984). Molecular genetic studies on transgenic lines of mice expressing the radial glial gene Blbp (Anthony et al., 2004; Feng et al., 1994) indicate that radial glia are neurogenic in all brain regions, including the cerebellum (Anthony et al., 2004). Biochemical and molecular genetic studies by Heintz and colleagues further demonstrate that Blbp is a target of Notch signaling (Anthony et al., 2005). Recent genetic studies on mice lacking MycN, enhanced proliferation of GCPs and impaired differentiation of cerebellar glial cells (D'Arca et al., 2010). These studies raise the question as to whether neuron–glial interactions regulate cerebellar neurogenesis, especially granule cell neurogenesis, as well as the formation and growth of medulloblastoma and glioblastoma.
Figure 8.6.
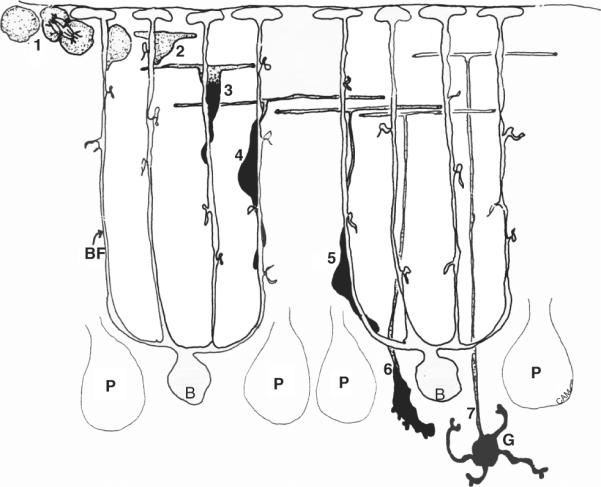
GCP migration and development. GCPs divide in the superficial layer of the neonatal cerebellar cortex (1), after which postmitotic GCPs begin to extend axons (2) and a descending, leading process (black) (3) which migrates along the radial fibers of Bergmann glia (B) by closely opposing the cell soma to the glial fiber (4,5). At the level of the Purkinje neurons (P) GCPs detach from the glial fiber (6) and move into a deeper layer where they differentiate into mature granule neurons (G) with short, claw-like dendrites that form connections with mossy fiber afferents (not shown). Drawing kindly provided by Dr. Carol A. Mason.
2.4. Postnatal cerebellar development: GCP proliferation and migration
After birth, between the second and fourth days postnatal (P2–P4), a number of signaling pathways promote GCP proliferation (Wechsler-Reya and Scott, 1999). At peak periods of proliferation (P5–8) more than a million GCPs can be isolated from a single mouse, providing an unparalleled resource for studies of primary neural progenitor populations (Hatten, 1985). Between birth and the end of the second postnatal week, GCPs exit the cell cycle and move into the inner regions of the EGL where they extend parallel fibers and migrate along the radial fibers of Bergmann glial cells (Edmondson and Hatten, 1987; Rakic, 1971) to a position beneath the Purkinje cells, where they form the inner granule layer (IGL). The spatiotemporal dynamics of GCP proliferation in the superficial zone of the EGL, cell cycle exit, formation of parallel fibers and migration into the IGL are potentially relevant to tumorigenesis. Classical [3H]-thymidine labeling studies by Fujita (Fujita, 1967; Fujita et al., 1966) show temporal regulation of the step-wise progression of GCPs through phases of rapid proliferation to cell cycle exit and differentiation. In the mouse, the transit time of postmitotic GCPs across the EGL and the molecular layer (ML) is 21 and 4 h, respectively. As GCPs exit the cell cycle, they down regulate expression of the bHLH gene Atoh1/Math1 (Ben-Arie et al., 1996; Helms et al., 2000) and upregulate expression of NeuroD1 (Lee et al., 2000; Pan et al., 2009), Zic1,3 (Aruga, 2004; Aruga et al., 1996), and the tumor suppressor cyclin-dependent kinase inhibitory protein p27Kip1 (Ayrault et al., 2009). During their movement from the mantle of the EGL into the deeper layers of the EGL, GCPs extend long, parallel fiber axons that express the GPI-linked axonal glycoprotein TAG1 (Furley et al., 1990), among others. Recent studies by Luo et al. (Espinosa and Luo, 2008), discussed below, suggest that clones of GCPs stack their parallel fibers in a chronological order that relates to the timing of their penultimate cell division. The importance of correlated timing of axon outgrowth and gene expression is underscored by the anatomical studies of Rivas, who demonstrated TAG1 immunoreactive of parallel fibers peaks during the first 3 days after the GCPs become postmitotic. TAG1 levels decrease dramatically with the differentiation of Purkinje into the ML and formation of parallel fiber synapses with Purkinje cell dendrites (Stottmann and Rivas, 1998).
The timing of GCP migration away from the EGL is another important determinant of cerebellar patterning. GCPs first begin to exit the cell cycle and migrate across the ML into the IGL in the late embryonic period. Several classes of genes that modify the timing of glial-guided GCP migration out of the EGL have potential importance to medulloblastoma. First, in mice lacking the Gi Protein-coupled chemokine CXCR4, which is broadly expressed in both the CNS and immune system, GCPs migrate along Bergmann glial fibers prematurely, with large numbers of cells pouring down the Bergmann glia in the late embryonic period. The premature migration of GCPs from the EGL into the cerebellar anlagen, prevents the massive expansion of GCPs that normally occurs in the postnatal period. Several studies indicate a potential relevance of CXCR4 to medulloblastoma, as blocking CXCR4-mediated cyclin AMP suppression inhibits tumor growth (Yang et al., 2007). Further support for the idea that CXCR4 is critical for the progression of brain tumors comes from studies showing that small molecule inhibitors of CXCR4 inhibit intracranial tumor growth (Rubin et al., 2003).
Molecular genetic studies on genes involved in glial-guided migration, including Astn1 and Pex5, raise the possibility that slowed migration or altered patterns of GCP survival and differentiation cause ectopic zones of GCPs that fail to migrate into the IGL. In the case of mice lacking Astn1 or Pex5, GCP migration along Bergmann glial fibers is slowed markedly, causing the dendritic arbors of young Purkinje neurons to tilt out of the sagittal plane (Adams et al., 2002; Faust, 2003; Faust and Hatten, 1997). In addition, the slowed migration results in ectopic zones of GCPs in the EGL that persist into late childhood, a time by which the EGL has normally dissipated. Since Purkinje neurons provide the mitogen sonic hedgehog (SHH) for GCP expansion in the EGL, it is possible that the growth rates of GCPs depend on the spatiotemporal dynamics of GCP and Purkinje neuron maturation and the assembly of the cerebellar circuit. Finally, it is worth noting that Purkinje cells also provide BDNF, a neurotrophin that promotes the stepwise differentiation of postmitotic GCPs, including migration along Bergmann fibers (Borghesani et al., 2002). Indeed, mice lacking BDNF also have defects in cerebellar patterning that include ectopic zones of immature GCPs in the superficial aspect of the cerebellar cortex (Schwartz et al., 1997). All of these perturbations of the normal spatiotemporal program of GCP expansion and differentiation could be relevant to medulloblastoma formation in childhood.
Although a number of studies have demonstrated that the EGL gives rise to a single class of neurons, the granule cells (Alder et al., 1996, 1999; Hallonet et al., 1990; Zhang and Goldman, 1996), recent studies by Luo and colleagues using the mosaic analysis with double markers (MADM) show that GCPs undergo predominantly symmetric divisions during EGL development and that clonally related granule cells exit the cell cycle within a narrow time frame (Espinosa and Luo, 2008). In agreement with the earlier studies by Fujita (Fujita, 1967; Fujita et al., 1966), these studies showed a progressive slowing of GCP proliferation just before birth, and a rapid expansion of clonally related GCPs just prior to cell cycle exit in the postnatal period (Espinosa and Luo, 2008). These studies show that GCP expansion produces distinct clones of granule cells (GCs). Studies on a large number of BAC transgenic lines of mice in the GENSAT Project also provide molecular genetic evidence for subpopulations of GCPs and of cerebellar granule cells (Hatten and Heintz, unpublished; www.gensat.org). These subpopulations of GCPs may be relevant to some of the subtypes of medulloblastomas (Gilbertson and Ellison, 2008).
2.5. Differentiation of ES cells toward a granule cell identity
As a proof of principle of the role of key developmental regulators in granule cell development (Hatten and Heintz, 1995), we treated E14 embryonic stem cells, as well as ES cell lines derived from Tg(Pde1c–Egfp) transgenic mice generated using BAC methodology (Gong et al., 2003), with “cerebellar organizers” (Fgf8, RA), followed by the local signals that “dorsalize” the cerebellar anlagen (Wnt1, Wnt3a, Bmp7, Gdf7, Bmp6), and mitogens that expand the cerebellar granule cell progenitor cell population (Shh, Jag1) (Salero and Hatten, 2007). To achieve terminal differentiation, we cultured differentiated ES cells in medium conditioned by purified cerebellar glial cells. After these treatments, which parallel known steps of GCP differentiation in vivo, treated ES cells expressed multiple markers of granule neurons (En1, Math1, Zic1, Zic2, Pax6, and GABAa6). To test the behavior of these differentiated Tg(Pde1c–Egfp) ES cells into the EGL of the cerebella of living, neonatal mice, using a stereotaxic injection system, and followed the migration and development of EGFP-positive ES cells in vivo. After 3 days to 2 weeks, the cells migrated into the IGL, the position where mature granule neurons function in the cerebellar cortex. Thus, ES cell differentiation confirms the role of local signals in normal cerebellar neuron specification (Salero and Hatten, 2007), and provides an initial strategy for CNS cell replacement therapy.
2.6. Mitogenic pathways that promote GCP proliferation
A number of mitogenic pathways maintain the rapid expansion of the pool of GCPs in the EGL. A role for Shh as a GCP mitogen was suggested by studies of Patched (Ptch), a gene that encodes a Shh-binding protein that functions as an antagonist of Shh signaling (Fig. 8.7). Since mutations in Ptch occur in sporadic human medulloblastoma and promote medulloblastoma in mouse models, Wechsler-Reya and Scott tested the idea that Shh signaling regulates cerebellar growth. Their studies identified Shh as a key mitogen for GCP expansion (Wechsler-Reya and Scott, 1999), and numerous subsequent studies indicate that Shh signaling is defective in ~25% of medulloblastoma (reviewed in Gilbertson and Ellison, 2008). Activation of the SHH signaling pathway leads to increased transcription of the primary downstream mediator of Shh signaling, the zinc finger protein Gli. In the embryonic cerebellar development, Shh signaling regulates midbrain/hindbrain development through positive regulation of the Gli activators (GliA) and inhibition of the Gli3 repressor (Gli3R). The levels of Gli3R control the overall growth of the midbrain/hindbrain territory and play a critical role in sustaining the activity of the anterior posterior (AP) organizer Fgf8 in the isthmus region (Blaess et al., 2006). In the early postnatal period, Shh signaling is a primary driver of the dramatic expansion of the GCP precursor pool. Analysis of the role of Gli family members in medulloblastoma, by inactivation of Gli alleles in Pcth1(+/−) mice suggests that Gli1 both provides a marker of Shh pathway activation and functions in medulloblastoma formation from GCPs (Kimura et al., 2005).
Figure 8.7.
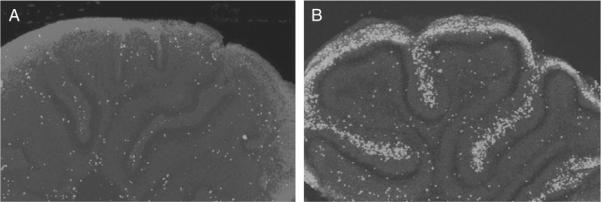
Mitogenic effect of sonic hedgehog (Shh) on the proliferation of granule cell progenitors (GCPs) of the developing cerebellum. The addition of SHH to early postnatal cerebellar cortex (B) results in a large increase in the number of dividing cells, as measured by incorporation of bromodeoxyuridine (BrdU), a deoxythymidine analog (light gray). Adapted from Wechsler-Reya and Scott, (1999).
Shh appears to regulate GCP proliferation by several mechanisms (Fig. 8.8). First, Shh signaling regulates the expression of the cell cycle regulators cyclin D1 (Ccnd1), cyclin D2 (Ccnd2), and cyclin E (Ccne) (Kenney and Rowitch, 2000). During cerebellar development, early post-natal GCPs express cyclin D1 while GCPs generated during the peak of GCP neurogenesis express both Ccnd1 and Ccnd2. Mice lacking Ccnd1 have slowed GCP proliferation and cerebellar development (Pogoriler et al., 2006). Interestingly mice lacking both Ccnd1 and Ptc1 had a reduced incidence of medulloblastoma, suggesting that a loss of Ccnd1 may suppress medulloblastoma formation (Pogoriler et al., 2006). Second, activation of the SHH pathway upregulates the expression of the proto-oncogene MycN that when overexpressed, promotes cell autonomous upregulation of Ccnd1 mRNA and protein independently of Shh signaling (Kenney et al., 2003). Thus, Shh controls cerebellar growth and GCP proliferation by multiple mechanisms.
Figure 8.8.
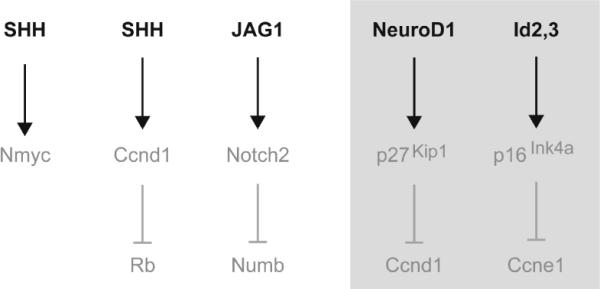
Mitogenic pathway and their inhibitors in GCPs of the developing cerebellum. Schema of the primary mitogens (Shh and JAG1) in the early postnatal cerebellum.
The importance of SHH to cerebellar histogenesis is also underscored by elegant genetic analyses of Joyner and colleagues, who demonstrated that the levels of Shh signaling control the foliation patterning of the cerebellar cortex (Corrales et al., 2006). At present, the regulation of Purkinje cell expression and release of Shh is not as well understood as the mitogenic Shh signaling pathways in GCP neurogenesis. Gene expression studies show that Purkinje neurons are the primary source of Shh in the neonatal cerebellum (Wechsler-Reya and Scott, 1999). The role of the Purkinje neuron in GCP expansion is of particular interest, given the increasing ratio of granule cells to Purkinje cells during evolution (750:1 in the mouse; 3300:1 in humans; Lange, 1975; Fig. 8.9). This suggests the possibility that the PC as the primary output neuron of the cerebellar cortex, regulates the production of interneurons in the cerebellar circuitry (Hatten, 1999) by a feedback loop that involves Shh.
Figure 8.9.
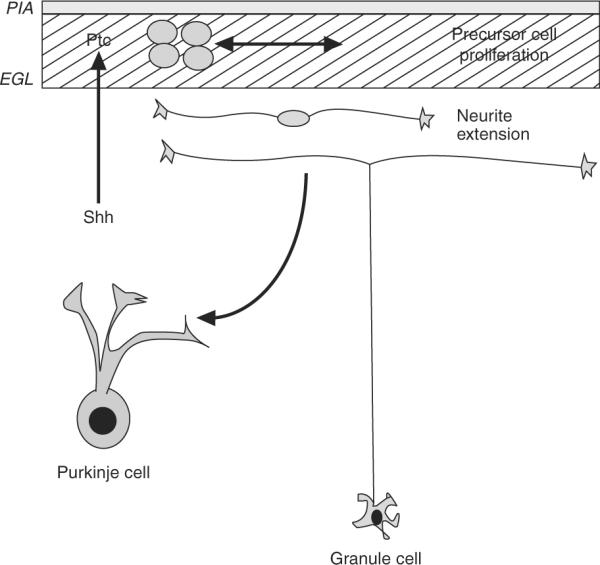
Model for the action of sonic hedgehog in cerebellar development. Sonic hedgehog (Shh) is produced by Purkinje neurons during the developmental stage when they are extending their dendritic tree and beginning to form synaptic connections with the axons of granule neurons. Granule cell precursors bind Shh via the protein Patched (Ptc), which releases Ptc's inhibition of Smoothened and induces a cascade of signals leading to proliferation. Granule cells later exit the cell cycle, extend neurites, and form synaptic connections with Purkinje cells. Thus, Purkinje cells appear to control the number of granule cells entering the cerebellar circuit. Adapted from Hatten (1999).
A number of studies have defined a role for Shh pathway activation in desmoplastic medulloblastoma that represent ~25% of all medulloblastoma in children. Major effort has been spent identifying genes that are specifically expressed in embryonal tumors and medulloblastoma (Pomeroy et al., 2002; Thompson et al., 2006), especially the targets of the Gli1 expressed in medulloblastoma (Yoon et al., 2009). Their studies suggest that Gli1 affects medulloblastoma growth and survival via targets that include p53, SGK1, MGMT, and NTRK2.
Notch2 signaling also stimulates granule cell proliferation (Solecki et al., 2001), as activated Notch2 maintains GCP proliferation and inhibits granule neuron differentiation. Exposure of GCPs to the Notch2 ligand, JAG1, or a constitutively activated form of the Notch2 receptor results in increased (3–5×) cell proliferation. In these cells, expression of the downstream transcription factor, HES1, is upregulated and HES1 overexpression has a similar ability to maintain proliferation in granule cell progenitor populations. HES1 expression is also induced by the SHH pathway, suggesting that it is a common downstream effector of these two pathways (Solecki et al., 2001). This hypothesis is consistent with gene expression studies in medulloblastoma from Ptch1(+/−) mice with elevated Shh signaling, showing increased expression of components of the Notch and Wnt pathways. In addition, genetic studies on mice with reduced Shh signaling report a downregulation of expression of Notch2, Jag1, and Hes1 (Dakubo et al., 2006). Studies on the expression of Notch family members in medulloblastomas and other tumors also report a link between Notch2 activation and tumorigenesis. Immunocytochemical studies indicate expression of Notch2, but not Notch 1,3,4, is detected in medulloblastoma, with increasing numbers of immunopositive cells with tumor (Xu et al., 2009). Direct studies on the effect of expressing truncated, constitutively active forms of Notch1 or Notch2 support the idea that activation of Notch2, but not Notch1, has oncogenic effects on tumor formation. Examination of the levels of Notch2, and its downstream target Hes1, mRNAs in 40 embryonal tumors showed enhanced expression in 15% of the tumors. Taken together, these findings suggest that Notch1 and Notch2 have different effects on GCP proliferation, as well as on human medulloblastoma (Fan et al., 2004).
Cell cycle regulators are also important regulators of normal GCP proliferation. In the postnatal EGL, p18Ink4c is transiently expressed in GCPs during cell cycle exit (Uziel et al., 2005). N-Myc is also required for the rapid expansion of GCP proliferation in the postnatal cerebellum (Knoepfler et al., 2002). Loss of MycN increases expression of two cyclin-dependent kinase inhibitory proteins, p27Kip1 and p18Ink4c in the cerebel lum, and the D-type cyclins dramatically expand the GCP population in the EGL. Studies by Zindy et al. (2006) further demonstrate that MycN expression and downregulation of p18Ink4c and p27Kip1 are both critical for GCP expansion during cerebellar development.
2.7. Negative regulators of GCP proliferation
In addition to signaling pathways that promote growth, GCP cell cycle exit and differentiation depend on signaling pathways that provide negative growth regulation. Recent studies have described several signaling molecules that antagonize the SHH-mediated proliferation of GCPs, including bFGF (Fogarty et al., 2007) and members of the BMP family. Among the latter, BMP2 and BMP4 are expressed in granule cell progenitors, and in postmitotic, differentiating GCPs in the EGL. In vitro assays indicate that Bmp2 and Bmp4, but not Bmp7, inhibit Shh-induced GCP proliferation (Rios et al., 2004) via the Smad signaling pathway (Zhao et al., 2008). Recent studies showed that Bmp4 antigonizes Shh signaling and induce differentiation of GCPs by rapid posttranscriptional turnover of Math1/Atoh1 (Ayrault et al., 2010). Recently, we have used a combination of genetic, cell, molecular, and biochemical methods to identify Wnt3 as a novel negative regulator of GCP proliferation (Kim et al., 2010, submitted). Our experiments show that Wnt3, which is provided by Bergmann glia in the developing cerebellum, blocks SHH-induced stimulation of GCP growth and slows medulloblastoma growth through a noncanonical Wnt signaling pathway.
In addition to changes in the regulation of cell cycle regulator RNAs and proteins, cell-cycle transitions are driven by Ubiquitin-dependent degradation of key cell-cycle regulators. SCF (Skp1/Cullin/F-box protein) complexes and anaphase-promoting complexes (APC) represent two major classes of ubiquitin ligases whose activities are thought to regulate the G1/S and metaphase/anaphase cell-cycle transitions, respectively (Ayad et al., 2005; Rankin et al., 2005; Wei et al., 2004). The APC complex is an E3 ubiquitin ligase required for the metaphase-to-anaphase transition and mitotic exit. GCPs provide a unique model for understanding cell cycle transitions. Recent studies by Harmey et al. (2009) indicate that the APC is essential for GCP proliferation and differentiation. Inhibition of APC activity by either a dominant negative approach or shRNAi depletion of the APC activator Cdh1 reduced GCP neurite outgrowth and migration in organotypic slice cultures. Subsequent studies using high-throughput over-expression screens identified ubiquitin ligases, which are similar to Cdh1, in controlling GCP migration (Simanski et al., submitted). A clearer understanding of the role of ubiquitin-dependent degradation of GCP cell cycle regulators in normal cerebellar development could have implications for medulloblastoma growth and treatment.
2.8. Addendum: Timing of human cerebellar development
In humans, between 24 and 40 weeks of gestation, the cerebellum undergoes a remarkable rate of growth as reviewed by Volpe (2009). During this period, as assessed by 3-D volumetric ultrasound, the volume of the cerebellar territory increases fivefold (Limperopoulos et al., 2005). In humans, the major embryonic histogenic events occur by 20 weeks gestation. Between 20 and 30 weeks, the EGL forms and development accelerates as evidenced by surface foliation (Volpe, 2009). At 25 weeks, the EGL reaches peak thickness, 6–8 cells deep. GCP proliferation and migration continue between 30 and 40 weeks. A number of features of GCP expansion, including a role for Shh, provided by Purkinje cells have been documented in human cerebellar development (Carletti and Rossi, 2008). During this period, although the thickness of the EGL remains constant, the surface expands horizontally via a more than 30-fold increase in the surface area of the cerebellar cortex. In human infants, the granule cell population accounts for more than 95% of the neurons of cerebellar cortex (Andersen et al., 1992), with the ratio of granule cells to Purkinje neurons being 3300:1. During the first postnatal year, as the EGL dissipates, the IGL continues to increase in size as the cerebellar circuitry matures (Volpe, 2009).
3. Medulloblastoma
Medulloblastoma is the most common malignant pediatric brain tumor with about 1000 new cases every year worldwide and a mean age between 3 and 7 years (Fogarty et al., 2005). Medulloblastoma, a cancer of the cerebellum, (Fig. 8.10), is a heterogenous class of embryonal tumors, that include subgroups with genetic anomalies in developmental pathways that are critical for normal cerebellar development. In the last 10 years a variety of studies, including analyses of primary human patient medulloblastoma samples as well as cell culture and mouse models have identified signaling pathways that promote or suppress medulloblastoma. As we refine our understanding of the disease at the molecular level, we will no doubt improve diagnostic tools and develop new and improved targeted therapies.
Figure 8.10.
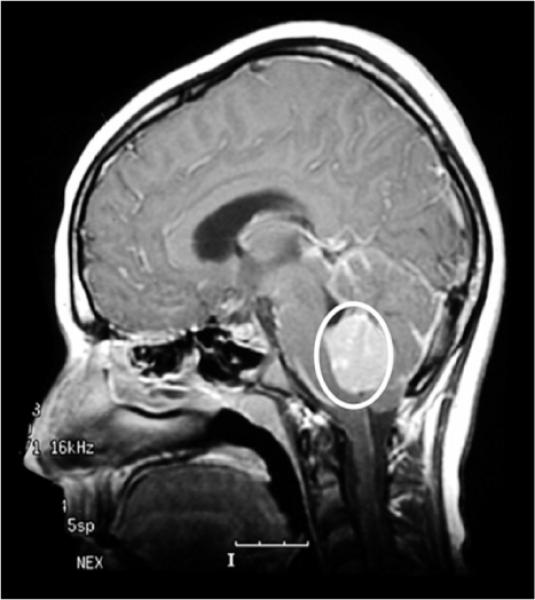
Medulloblastoma. Sagittal MRI scan following gadolinium. The tumor appears dense and is outlined by a circle. Image provided by Dr. Richard Gilbertson.
3.1. Current therapy and its consequences and targeted therapy
Current therapies for all subgroups of MB regardless of the subgroup include surgical resection, radiation therapy, and chemotherapy. Although advances in the radiation oncology and chemotherapy have led to dramatic increases in survival rates, ~70% of patients with average risk MBs, current treatments have significant morbidity and adverse secondary effects including loss of hearing, due mainly to the use of ototoxic drugs, and cognitive impairment (Gajjar et al., 2006). Such toxicities have profound effects in children that even if cured, face many years of posttreatment disabilities. High risk patients fair less well and ~30% of children die of advanced disease High risk MBs having the large cell anaplastic variants with MYC amplification and those associated with metastasis are associated with a poor outcome (Finlay et al., 2007; Gajjar et al., 2006). In contrast, a subgroup of MBs with mutations in the Wnt signaling pathway (see below), found primarily in adolescents and invariably are associated with high survival rates.
Our understanding of Shh/Ptch signaling has led to the discovery of small molecule inhibitors of Smoothened function that worked remarkably well to suppress MBs in allografts and mouse models of medulloblastoma (Berman et al., 2002; Romer and Curran, 2005). These compounds are currently in clinical trials. Although these small molecule inhibitors were very effective in reducing and eliminating tumor cells in mice, a case report indicates that one patient with remarkable remission of the disease later had a recurrence of his MB associated with a mutation in Smo, which made the tumor cells insensitive to the drug (Rudin et al., 2009). This emphasizes the need to discover novel targets and small molecule antagonists to the Shh pathway. A nasal antifungal, itracodazole, was recently found to block Smo function while arsenic trioxide was shown to antagonize Shh signaling by targeting Gli2 rather than Smoothened. Like cyclopamine and analogs, itracodazole and arsenic block proliferation of tumor cells. However, arsenic has the advantage to be active against MBs in which Smo is mutated, suggesting that it could be used therapeutically in patients in which tumors express a mutated Smo protein (Kim et al., 2010, Phillip Beachy, personal communication). Ongoing molecular analysis of primary MBs combined with the discovery of new biomarkers for histopathology studies should provide physicians with better diagnostic and therapeutic tools. In patients with MBs with a good prognosis, including those with mutations in the Wnt pathway, the treatment could be less aggressive thus lessening the negative secondary effects of the treatment. Such approaches are currently being tested in clinical trials.
3.2. Histopathology
The 2007 WHO classification of medulloblastomas defines five histopathologic subgroups or variants, including the classic subgroup with some differentiated neurons, the desmoplastic variant in which tumor cells show some differentiation and are surrounded by extracellular matrix, MBs with nodularity, and the anaplastic and large cell anaplastic forms, the most aggressive of the disease that invariably connotes poor prognosis (Fig. 8.11; Louis et al., 2007; reviewed in Gilbertson and Ellison, 2008; Northcott et al., 2009). Molecular genetic analyses have identified only four subgroups that overlap the WHO pathological classification (Kool et al., 2008; Thompson et al., 2006) Whereas medulloblastoma with mutations in the Shh/Ptch pathway are characterized as desmoplastic and represent ~25% of all MBs and the classic type is the most represented while the aggressive large cell anaplastic MBs are found within each molecularly distinct subgroup and represent ~10% of all MBs (Thompson et al., 2006). MBs with a good prognosis include desmoplastic MBs and those with increased nodularity characterized with monosomy 6, mutations in the Wnt pathway, and high TrkC expression. In contrast, high risk patients with medulloblastoma often have metastases and amplification or high expression of C-MYC or MYCN which is associated with the large cell anaplastic variants, 17p loss and 1q gain (Eberhart et al., 2004; von Hoff et al., 2010). MBs also can spread within the cerebrospinal fluid and outside the cerebral nervous system (CNS) in bone and bone marrow in very few cases conferring an adverse prognosis (reviewed in Pizer and Clifford, 2009). The identification of new biomarkers that define subgroups of MB's and the development of antibodies to these markers will likely lead to better diagnostic tools.
Figure 8.11.
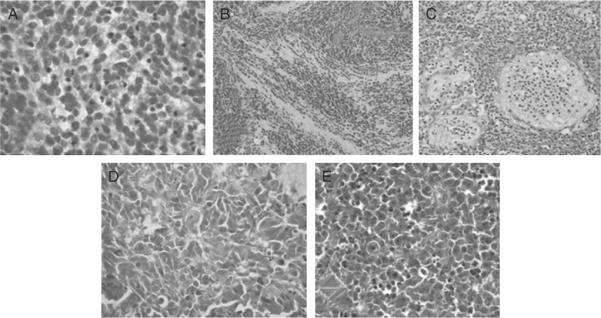
Histopathology of the five medulloblastoma variants. (A) Classic medulloblastoma marked here by a syncytial arrangement of densely packed small uniform cells with hyperchromatic nuclei. (B) Desmoplastic/nodular medulloblastoma showing nodules of differentiated neurocytic cells separated by regions containing moderately pleomorphic cells with a high growth fraction. (C) Medulloblastoma with extensive nodularity with a large irregularly shaped nodules of neurocytic cells often forming linear patterns interspersed with small collections of pleomorphic cells. (D) Anaplastic medulloblastoma characterized with a large markedly pleomorphic cells with a high mitotic count and nuclear:cytoplasmic ratio mold to each other. (E) Large cell medulloblastoma showing large cells with a single nucleolus among more pleomorphic cells with an anaplastic phenotype.
3.3. Origin of medulloblastomas
Like other embryonal tumors in children, MBs are thought to arise from neuronal progenitors with defects in gene regulation or genetic anomalies in genes and proteins that regulate normal growth and development. To date, one subgroup of MBs with constitutive activation of the Shh/Ptch pathway has been shown to originate from granule cerebellar progenitors (GCPs) in the external granule layer (EGL) (reviewed in Behesti and Marino, 2009; Gilbertson and Ellison, 2008). The question remains as to whether the other subgroups of MBs arise from subpopulations of GCPs in the cerebellar EGL, from ectopic GCPs at developmental stages after the EGL dissipates, or from other types of cerebellar neurons. Recently, Gilbertson and colleagues developed a mouse model for MBs that recapitulate human MBs from the Wnt subgroup by targeting neuronal progenitors in the ventricular zone distinct from the GCPs using a mouse line in which the Cre recombinase is expressed in neuronal progenitors located in the ventricular zone (Blbp-Cre). By breeding this mouse line with a conditional form of a constitutively activated β-Catenin protein in a p53-null background, they were able to generate a mouse model that mirror the human subgroup with a Wnt signature (Gibson et al., in revision). This is especially interesting because progenitors that express the glial gene Blbp give rise to both neurons and glial cells (Anthony and Heintz, 2008) and Blbp is a direct target of Notch1 in radial glial cells (Anthony et al., 2005). Thus, different subgroups of MB may derive from different subpopulations of cerebellar progenitors, including embryonic radial glial cells, although animal models will be required to test this hypothesis.
3.4. Molecular characterization of MBs
Transcriptional profiling of messenger mRNAs in human medulloblastomas (Kool et al., 2008; Thompson et al., 2006) and more recently microRNAs profiling (Ferretti et al., 2008; Northcott et al., 2009) have identified four subgroups of medulloblastoma with distinct RNA signatures. It is likely that more tumor suppressors and possibly oncogenic mutations and amplifications will be identified as we continue to analyze an increasing number of tumors (reviewed in Lindsey et al., 2005). Genetic and molecular genetic analyses of medulloblastoma have identified changes in the regulation of signaling pathways that are important for cerebellar progenitor cell neuro-genesis, including pathways that promote growth (Shh/Patched, the Wnt, Notch, IGF/PTEN/mTOR) and negative growth regulators (BMP). Molecular changes in these pathways associated with MB include both loss of function and gain of function mutations, as well as alterations in the mRNA or protein levels of regulators of these pathways.
3.4.1. Shh/Patched signaling
The Shh/Patched signaling pathway is the most studied in medulloblastoma (Fig. 8.12). The mitogen Sonic hedgehog (Shh) drives proliferation of granule cerebellar progenitors (GCPs) by binding to the 12 transmembrane receptor Patched (Ptch) that exists in two forms, Ptch1 and Ptch2 (Hatten, 1999). In the absence of Shh, Ptch represses the function of Smoothened (Smo), a seven transmembrane G-protein-coupled receptor-like protein that activates the Gli1 and Gli2 transcription factors and inactivates the transcriptional repressor Gli3 that together regulate the transcriptional program in the cell nucleus (reviewed by Wechsler-Reya and Scott, 2001). The primary cilium, a cellular structure essential to cell proliferation and function, was recently found to concentrate components of Shh signaling in cerebellar GCPs and to be required for Shh signaling in GCP expansion (Fig. 8.13; Rohatgi et al., 2007; Spassky et al., 2008). While the three Gli transcription factors and their partner SuFu are present in the cilia together with Ptch1 in the absence of Shh stimulation, in the presence of the mitogen, Smo is recruited to the cilia while Ptch is degraded in the cytoplasm (Wen et al., 2010).
Figure 8.12.
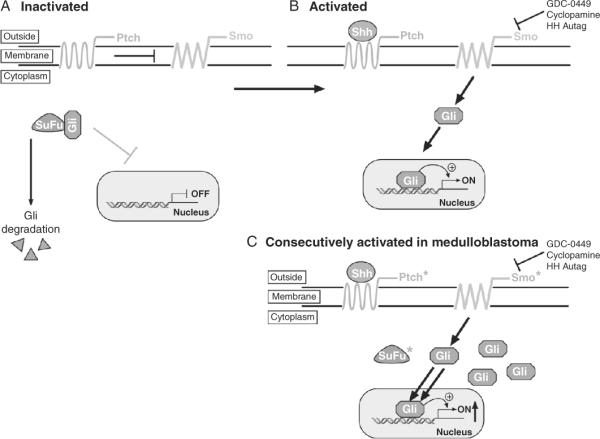
Schematic of the sonic hedgehog signaling pathway. (A) In the absence of Sonic hedgehog (Shh), the 12 transmembrane receptor Patched (Ptch) suppresses the seven transmembrane G-coupled receptor Smoothened (Smo) and the transcription factor Gli is sequestered into the cytoplasm by the Suppressor-of-Fuse (SuFu) preventing it from translocating to the nucleus and is degraded leading to a block of transcription. (B) In the presence of Shh, the suppression by Ptch is relieved and Smo activates Gli1 that is translocated to the nucleus and induces the transcription of Gli targets, including many proproliferative genes. (C) In medulloblastoma, loss of Ptch and mutations in Smo or SuFu lead to the constitutive activity of Smo, accumulation of Gli and increased transcription of proproliferative Gli-dependent targets. Asterisk (*) identifies proteins mutated in medulloblastoma.
Figure 8.13.
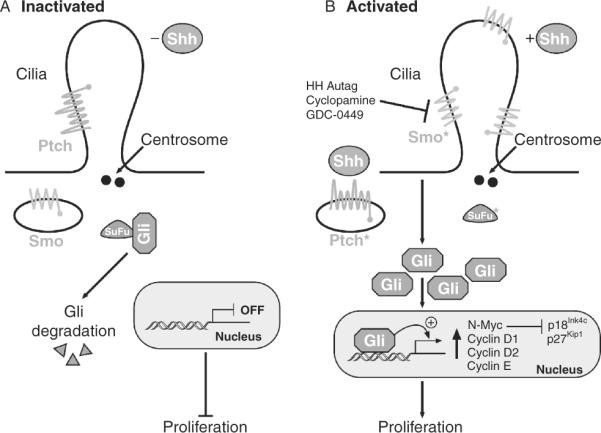
Schematic of the sonic hedgehog signaling pathway in the primary cilium. (A) In the absence of Shh, Ptch resides at the base of the cilium whereas Smo is in the cytoplasm and the pathway is off. (B) In the presence of Shh or in medulloblastoma, Smo becomes located to the cilium. In the absence of Ptch or when Smo or SuFu are mutated, Gli and the levels of targets increase. Asterisk (*) identifies proteins mutated in medulloblastoma.
Patients with Gorlin syndrome (also called Nevoid Basal Cell Carcinoma Syndrome) sustain germline mutations in PTCH1 and SUFU that predisposes them to multiple cancers including medulloblastomas (Hahn et al., 1996; Johnson et al., 1996; Pastorino et al., 2009). Loss of function mouse models in which Ptch1 is deleted in the germline or conditionally demonstrates that loss of Ptch1 induces medulloblastoma with histopathologic features of desmoplastic human MBs (Goodrich et al., 1997; Lee et al., 2003; Marino et al., 2000; Oliver et al., 2005). Molecular analysis of sporadic human MB primary samples revealed activation of the SHH/PTCH pathway from the loss of PTCH, and mutations in SUFU (Brugieres et al., 2010), and SMO (reviewed by Rubin and Rowitch, 2002; Thompson et al., 2006). Mice lacking Sufu develop medulloblastomas in conjunction with p53 loss, demonstrating that Sufu functions as a tumor suppressor gene (Lee et al., 2007).
The two Shh-dependent transcription factors, Gli1 and Gli2, activate the transcription of several proproliferative genes, including MycN, cyclins D1, and D2, and cyclin E (Fig. 8.5; Ciemerych et al., 2002; Hatton et al., 2006; Kenney et al., 2003; reviewed in Katoh and Katoh, 2009). In turn, MycN, a b-HLH-transcription factor induces the expression of many genes and microRNAs, among which is the miR-17~92 cluster (see below). Conversely, MycN suppresses the expression of two cyclin-dependent kinase inhibitory proteins, p18Ink4c and p27Kip1 to induce Rb and p107 phosphorylation and cell cycle progression (Fig. 8.14; Knoepfler et al., 2002). While Rb and p107 are required for normal cerebellar development and GNP survival, their loss in mice in conjunction with loss of p53 induces MBs (Marino et al., 2003). We found that in mice p18Ink4c is transiently expressed in GCPs to time their exit from the cell cycle, whereas p27Kip1 is turned on in postmitotic GCPs in the inner EGL and in the IGL (Uziel et al., 2005). Loss of Ink4c or Kip1 indeed collaborates with the loss of Ptch1 to induce medulloblastoma in mice (Uziel et al., 2005). Similarly, Ink4c and the tumor suppressor p53 co-repress the induction of tumors (Zindy et al., 2003). Although TP53 mutations are seldom found in human MBs that represent only ~10%, Li-Fraumeni patients with familial TP53 mutations are prone to medulloblastoma development (Malkin et al., 1990; Shrivastava et al., 1990). For reasons that are not entirely clear, GCPs are exquisitely sensitive to DNA damage and rely heavily on an intact p53 pathway to eliminate cells that have acquired mutations during their division cycle. Several experiments illustrate this point: first, mouse models of medulloblastoma with a Shh signature were generated by mutations of genes in the DNA repair pathways, including Ligase 4, PARP1, XRCC4 in combination with p53 loss (Lee and McKinnon, 2002; Tong et al., 2003; Yan et al., 2006); second, 5 days old mice irradiated with nonlethal doses (~4 Gy) all develop medulloblastoma by 3 months of age (Zindy et al., 2007).
Figure 8.14.
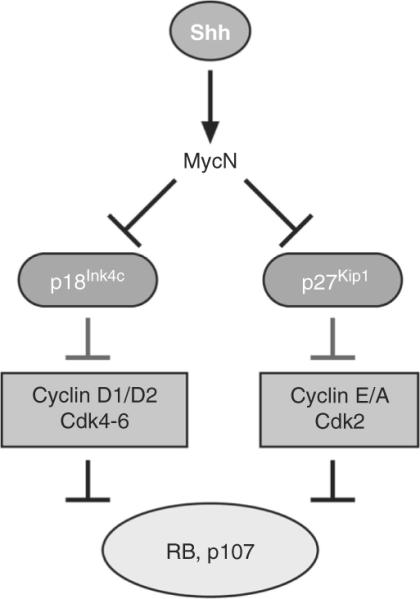
Regulation of MycN and cell cycle proteins by sonic hedgehog signaling. Shh signaling directly induces the expression of MycN and Cyclins D1, D2, and E. MycN in turn suppresses the expression of two cyclin-dependent kinase (Cdk) inhibitory proteins, p18Ink4c and p27Kip1. While p18Ink4c inhibits the activity of Cdk4 and Cdk6 in complex with cyclins D1 and D2, p27Kip1 suppresses the activity of the Cyclin E/ and A/Cdk2 complexes. Cyclin/Cdk complexes phosphorylate Rb and p107 to allow the cells to progress through the first gap (G1) phase of the cell cycle and enter the DNA synthetic (S) phase when they replicate their DNA.
Initial karyotyping and recent CGH analysis revealed that the genes of the MYC family, c-MYC on chromosome 8q24, MYCN on chromosome 2p, and MYCL1 on chromosome 1p are frequently amplified in medulloblastomas (reviewed in Northcott et al., 2010). Indeed, enforced expression of C-Myc or MycN in GCPs either from p53-null or Ptch1+/− mice induce medulloblastoma with full penetrance (100%) after orthotopic transplants in naïve recipient animals (Zindy et al., 2007).
3.4.2. Collaboration between Shh and insulin-like growth factor
Shh and IGF promote GCP growth and constitutive activation of the Shh/Ptch and IGF pathway are found in medulloblastomas in mouse and man (Kenney et al., 2004). IGF-I and IGF-II are potent survival factors that bind to IGF-receptors (IGF-R). IGFRs in turn activate PI3K, Akt, and mTOR, to promote mRNA translation (Fig. 8.15). Most growth factors activate mTOR by suppressing the tuberous sclerosis complex (TSC) that include TSC1 (hamartin) and TSC2 (tuberin) that normally restrain mTOR activity. TSC1 stabilizes TSC2, and TSC2 upregulates p27Kip1 levels by preventing its degradation, promoting its nuclear localization and preventing cell cycle progression. Although TSC inactivation alone has no effects, in collaboration with Ptch1 mutation, it drives MB formation in mice by increasing translation of mRNAs and possibly affecting p27Kip1 localization (Bhatia et al., 2009, 2010). Similar results were found using an IGF-1 transgene which expression was driven in GNPs and in a Ptch1+/− background (Tanori et al., 2010). Several inhibitors of the mTOR pathway (rapamycin) are currently being tested as a treatment for MBs.
Figure 8.15.
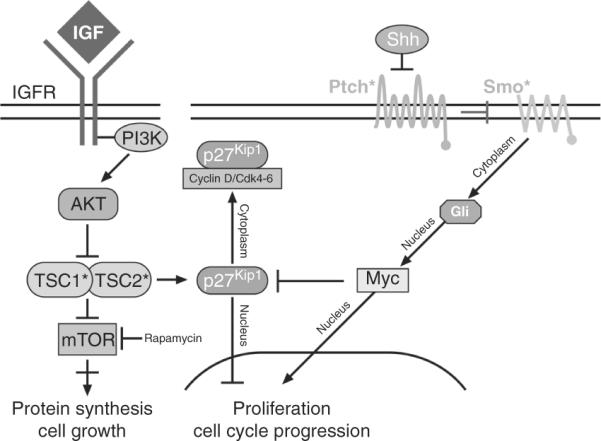
Collaboration between insulin-like growth factor and Shh signaling. IGF activates the insulin-like growth factor (IGF) receptor (IGFR) which in turns activates the phosphoinositol 3-kinase (PI3K), the AKT kinase which suppresses the tumor suppressor TSC1 and TSC2 proteins. TSC1 and TSC2 proteins induce p27Kip1 that prevent G1 progression and block mTOR which is required for protein synthesis and cell growth. Rapamycin blocks mTOR activity. Shh signaling in turn activates Mycn that suppresses p27Kip1. Asterisk (*) identifies proteins mutated in medulloblastoma. Adapted from Bathia et al. (2010).
The 3.4.3. Collaboration between Shh and Hippo pathways
Hippo pathway is thought to limit cell proliferation and promote apoptosis in differentiating cells (reviewed in Saucedo and Edgar, 2007). Moreover, mutations in this pathway have been proposed to provide cancer cells with a competitive advantage in genetically mosaic tumors, thereby promoting proliferation. The Hippo pathway contains two serine-threonine kinases Mst1 and Mst2 that redundantly phosphorylate the serine-threonine kinases Lats1 or Lats2. Activity of these kinases is enhanced by scaffolding proteins Sav1 (Ww45) for Mst 1 and 2 and Mob1–4 for Lats proteins. Lats proteins phosphorylate two related transcriptional adaptor proteins Yes-associated protein (YAP) and TAZ. Phosphorylation of YAP and TAZ induces their interaction with the chaperone protein 14-3-3 and retention into the cytoplasm thus inhibiting their function. In the absence or the presence of reduced Hippo signaling, YAP/TAZ phosphorylation is absent or low, respectively, inducing their translocation to the nucleus where they interact with DNA binding TEA domain transcription factor TEAD (1–4), as well as several other transcription factors including RUNX2, Smad7, p73, and p53BP2 to activate the transcription of genes involved in cell proliferation and survival (reviewed in Saucedo and Edgar, 2007).
Recent studies implicate the Hippo pathway in medulloblastomas in concert with the Shh/Ptch signaling pathway (Fernandez et al., 2009). Fernandez and collaborators found that YAP expression is regulated by Shh in granule neuron progenitors, that its translocation is mediated by Shh and that it regulates GCPs proliferation (Fig. 8.16). Consistent with its expression in GCPs, YAP is upregulated by overexpression and amplification of the locus on chromosome 11q22 in human medulloblastomas in which the Shh signaling pathway is activated. YAP is also highly overexpressed in human medulloblastomas with activation of the Wnt signaling pathway. TEAD, the major partner of YAP1 is similarly overexpressed in human MBs with constitutive activation of the Shh or Wnt signaling pathways. In a mouse medulloblastoma model that recapitulates the human tumors with a Shh “signature,” YAP is expressed in the perivascular niche, a region of the tumor surrounding blood vessels, in which cancer stem cells reside. These YAP expressing cells were resistant to irradiation that otherwise eradicated all other cell types (Fernandez et al., 2009). Since YAP may maintain “stemness” and protect tumor cells from DNA damage-induced apoptosis, it is a potential new therapeutic target in the treatment of medulloblastomas with Shh and Wnt pathway activation. However, experiments in animal models will be required to validate the function of YAP1 in MBs.
Figure 8.16.
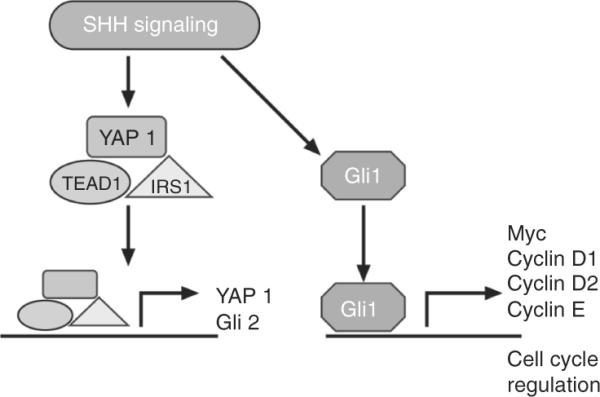
Collaboration between Shh and Hippo signaling. Shh regulates YAP1 in complex with TEAD and IRS1 that is translocated to the nucleus where it regulates YAP1 itself and Gli2. Shh signaling regulates Gli1 and its direct targets. Adapted from Fernandez et al. (2009).
3.4.4. The Wnt signaling pathway
The canonical Wnt signaling pathway is activated when Wnt binds to the seven transmembrane receptor Frizzled that activates the Disheveled protein which has two functions: one to block β-Catenin activation and nuclear localization and the other to regulate actin stress fibers (Fig. 8.17). β-Catenin protein is part of a complex of proteins containing the kinase GSK-3β, Axins 1 and 2, the adenomatous polyposis coli (APC) and Caseine kinase a, CK1a. β-Catenin levels are normally regulated by phosphorylation by GSK-3β leading to ubiquitination and proteasome-dependent degradation. Mutations in APC or in the phosphorylation site of β-Catenin that can no longer be degraded by the proteasome, induces the constitutive activation of nuclear β-Catenin that by binding to the transcription factors TCF/LEF induces unabated transcription of proproliferative target genes including cyclin D1 and c-Myc.
Figure 8.17.
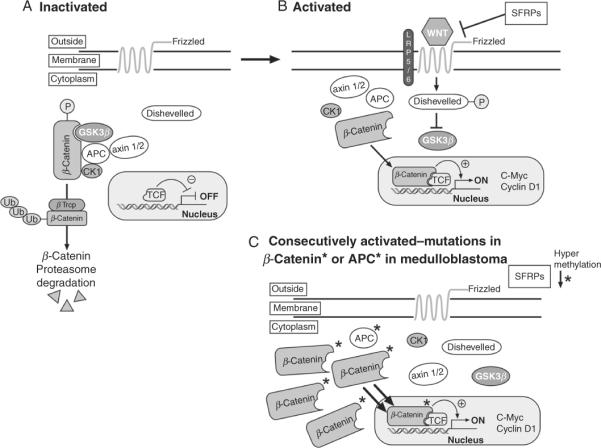
The Wnt signaling pathway. (A) In the absence of ligand, the transmembrane frizzled receptor is inactive and β-Catenin is in complex with APC, axin 1 and 2, caseine kinase 1 (CK1), and GSK-3beta. This enables GSK-3beta to phosphorylate β-Catenin that is then ubiquitinaed by the E3 ligase β-Trcp and degraded by the proteasome. (B) Upon binding of Wnt ligands, lrp5/6 and Frizzell are activated, disheveled is phosphorylated and blocks GSK-3β kinase activity. β-Catenin is no longer in a complex with APC, Axin, and CK1 and β-Catenin is translocated to the nucleus where it transcribed cyclin D1 and C-Myc. SFRPs prevent Wnt from binding to Frizzell and inhibit Wnt signaling. (C) In medulloblastomas, the Wnt pathway is constitutively activated leading to the accumulation of β-Catenin levels and increase transcription.
Deregulated Wnt signaling occurs in ~10–15% of human medulloblastomas. It was first identified in patients with Turcot's syndrome in whom colon cancer and malignant neuroepithelial brain tumors including medulloblastomas were associated with mutations in APC (reviewed in Gilbertson and Ellison, 2008; Marino, 2005). Subsequently mutations in BETA-CATENIN/CTNNB1 were also found in human medulloblastomas (Thompson et al., 2006). Besides activated mutations of B-CATENIN/CTNNB1, these MBs harbor a single copy loss of chromosome 6, also called monosomy 6. Monosomy 6 represents the most significant prognostic factor for the WNT subgroup of tumors that correlates with a good outcome (Gajjar et al., 2006). Finally, epigenetic silencing of the secreted frizzled-related proteins (SFRP) family of Wnt inhibitors, SFRP-1,-2, and -3 is thought to elevate Wnt signaling in MBs (Fig. 8.17). In addition, forced expression of these SFRP proteins reduces the proliferation and anchorage-independent growth of medulloblastoma cells, limits tumor burden, and prolongs survival in xenografts of MB in mice (Kongkham et al., 2010). The recent mouse model that recapitulates the WNT MB human variant will provide a preclinical model for potential novel therapeutic intervention in patients afflicted with this subgroup of MB (Gibson et al., 2010).
3.4.5. The Notch signaling pathway
Notch signaling is involved in many cellular processes, including stem cell renewal, cell fate specification, and proliferation. Intercellular Notch signaling is mediated by expression of the membrane-bound ligands Delta (1–4) and Jagged ( Jag1, 2) on one cell and of the transmembrane receptors Notch (1–4) on the other cell (Fig. 8.18). Upon ligand binding, the intracellular domain (ICD) is clipped by the γ-secretase and translocated to the nucleus where it enters in complexes to activate the transcription of several targets, in particular the basic-helix-loop-helix (bHLH) transcription factors, Hes1 and Hes5. Activated Notch 2 stimulates GCP proliferation (Solecki et al., 2001). Consistent with a role for Notch2 in GCP proliferation, high levels of Notch 2 expression, but not of other Notch family members, was detected in human medulloblastoma (Fan et al., 2004). In addition to Notch2, Hes1 and Jag1 were found to be highly expressed in mouse medulloblastoma models by several groups (Dakubo et al., 2006; Hallahan et al., 2004). In addition, genetic studies do not support a role for Notch1 in medulloblastoma formation in collaboration with the Shh/Ptch pathway (Julian et al., 2010) Thus, the components of the Notch1 and Notch2 signaling pathways may delineate important differences among subgroups of MBs
Figure 8.18.
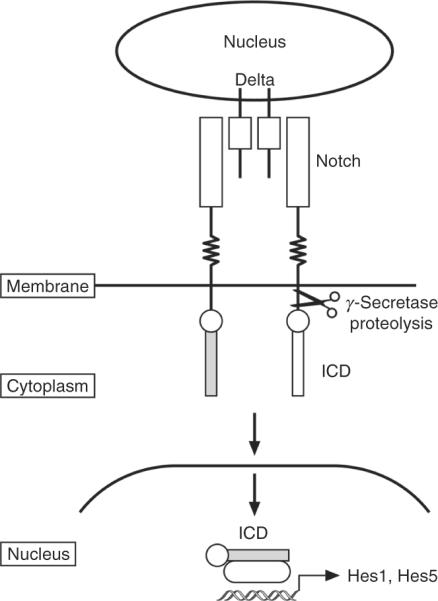
The Notch signaling pathway. Notch signaling is mediated by two cells; one providing the transmembrane ligands Delta and Jagged, the other the single transmembrane receptor, Notch, that transmits the signals. Upon ligand binding, the intracellular domain (ICD) is clipped by the γ-secretase at the membrane, is translocated to the nucleus where it enters in a complex with other proteins, and induces transcription of specific targets including the basic-helix-loop-helix transcription factors, Hes1 and Hes5.
3.4.6. The BMP2/4 signaling pathway
Unlike the growth promoting signaling pathways described above, the BMP signaling pathway inhibits proliferation and induces the differentiation of GCPs postnatally and GCP-like tumor cells. BMPs, 1–7, belong to the TGF-β family of cytokines (reviewed in Massague, 1996; Massague et al., 2005). BMP-2,-4, and 7 bind as dimers to promote the heterodimerization and transphosphorylation of serine/threonine BMP receptors type I (Alk3) and II (Alk6) (Fig. 8.19). Upon receptor activation, Smad1is recruited to and phosphorylated by the receptor type II (Alk6), inducing its conformational change and heterodimerization with Smad4. The heterodimer is then translocated to the nucleus where it directs the transcription of several genes including the Inhibitors of division (Ids), Id1-4, and TGF-B-early inducible gene 1 (TEIG-1) (Fig. 8.19).
Figure 8.19.
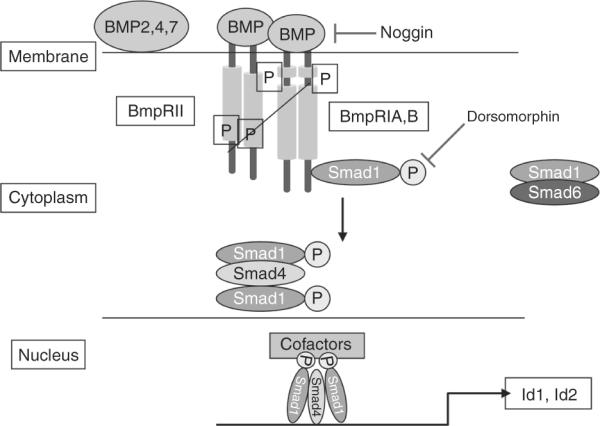
Bone morphogenic protein (BMP) signaling. BMP-2, 4, 7 bind to heterodimeric serine/threonine kinase receptors BMP-RII and BMP-RIA/B to induce their activation and trans-phosphorylation. This induces the recruitment of Smad1 that is sequestered by Smad6 in the cytoplasm in the absence of BMPs, and its phosphorylation. Phosphorylated Smad1 enters in complexes with Smad4 and is translocated to the nucleus where is induces the transcription of several genes including the inhibitors of DNA binding, Id1 and Id2.
Several studies found that in postnatal cerebellum development and in MBs with a Shh signature BMPs oppose Shh-induced proliferation and induce irreversible neuronal differentiation. In 2004, Rios and collaborators showed that BMP2 antigonizes Shh-dependent proliferation of GCPs (Rios et al., 2004). In 2007, the same group found that BMP2 antigonizes Shh-dependent proliferation by expressing TIEG-1 which directly represses MycN transcription. Conversely, they found that TIEG-1 overexpression promotes cell cycle arrest and apoptosis of GCPs in the absence of other differentiation signals (Alvarez-Rodriguez et al., 2007). In 2008, we found that cell cycle exit and terminal differentiation of GCPs and GCP-like tumor cells is mediated by the rapid posttranscriptional downregulation of Math1/Atoh1, a b-HLH- transcription factor that expression marks proliferating GNPs. Enforced Math1/Atoh1 expression in turn prevented differentiation (Fig. 8.20). Finally, Math1/Atoh1 was recently demonstrated to be essential for MB development (Flora et al., 2010) and to collaborate with Gli1 to transform normal GCPs into tumor-initiating cells (Ayrault et al., 2010).
Figure 8.20.
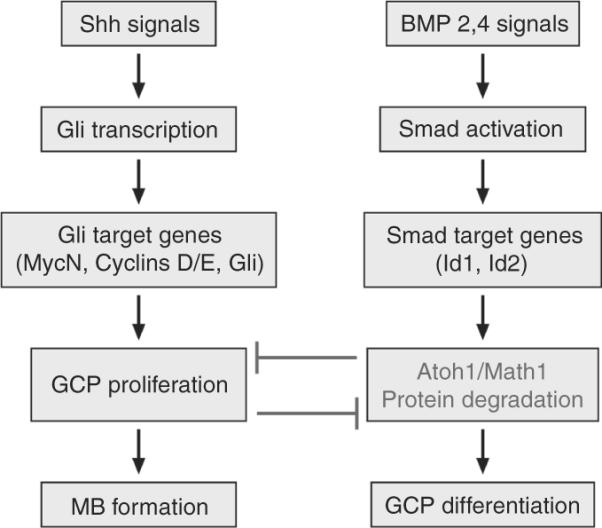
Shh and BMP signaling. Both signaling pathways antagonize each other to regulate proliferation of GCPs by regulating the levels of the bHLH transcription factor Atoh1/Math1. Whereas Shh induces proliferation, BMP inhibits proliferation and induces the irreversible differentiation of GCPs. BMP induces cell growth arrest by the rapid posttranscriptional downregulation of Atoh1/Math1 protein by the proteasome. In medulloblastomas, Atoh1/Math1 levels are high whereas the genes regulating the BMP pathway are downregulated.
Gene profiling of mouse and human MBs with a constitutively activated SHH pathway revealed that most genes within the BMP signaling pathway are downregulated in tumors compared to proliferating GCPs and neurons, whereas Atoh1 levels are high (Zhao et al., 2008). Epigenetic studies of human medulloblastoma samples identified a tumor suppressor called HIC1, for hypermethylated in cancer-1, the normal function of which is to suppress ATOH1/MATH1. In human medulloblastoma, the promoter of HIC1 is hypermethylated leading to inhibition of its expression and to the induction of high ATOH1/MATH1 protein levels (Lindsey et al., 2004; reviewed by Briggs et al., 2008a,b). Current CGH analysis confirmed that the most frequently affected chromosome in MBs with a SHH signature is 17 that encodes the tumor suppressor HIC1 (Ferretti et al., 2005). However, it is likely that other genes are affected as the result of the frequent loss of heterozygocity on 17p.
3.5. Epigenetic silencing in MBs
Besides loss of function by gene deletions or mutations, epigenetic silencing also plays an important role in the development of MBs (reviewed in Lindsey et al., 2005). Survey of the methylation status of tumor suppressors or oncogenes in human MBs has revealed several genes the disregulation of which is associated with MBs. They include SFRPs that negatively regulate the Wnt pathway (Kongkham et al., 2010), the S100 gene family (Lindsey et al., 2007), the Kruppel-like factor 4 (Nakahara et al., 2010), and HIC1 that regulates Atoh1/Math1 expression (Briggs et al., 2008a,b). As these studies are expanded to an increasing number of primary tumors in the future, it is likely that more genes will be found to be regulated epigenetically and to be relevant to MB formation.
3.6. Other genes involved in medulloblastoma
A number of other genes have been associated with human medulloblastoma. However, the mechanism of action for these genes in MB or their association with specific subgroups of MBs remains to be determined. Among these, high expression of the neurotrophin TrkC receptor is associated with a good prognostic and a favorable outcome in patients with medulloblastoma. TrkC is found in desmoplastic nodular medulloblastoma and is suggested to be responsible for neuronal differentiation. Unlike TrkC, other proteins are correlated with poor prognosis and metastasis. These include ERBB2, a member of the epidermal growth factor receptor, the platelet-derived growth factor receptor, PDFGR and C-Met, the receptor tyrosine kinase for the hepatocyte growth factor (reviewed in Guessous et al., 2008).
In addition, RENKCTD11 that maps to 17p13.2 and is deleted in human medulloblastoma with a SHH “signature” is a suppressor of Hedgehog signaling and was shown by the same group to regulate proliferation of GCPs (Argenti et al., 2005; Di Marcotullio et al., 2006). On the other hand, expression of OTX1 and OTX2, two developmentally regulated transcription factors was found to correlate with two subgroups of medulloblastoma; whereas overexpression of OTX1 was found in nodular/desmoplastic human tumors, OTX2 corresponded to the classic subgroup in man and mice (Adamson et al., 2010; de Haas et al., 2006).
3.7. MicroRNAs and medulloblastoma
In the last 10 years, microRNAs and long noncoding RNAs have emerged as novel regulators of many aspects of cellular biology and development. MicroRNAs are small noncoding 22 nucleotides long RNAs the sequence of which is complementary to those in the 3′-UTR of targeted messenger RNAs. MicroRNAs are thought to regulate hundreds of mRNA targets (reviewed in Bartel, 2004).
MicroRNAs are encoded on human and mouse chromosomes as long pri-miRNAs that are processed in the nucleus as pre-miRNAs by a processing enzyme called Drosha (Fig. 8.21). Pre-miRNAs are processed by Dicer in the cytoplasm as mature microRNAs that form complexes, called RISC with argonaute proteins. The RISC complex recognizes the complementary sequences on the messenger RNA targets within its 3′ end, 5′ end and often coding sequences. Several microRNAs were found to be overexpressed and possibly act as oncogenes in tumors whereas others were shown to be underexpressed in tumors suggesting that their expression might suppress tumor development (reviewed in Garzon et al., 2006). As such, microRNAs are being considered as targets for therapy.
Figure 8.21.
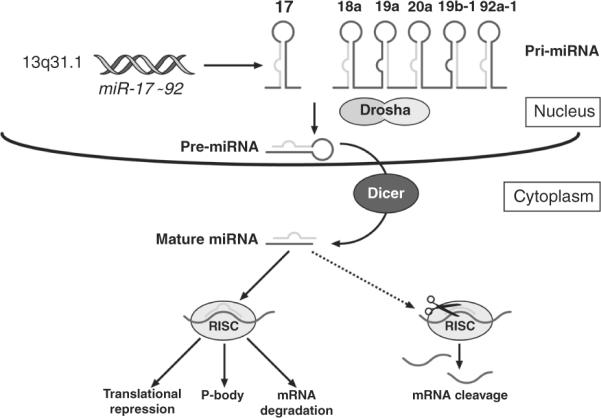
MicroRNA biogenesis and miR-17~92 processing. MicroRNAs (miRs) are encoded by the genome on many chromosomes but do not encode proteins. The miR-17~92 cluster is encoded on human chromosome 13q31.1 as a long pri-miRNA and processed by the processing enzyme Drosha. Pre-miRNAs are further processed in mature miRs where they enter in a RISC complex with argonaute proteins. RISC complexes recognize the complementary sequence on the 3′ end, 5′ end or even the coding region of the messenger mRNA targets. This leads mainly to the inhibition of protein synthesis but the mRNA target could also be trapped in P bodies or degraded. Ultimately, gene expression is suppressed. Figure provided by Dr. Zindy.
Two groups identified the microRNA miR-17~92 cluster as an oncogene in MBs with a Shh signature (Northcott et al., 2009; Uziel et al., 2009). The mir-17~92 cluster belongs to a family of a three clusters encoded by different chromosomes in mouse and man (Ventura et al., 2008) (Fig. 8.22). It encodes six unique microRNAs, miR-17, 18, 19, 20, and 92, that share common seed sequences with microRNAs encoded by the two other clusters of the family, miR-106b~25 and miR-106a-363. MiR-17~92 was found to be a direct target of C-Myc and MycN (O'Donnell et al., 2005). Consistent with this finding, the miR-17~92 cluster is found at high levels in human medulloblastoma with high expression or amplification of C-MYC or MYCN. Although several important targets of the miR-17~92 cluster were found in B cells including E2F-1, PTEN, and Bim that all induce apoptosis (Fig. 8.23), specific bona fide targets in MBs have not yet been identified. Whereas ectopic expression of miR-17~92 in GCPs is not sufficient to drive tumorigenesis on its own, it does so in collaboration with Ptch1 loss in an orthotopic transplant model in mice (Uziel et al., 2009). If this cluster is required for MB development, anti-gomiRs to this cluster may prove to be an efficacious and novel therapeutic approach for MBs in which this cluster is overexpressed.
Figure 8.22.
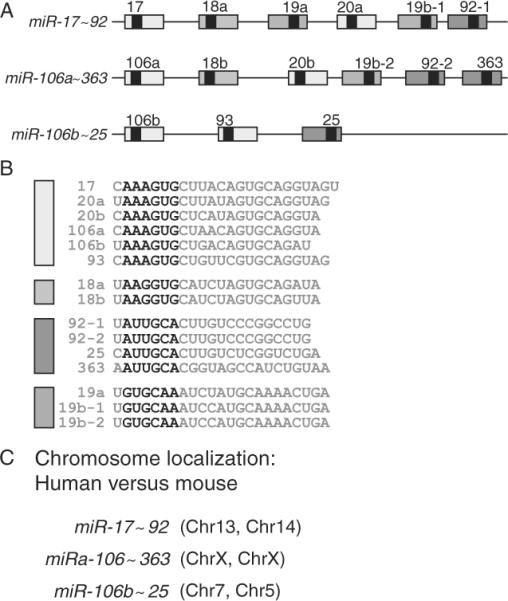
The miR-17~92 cluster family. The miR-17~92 cluster family comprises three members (A) encoding unique miRs that share sequence homology in the 6 mer “seed” sequence labeled in bold letters (B) and encoded in different chromosomes in human and mice (C). Adapted from Ventura et al. (2008).
Figure 8.23.
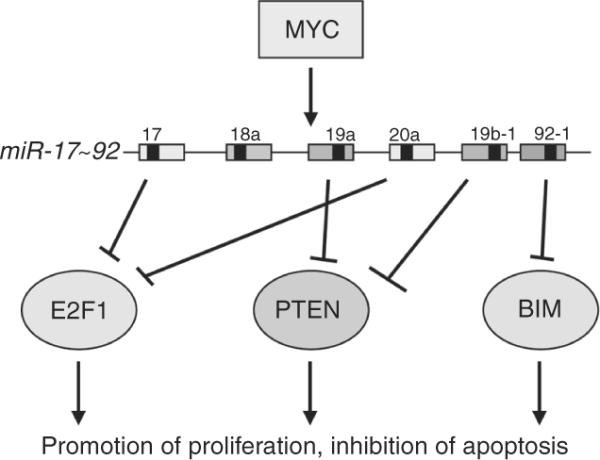
The miR-17~92 cluster is an MYC target. Myc directly binds to the promoter of the cluster to induce its expression. The miR-17~92 encodes six individual miRs that target different cell death-inducing proteins, including E2F-1, PTEN, and BIM. Figure provided by Dr. Zindy.
3.8. Mouse models of medulloblastoma and preclinical testing of novel targeted therapies
Remarkably, all mouse models of medulloblastoma that have been developed to date mirror only one subgroup of human MBs, with a Shh signature, even though they were generated with different founding mutations (Fig. 8.24; Lee et al., 2003). These include mutations in Sufu, Smo, loss of Ptch1 alone or together with Ink4c or Kip1, Rb, hypermethylation of the HIC1 promoter, loss of Ligase 4, XRCC4, Brca2, and PARP1, often in combination with p53 loss (reviewed in Behesti and Marino, 2009). In contrast, forced expression of MycN, C-Myc, and miR-17~92, induced MB development in combination with loss of Ptch1 or loss of p53 (Uziel et al., 2009; Zindy et al., 2007). Comparison of the gene profile of GCPs and tumors with that of differentiated neurons revealed that in all these mouse models, the cell of origin is a GCP (Lee et al., 2003). Although p53 mutations are not common in human MBs (~10%), most of the mouse models, except those from Ptch1+/−, Ink4c−/− and Ptch1+/−, Kip1−/−, required the loss of p53. This may reflect the extreme sensitivity of GCPs to DNA damage that must rely on the p53 checkpoint to eliminate all cells that fail to properly replicate their DNA. It is plausible that mutations upstream of downstream of p53 affect its function without a requirement for its elimination in human tumors.
Figure 8.24.
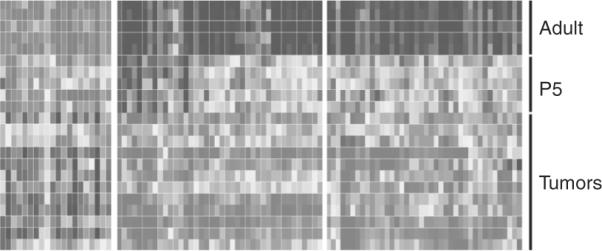
Gene profile of mouse genes in the cerebellum and medulloblastoma. Upregulated (light gray) and downregulated (dark gray) gene expressions are compared between adult mouse cerebella, GCPs purified at postnatal day P5 and in medulloblastomas spontaneously arisen from genetically engineered mice, including Ligase4−/−, p53−/−, Ptch1+/−, Ptch1+/−, P53−/−, P53−/−, Ink4c−/−. Note that the profile of gene expression is similar between GCPs from P5 cerebella and tumors consistent with the GCPs as the cell of origin for this MB variant. Adapted from Lee et al. (2003).
These recent findings suggest that successful development of mouse models that recapitulate the two other subgroups of human MBs, will require a better understanding of the location and identity of the GCPs as well as the alterations in gene or/and microRNAs associated with these specific tumor subgroups. This will enable the targeting of the right mutations into the right cell type at the right time.
3.9. What does the future hold?
An understanding of the Shh signaling pathway and the development of a mouse model that recapitulates the human disease has led to a therapeutic drug targeting the Shh pathway currently in clinical trial. A better understanding of the signaling pathways that are disrupted in each subgroup of medulloblastoma will enable the development of mouse models for all subgroups of human medulloblastomas and the screening of small molecules that ultimately will be used as novel therapeutic drugs. The complete sequencing of human primary medulloblastoma samples and the complete molecular analysis of these tumors will provide additional information that hopefully will lead to better targeted therapies.
ACKNOWLEDGMENTS
We thank Joshua Stokes from Biomedical Communication for the drawing of the figures; Dr. Eve-Ellen Govek for reading the chapter; and Drs. David Ellison, Richard Gilbertson, Frederique Zindy, Jane Johnson, and Carol A. Mason for providing illustrations used in this chapter. This work was funded by NIH grants CA-096832 (MFR), NINDS R01 NS051778-05 (M. E. H.), the Children's Brain Tumor Foundation (MFR), the Pediatric Brain Tumor Foundation (M. F. R.), the American Brain Tumor Foundation (MFR), the Starr Cancer Consortium (M. E. H.), and the American Lebanese-Syrian Associated Charities (ALSAC) of St. Jude Children's Research Hospital (MFR).
REFERENCES
- Adams NC, Tomoda T, Cooper M, Dietz G, Hatten ME. Mice that lack astrotactin have slowed neuronal migration. Development. 2002;129:965–972. doi: 10.1242/dev.129.4.965. [DOI] [PubMed] [Google Scholar]
- Adamson DC, Shi Q, Wortham M, Northcott PA, et al. OTX2 is critical for the maintenance progression of Shh-independent medulloblastoma. Cancer Res. 2010;70:181–191. doi: 10.1158/0008-5472.CAN-09-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder J, Cho NK, Hatten ME. Embryonic precursor cells from the rhombic lip are specified to a cerebellar granule neuron identity. Neuron. 1996;17:389–399. doi: 10.1016/s0896-6273(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Alder J, Lee K, Jessell T, Hatten ME. Generation of cerebellar granule neurons in vivo by transplantation of BMP-treated neural progenitor cells. Nat. Neurosci. 1999;2:535–540. doi: 10.1038/9189. [DOI] [PubMed] [Google Scholar]
- Aldinger KA, Elsen GE. Ptf1a is a molecular determinant for both glutamatergic and GABAergic neurons in the hindbrain. J. Neurosci. 2008;28:338–339. doi: 10.1523/JNEUROSCI.5139-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Rodriguez R, Barzi M, Berenguer J, Pons S. Bone morphogenetic protein 2 opposes Shh-mediated proliferation in cerebellar granule cells through a TIEG-1-based regulation of Nmyc. J. Biol. Chem. 2007;282:37170–37180. doi: 10.1074/jbc.M705414200. [DOI] [PubMed] [Google Scholar]
- Andersen BB, Korbo L, Pakkenberg B. A quantitative study of the human cerebellum with unbiased stereological techniques. J. Comp. Neurol. 1992;326:549–560. doi: 10.1002/cne.903260405. [DOI] [PubMed] [Google Scholar]
- Anthony TE, Heintz N. Genetic lineage tracing defines distinct neurogenic. Neural Dev. 2008;5:3–30. doi: 10.1186/1749-8104-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- Anthony TE, Mason HA, Gridley T, Fishell G, Heintz N. Brain lipid-binding protein is a direct target of Notch signaling in radial glial cells. Genes Dev. 2005;19:1028–1033. doi: 10.1101/gad.1302105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argenti B, Gall R, Di Marcotulllio L, et al. Hedgehog antagonist RENKCTD11 regulated proliferation and apoptosis of developing granule cell progenitors. J. Neurosci. 2005;25:8338–8346. doi: 10.1523/JNEUROSCI.2438-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruga J. The role of Zic genes in neural development. Mol. Cell. Neurosci. 2004;26:205–221. doi: 10.1016/j.mcn.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Aruga J, Nagai T, Tokuyama T, Hayashizaki Y, Okazaki Y, Chapman VM, Mikoshiba K. The mouse zic gene family. Homologues of the Drosophila pair-rule gene odd-paired. J. Biol. Chem. 1996;271:1043–1047. doi: 10.1074/jbc.271.2.1043. [DOI] [PubMed] [Google Scholar]
- Ayad NG, Rankin S, Ooi D, Rape M, Kirschner MW. Identification of ubiquitin ligase substrates by in vitro expression cloning. Methods Enzymol. 2005;399:404–414. doi: 10.1016/S0076-6879(05)99028-9. [DOI] [PubMed] [Google Scholar]
- Ayrault O, Zindy F, Rehg J, Sherr CJ, Roussel MF. Two tumor suppressors, p27Kip1 and patched-1, collaborate to prevent medulloblastoma. Mol. Cancer Res. 2009;7:33–40. doi: 10.1158/1541-7786.MCR-08-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayrault O, Zhao H, Zindy F, Qu C, Sherr CJ, Roussel MF. Atoh1 inhibits neuronal differentiation and collaborates with Gli2 to generate medulloblastoma-initiating cell. Cancer Res. 2010;70:5618–5627. doi: 10.1158/0008-5472.CAN-09-3740. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey P, Cushing H. Medulloblastoma cerebelli: A common type of midcerebellar glioma of childhood. Arch. Neurol. Psychiatry. 1925;14:192–223. [Google Scholar]
- Bailey P, Cushing H. A Classification of the Tumors of the Glioma Group on a Histogenetic Basis with a Correlated Study of Prognosis. Lippincott; Philadelphia: 1926. [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanisms and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Behesti H, Marino S. Cerebellar granule cells: Insights into proliferation, differentiation, and role in medulloblastoma pathogenesis. Int. J. Biochem. Cell Biol. 2009;41:435–445. doi: 10.1016/j.biocel.2008.06.017. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, McCall A, Berkman S, Eichele G, Bellen H, Zoghbi H. Evolutionary conservation of sequence and expression of the bHLH protein atonal suggests a conserved role in neurogenesis. Hum. Mol. Genet. 1996;5:1207–1216. doi: 10.1093/hmg/5.9.1207. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Berman DM, Karhadkar SS, Hallahan AR, Pritchard JI, Eberhart CG, Watkins DN, Chen JK, Cooper MK, Taipale J, Olson JM, Beachy PA. Medulloblastoma growth inhibition by hedgehog pathway blockade. Science. 2002;297:1559–1561. doi: 10.1126/science.1073733. [DOI] [PubMed] [Google Scholar]
- Bhatia B, Northcott PA, Hambardzumyan D, Govindarajan B, Brat DJ, Arbiser JL, Holland EC, Taylor MD, Kenney AM. Tuberous sclerosis complex suppression in cerebellar development and medulloblastoma: Separate regulation of mammalian target of rapamycin activity and p27 Kip1 localization. Cancer Res. 2009;69:7224–7234. doi: 10.1158/0008-5472.CAN-09-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia B, Nahle Z, Kenney AM. Double trouble: When sonic hedgehog signaling meets TSC inactivation. Cell Cycle. 2010;9:456–459. doi: 10.4161/cc.9.3.10532. [DOI] [PubMed] [Google Scholar]
- Blaess S, Corrales JD, Joyner AL. Sonic hedgehog regulates Gli activator and repressor functions with spatial and temporal precision in the mid/hindbrain region. Development. 2006;133:1799–1809. doi: 10.1242/dev.02339. [DOI] [PubMed] [Google Scholar]
- Borghesani PR, Peyrin JM, Klein R, Rubin J, Carter AR, Schwartz PM, Luster A, Corfas G, Segal RA. BDNF stimulates migration of cerebellar granule cells. Development. 2002;129:1435–1442. doi: 10.1242/dev.129.6.1435. [DOI] [PubMed] [Google Scholar]
- Briggs KJ, Corcoran-Schwartz IM, Zhang W, Harcke T, Devereux WL, Baylin SB, Eberhart CG, Watkins DN. Cooperation between the Hic1 and Ptch1 tumor suppressors in medulloblastoma. Genes Dev. 2008a;22:770–785. doi: 10.1101/gad.1640908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs KJ, Eberhart CG, Watkins DN. Just say no to ATOH: How HIC1 methylation might predispose medulloblastoma to lineage addiction. Cancer Res. 2008b;68:8654–8656. doi: 10.1158/0008-5472.CAN-08-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugieres L, Pierron G, Chompret A, Paillerets BB, Di Rocco F, Varlet P, Pierre-Kahn A, Caron O, Grill J, Delattre O. Incomplete penetrance of the predisposition to medulloblastoma associated with germ-line SUFU mutations. J. Med. Genet. 2010;47:142–144. doi: 10.1136/jmg.2009.067751. [DOI] [PubMed] [Google Scholar]
- Carletti B, Rossi F. Neurogenesis in the cerebellum. Neuroscientist. 2008;14:91–100. doi: 10.1177/1073858407304629. [DOI] [PubMed] [Google Scholar]
- Chi CL, Martinez S, Wurst W, Martin GR. The isthmic organizer signal FGF8 is required for cell survival in the prospective midbrain and cerebellum. Development. 2003;130:2633–2644. doi: 10.1242/dev.00487. [DOI] [PubMed] [Google Scholar]
- Ciemerych MA, Kenney AM, Sicinska E, Kalaszczynska I, Bronson RT, Rowitch DH, Gardner H, Sicinski P. Development of mice expressing a single D-type cyclin. Genes Dev. 2002;16:3277–3289. doi: 10.1101/gad.1023602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales JD, Blaess S, Mahoney EM, Joyner AL. The level of sonic hedgehog signaling regulates the complexity of cerebellar foliation. Development. 2006;133:1811–1821. doi: 10.1242/dev.02351. [DOI] [PubMed] [Google Scholar]
- Crossley PH, Martinez S, Martin GR. Midbrain development induced by FGF8 in the chick embryo. Nature. 1996;380:66–68. doi: 10.1038/380066a0. [DOI] [PubMed] [Google Scholar]
- Dakubo GD, Mazerolle CJ, Wallace VA. Expression of Notch and Wnt pathway components and activation of Notch signaling in medulloblastomas from heterozygous patched mice. J. Neurooncol. 2006;79:221–227. doi: 10.1007/s11060-006-9132-2. [DOI] [PubMed] [Google Scholar]
- D'Arca D, Zhao X, Xu W, Ramirez-Martinez NC, Iavarone A, Lasorella A. Huwe1 ubiquitin ligase is essential to synchronize neuronal and glial differentiation in the developing cerebellum. Proc. Natl. Acad. Sci. USA. 2010;107:5875–5880. doi: 10.1073/pnas.0912874107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Haas T, Oussoren E, Grojkowska W, et al. OTX1 and OTX2 expression correlated with the clinical pathologic classification of medulloblastoma. J. Neuropathol. Exp. Neurol. 2006;65:176–186. doi: 10.1097/01.jnen.0000199576.70923.8a. [DOI] [PubMed] [Google Scholar]
- De Zeeuw CI, Yeo CH. Time and tide in cerebellar memory formation. Curr. Opin. Neurobiol. 2005;15:667–674. doi: 10.1016/j.conb.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Di Marcotullio L, Ferretti E, De Smaele E, et al. RENKTCD11 is a suppressor of hedgehog signaling and is delted in human medulloblastoma. Proc. Natl. Acad. Sci. USA. 2006;29:10833–10838. doi: 10.1073/pnas.0400690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dino MR, Schuerger RJ, Liu Y, Slater NT, Mugnaini E. Unipolar brush cell: A potential feedforward excitatory interneuron of the cerebellum. Neuroscience. 2000;98:625–636. doi: 10.1016/s0306-4522(00)00123-8. [DOI] [PubMed] [Google Scholar]
- du Lac S, Raymond JL, Sejnowski TJ, Lisberger SG. Learning and memory in the vestibulo-ocular reflex. Annu. Rev. Neurosci. 1995;18:409–441. doi: 10.1146/annurev.ne.18.030195.002205. [DOI] [PubMed] [Google Scholar]
- Dymecki SM, Tomasiewicz H. Using Flp-recombinase to characterize expansion of Wnt1-expressing neural progenitors in the mouse. Dev. Biol. 1998;201:57–65. doi: 10.1006/dbio.1998.8971. [DOI] [PubMed] [Google Scholar]
- Eberhart CG, Kratz J, Wang Y, Summers K, et al. Histopatholical and molecular prognostic markers in medulloblastoma: c-Myc, N-Myc, TrkC, and anaplasia. J. Neuropathol. Exp. Neurol. 2004;63:441–449. doi: 10.1093/jnen/63.5.441. [DOI] [PubMed] [Google Scholar]
- Ebert PJ, Timmer JR, Nakada Y, Helms AW, Parab PB, Liu Y, Hunsaker TL, Johnson JE. Zic1 represses Math1 expression via interactions with the Math1 enhancer and modulation of Math1 autoregulation. Development. 2003;130:1949–1959. doi: 10.1242/dev.00419. [DOI] [PubMed] [Google Scholar]
- Edmondson JC, Hatten ME. Glial-guided granule neuron migration in vitro: A high-resolution time-lapse video microscopic study. J. Neurosci. 1987;7:1928–1934. doi: 10.1523/JNEUROSCI.07-06-01928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa JS, Luo L. Timing neurogenesis and differentiation: Insights from quantitative clonal analyses of cerebellar granule cells. J. Neurosci. 2008;28:2301–2312. doi: 10.1523/JNEUROSCI.5157-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Mikolaenko I, Elhassan I, Ni X, Wang Y, Ball D, Brat DJ, Perry A, Eberhart CG. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004;64:7787–7793. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- Faust PL. Abnormal cerebellar histogenesis in PEX2 Zellweger mice reflects multiple neuronal defects induced by peroxisome deficiency. J. Comp. Neurol. 2003;461:394–413. doi: 10.1002/cne.10699. [DOI] [PubMed] [Google Scholar]
- Faust PL, Hatten ME. Targeted deletion of the PEX2 peroxisome assembly gene in mice provides a model for Zellweger syndrome, a human neuronal migration disorder. J. Cell Biol. 1997;139:1293–1305. doi: 10.1083/jcb.139.5.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Hatten ME, Heintz N. Brain lipid-binding protein (BLBP): A novel signaling system in the developing mammalian CNS. Neuron. 1994;12:895–908. doi: 10.1016/0896-6273(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Fernandez LA, Northcott PA, Dalton J, Fraga C, Ellison D, Angers S, Taylor MD, Kenney AM. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23:2729–2741. doi: 10.1101/gad.1824509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti E, De Smaele E, Di Marcotullio L, Screpanti I, Gulino A. Hedgehog checkpoints in medulloblastoma: The chromosome 17p deletion paradigm. Trends Mol. Med. 2005;11:537–545. doi: 10.1016/j.molmed.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Ferretti E, De Smaele E, Miele E, Lavene P, et al. Concerted microRNA control of Hedeghog signaling in cerebellar neuronal progenitor and tumour cell. EMBO J. 2008;27:2616–2627. doi: 10.1038/emboj.2008.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink AJ, Englund C, Daza RA, Pham D, Lau C, Nivison M, Kowalczyk T, Hevner RF. Development of the deep cerebellar nuclei: Transcription factors and cell migration from the rhombic lip. J. Neurosci. 2006;26:3066–3076. doi: 10.1523/JNEUROSCI.5203-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay JL, Erdreich-Epstein A, Parker RJ. Progress in the treatment of childhood brain tumors: No room for complacency. Pediatr. Hematol. Oncol. 2007;24:79–84. doi: 10.1080/08880010601001073. [DOI] [PubMed] [Google Scholar]
- Flora A, Klisch TJ, Schuster G, Zoghbi HY. Deletion of Atoh1 disrupts sonic hedgehog signaling in the developing cerebellum and prevents medulloblastoma. Science. 2010;326:1424–1427. doi: 10.1126/science.1181453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty MP, Kessker JD, Wechsler-Reya RJ. Morphing into cancer: The role of developmental signaling pathways in brain tumor formation. J. Neurobiol. 2005;64:458–475. doi: 10.1002/neu.20166. [DOI] [PubMed] [Google Scholar]
- Fogarty MP, Emmenegger BA, Grasfeder LL, Oliver TG, Wechsler-Reya RJ. Fibroblast growth factor blocks Sonic hedgehog signaling in neuronal precursors and tumor cells. Proc. Natl. Acad. Sci. USA. 2007;104:2973–2978. doi: 10.1073/pnas.0605770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S. Quantitative analysis of cell proliferation and differentiation in the cortex of the postntal mouse cerebellum. J. Cell Biol. 1967;32:277–287. doi: 10.1083/jcb.32.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S, Shimada M, Nakanuna T. 3H-thymidine autoradiographic studies on the cell proliferation and differentiation in the external and internal granular layers of the mouse cerebellum. J. Comp. Neurol. 1966;128:191–209. doi: 10.1002/cne.901280206. [DOI] [PubMed] [Google Scholar]
- Furley AJ, Morton SB, Manalo D, Karagogeos D, Dodd J, Jessell TM. The axonal glycoprotein TAG-1 is an immunoglobulin superfamily member with neurite outgrowth-promoting activity. Cell. 1990;61:157–170. doi: 10.1016/0092-8674(90)90223-2. [DOI] [PubMed] [Google Scholar]
- Gajjar A, Chintagumpala M, Ashley D, Kellie S, Kun LE, Merchant TE, Woo S, Wheeler G, Ahern V, Krasin MJ, Fouladi M, Broniscer A, et al. Risk-adapted craniospinal radiotherapy followed by high-dose chemotherapy and stem-cell rescue in children with newly diagnosed medulloblastoma (St Jude Medulloblastoma-96): Long-term results from a prospective, multicentre trial. Lancet Oncol. 2006;7:813–820. doi: 10.1016/S1470-2045(06)70867-1. [DOI] [PubMed] [Google Scholar]
- Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol. Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Gibson P, Tong Y, Robinson G, Thompson M, Currle DS, Eden C, Hogg T, Poppleton H, Martin J, Finkelstein D, Pounds S, Patay Z, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010 doi: 10.1038/nature09587. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson RJ. Medulloblastoma: Signalling a change in treatment. Lancet Oncol. 2004;5:209–218. doi: 10.1016/S1470-2045(04)01424-X. [DOI] [PubMed] [Google Scholar]
- Gilbertson RJ, Ellison DW. The origins of medulloblastoma subtypes. Annu. Rev. Pathol. 2008;3:341–365. doi: 10.1146/annurev.pathmechdis.3.121806.151518. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Goodrich LV, Milenkovic L, Higgins KM, Scott MP. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277:1109–1113. doi: 10.1126/science.277.5329.1109. [DOI] [PubMed] [Google Scholar]
- Guessous F, Li Y, Abounader R. Signaling pathways in medulloblastoma. J. Cell. Physiol. 2008;217:577–583. doi: 10.1002/jcp.21542. [DOI] [PubMed] [Google Scholar]
- Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, Vorechovsky I, Holmberg E, Unden AB, Gillies S, Negus K, Smyth I, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, Russell TL, Ellenbogen RG, Bernstein ID, Beachy PA, Olson JM. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- Hallonet ME, Teillet MA, Le Douarin NM. A new approach to the development of the cerebellum provided by the quail-chick marker system. Development. 1990;108:19–31. doi: 10.1242/dev.108.1.19. [DOI] [PubMed] [Google Scholar]
- Harmey D, Smith A, Simanski S, Moussa CZ, Ayad NG. The anaphase promoting complex induces substrate degradation during neuronal differentiation. J. Biol. Chem. 2009;284:4317–4323. doi: 10.1074/jbc.M804944200. [DOI] [PubMed] [Google Scholar]
- Hatten ME. Neuronal regulation of astroglial morphology and proliferation in vitro. J. Cell Biol. 1985;100:384–396. doi: 10.1083/jcb.100.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME. Expansion of CNS precursor pools: A new role for Sonic Hedgehog. Neuron. 1999;22:2–3. doi: 10.1016/s0896-6273(00)80668-6. [DOI] [PubMed] [Google Scholar]
- Hatten ME. New directions in neuronal migration. Science. 2002;297:1660–1663. doi: 10.1126/science.1074572. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Heintz N. Mechanisms of neural patterning and specification in the developing cerebellum. Annu. Rev. Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Mason CA, Liem RK, Edmondson JC, Bovolenta P, Shelanski ML. Neuron-astroglial interactions in vitro and their implications for repair of CNS injury. Cent. Nerv. Syst. Trauma. 1984;1:15–27. doi: 10.1089/cns.1984.1.15. [DOI] [PubMed] [Google Scholar]
- Hatton BA, Knoepfler PS, Kenney AM, Rowitch DH, de Alboran IM, Olson JM, Eisenman RN. N-myc is an essential downstream effector of Shh signaling during both normal and neoplastic cerebellar growth. Cancer Res. 2006;66:8655–8661. doi: 10.1158/0008-5472.CAN-06-1621. [DOI] [PubMed] [Google Scholar]
- Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- Hogan BL. Bone morphogenetic proteins: Multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, Watanabe M, Bito H, et al. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47:201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Ibanez-Tallon I, Miwa JM, Wang HL, Adams NC, Crabtree GW, Sine SM, Heintz N. Novel modulation of neuronal nicotinic acetylcholine receptors by association with the endogenous prototoxin lynx1. Neuron. 2002;33:893–903. doi: 10.1016/s0896-6273(02)00632-3. [DOI] [PubMed] [Google Scholar]
- Ito M. Cerebellar circuitry as a neuronal machine. Prog. Neurobiol. 2006;78:272–303. doi: 10.1016/j.pneurobio.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, Quinn AG, Myers RM, Cox DR, Epstein EH, Jr., Scott MP. Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science. 1996;272:1668–1671. doi: 10.1126/science.272.5268.1668. [DOI] [PubMed] [Google Scholar]
- Joyner AL. Engrailed, Wnt and Pax genes regulate midbrain–hindbrain development. Trends Genet. 1996;12:15–20. doi: 10.1016/0168-9525(96)81383-7. [DOI] [PubMed] [Google Scholar]
- Joyner AL, Skarnes WC, Rossant J. Production of a mutation in mouse En-2 gene by homologous recombination in embryonic stem cells. Nature. 1989;338:153–156. doi: 10.1038/338153a0. [DOI] [PubMed] [Google Scholar]
- Joyner AL, Liu A, Millet S. Otx2, Gbx2 and Fgf8 interact to position and maintain a mid-hindbrain organizer. Curr. Opin. Cell Biol. 2000;12:736–741. doi: 10.1016/s0955-0674(00)00161-7. [DOI] [PubMed] [Google Scholar]
- Julian E, Dave RK, Robson JP, Hallahan AR, Wainwright BJ. Canonical Notch signaling is not required for the growth of Hedgehog pathway-induced medulloblastoma. Oncogene. 2010;29:3465–3476. doi: 10.1038/onc.2010.101. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Katoh M. Hedgehog target genes: Mechanisms of carcinogenesis induced by aberrant hedgehog signaling activation. Curr. Mol. Med. 2009;9:873–886. doi: 10.2174/156652409789105570. [DOI] [PubMed] [Google Scholar]
- Kenney AM, Rowitch DH. Sonic hedgehog promotes G(1) cyclin expression and sustained cell cycle progression in mammalian neuronal precursors. Mol. Cell. Biol. 2000;20:9055–9067. doi: 10.1128/mcb.20.23.9055-9067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney AM, Cole MD, Rowitch DH. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- Kenney AM, Widlund HR, Rowitch DH. Hedgehog and PI-3 kinase signaling converge on Nmyc1 to promote cell cycle progression in cerebellar neuronal precursors. Development. 2004;131:217–228. doi: 10.1242/dev.00891. [DOI] [PubMed] [Google Scholar]
- Kim J, Tang JY, Gong R, Lee JJ, Clemons KV, Chong CR, Chang KS, Fereshteh M, Gardner D, Reya T, Liu JO, Epstein EH, et al. Itraconazole, a commonly used antifungal that inhibits Hedgehog pathway activity and cancer growth. Cancer Cell. 2010;17:388–399. doi: 10.1016/j.ccr.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H, Stephen D, Joyner A, Curran T. Gli1 is important for medulloblastoma formation in Ptc1+/− mice. Oncogene. 2005;24:4026–4036. doi: 10.1038/sj.onc.1208567. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kongkham PN, Northcott PA, Croul SE, Smith CA, Taylor MD, Rutka JT. The SFRP family of WNT inhibitors function as novel tumor suppressor genes epigenetically silenced in medulloblastoma. Oncogene. 2010;29:3017–3024. doi: 10.1038/onc.2010.32. [DOI] [PubMed] [Google Scholar]
- Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, Troost D, Meeteren NS, Caron HN, Cloos J, Mrsic A, Ylstra B, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS ONE. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhar SG, Feng L, Vidan S, Ross ME, Hatten ME, Heintz N. Changing patterns of gene expression define four stages of cerebellar granule neuron differentiation. Development. 1993;117:97–104. doi: 10.1242/dev.117.1.97. [DOI] [PubMed] [Google Scholar]
- Laine J, Axelrad H. Extending the cerebellar Lugaro cell class. Neuroscience. 2002;115:363–374. doi: 10.1016/s0306-4522(02)00421-9. [DOI] [PubMed] [Google Scholar]
- Lange W. Cell number and cell density in the cerebellar cortex of man and some other mammals. Cell Tissue Res. 1975;157:115–124. doi: 10.1007/BF00223234. [DOI] [PubMed] [Google Scholar]
- Lee Y, McKinnon PJ. DNA ligase IV suppresses medulloblastoma formation. Cancer Res. 2002;62:6395–6399. [PubMed] [Google Scholar]
- Lee JK, Cho JH, Hwang WS, Lee YD, Reu DS, Suh-Kim H. Expression of neuroD/BETA2 in mitotic and postmitotic neuronal cells during the development of nervous system. Dev. Dyn. 2000;217:361–367. doi: 10.1002/(SICI)1097-0177(200004)217:4<361::AID-DVDY3>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Lee Y, Miller HL, Jensen P, Hernan R, Connelly M, Wetmore C, Zindy F, Roussel MF, Curran T, Gilbertson RJ, McKinnon PJ. A molecular fingerprint for medulloblastoma. Cancer Res. 2003;63:5428–5437. [PubMed] [Google Scholar]
- Lee Y, Kawagoe R, Sasai K, Li Y, Russell HR, Curran T, McKinnon PJ. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene. 2007;26:6442–6447. doi: 10.1038/sj.onc.1210467. [DOI] [PubMed] [Google Scholar]
- Levisohn L, Cronin-Golomb A, Schmahmann JD. Neuropsychological consequences of cerebellar tumour resection in children: Cerebellar cognitive affective syndrome in a paediatric population. Brain. 2000;123(Pt 5):1041–1050. doi: 10.1093/brain/123.5.1041. [DOI] [PubMed] [Google Scholar]
- Limperopoulos C, Soul JS, Gauvreau K, Huppi PS, Warfield SK, Bassan H, Robertson RL, Volpe JJ, du Plessis AJ. Late gestation cerebellar growth is rapid and impeded by premature birth. Pediatrics. 2005;115:688–695. doi: 10.1542/peds.2004-1169. [DOI] [PubMed] [Google Scholar]
- Lindsey JC, Lusher ME, Anderton JA, Bailey S, Gilbertson RJ, Pearson AD, Ellison DW, Clifford SC. Identification of tumour-specific epigenetic events in medulloblastoma development by hypermethylation profiling. Carcinogenesis. 2004;25:661–668. doi: 10.1093/carcin/bgh055. [DOI] [PubMed] [Google Scholar]
- Lindsey JC, Anderton JA, Lusher ME, Clifford SC. Epigenetic events in medulloblastoma development. Neurosurg. Focus. 2005;15:E10. doi: 10.3171/foc.2005.19.5.11. [DOI] [PubMed] [Google Scholar]
- Liu W, Chao Y-H, Chao T-F, et al. Identification of differentially expressed microRNAs by microarray: A possible role for microRNAs gene in medulloblastomas. Chin. Med. J. 2009;122:2405–2411. [PubMed] [Google Scholar]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Machold RP, Kittell DJ, Fishell GJ. Antagonism between Notch and bone morphogenetic protein receptor signaling regulates neurogenesis in the cerebellar rhombic lip. Neural Dev. 2007;2:5. doi: 10.1186/1749-8104-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin D, Li FP, Strong LC, Fraumeni JF, Jr., Nelson CE, Kim DH, et al. Germline p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- Marino S. Medulloblastoma: Developmental mechanisms out of control. Trends Mol. Med. 2005;11:17–22. doi: 10.1016/j.molmed.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- Marino S, Hoogervoorst D, Brandner S, Berns A. Rb and P107 are required for normal cerebellar development and granule cell survival but nor Purkinje cell persistence. Development. 2003;130:3359–3368. doi: 10.1242/dev.00553. [DOI] [PubMed] [Google Scholar]
- Martinez S, Crossley PH, Cobos I, Rubenstein JL, Martin GR. FGF8 induces formation of an ectopic isthmic organizer and isthmocerebellar development via a repressive effect on Otx2 expression. Development. 1999;126:1189–2000. doi: 10.1242/dev.126.6.1189. [DOI] [PubMed] [Google Scholar]
- Massague J. TGFbeta signaling: Receptors, transducers, and Mad proteins. Cell. 1996;85:947–950. doi: 10.1016/s0092-8674(00)81296-9. [DOI] [PubMed] [Google Scholar]
- Massague J, Seoane J, Wotton D. Smad transcripton factors. Genes Dev. 2005;19:2783–2810. doi: 10.1101/gad.1350705. [DOI] [PubMed] [Google Scholar]
- McMahon AP, Bradley A. The Wnt-1 (int-1) proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- Miale I, Sidman RL. An autoradiographic analysis of histogenesis in the mouse cerebellum. Exp. Neurol. 1961;4:277–296. doi: 10.1016/0014-4886(61)90055-3. [DOI] [PubMed] [Google Scholar]
- Miwa JM, Ibanez-Tallon I, Crabtree GW, Sanchez R, Sali A, Role LW, Heintz N. lynx1, an endogenous toxin-like modulator of nicotinic acetylcholine receptors in the mammalian CNS. Neuron. 1999;23:105–114. doi: 10.1016/s0896-6273(00)80757-6. [DOI] [PubMed] [Google Scholar]
- Miwa JM, Stevens TR, King SL, Caldarone BJ, Ibanez-Tallon I, Xiao C, Fitzsimonds RM, Pavlides C, Lester HA, Picciotto MR, Heintz N. The prototoxin lynx1 acts on nicotinic acetylcholine receptors to balance neuronal activity and survival in vivo. Neuron. 2006;51:587–600. doi: 10.1016/j.neuron.2006.07.025. [DOI] [PubMed] [Google Scholar]
- Morales D, Hatten ME. Molecular markers of neuronal progenitors in the embryonic cerebellar anlage. J. Neurosci. 2006;26:12226–12236. doi: 10.1523/JNEUROSCI.3493-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara Y, Northcott PA, Li M, Kongkham PN, et al. Genetic and epigentic inactivation of Kruppel-like factor 4 in medulloblastoma. Neoplasia. 2010;12:20–27. doi: 10.1593/neo.91122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Fernandez LA, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, Grundy R, Van Meter T, Rutka JT, Croce CM, Kenney AM, Taylor MD. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69:3249–3255. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Rutka JT, Taylor MD. Genomics of medulloblastoma: From Giemsa-banding to next-generation sequencing in 20 years. Neurosurg. Focus. 2010;28(E6):1–20. doi: 10.3171/2009.10.FOCUS09218. [DOI] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. C-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Oliver TG, Read TA, Kessler JD, Mehmeti A, Wells JF, Huynh TT, Lin SM, Wechsler-Reya RJ. Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medulloblastoma. Development. 2005;132:2425–2439. doi: 10.1242/dev.01793. [DOI] [PubMed] [Google Scholar]
- Packer RJ, Cogen P, Vezina G, Rorke LB. Medulloblastoma: Clinical and biologic aspects. Neuro-oncology. 1999;1:232–250. doi: 10.1215/15228517-1-3-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palay SL, Chan-Palay S. The Cerebellar Cortex: Cytology and Organization. Springer-Verlag; New York: 1974. [Google Scholar]
- Pan N, Jahan I, Lee JE, Fritzsch B. Defects in the cerebella of conditional Neurod1 null mice correlate with effective Tg(Atoh1-cre) recombination and granule cell requirements for Neurod1 for differentiation. Cell Tissue Res. 2009;337:407–428. doi: 10.1007/s00441-009-0826-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M, Abasolo I, Mingorance-Le Meur A, Martinez A, Del Rio JA, Wright CV, Real FX, Soriano E. Cerebellar GABAergic progenitors adopt an external granule cell-like phenotype in the absence of Ptf1a transcription factor expression. Proc. Natl. Acad. Sci. USA. 2007;104:5193–5198. doi: 10.1073/pnas.0605699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino L, Ghiorzo P, Nasti S, Battistuzzi L, Cusano R, Marzocchi C, Garre ML, Clementi M, Scarra GB. Identification of a SUFU germline mutation in a family with Gorlin syndrome. Am. J. Med. Genet. A. 2009);149A:1539–1543. doi: 10.1002/ajmg.a.32944. [DOI] [PubMed] [Google Scholar]
- Pizer BL, Clifford SC. The potential impact of tumour biology on improved clinical practice for medulloblastoma: Progress towards biologically driven clinical trials. Br. J. Neurosurg. 2009;23:364–375. doi: 10.1080/02688690903121807. [DOI] [PubMed] [Google Scholar]
- Pogoriler J, Millen K, Utset M, Du W. Loss of cyclin D1 impairs cerebellar development and suppresses medulloblastoma formation. Development. 2006;133:3929–3937. doi: 10.1242/dev.02556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, Kim JY, Goumnerova LC, Black PM, Lau C, Allen JC, Zagzag D, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- Qin L, Wine-Lee L, Ahn KJ, Crenshaw EB., 3rd Genetic analyses demonstrate that bone morphogenetic protein signaling is required for embryonic cerebellar development. J. Neurosci. 2006;26:1896–1905. doi: 10.1523/JNEUROSCI.3202-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Neuron-glia relationship during granule cell migration in developing cerebellar cortex. A Golgi and electronmicroscopic study in Macacus Rhesus. J. Comp. Neurol. 1971;141:283–312. doi: 10.1002/cne.901410303. [DOI] [PubMed] [Google Scholar]
- Ramon y Cajal S. Histology of the Nervous System of Man and Vertebrates. Oxford University Press; Oxford: 1995. [Google Scholar]
- Rankin S, Ayad NG, Kirschner MW. Sororin, a substrate of the anaphase-promoting complex, is required for sister chromatid cohesion in vertebrates. Mol. Cell. 2005;18:185–200. doi: 10.1016/j.molcel.2005.03.017. [DOI] [PubMed] [Google Scholar]
- Rios I, Alvarez-Rodriguez R, Marti E, Pons S. Bmp2 antagonizes sonic hedgehog-mediated proliferation of cerebellar granule neurones through Smad5 signalling. Development. 2004;131:3159–3168. doi: 10.1242/dev.01188. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Milenkovic L, Scott MP. Patched 1 regulates hedgehog signaling at the primary cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- Romer J, Curran T. Targeting medulloblastoma: Small-molecule inhibitors of the Sonic Hedgehog pathway as potential cancer therapeutics. Cancer Res. 2005;65:4975–4978. doi: 10.1158/0008-5472.CAN-05-0481. [DOI] [PubMed] [Google Scholar]
- Rubin JB, Rowitch DH. Medulloblastoma: A problem of developmental biology. Cancer Cell. 2002;2:7–8. doi: 10.1016/s1535-6108(02)00090-9. [DOI] [PubMed] [Google Scholar]
- Rubin JB, Kung AL, Klein RS, Chan JA, Sun Y, Schmidt K, Kieran MW, Luster AD, Segal RA. A small-molecule antagonist of CXCR4 inhibits intracranial growth of primary brain tumors. Proc. Natl. Acad. Sci. USA. 2003;100:13513–13518. doi: 10.1073/pnas.2235846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Hann CL, Laterra J, Yauch RL, Callahan CA, Fu L, Holcomb T, Stinson J, Gould SE, Coleman B, LoRusso PM, Von Hoff DD, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N. Engl. J. Med. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salero E, Hatten ME. Differentiation of ES cells into cerebellar neurons. Proc. Natl. Acad. Sci. USA. 2007;104:2997–3002. doi: 10.1073/pnas.0610879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo LJ, Edgar BA. Filling out the Hippo pathway. Nat. Rev. Mol. Cell Biol. 2007;8:613–621. doi: 10.1038/nrm2221. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD, Caplan D. Cognition, emotion and the cerebellum. Brain. 2006;129:290–292. doi: 10.1093/brain/awh729. [DOI] [PubMed] [Google Scholar]
- Schwartz PM, Borhesani PR, Levy RL, Pomeroy SL, Segal R. Abnormal cerebellar development and foliation in BDNF−/− mice reveals a role for neurotrophins in CNS patterning. Neuron. 1997;19:269–281. doi: 10.1016/s0896-6273(00)80938-1. [DOI] [PubMed] [Google Scholar]
- Shrivastava S, Zou ZQ, Pirollo K, Blattner W, Chang EH. Germline transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature. 1990;348:747–749. doi: 10.1038/348747a0. [DOI] [PubMed] [Google Scholar]
- Solecki DJ, Liu XL, Tomoda T, Fang Y, Hatten ME. Activated Notch2 signaling inhibits differentiation of cerebellar granule neuron precursors by maintaining proliferation. Neuron. 2001;31:557–568. doi: 10.1016/s0896-6273(01)00395-6. [DOI] [PubMed] [Google Scholar]
- Spassky N, Han YG, Aguilar A, Strehl L, Besse L, Laclef C, Ros MR, Garcia-Verdugo JM, Alvarez-Buylla A. Primary cilia are required for cerebellar development and Shh-dependent expansion of progenitor pool. Dev. Biol. 2008;317:246–259. doi: 10.1016/j.ydbio.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stottmann RW, Rivas RJ. Distribution of TAG-1 and synaptophysin in the developing cerebellar cortex: Relationship to Purkinje cell dendritic development. J. Comp. Neurol. 1998;395:121–135. [PubMed] [Google Scholar]
- Tanori M, Santone M, Mancuso M, Pasquali E, Leonardi S, Di Majo V, Rebessi S, Saran A, Pazzaglia S. Developmental and oncogenic effects of insulin-like growth factor-I in Ptc1+/− mouse cerebellum. Mol. Cancer. 2010;9:53. doi: 10.1186/1476-4598-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MC, Fuller C, Hogg TL, Dalton J, Finkelstein D, Lau CC, Chintagumpala M, Adesina A, Ashley DM, Kellie SJ, Taylor MD, Curran T, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J. Clin. Oncol. 2006;24:1924–1931. doi: 10.1200/JCO.2005.04.4974. [DOI] [PubMed] [Google Scholar]
- Timmann D, Watts S, Hore J. Failure of cerebellar patients to time finger opening precisely causes ball high-low inaccuracy in overarm throws. J. Neurophysiol. 1999;82:103–114. doi: 10.1152/jn.1999.82.1.103. [DOI] [PubMed] [Google Scholar]
- Tong WM, Ohgati H, Hunag H, Granier C, Kleihues P, Wang ZQ. Null mutation of DNA strand break-binding molecule poly(ADP-ribose) polymerase causes medulloblastoms in p53−/− mice. Am. J. Pathol. 2003;162:343–352. doi: 10.1016/S0002-9440(10)63825-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel T, Zindy F, Xie S, Lee Y, Forget A, Magdaleno S, Rehg JE, Calabrese C, Solecki D, Eberhart CG, Sherr SE, Plimmer S, et al. The tumor suppressors Ink4c and p53 collaborate independently with Patched to suppress medulloblastoma formation. Genes Dev. 2005;19:2656–2667. doi: 10.1101/gad.1368605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uziel T, Karginov FV, Xie S, Parker JS, Wang Y-D, Gajjar A, He L, Ellison D, Gilbertson RG, Hannon G, Roussel MF. The miR-17 92 cluster collaborates with the sonic hedgehog pathway in medulloblastoma. Proc. Natl. Acad. Sci. USA. 2009;106:2812–2817. doi: 10.1073/pnas.0809579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, et al. Targeted deletion revelas essentail and overlapping functions af the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe JJ. Cerebellum of the premature infant: Rapidly developing, vulnerable, clinically important. J. Child Neurol. 2009;24:1085–1104. doi: 10.1177/0883073809338067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hoff K, Hartmann W, von Bueren AO. Large cell anaplastic medulloblastoma: Outcome according to Myc status, histopatholical and clinical risk factors. Pediatr. Blood Cancer. 2010;54:369–376. doi: 10.1002/pbc.22339. [DOI] [PubMed] [Google Scholar]
- Wang VY, Rose MF, Zoghbi HY. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron. 2005;48:31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya RJ, Scott MP. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- Wechsler-Reya R, Scott MP. The developmental biology of brain tumors. Annu. Rev. Neurosci. 2001;24:385–428. doi: 10.1146/annurev.neuro.24.1.385. [DOI] [PubMed] [Google Scholar]
- Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG., Jr Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194–198. doi: 10.1038/nature02381. [DOI] [PubMed] [Google Scholar]
- Wen X, Lai CK, Evangelista M, Hongo JA, de Sauvage FJ, Scales SJ. Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Mol. Cell. Biol. 2010;30:1910–1922. doi: 10.1128/MCB.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingate RJ. The rhombic lip and early cerebellar development. Curr. Opin. Neurobiol. 2001;11:82–88. doi: 10.1016/s0959-4388(00)00177-x. [DOI] [PubMed] [Google Scholar]
- Wingate RJ, Hatten ME. The role of the rhombic lip in avian cerebellum development. Development. 1999;126:4395–4404. doi: 10.1242/dev.126.20.4395. [DOI] [PubMed] [Google Scholar]
- Xu P, Yu S, Jiang R, Kang C, Wang G, Jiang H, Pu P. Differential expression of Notch family members in astrocytomas and medulloblastomas. Pathol. Oncol. Res. 2009;15:703–710. doi: 10.1007/s12253-009-9173-x. [DOI] [PubMed] [Google Scholar]
- Yan CT, Kaushal D, Murphy M, et al. XRCC4 suppresses medulloblastomas with recurrent translocations in p53-deficient mice. Proc. Natl. Acad. Sci. USA. 2006;103:7378–7383. doi: 10.1073/pnas.0601938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Jackson E, Woerner BM, Perry A, Piwnica-Worms D, Rubin JB. Blocking CXCR4-mediated cyclic AMP suppression inhibits brain tumor growth in vivo. Cancer Res. 2007;67:651–658. doi: 10.1158/0008-5472.CAN-06-2762. [DOI] [PubMed] [Google Scholar]
- Yoon JW, Gilbertson R, Iannaccone S, Iannaccone P, Walterhouse D. Defining a role for Sonic hedgehog pathway activation in desmoplastic medulloblastoma by identifying GLI1 target genes. Int. J. Cancer. 2009;124:109–119. doi: 10.1002/ijc.23929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Goldman JE. Developmental fates and migratory pathways of dividing progenitors in the postnatal rat cerebellum. J. Comp. Neurol. 1996;370:536–550. doi: 10.1002/(SICI)1096-9861(19960708)370:4<536::AID-CNE9>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Zhao H, Ayrault O, Zindy F, Kim JH, Roussel MF. Post-transcriptional down-regulation of Atoh1/Math1 by bone morphogenic proteins suppresses medulloblastoma development. Genes Dev. 2008;22:722–727. doi: 10.1101/gad.1636408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Heintz N, Hatten ME. CNS gene encoding astrotactin, which supports neuronal migration along glial fibers. Science. 1996;272:417–419. doi: 10.1126/science.272.5260.417. [DOI] [PubMed] [Google Scholar]
- Zindy F, Nilsson LM, Nguyen L, Meunier C, Smeyne RJ, Rehg JE, Eberhart C, Sherr CJ, Roussel MF. Hemangiomas, Medulloblastomas, and other tumors in Ink4c/p53-null mice. Cancer Res. 2003;63:5420–5437. [PubMed] [Google Scholar]
- Zindy F, Knoepfler PS, Xie S, Sherr CJ, Eisenman RN, Roussel MF. N-Myc and the cyclin-dependent kinase inhibitors p18Ink4c and p27Kip1 coordinately regulate cerebellar development. Proc. Natl. Acad. Sci. USA. 2006;103:11579–11583. doi: 10.1073/pnas.0604727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Uziel T, Ayrault O, Calabrese C, Valentine M, Rehg G, Gilbertson R, Sherr CJ, Roussel MF. Genetic alterations in mouse medulloblastoma and generation of tumors de novo from primary cerebellar granule neuron precursors. Cancer Res. 2007;67:2676–2684. doi: 10.1158/0008-5472.CAN-06-3418. [DOI] [PubMed] [Google Scholar]


