Abstract
Context:FGFR1 mutations cause isolated hypogonadotropic hypogonadism (IHH) with or without olfactory abnormalities, Kallmann syndrome, and normosmic IHH respectively. Recently, missense mutations in FGF8, a key ligand for fibroblast growth factor receptor (FGFR) 1 in the ontogenesis of GnRH, were identified in IHH patients, thus establishing FGF8 as a novel locus for human GnRH deficiency.
Objective: Our objective was to analyze the clinical, hormonal, and molecular findings of two familial IHH patients due to FGF8 gene mutations.
Methods and Patients: The entire coding region of the FGF8 gene was amplified and sequenced in two well-phenotyped IHH probands and their relatives.
Results: Two unique heterozygous nonsense mutations in FGF8 (p.R127X and p.R129X) were identified in two unrelated IHH probands, which were absent in 150 control individuals. These two mutations, mapped to the core domain of FGF8, impact all four human FGF8 isoforms, and lead to the deletion of a large portion of the protein, generating nonfunctional FGF8 ligands. The p.R127X mutation was identified in an 18-yr-old Kallmann syndrome female. Her four affected siblings with normosmic IHH or delayed puberty also carried the p.R127X mutation. Additional developmental anomalies, including cleft lip and palate and neurosensorial deafness, were also present in this family. The p.R129X mutation was identified in a 30-yr-old man with familial normosmic IHH and severe GnRH deficiency.
Conclusions: We identified the first nonsense mutations in the FGF8 gene in familial IHH with variable degrees of GnRH deficiency and olfactory phenotypes, confirming that loss-of-function mutations in FGF8 cause human GnRH deficiency.
Two novel nonsense FGF8 gene mutations were identified in two families with isolated hypogonadotropic hypogonadism, confirming that its loss-of-function causes human GnRH deficiency.
The human fibroblast growth factors (FGFs) are encoded by 18 distinct genes (FGF1-FGF10 and FGF16-FGF23) that play pleiotropic roles in human development and metabolism (1). The biological activities of the FGFs are mediated by FGF receptor tyrosine kinases (FGFRs) encoded by four distinct genes (FGFR1–FGFR4) in mammals (1). Extracellular interaction between the FGFs, the receptor and heparan sulfate (HS) lead to formation of a 2:2:2 FGF-FGFR-HS symmetric dimer, which enables the intracellular tyrosine kinase domains to trans-autophosphorylate each other and become activated (2). Aberrant FGF-FGFR signaling due to gain- or loss-of-function mutations or misexpression has been implicated in a multitude of human diseases, including craniosynostosis and chondrodysplasia syndromes, cancer, and isolated hypogonadotropic hypogonadism (IHH) (1,3,4).
Congenital IHH is characterized by absent or partial puberty due to GnRH deficiency, with otherwise normal pituitary function and normal imaging of the hypothalamus-pituitary area (5). IHH may occur in association with olfactory abnormalities (Kallmann syndrome), whereas a normal sense of smell defines normosmic IHH (5). In addition, it is occasionally associated with other developmental anomalies, including renal agenesis, cleft lip and/or palate, selective tooth agenesis, and bimanual synkinesis (4,6). Loss-of-function mutations in FGFR1, KAL1, GNRHR/GNRH1, PROKR2/PROK2, GPR54, TACR3/TAC3, and CHD7 genes have been reported in IHH patients (4,5,6,7,8). However, the genetic defects in nearly 70% of IHH cases remain to be discovered.
Research on the functional impact of a FGFR1 mutation (p.L342S), identified in a Kallmann syndrome patient, has shed light on the identity of the cognate FGFs of FGFR1 that partake in the GnRH neuron’s ontogenesis (9). It was shown that the p.L342S mutation eliminates a key binding site for FGF8 on FGFR1, thereby causing a specific reduction in FGF8’s ability to bind and signal through FGFR1. Subsequently, Falardeau et al. (10) identified several heterozygous and one homozygous missense mutation in the FGF8 gene in patients with IHH and variable sense of smell. In support of these clinical studies, mice homozygous for Fgf8 hypomorphic allele have a small telencephalon with no olfactory bulbs as well as other cardiac, craniofacial, forebrain, midbrain, and cerebellar developmental abnormalities including absent GnRH neurons (11). In contrast to knockout Fgfr1 mice, Fgf8 hypomorphic mice had also a severe renal hypoplasia, an abnormality frequently observed in KAL1 but not in FGFR1 Kallmann syndrome patients (4,12,13).
In this report, we provide clinical and hormonal findings concerning two familial cases of IHH, one with Kallmann syndrome and one with normosmic IHH, carrying distinct FGF8 nonsense mutations, supporting the pathogenic role of impaired FGF8 signaling in human GnRH deficiency.
Patients and Methods
Two unrelated patients, one with Kallmann syndrome (case 1) and one with normosmic IHH (case 2), are presented. These patients were referred to the Endocrine Unit at the Clinical Hospital of University of Sao Paulo and University of Campinas (Unicamp), respectively. Both had a family history of IHH. Olfactory acuity was evaluated by the University of Pennsylvania Smell Identification Test (UPSIT) or Alcohol Sniff Test http://drdavidson.ucsd.edu/Default.aspx?tabid=144. Both patients had been previously screened for KAL1, FGFR1, PROKR2/PROK2, GnHR-R, and GPR54/KiSS1 and found to be negative for mutations (14,15).
One hundred fifty Brazilian adult individuals with normal sexual development at the appropriate chronological age and normal olfactory status were used as control group. This study was approved by the Ethical Committee of each institution, and written informed consent was obtained from the patients and their families for clinical and genetic studies.
Hormone assays
Gonadotropin levels were measured by immunofluorometric assays (IFMA) or RIA. The coefficient of variation was less than 5% for all assays. The lower limits of detection were 0.1 U/liter for LH, 1.0 U/liter for FSH, 13 pg/ml (47 pmol/liter) for estradiol, and 19 ng/dl (0.6 nmol/liter) for testosterone. Reference ranges are depicted in Table 1. Serum LH and FSH levels were measured at −15, 0, 15, 30, 45, and 60 min after the iv administration of 100 μg of GnRH. For RIA, LH peak after GnRH (100 μg gonadorelin iv) higher than 15 U/liter in females and higher than 25 U/liter in males were considered pubertal responses. For IFMA, GnRH-stimulated LH peak higher than 6.9 U/liter in girls and higher than 9.6 U/liter in boys were considered pubertal responses.
Table 1.
Clinical and hormonal features of individuals with familial GnRH deficiency due to nonsense mutations of the FGF8 gene
| Sex | Genotype | Hormonal evaluation (IFMA) |
Puberty | Olfactory status | Reproductive phenotypes | Other features | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LH (IU/liter) | FSH (IU/liter) | T (ng/dl) | E2 (pg/ml) | |||||||||
| Basal | Peak | Basal | Peak | |||||||||
| Family 1 | ||||||||||||
| II:1 | M | p.R127X/wt | 0.5 | <1.0 | <11 | IHH | Normosmic | Micropenis | Cleft lip and palate, deafness | |||
| II:2 | F | p.R127X/wt | Delay | Normosmic | Deafness | |||||||
| II:4 (case 1) | F | p.R127X/wt | 2.3 | 6.8 | <1.0 | 3.4 | <13 | IHH | Hyposmic | Primary amenorrhea | ||
| II:5 | F | p.R127X/wt | 0.2 | 1.6 | <13 | IHH | Normosmic | Primary amenorrhea | ||||
| II:7 | F | p.R127X/wt | 3.5 | 22.5 | 5.9 | 12.9 | 244 | Delay | Normosmic | Obesity, psychiatric disorder | ||
| Family 2 | ||||||||||||
| II:3 (case 2) | M | p.R129X/wt | <0.1 | 2.0 | <1.0 | 70 | IHH | Normosmic | Micropenis, cryptorchidism, azospermia | |||
| II:4 | F | IHH | Normosmic | Primary amenorrhea | ||||||||
Normal values measured by IFMA are as follows: testosterone (T), 271–965 ng/dl; estradiol (E2), prepubertal less than 21 pg/ml; basal LH, 1.0–8.4 U/liter; basal FSH, 1.0–10.5 U/liter; and GnRH-stimulated LH,12–29.7 U/liter, and FSH, 2.9–7.8 U/liter. F, Female; M, male.
RIA; normal values are as follows: testosterone, 12–38 nmol/liter; basal LH, 1.7–3.0 U/liter; basal FSH, 1.0–6.6 U/liter; and GnRH-stimulated LH, 25 U/liter, and FSH, 3–26 U/liter.
Moderate microsmia [University of Pennsylvania Smell Identification Test (UPSIT)].
Not available for evaluation.
Genetic analysis of FGF8 gene
Genomic DNA was extracted from peripheral blood leukocytes using standard procedures. All exons and adjacent intronic sequences for the FGF8 gene (NM 033163) were amplified in 20 μl reaction mixes containing 200–500 ng DNA, 0.2 mm dNTPs, 1.5 mm MgCl2, 0.6 pmol of each of the primers, 1× PCR buffer, and 1 U Taq polymerase (Amersham Pharmacia, Piscataway, NJ). Amplified products were pretreated with an enzymatic combination of exonuclease I and shrimp alkaline phosphatase (U.S. Biochemical Corp., Cleveland, OH) and directly sequenced using the BigDye terminator cycle sequencing ready reaction kit (PE Applied Biosystems, Foster City, CA) in an ABI PRISM 310 automatic sequencer (PerkinElmer Cetus, Foster City, CA). The identified mutations were confirmed by sequencing both strands of PCR products from three independent reactions. For primer sequences and annealing temperatures, see Supplemental Table 1 (published on The Endocrine Society’s Journals Online web site at http://jcem.endojournals.org).
Results
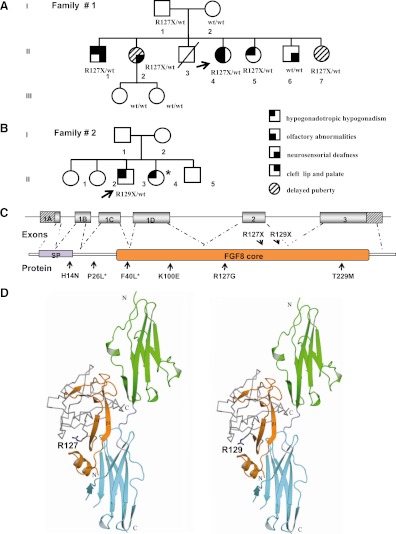
Automatic sequencing of the FGF8 gene revealed two heterozygous nonsense mutations, p.R127X and p.R129X in two familial cases of IHH due to C to T transitions at nucleotides 763 and 769 (c.763C→T and c.769C→T), respectively (Supplemental Fig. 1). The p.R127X was identified in the female patient with Kallmann syndrome (case 1; subject II:4, family 1) and the p.R129X in the male patient with normosmic IHH (case 2; subject II:3, family 2). Both mutations are located in the highly conserved FGF β-trefoil core domain and were absent in 150 unaffected control individuals (Fig. 1, C and D).
Figure 1.
The p.R127X and p.R129X mutations in the FGF8 gene. Panel A, Pedigree of family 1, a female proband with Kallmann syndrome and her four siblings carrying the p.R127X FGF8 mutation. Two of them reported delayed puberty, whereas the other two had normosmic IHH. They also had other additional abnormalities such as bilateral neurosensorial deafness and cleft lip and palate. Panel B, Pedigree of family 2, a male proband with normosmic IHH who harbors the p.R129X mutation of the FGF8. Arrows indicate proband; squares are males, and circles are females. Panel C, FGF8 genomic and protein structures showing the localization of the novel (top) and the previously described (bottom) FGF8 mutations. The hatched areas indicate the 5′ and 3′ untranslated region. SP, Signal peptide. *, These mutations affected only the isoforms e and f of FG8 protein. Panel D, Analysis of the impact of the p.R127Xand p.R129X mutations on the function of FGF8 ligand based on the FGF8b:FGFR2c (2FDB Protein Data Bank) (16). D2 and D3 of FGFR2c are green and cyan, respectively. The mutations map to the start of β5 strand and are predicted to truncated FGF8 ligands lacking a large portion of the protein that is essential for receptor and HS binding. The expressed regions of the mutant proteins are shown in orange ribbons. The amino acid residues predicted to be absent in both FGF8 mutants are shown in gray. The N terminus and C terminus of FGFR and FGF8b are denoted by the letters N and C, respectively.
Basal hormone evaluation showed inappropriately low gonadotropin levels together with prepubertal concentrations of sex steroids in both patients [case 1: LH 0.2 U/liter, FSH <1.0 U/liter (RIA), and estradiol <13 pg/ml (RIA); case 2: LH <0.1 IU/liter, FSH <1.0 IU/liter, and testosterone 70 ng/dl]. Both patients exhibited a poor response after a prolonged GnRH stimulation test (Table 1). Other tests of anterior pituitary function, as well as hypothalamus-pituitary imaging and an abdominal ultrasonography, were normal. Magnetic resonance imaging of the olfactory structure revealed normal olfactory bulbs and sulci. Reproductive phenotypes included primary amenorrhea (case 1) and micropenis and cryptorchidism (case 2). No other phenotypic stigmata, including dental agenesis, cleft lip/palate, nystagmus, hearing loss, bimanual synkinesis, and color blindness was observed. Patients’ characteristics and their hormonal profile are outlined in Table 1. Pedigree presentations of both cases are shown in Fig. 1, A and B, and all familial genotype-phenotype information available is described in Table 1. Further details are given in the Supplemental Data.
Discussion
FGF8 is expressed in many organizing centers during embryogenesis and controls patterning of many tissues/organs including brain, limbs, heart, ear, and eye (16,17). An alternative splicing event at the N terminus of FGF8 generates four isoforms (a, b, e, and f) that serves to modulate binding interactions of FGF8 with its cognate FGFRs including FGFR1c (16,17). Here, we have identified the first heterozygous nonsense FGF8 mutations, p.R127X and p.R129X, in two probands with familial IHH, one with Kallmann syndrome and one with normosmic IHH, respectively. Both mutations map to the core domain of FGFs (Fig. 1, C and D) and therefore should manifest in all four FGF8 isoforms. The mutations are predicted to give rise to truncated FGF8 ligands that lack key regions required for receptor and HS binding (16). Alternatively, mRNA transcripts that contain a premature termination codon may be destroyed by a nonsense mRNA decay mechanism leading also to FGF8 haploinsufficiency (18).
The first loss-of-function FGF8 mutation, a homozygous p.D73H, in humans was identified in 2007 by Riley et al. (19), in a male with a nonsyndromic cleft lip and palate. One year later, Falardeau et al. (10) reported FGF8 missense mutations in patients with Kallmann syndrome and normosmic IHH (10). Six missense mutations were detected in FGF8 in IHH probands with variable olfactory phenotypes and different degrees of GnRH deficiency, including the rare adult-onset form of IHH. Four of these mutations (p.H14N, p.K100E, p.R127G, and p.T229M) affected all four FGF8 splice isoforms, whereas two mutations (p.P26L and p.F40L) affected only e and f FGF8 isoforms. All FGF8 mutations were identified in the heterozygous state, except for p.F40L mutation. Biochemical analysis of mutated FGF8b and FGF8f revealed that mutations in the FGF8 gene impair the biological activity of ligand in vitro (10).
Interestingly, although mice homozygous for a Fgf8 hypomorphic allele lack GnRH neurons in the hypothalamus, heterozygous mice exhibited markedly decreased numbers of GnRH neurons, indicating an exquisite sensitivity of decreased FGF8 signaling for GnRH neuron development (10). In addition, experimental results of Chung et al. (20) clearly indicated that one Fgf8 hypomorphic allele is sufficient to significantly impair GnRH neuronal development. These authors suggested that FGF8 mutations in Kallmann syndrome may compromise human GnRH system function by abrogating the initial emergence of GnRH neurons from the olfactory epithelium (20).
Several nonreproductive phenotypes have been described in IHH patients. Some of these anomalies, such as bimanual synkinesis, midline defects, and dental agenesis, were largely attributed to KAL1 and FGFR1 mutations (Supplemental Table 2). Considerable phenotypic heterogeneity was evident in family 1, with affected members presenting variable degrees of GnRH deficiency, from severe IHH to delayed puberty and normal reproductive function. Furthermore, variable olfactory phenotypes, craniofacial abnormalities, and neurosensorial deafness also prevailed among them (Fig. 1A and Table 1). It is likely that the effect of the heterozygous FGF8 mutations is modified by other gene defects and/or the environment, explaining at least partially the variable expressivity and incomplete penetrance. In fact, a digenic model of inheritance has been proposed as a cause of the highly phenotypic heterogeneity observed in families with GnRH deficiency (9).
Recently, a missense mutation of FGF8, disturbing the same R127 residue (p.R127G), was reported to cause Kallmann syndrome associated with cleft lip/palate, deafness, flat nasal bridge, camplodactyly, and hyperlaxity (10). In family 1 (Fig. 1A), cleft lip and palate was present in one affected member (subject II:1) who harbored the p.R127X FGF8 mutation. These findings, together with the previous descriptions of patients with cleft lip palate due to FGFR1 or FGF8 mutations, strongly suggest that impaired FGF8 signaling contributes to cleft lip/palate (4,10,19). Neurosensorial deafness was also observed in several members of family 1 harboring the p.R127X, including one brother with normosmic IHH and cleft lip and palate (subject II:1) and two siblings with delayed puberty and deafness (subjects II:2 and II:7). Notably, neurosensorial deafness was also observed in one sibling without the p.R127X (subject II:6), suggesting that deafness segregates independently of the FGF8 mutation in this family.
In conclusion, we described novel molecular FGF8 gene defects and provided unequivocal evidence that FGF8 loss-of-function mutations are a cause of IHH in humans. In addition, the findings of FGF8 mutations in patients with and without olfactory abnormalities extend the repertoire of genes implicated in both Kallmann syndrome and normosmic IHH.
Footnotes
This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo [FAPESP Grants 07/50938-4 (to E.B.T.), 06/52583-6 (to A.P.A.), and 05/04726-0 (to A.C.L.)] and Conselho Nacional de Desenvolvimento Científico e Tecnológico [CNPq Grants 300209/2008-9 (to A.C.L.) and 300828/2005-5 (to B.B.M.)].
Disclosure Summary: The authors have nothing to disclose.
First Published Online May 12, 2010
Abbreviations: FGF, Fibroblast growth factor; FGFR, FGF receptor tyrosine kinase; HS, heparan sulfate; IFMA, immunofluorometric assays; IHH, isolated hypogonadotropic hypogonadism.
References
- Turner N, Grose R 2010 Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer 10:116–129 [DOI] [PubMed] [Google Scholar]
- Eswarakumar VP, Lax I, Schlessinger J 2005 Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev 16:139–149 [DOI] [PubMed] [Google Scholar]
- Marie PJ, Coffin JD, Hurley MM 2005 FGF and FGFR signaling in chondrodysplasias and craniosynostosis. J Cell Biochem 96:888–896 [DOI] [PubMed] [Google Scholar]
- Trarbach EB, Silveira LG, Latronico AC 2007 Genetic insights into human isolated gonadotropin deficiency. Pituitary 10:381–391 [DOI] [PubMed] [Google Scholar]
- Seminara SB, Hayes FJ, Crowley Jr WF 1998 Gonadotropin-releasing hormone deficiency in the human (idiopathic hypogonadotropic hypogonadism and Kallmann’s syndrome): pathophysiological and genetic considerations. Endocr Rev 19:521–539 [DOI] [PubMed] [Google Scholar]
- Bhangoo A, Jacobson-Dickman E 2009 The genetics of idiopathic hypogonadotropic hypogonadism: unraveling the biology of human sexual development. Pediatr Endocrinol Rev 6:395–404 [PubMed] [Google Scholar]
- Bouligand J, Ghervan C, Tello JA, Brailly-Tabard S, Salenave S, Chanson P, Lombès M, Millar RP, Guiochon-Mantel A, Young J 2009 Isolated familial hypogonadotropic hypogonadism and a GNRH1 mutation. N Engl J Med 360:2742–2748 [DOI] [PubMed] [Google Scholar]
- Chan YM, de Guillebon A, Lang-Muritano M, Plummer L, Cerrato F, Tsiaras S, Gaspert A, Lavoie HB, Wu CH, Crowley Jr WF, Amory JK, Pitteloud N, Seminara SB 2009 GNRH1 mutations in patients with idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci USA 106:11703–11708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitteloud N, Quinton R, Pearce S, Raivio T, Acierno J, Dwyer A, Plummer L, Hughes V, Seminara S, Cheng YZ, Li WP, Maccoll G, Eliseenkova AV, Olsen SK, Ibrahimi OA, Hayes FJ, Boepple P, Hall JE, Bouloux P, Mohammadi M, Crowley W 2007 Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J Clin Invest 117:457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falardeau J, Chung WC, Beenken A, Raivio T, Plummer L, Sidis Y, Jacobson-Dickman EE, Eliseenkova AV, Ma J, Dwyer A, Quinton R, Na S, Hall JE, Huot C, Alois N, Pearce SH, Cole LW, Hughes V, Mohammadi M, Tsai P, Pitteloud N 2008 Decreased FGF8 signaling causes deficiency of gonadotropin-releasing hormone in humans and mice. J Clin Invest 118:2822–2831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers EN, Lewandoski M, Martin GR 1998 An Fgf8 mutant allelic series generated by Cre- and Flp-mediated recombination. Nat Genet 18:136–141 [DOI] [PubMed] [Google Scholar]
- Perantoni AO, Timofeeva O, Naillat F, Richman C, Pajni- Underwood S, Wilson C, Vainio S, Dove LF, Lewandoski M 2005 Inactivation of FGF8 in early mesoderm reveals an essential role in kidney development. Development 132:3859–3871 [DOI] [PubMed] [Google Scholar]
- Poladia DP, Kish K, Kutay B, Hains D, Kegg H, Zhao H, Bates CM 2006 Role of fibroblast growth factor receptors 1 and 2 in the metanephric mesenchyme. Dev Biol 291:325–339 [DOI] [PubMed] [Google Scholar]
- Abreu AP, Trarbach EB, de Castro M, Frade Costa EM, Versiani B, Matias Baptista MT, Garmes HM, Mendonca BB, Latronico AC 2008 Loss-of-function mutations in the genes encoding prokineticin-2 or prokineticin receptor-2 cause autosomal recessive Kallmann syndrome. J Clin Endocrinol Metab 93:4113–4118 [DOI] [PubMed] [Google Scholar]
- Trarbach EB, Costa EM, Versiani B, de Castro M, Baptista MT, Garmes HM, de Mendonca BB, Latronico AC 2006 Novel fibroblast growth factor receptor 1 mutations in patients with congenital hypogonadotropic hypogonadism with and without anosmia. J Clin Endocrinol Metab 91:4006–4012 [DOI] [PubMed] [Google Scholar]
- Olsen SK, Li JY, Bromleigh C, Eliseenkova AV, Ibrahimi OA, Lao Z, Zhang F, Linhardt RJ, Joyner AL, Mohammadi M 2006 Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes Dev 20:185–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemel J, Gorry M, Ehrlich GD, MacArthur CA 1996 Structure and sequence of human FGF8. Genomics 35:253–257 [DOI] [PubMed] [Google Scholar]
- Maquat LE, Serin G 2001 Nonsense-mediated mRNA decay: insights into mechanism from the cellular abundance of human Upf1, Upf2, Upf3, and Upf3X proteins. Cold Spring Harb Symp Quant Biol 66:313–320 [DOI] [PubMed] [Google Scholar]
- Riley BM, Mansilla MA, Ma J, Daack-Hirsch S, Maher BS, Raffensperger LM, Russo ET, Vieira AR, Dodé C, Mohammadi M, Marazita ML, Murray JC 2007 Impaired FGF signaling contributes to cleft lip and palate. Proc Natl Acad Sci USA 104:4512–4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WC, Moyle SS, Tsai PS 2008 Fibroblast growth factor 8 signaling through fibroblast growth factor receptor 1 is required for the emergence of gonadotropin-releasing hormone neurons. Endocrinology 149:4997–5003 [DOI] [PMC free article] [PubMed] [Google Scholar]