SUMMARY
The antimycobacterial efficacy of the abyssomicin C family of natural products, in addition to a key synthetic intermediate, has been investigated given their reported inhibition of Bacillus subtilis p-aminobenzoate biosynthesis. The naturally occurring (−)-abyssomicin C and its atropisomer were found to exhibit low micromolar growth inhibition against the relatively fast-growing and non-virulent Mycobacterium smegmatis and the vaccine strain Mycobacterium bovis BCG, while their antipodes were slightly less active. (−)-Abyssomicin C and its atropisomer were particularly efficacious against Mycobacterium tuberculosis H37Rv, exhibiting MIC values of 3.6 and 7.2 μM, respectively. More specifically, (−)-abyssomicin C was bactericidal. This complex natural product and its analogs, thus, hold promise as chemical tools in the study of M. tuberculosis metabolism.
Keywords: Mycobacterium tuberculosis, Abyssomicin, p-Aminobenzoate metabolism
Tuberculosis is a global epidemic, claiming nearly 2 million lives and infecting more than 9 million people per annum.1 The current front-line therapy consists of isoniazid, pyrazinamide, rifampicin, and ethambutol - drugs that arose from discovery efforts more than forty years old. The challenges in tuberculosis drug discovery set by drug resistance, mycobacterial persistence, and latency are significant.2 The rates at which these hurdles, especially that of drug resistance,3 have grown have dramatically outpaced the development of new tuberculosis therapies. While drug candidates such as TMC2074 and PA-8245 are progressing through clinical trials, the need is evident for new molecules that modulate novel mycobacterial targets.
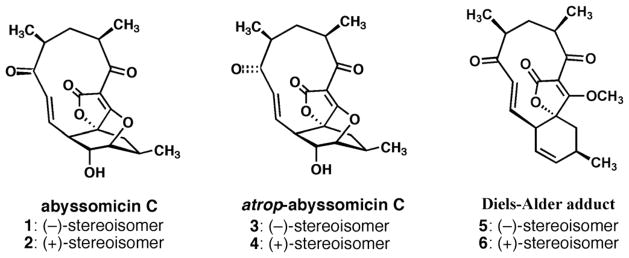
The natural product (−)-abyssomicin C is a structurally intriguing polyketide-like molecule initially discovered with activity against the para-aminobenzoate (pABA) biosynthetic pathway in Bacillus subtilis.6,7 More specifically, (−)-abyssomicin C (compound 1 in Figure 1) was found to inhibit the biosynthesis of pABA from chorismate. Further investigation by the authors demonstrated the antibacterial activity of (−)-abyssomicin C against a number of Gram-positive bacteria, including clinical isolates of multi drug-resistant Staphylococcus aureus. We have previously reported the total synthesis8 of (−)-abyssomicin C and what was eventually identified by Nicolaou and co-workers as (−)-atrop-abyssomicin C.9 It is interesting to note that (−)-atrop-abyssomicin C was later shown to not only be present in the original isolates from Verrucosispora strain AB-18-032, but to be the predominant abyssomicin by quantity.10 We have tested these compounds to determine their efficacy against Mycobacterium tuberculosis (Mtb). The genes involved in the mycobacterial pABA biosynthesis from chorismate had been located,11 and data existed to support the essential nature of pABA biosynthesis.12–14 Importantly, human biosynthesis of pABA is non-existent. Intriguingly, no reported efforts to specifically target mycobacterial pABA biosynthesis had been disclosed in the literature. Thus, the potential for a small molecule that inhibited mycobacterial pABA biosynthesis appeared to hold significant promise in further probing Mtb metabolism, with a long-term goal of seeding the discovery of novel antituberculars.
Figure 1.
Chemical structures of the studied abyssomicin C family.
We set out to examine the antimycobacterial efficacy (Table 1) of not only (−)-abyssomicin C, but a larger “abyssomicin family.” This family (Figure 1) includes the naturally occurring (−)-abyssomicin C (1) and its (+)-antipode (2), (−)-atrop-abyssomicin C (3) and its (+)-antipode 4, and the so-called Diels–Alder adduct precursors 5 and 6 that were central to our first reported synthesis of the natural product,8 and have also been reported in the literature by Snider15 and Couladouros.16 Initially, these compounds were tested against the non-virulent, faster-growing Mycobacterium smegmatis mc2155.17 Experiments with this strain and other mycobacteria were performed with the microplate Alamar Blue assay, as reported by Franzblau.18 (−)-Abyssomicin C and its atropisomer 3, in particular, showed modest activities with liquid culture MIC values of 29 and 58 μM, respectively. The respective antipodes 2 and 4 displayed diminished efficacy (MIC = 110 μM). The overall activity trends in M. smegmatis held when examining the compound family against Mycobacterium bovis BCG. (−)-Abyssomicin C was the most potent compound, with a liquid culture MIC of 7.2 μM and its atropisomer was twofold less potent. The corresponding antipodes of 1 and 3 are 8- and 2-fold less efficacious, respectively, perhaps pointing to some specificity in mode of action. The Diels–Alder adducts 5 and 6 were again much less potent, connoting the importance of structural features other than the presence of the α,β-unsaturated carbonyl systems in driving whole-cell efficacy.
Table 1.
Minimum inhibitory concentration (MIC) data for (−)-abyssomicin C family and drug controls.
| Compound | MIC* in M. smegmatis mc2155 in μg/ml (μM) | MIC* in M. bovis BCG in μg/ml (μM) | MIC* in M. tuberculosis in μg/ml (μM) |
|---|---|---|---|
| rifampicin | 1.2 (1.5) | 0.00064 (0.00078) | ND |
| isoniazid | 2.5 (18) | 0.16 (1.2) | 0.040 (0.29) |
| (−)-abyssomicin C | 10 (29) | 2.5 (7.2) | 1.2 (3.6) |
| (+)-abyssomicin C | 40 (110) | 20 (58) | ND |
| (−)-atrop-abyssomicin C | 20 (58) | 5.0 (14) | 2.5 (7.2) |
| (+)-atrop-abyssomicin C | 40 (110) | 10 (29) | ND |
| (−)-Diels-Alder adduct | >80 (>230) | 40 (120) | ND |
| (+)-Diels-Alder adduct | >80 (>230) | 40 (120) | ND |
The values reported represent the mean of three or more independent determinations.
The structure–activity relationships (SAR) against M. smegmatis and M. bovis BCG are indicative of definite structural requirements for efficacy. Clearly, abyssomicins 1 and 2 and their respective atropisomers 3 and 4 are significantly more active than the Diels–Alder adducts 5 and 6. This points toward the importance of the oxabicyclic ring, which may serve as a conformational constraint, and the secondary hydroxyl. We observe twofold differences in MIC values between 1 and atropisomer 3, which are small but may be attributed to the positioning of the potentially reactive α,β-unsaturated enone in the 11-membered macrocyclic ring. Nicolaou and co-workers reported 3 to be a better Michael acceptor (under 1,4-reduction conditions with hydride reagent) than 1, which they proposed to be responsible for a small (15 μM vs. 20 μM) MIC differential against methicillin-resistant S. aureus.19 Finally, it is noteworthy that the naturally occurring enantiomer of (−)-abyssomicin 1 is 4–8 times more active than its antipode 2, supporting inhibition through a specific binding event.
The Mtb H37Rv strain was utilized in liquid cultures to assess the MIC values of the two most potent compounds 1 and 3. The MIC of (−)-abyssomicin C was 3.6 μM and the MIC of its atropisomer 3 was 7.2 μM. Cultured Mtb H37Rv (OD600 ~ 0.5) were exposed to ca. 4×, 40×, and 80× multiples of the MIC of 1, incubated for 18 h, and then plated and serially diluted in duplicate. Incubation with the compound resulted in bacterial death (data not shown). Bacterial outgrowth after ca. 6 weeks was only observed with the first dilution of the lowest concentration of 1 (4×; 14 μM), and at a visibly lower number of colony-forming units as compared to the control. At higher concentrations of 1, no visible bacterial outgrowth at any of the dilutions was noted, in stark contrast to the control. While there was not enough compound available to test the MBC formally, we did observe bactericidal activity.
While these abyssomicins are ca. 10–20 fold less active against Mtb H37Rv than the front-line drug isoniazid on a molar basis, they compare favorably to second-line drugs such as ethionamide (MIC = 18 μM).20 With very limited amounts remaining of these abyssomicins, we were unable to determine if this promising efficacy is extended to other members of the Mycobacterium genus. It should be noted that Bister and colleagues reported a lack of efficacy of 1 against Mycobacterium phlei DSM 750 on complex medium.7
In liquid culture, we investigated whether antimycobacterial activity is rescued by added pABA. We began by examining whether pABA added to liquid cultures of M. bovis BCG could reverse growth inhibition of the abyssomicins. It has been reported that the efficacy of (−)-abyssomicin C versus B. subtilis was reversed by a 5 mM solution of pABA.7 At concentrations ranging from 2.5 to 20 mM, pABA was observed, through the Alamar Blue assay, to not be toxic to the mycobacteria. As a control experiment, pABA (at ca.1.5 times the concentration of drug by weight) was found to reverse the inhibition of M. bovis BCG by p-aminosalicylate (PAS),21 in sync with the original experiments with Mtb by Youmans and colleagues.22 In contrast, upon adding pABA (at seven concentrations from 27 μM to 5 mM) to M. bovis BCG exposed to (−)-abyssomicin C (at 7.2, 14, or 29 μM), we failed to observe reversal of mycobacterial growth inhibition.
The antimycobacterial activity of the abyssomicin C family has been established through liquid culture MIC determinations. In particular, bactericidal (−)-abyssomicin C and its atropisomer have demonstrated single-digit micromolar MICs versus M. tuberculosis H37Rv. The biological data encourage studies to explore the SAR around this family of molecules while further probing the mechanism of action of these natural products in order to better understand M. tuberculosis metabolism.
1. Materials and methods
1.1. Synthesis of abyssomicin C family members
The syntheses of the family members 1–6 have been described previously.8,23
1.1.1. Determination of MIC values for abyssomicin C family members versus M. smegmatis mc2155, M. bovis BCG, and M. tuberculosis H37Rv
A drug susceptibility assay in 96-well plate format by Alamar Blue was modified from Franzblau and co-workers.18 The respective bacteria were grown to mid-log phase (OD600 = 0.5) and diluted to OD600 = 0.001 or 0.003 for M. smegmatis or M. bovis BCG/M. tuberculosis, respectively. 200 μL of sterile deionized water was added to all outer-perimeter wells of sterile 96-well plates to minimize evaporation of the medium in the test wells during incubation. Compounds were dissolved in DMSO and subsequent serial dilutions were performed in 7H9 complete medium.100 μL of serial diluted drug solutions were added to the wells in columns 3–10, and 100 μL 7H9 medium were added in columns 2 and 11. 100 μL of mycobacterial inoculum was added to the wells in rows B to G in columns 2–9 by using a multichannel pipette (yielding a final volume of 200 μL per well). Thus, the wells in column 2 served as drug-free (inoculum-only) controls, the wells in column 10 served as drug-only controls and the wells in column 11 served as medium-only controls. The plates were sealed with breathable film (VWR) and were incubated at 37 °C for 24 h for M. smegmatis or 4 days for M. bovis BCG/M. tuberculosis. Alamar Blue (Biosource) reagent was added to every well. The plates were reincubated at 37 °C for 6 h for M. smegmatis or 24 h for M. bovis BCG/M. tuberculosis, and the colors of all wells were recorded. A blue color in the well was interpreted as no growth, and a pink color was scored as growth. A few wells appeared violet after 24 h of incubation, but they invariably changed to pink after another day of incubation and thus were scored as growth (while the adjacent blue wells remained blue). The MIC was defined as the lowest drug concentration that prevented a color change from blue to pink.
1.1.2. Determination of bactericidal activity of (−)-abyssomicin C versus M. tuberculosis H37Rv
M. tuberculosis H37Rv was grown at 37 °C to log phase in Middlebrook 7H9 broth (Difco) supplemented with 0.2% glycerol, 0.05% Tween-80, 0.5% bovine serum albumin, 0.2% dextrose, and 0.085% sodium chloride. A 1 mL aliquot was removed and incubated in the presence of (−)-abyssomicin C, at concentrations of 5, 50, and 100 μg/mL, or in the absence of compound as a control, for 18 h. Post incubation, the cultures were serially diluted to 10−1, 10−2, 10−3, 10−4, and 10−5, and plated in duplicate. The plates were 7H10 agar (Difco) supplemented with oleic acid, albumin, dextrose and catalase. The plates were examined for the outgrowth of bacterial colonies after 21 days and then again after a total of ca. 6 weeks.
1.1.3. Evaluation of pABA effect on PAS treated M. bovis BCG
The Alamar blue assays were conducted as described above. pABA solutions, at concentrations ranging from 0.1 to 6 μg/mL, in ethanol/media and PAS (MIC ~ 0.03 μg/mL; concentrations ranging from 0.045 to 4 μg/mL) were added to M. bovis BCG. Bacteria were cultured for 4 days and followed by addition of Alamar blue to evaluate their growth.
1.1.4. Evaluation of pABA effect on (−)-abyssomicin C treated M. bovis BCG
A similar procedure was utilized as described for the rescue of PAS treated M. bovis BCG by pABA. At concentrations ranging from 2.5 to 20 mM, pABA solutions in ethanol/media were added to cultured M. bovis BCG and evaluated after 4 days by the addition of Alamar blue to check for pABA toxicity. Similarly, pABA solutions, at concentrations ranging from 27 μM to 5 mM, in ethanol/media and (−)-abyssomicin C, at the concentration of 7.2, 14, or 29 μM, were added to M. bovis BCG culture. Bacteria were cultured for 4 days and followed by addition of Alamar blue to evaluate their growth.
Acknowledgments
J.S.F. and E.J.S. acknowledge support from the Princeton University Grand Challenges in Global Health and Carmen F. Drahl for supplying samples of the abyssomicin C family members.
Funding: This work was supported by the Bill and Melinda Gates Foundation (grant 42844). J.C.S. acknowledges the Robert A. Welch Foundation (grant A-0015).
Footnotes
Competing interests: None declared.
Ethical approval: Not required.
References
- 1.World Health Organization. Global tuberculosis control report. 2009. [Google Scholar]
- 2.Sacchettini JC, Rubin EJ, Freundlich JS. Drugs versus bugs: in pursuit of the persistent predator Mycobacterium tuberculosis. Nat Rev Microbiol. 2008;6:41–52. doi: 10.1038/nrmicro1816. [DOI] [PubMed] [Google Scholar]
- 3.Jassal M, Bishai WR. Extensively drug-resistant tuberculosis. Lancet Infect Dis. 2009;9:19–30. doi: 10.1016/S1473-3099(08)70260-3. [DOI] [PubMed] [Google Scholar]
- 4.Andries K, Verhasselt P, Guillemont J, Gohlmann HWH, Neefs J-M, Winkler H, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–7. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 5.Stover CK, Warrener P, VanDevanter DR, Sherman DR, Arain TM, Langhorne MH, et al. A small-molecule nitroimidazopyran drug candidate for the treatment of tuberculosis. Nature. 2000;405:962–6. doi: 10.1038/35016103. [DOI] [PubMed] [Google Scholar]
- 6.Bister B, Bischoff D, Ströbele M, Riedlinger J, Reicke A, Wolter F, et al. Abyssomicin C-A polycyclic antibiotic from a marine Verrucosispora strain as an inhibitor of the p-aminobenzoic acid/tetrahydrofolate biosynthesis pathway. Angew Chem Int Ed Engl. 2004;43:2574–6. doi: 10.1002/anie.200353160. [DOI] [PubMed] [Google Scholar]
- 7.Riedlinger J, Reicke A, Zähner H, Krismer B, Bull AT, Maldonado LA, et al. Abyssomicins, inhibitors of the para-aminobenzoic acid pathway produced by the marine Verrucosispora strain AB-18-032. J Antibiot. 2004;57:271–9. doi: 10.7164/antibiotics.57.271. [DOI] [PubMed] [Google Scholar]
- 8.Zapf CW, Harrison BA, Drahl C, Sorensen EJ. A Diels-Alder macrocyclization enables an efficient asymmetric synthesis of the antibacterial natural product abyssomicin C. Angew Chem Int Ed Engl. 2005;44:6533–7. doi: 10.1002/anie.200502119. [DOI] [PubMed] [Google Scholar]
- 9.Nicolaou KC, Harrison ST. Total synthesis of abyssomicin C and atrop-abyssomicin C. Angew Chem Int Ed Engl. 2006;45:3256–60. doi: 10.1002/anie.200601116. [DOI] [PubMed] [Google Scholar]
- 10.Keller S, Nicholson G, Drahl C, Sorensen E, Fielder H-P, Süssmuth RD. Abyssomicins G and H and atrop-abyssomicin C from the marine Verrucosispora strain AB-18-032. J Antibiot (Tokyo) 2007;60:391–4. doi: 10.1038/ja.2007.54. [DOI] [PubMed] [Google Scholar]
- 11.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 12.Lamichhane G, Zignol M, Blades NJ, Geiman DE, Dougherty A, Grosset J, et al. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2003;100:7213–8. doi: 10.1073/pnas.1231432100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sassetti CM, Boyd DH, Rubin EJ. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci USA. 2001;2001:12712–7. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 15.Snider BB, Zou Y. Synthesis of the carbocyclic skeleton of abyssomicins C and D. Org Lett. 2005;7:4939–41. doi: 10.1021/ol0518941. [DOI] [PubMed] [Google Scholar]
- 16.Couladouros EA, Bouzas EA, Magos AD. Formal synthesis of abyssomicin C. Tetrahedron. 2006;62:5272–9. [Google Scholar]
- 17.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4:1911–9. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 18.Franzblau SG, Witzig RS, McLaughlin JC, Torres P, Madico G, Hernandez A, et al. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J Clin Microbiol. 1998;36:362–6. doi: 10.1128/jcm.36.2.362-366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicolaou KC, Harrison ST. Total synthesis of abyssomicin C, atrop-abyssomicin C, and abyssomicin D: implications for natural origins of atrop-abyssomicin C. J Am Chem Soc. 2007;129:429–40. doi: 10.1021/ja067083p. [DOI] [PubMed] [Google Scholar]
- 20.Vilchèze C, Wang F, Arai M, Hazbon MH, Colangeli R, Kremer L, et al. Transfer of a point mutation in Mycobacterium tuberculosis inhA resolves the target of isoniazid. Nat Med. 2006;12:1027–9. doi: 10.1038/nm1466. [DOI] [PubMed] [Google Scholar]
- 21.Rengarajan J, Sassetti CM, Naroditskaya V, Sloutsky A, Bloom BR, Rubin EJ. The folate pathway is a target for resistance to the drug para-aminosalicylic acid (PAS) in mycobacteria. Mol Microbiol. 2004;53:275–82. doi: 10.1111/j.1365-2958.2004.04120.x. [DOI] [PubMed] [Google Scholar]
- 22.Youmans GP, Raleigh GW, Youmans AS. The tuberculostatic action of para-aminosalicyclic acid. J Bacteriol. 1947;54:409–16. doi: 10.1128/jb.54.4.409-416.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drahl CF. Chemical synthesis and activity-based proteomic studies of the abyssomicins, protein-reactive natural products. Princeton University; 2007. [Google Scholar]