Abstract
Eutypine (4-hydroxy-3-[3-methyl-3-butene-1-ynyl] benzaldehyde) is a toxin produced by Eutypa lata, the causal agent of eutypa dieback in the grapevine (Vitis vinifera). Eutypine is enzymatically converted by numerous plant tissues into eutypinol (4-hydroxy-3-[3-methyl-3-butene-1-ynyl] benzyl alcohol), a metabolite that is nontoxic to grapevine. We report a four-step procedure for the purification to apparent electrophoretic homogeneity of a eutypine-reducing enzyme (ERE) from etiolated mung bean (Vigna radiata) hypocotyls. The purified protein is a monomer of 36 kD, uses NADPH as a cofactor, and exhibits a Km value of 6.3 μm for eutypine and a high affinity for 3- and 4-nitro-benzaldehyde. The enzyme failed to catalyze the reverse reaction using eutypinol as a substrate. ERE detoxifies eutypine efficiently over a pH range from 6.2 to 7.5. These data strongly suggest that ERE is an aldehyde reductase that could probably be classified into the aldo-keto reductase superfamily. We discuss the possible role of this enzyme in eutypine detoxification.
Many pathogenic bacteria and fungi produce toxins that interfere with various functions of plant cells and may affect plant defense mechanisms (Durbin, 1981). Toxin production is commonly associated with disease severity and can be involved in colonization or systemic invasion by the pathogen (Schäfer, 1994). Toxin resistance has been shown in most cases to be based on the ability of the plant to metabolically detoxify pathogen toxins (Meeley and Walton, 1991; Zhang and Birch, 1997; Zweimuller et al., 1997). Few cloned toxin-resistance genes that encode proteins involved in detoxification mechanisms have been described (Utsumi et al., 1988; Johal and Briggs, 1992; Zhang and Birch, 1997). In many cases a relationship exists between toxin tolerance and resistance to the disease (Anzai et al., 1989; Meeley et al., 1992). The availability of toxin-resistance genes will permit a greater understanding of the mechanisms causing plant disease and will also set the stage for engineering resistance to plant disease (Keen, 1993).
Eutypine (4-hydroxy-3-[3-methyl-3-butene-1-ynyl] benzaldehyde) is a toxin produced by the ascomycete fungus Eutypa lata (Pers.: Fr.) Tul., the causal agent of eutypa dieback (Tey-Rulh et al., 1991). This disease is responsible for considerable loss in yield and is the most devastating disease of grapevine (Vitis vinifera) in many countries (Moller and Kasamitis, 1981; Munkvold et al., 1994). The fungus infects the stock through pruning wounds and is present in the xylem and phloem of the vine trunk and branches (Moller and Kasamitis, 1978; Duthie et al., 1991). After a long incubation period, a canker forms around the infected wound. The toxin synthesized by the fungus in the trunk is believed to be transported by the sap to the herbaceous parts of the vine (Fallot et al., 1997). Eutypine penetrates grapevine cells through passive diffusion and its accumulation in the cytoplasm has been explained by an ion-trapping mechanism related to the ionization state of the molecule (Deswarte et al., 1996b). In the cell the effects of eutypine include reduction of adenylated nucleotide content, inhibition of succinate dehydrogenase, uncoupling of oxidative phosphorylation, and mitochondrial swelling (Deswarte et al., 1996a).
Symptoms of eutypa dieback in the herbaceous part of the plant lead to dwarfed and withered new growth of branches, marginal necrosis of the leaves, dryness of the inflorescence, and, finally, death of one or more branches (Moller and Kasamitis, 1981). The toxin appears to be an important virulence factor involved in symptom development of the disease (Deswarte et al., 1996a). However, the absence of toxin-deficient mutants of the fungus and its long incubation period in the trunk before symptom development have prevented a critical study of the toxin in vine plants. Determining the gene responsible for eutypine resistance would therefore be an important critical tool in determining the role of eutypine toxin in symptom development in the disease; and it has the potential to confer resistance to transgenic grapevines.
Recently, Colrat et al. (1998) found detoxification to occur in grapevine cells through the enzymatic reduction of eutypine into its corresponding alcohol, eutypinol (4-hydroxy-3-[3-methyl-3-butene-1-ynyl] benzyl alcohol). We have determined that this derivative of the toxin is nontoxic for grapevine tissues. Furthermore, we have established a relationship between the susceptibility of grapevine to eutypa dieback and the ability of tissues to inactivate eutypine, suggesting that the detoxification mechanism plays an important role in defense reactions. Eutypine is enzymatically detoxified in numerous plant species and, among them, we found that the tissues of mung bean (Vigna radiata), a nonhost plant for the pathogen, exhibit an efficient detoxification activity. As a prerequisite for demonstrating the involvement of eutypine toxin in eutypa dieback, we report here the purification to homogeneity and the characterization of an ERE from etiolated mung bean hypocotyls.
MATERIALS AND METHODS
Chemicals
Nonradioactive eutypine and [14C]-labeled eutypine (1.36 GBq mmol−1) were synthesized according to previously described procedures (Defrancq and Tabacchi, 1992; Defrancq et al., 1993). Benzaldehyde and benzyl alcohol were purchased from Sigma, cinnamaldehyde from Fluka, and all other chemicals from Aldrich.
Plant Material
Seeds of mung bean (Vigna radiata [L.] R. Wilcz) were purchased from Cereal Wander Nutrition Co. (Annonay, France). They were allowed to imbibe overnight in running tap water under continuous aeration, and then sown in vermiculite. Seedlings were harvested after 5 d in the dark at 23°C, and hypocotyls (approximately 2 cm long) were cut for enzyme extraction.
Assay of ERE Activity
ERE activity was assayed spectrophotometrically at 25°C by measuring the rate of enzyme-dependent decrease of NADPH absorption at 340 nm. The reaction mixture consisted of 200 mm Na2HPO4/100 mm citric acid, pH 6.5, 100 μm NADPH, 100 μm eutypine or other aldehyde derivatives (listed in Table II), and 10 to 100 μL of proteins in a total volume of 500 μL. To verify the identity of the reaction product, eutypine was substituted by [14C]eutypine. After a 15-min incubation the reaction products were extracted three times with diethyl ether. The ether phases were evaporated under a stream of nitrogen, and the extracted compounds were suspended in 20 μL of ethanol. The samples were cochromatographed with labeled eutypinol on silica-gel TLC using dichloromethane as a solvent, and the plates were exposed to radiographic film (Kodak XAR-5) for several days.
Table II.
Substrate specificity of the ERE of mung bean
| Substrate | Km | kcat | kcat/Km |
|---|---|---|---|
| μm | min−1 | 105 min−1/m−1 | |
| Eutypine | 6.3 | 50 | 79 |
| Benzaldehyde | 33 | 5.1 | 1.5 |
| 2-Nitro-benzaldehyde | 376 | 10 | 0.3 |
| 3-Nitro-benzaldehyde | 2.9 | 11 | 40 |
| 4-Nitro-benzaldehyde | 5.3 | 11 | 23 |
| 2-Fluoro-benzaldehyde | 702 | 42 | 6.0 |
| 3-Fluoro-benzaldehyde | 5.4 | 9.0 | 16 |
| 4-Fluoro-benzaldehyde | 40 | 5.7 | 1.4 |
| 2-Methyl-benzaldehyde | 125 | 4.4 | 0.4 |
| 3-Methyl-benzaldehyde | 20 | 7.2 | 3.6 |
| 4-Methyl-benzaldehyde | 41 | 5.7 | 1.4 |
| 2-Methoxy-benzaldehyde | 49 | 7.8 | 1.5 |
| 3-Methoxy-benzaldehyde | 212 | 10 | 49 |
| 4-Methoxy-benzaldehyde | 150 | 7.2 | 0.5 |
| Propionaldehyde | 87 | 3.2 | 37 |
| Hexanaldehyde | 15 | 5.2 | 3.4 |
| Decylaldehyde | 3.3 | 18 | 54 |
| Sinapylaldehyde | 126 | 26 | 21 |
| Coniferylaldehyde | 60 | 36 | 5.9 |
| Cinnamaldehyde | 99 | 9.0 | 0.9 |
| Salicylaldehyde | 66 | 14 | 2.1 |
| Furaldehyde | 207 | 15 | 0.7 |
| 4-Pyridine carbaldehyde | 4.5 | 14 | 32 |
We determined the pH optimum for ERE activity in preliminary experiments by assaying activity in crude protein extracts. It was later confirmed with pure protein using a 0.1 m citric acid/0.2 m Na2HPO4 buffer solution ranging from pH 5.5 to 8.0. Protein content was determined using a bicinchoninic acid dye reagent (Pierce) and BSA as a standard.
Km values were determined from initial velocities by varying the concentration of the substrates (3–25 μm) and the cofactor (5–100 μm). Kinetic parameters were calculated assuming Michaelis-Menten enzyme performance. All kinetic mesurements were performed at least three times, and the mean values were used for the subsequent calculations.
Extraction and Purification of ERE
All purification steps were carried out at 4°C. Mung bean hypocotyls (400 g) were homogenized in 2 volumes (w/w) of extraction buffer consisting of 100 mm sodium borate buffer, pH 8.0, 10% (v/v) glycerol, 1% (w/v) polyvinylpyrrolidone (Mr 40,000), and 4 mm DTT. The resulting homogenate was centrifuged at 48,000g for 30 min. The clear supernatant was subjected to (NH4)2SO4 precipitation and the fraction precipitating between 30% and 70% saturation was resuspended in the same volume of 25 mm potassium-phosphate buffer, pH 8.0, and 10% glycerol (buffer A). Any material that was not readily solubilized was removed by centrifugation at 48,000g for 10 min and discarded. ERE activity was measured after desalting a 2.5-mL aliquot of the supernatant on a PD10 column (Pharmacia). The extract recovered from (NH4)2SO4 precipitation was adjusted to 1 m (NH4)2SO4 and loaded onto a phenyl Sepharose CL-4B column (1.6 × 30 cm; Pharmacia) preequilibrated with buffer A adjusted to 1 m (NH4)2SO4.
The enzyme was eluted from the column with a linear decreasing gradient of 1 to 0 m (NH4)2SO4 at a flow rate of 1.5 mL min−1. Fractions of 5 mL were collected and those exhibiting ERE activity were pooled and desalted on a PD10 column as previously described. To maximize the elimination of contaminants, only the fractions containing the highest levels of ERE activity were pooled after each step for use in the subsequent steps. The desalted fractions from the phenyl Sepharose column were loaded onto a hydroxyapatite column (Econo-Pac CHT II, Bio-Rad) preequilibrated with buffer A. Proteins were eluted with an increasing linear gradient of 25 to 150 mm potassium-phosphate buffer, pH 8.0. Active fractions were pooled, desalted as outlined above, concentrated to a volume of 300 μL by ultrafiltration on a Centrisart C4 membrane (Sartorius, Goettingen, Germany), and injected into a fast protein liquid chromatography column (30 × 1 cm) of Superose 12 HR (Pharmacia) equilibrated with buffer A. Proteins were eluted with the same buffer at a flow rate of 0.3 mL min−1, and fractions of 300 μL were collected.
Fractions containing ERE activity were pooled and desalted on a NAP10 (Pharmacia) column preequilibrated in Tris-HCl buffer, pH 8.0, and 10% glycerol (buffer B). The active, desalted Superose 12 HR fractions were loaded onto a Mono-Q HR5/5 column (Pharmacia) equilibrated with buffer B. The column was then rinsed with 10 mL of buffer B, and the ERE activity was eluted with an increasing linear gradient of 0 to 500 mm KCl at a flow rate of 1 mL min−1. Purified ERE fractions were stored at −80°C.
Determination of Molecular Mass of ERE
To determine the molecular mass of ERE, partially purified enzyme (post hydroxyapatite) was subjected to Superose 12 HR gel filtration. Calibration was performed using the following molecular mass markers (Sigma): BSA, 66 kD; ovalbumin, 45 kD; carbonic anhydrase, 29 kD; Cyt c, 12.5 kD. All proteins were loaded in a total volume of 300 μL, and elution was monitored by A280.
Electrophoretic Analysis
Denaturing SDS-PAGE electrophoresis was performed according to the method of Laemmli (1970) in gels containing 12% acrylamide. Proteins were stained with silver nitrate (Damerval et al., 1987). The pI of the enzyme was determined by rapid IEF according to the method of Robertson et al. (1987).
RESULTS
ERE Purification
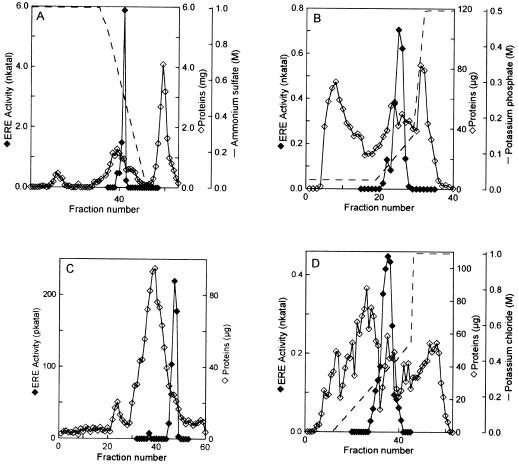
We achieved purification of ERE to apparent homogeneity by using a five-step protocol that included (NH4)2SO4 precipitation and four successive chromatographic steps (Table I). The corresponding chromatograms are presented in Figure 1. A second minor peak of ERE activity was found on the hydroxyapatite column. We focused on the major peak for the subsequent purification steps. After the last chromatographic step, an overall 1563-fold purification was obtained, with a recovery of 3.0%. The pure ERE had a specific activity of 891 nkat mg−1 using eutypine as a substrate. It was verified by TLC that the product from a reaction catalyzed by pure ERE (post Mono-Q) was a single spot that comigrated with authentic eutypinol (data not shown).
Table I.
Purification of ERE from etiolated mung bean hypocotyls
| Chromatography | Total Protein | Total Activity | Specific Activity | Purification Factor | Yield |
|---|---|---|---|---|---|
| mg | nkat | nkat/mg | % | ||
| Crude extract | 613 | 350.0 | 0.57 | — | 100 |
| (NH4)2SO4 | 86 | 108.0 | 1.25 | 2.2 | 30.8 |
| Phenyl Sepharose | 9.92 | 86.5 | 8.72 | 15.3 | 24.7 |
| Hydroxyapatite | 0.43 | 22.4 | 51.5 | 90.3 | 6.4 |
| Superose 12 | 0.03 | 12.5 | 403 | 707.0 | 3.5 |
| Mono-Q | 0.01 | 10.7 | 891 | 1 563.1 | 3.1 |
The results presented are for a typical purification starting from 400 g of tissue
Figure 1.
Chromatograms of ERE purification. Elution profiles of proteins (⋄) and ERE activity (♦) from Phenyl Sepharose (A), hydroxylapatite (B), Superose 12 (C), and Mono-Q (D) columns.
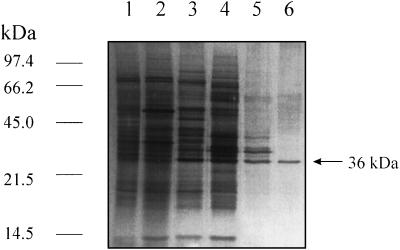
SDS-PAGE analysis of fractions from ERE sequential purification showed, after Mono-Q, a dominant polypeptide with an apparent molecular mass of approximately 36 kD (Fig. 2, lane 6). The purity of this band was verified by two-dimensional electrophoresis (data not shown).
Figure 2.
Silver-stained SDS-PAGE documenting the progress of purification of ERE. Fractions include: Lane 1, Crude extract; lane 2, (NH4)2SO4; lane 3, Phenyl-Sepharose chromatography; lane 4, hydroxyapatite chromatography; lane 5, Superose 12 chromatography; lane 6, Mono-Q chromatography. The positions of molecular mass markers are indicated on the left in kD.
Physicochemical Properties of ERE
The apparent molecular mass of the native ERE determined by gel filtration on Superose 12 corresponded to 36 kD. The fact that the protein size on SDS-PAGE-denaturing electrophoresis and native molecular mass estimation on Sepharose 12 gel filtration were both approximately the same suggested that ERE is monomeric. The pI of the ERE protein was found to be 7.2 (data not shown). The pH optimum of ERE activity was determined on the pure ERE (post Mono-Q) in citric acid/Na2HPO4 buffer over a pH range of 5.5 to 8.0 under saturating substrate conditions. Enzymatic activity was optimal between pH 6.2 and 7.5, with a maximum at 6.5. Activity was maximum at 45°C, with an Arrhenius activation energy of 49.8 kJ mol−1 over the range of 25°C to 45°C.
Substrate Specificity of ERE
We determined the apparent Km for eutypine to be 6.3 μm, indicating that ERE exhibited a high affinity toward the toxin (Table II). The Km value for NADPH was 8.4 μm. NADH could not substitute for NADPH as cofactor. When ERE was incubated in the presence of NADP+ and eutypinol, no dehydrogenase activity was detected, even at high protein concentrations.
To identify other potential substrates of ERE, we examined its affinity toward various aromatic and aliphatic aldehydes. The data presented in Table II show that the enzyme had a relatively broad range of substrates. ERE had the highest specificity constant (kcat/Km) for eutypine and the lowest Km values for 3-nitro-benzaldehyde and 4-pyridine carbaldehyde. Benzaldehyde and benzaldehyde derivatives containing substituents at position 3 on the aromatic ring (like eutypine) were reduced by ERE, but the enzyme showed higher Km values for 3-methoxy-, 3-methyl-, and 3-fluoro-benzaldehyde than for 3-nitro-benzaldehyde.
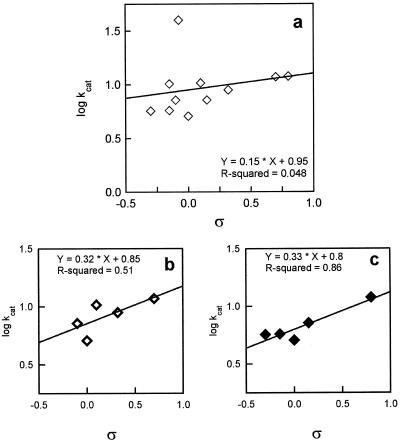
To determine the influence of the electronic nature of the substituents (represented by the constant ς) on the rate of the ERE-catalyzed reaction, the log (apparent kcat) values of ERE for different substrates were plotted against their ς values. The ς constant represents the electrical effect for a group R attached to an aromatic ring (March, 1985). The data showed that the electronic nature of the substituents weakly influenced the velocity of the enzyme-catalyzed reaction (the slope was only 0.15 ± 0.06), suggesting that electron-withdrawing groups slightly accelerated the reaction, whereas electron-donating groups apparently decreased ERE activity (Fig. 3a). To determine whether steric hindrance plays a role in the reduction rates, the kcat values of 3- and 4-substituted compounds were plotted separately (Fig. 3, b and c). The two plots were essentially the same (i.e. they had very similar slopes, 0.32 ± 0.04 and 0.33 ± 0.06 for 3- and 4-substituted compounds, respectively). The data indicate that the steric hindrance at position 3 is similar to that at position 4.
Figure 3.
Correlation of kcat with ς, dependent on the electronic nature of the substituent R, for ERE. a, Variation of log (apparent kcat) with ς for substituted benzaldehydes. b, Variation of log (apparent kcat) with ς for 3-substituted benzaldehydes. c, Variation of log (apparent kcat) with ς for 4-substituted benzaldehydes.
Various other aromatic aldehydes, such as cinnamaldehyde, coniferaldehyde, and sinapaldehyde, were converted much less efficiently than eutypine. Dihydroquercitin and dihydromyricetin, the two major dihydroflavonols found by Mato and Ishikura (1993) in mung bean, were not converted at all. Furthermore, ERE converted the reduction of various aliphatic aldehydes, and the efficiency of the reduction increased with the length of their hydrocarbon chains. We found the Km value for decylaldehyde to be close to that of eutypine.
DISCUSSION
Our results show that the capacity of mung bean to detoxify eutypine is determined by ERE. This protein was purified to apparent electrophoretic homogeneity from etiolated hypocotyls, and the purified enzyme catalyzed the reduction of eutypine into eutypinol, a nontoxic compound. We found ERE to be a NADPH-dependent oxidoreductase with a molecular mass of 36 kD and a monomeric active form. ERE exhibited a high affinity for eutypine and a broad substrate specificity but failed to catalyze the reverse reaction using eutypinol as a substrate. It showed a preference for 4-nitro-benzaldehyde, as do certain aldehyde reductases (Vander Jagt et al., 1990; Bohren et al., 1991). Furthermore, the pI of the purified enzyme on IEF gel electrophoresis was similar to the pI values of several aldehyde reductases (Bohren et al., 1989; Inoue et al., 1993). ERE is therefore probably a member of the aldo-keto reductase superfamily (Jez et al., 1997).
Aldo-keto reductases catalyze the NADPH-dependent reduction of a variety of biogenic and xenobiotic aldehydes to their corresponding alcohols in mammalian and plant tissues. In mammalian cells aldehyde or aldose reductases play an important role in the biological inactivation of various toxic aldehydes such as chlordecone (Winters et al., 1990), N-acetyl-leucyl-leucyl-norleucinal (Inoue et al., 1993), and acreoline (Kolb et al., 1994). Similarly, aflatoxin B1, a mycotoxin secreted by Aspergillus flavus, can be metabolized to aflatoxin B1 dihydrodiol by an aldo-keto reductase of 36 kD isolated from rat liver (Hayes et al., 1993). In plants few reductases involved in toxin inactivation have been described. The first to be well characterized was a NADPH-dependent carbonyl reductase able to metabolize HC toxin, a cyclic tetrapeptide from Cochliobolus carbonum race 1 that parasitizes sensitive maize cultivars (Meeley et al., 1992). The characterization of ERE contributes to an increased understanding of detoxification mechanisms for fungal toxins.
The analysis of purified ERE activity revealed that it reduced a broad range of aromatic and aliphatic aldehydes. It converted benzaldehyde and diverse substituted benzaldehydes, but the position and the nature of the substituents greatly affected the efficiency of the reduction. It was shown that electron-withdrawing groups accelerated the reaction, whereas electron-donating groups decreased ERE activity. This suggests that ERE differs from the benzyl alcohol dehydrogenases, because in the benzaldehyde reduction reaction these enzymes are known to be independent of the electronic nature of the benzaldehyde substituents (Shaw et al., 1992, 1993). Furthermore, because nitro groups are larger than methyl or fluoro groups, the size of the substituent at position 3 or 4 on the aromatic ring may not affect structural hindrance.
Because ERE was purified from mung bean, a nonhost plant for E. lata, ERE may have a physiological role in mung bean. The apparent substrate specificity is distinct from that of cinnamyl-alcohol dehydrogenases (Wyrambik and Grisebach, 1975; Goffner et al., 1992), dihydroflavonol-4-reductases (Heller et al., 1985), and aromatic alcohol:NADP+ oxidoreductase such as the Arabidopsis defense-related protein ELI3 (Somssich et al., 1996). Even though ERE is probably involved in the generation of benzyl or aliphatic alcohol derivates, the physiological function of the enzyme remains unclear.
The enzymatic degradation of eutypine was previously described in grapevine cells cultured in vitro, and a 54-kD protein able to biologically inactivate the toxin has been partially characterized (Colrat et al., 1998). The ERE protein purified from mung bean seems to be different from the enzyme described in the grapevine, suggesting that various aldehyde reductases could reduce eutypine in plants. However, the high affinity of ERE for the toxin and the fact that purified ERE effectively detoxified eutypine across a broad range of temperatures (25°C–45°C) and pH values (6.2–7.5) make the corresponding gene an attractive candidate to confer resistance to vinestock and to determine the role of eutypine in the symptom development of eutypa dieback. Cloning of the mung bean ERE cDNA is currently underway.
ACKNOWLEDGMENTS
The authors would like to thank R. Tabacchi for providing entypine and labeled entypine, P. Winterton for reviewing the English manuscript and H. Mondiès for her skillful technical assistance.
Abbreviation:
- ERE
eutypine-reducing enzyme
Footnotes
This work was supported by the Martell Company (Cognac, France). M.G. and S.C. hold grants from the Ministère de l'Enseignement Supérieur et de la Recherche (France).
LITERATURE CITED
- Anzaï H, Yoneyama K, Yamaguchi I. Transgenic tobacco resistant to a bacterial disease by detoxification of a pathogenic toxin. Mol Gen Genet. 1989;219:492–494. [Google Scholar]
- Bohren KM, Bullock B, Wermuth B, Gabbay KH. The aldo-keto reductase superfamily. J Biol Chem. 1989;264:9547–9551. [PubMed] [Google Scholar]
- Bohren KM, Page JL, Shankar R, Henry SP, Gabbay KH. Expression of human aldose and aldehyde reductases. J Biol Chem. 1991;266:24031–24037. [PubMed] [Google Scholar]
- Colrat S, Deswarte C, Latché A, Klaébé A, Bouzayen M, Fallot J, Roustan JP (1998) Enzymatic detoxification of eutypine, a toxin from Eutypa lata, by Vitis vinifera cells: partial purification of an NADPH-dependent aldehyde reductase. Planta (in press)
- Damerval C, LeGuilloux M, Blaisonneau J, DeVienne D. A simplification of Heukeneneshoven and Dermick's silver staining of proteins. Electrophoresis. 1987;8:158–159. [Google Scholar]
- Defrancq E, Tabacchi R (1992) The synthesis of (14C) labelled eutypine. J Labelled Compd Radiopharm 31: 1057–1063
- Defrancq E, Zeziger T, Tabacchi R. The synthesis of natural acetylenic compounds from Eutypa lata (Pers:) Tul. Helv Chim Acta. 1993;76:425–429. [Google Scholar]
- Deswarte C, Canut H, Klaebe A, Roustan JP, Fallot J. Transport, cytoplasmic accumulation and mechanism of action of the toxin eutypine in Vitis vinifera cells. Arch Biochem Biophys. 1996a;149:336–342. [Google Scholar]
- Deswarte C, Eychenne J, Davy de Virville J, Roustan JP, Moreau F, Fallot J. Protonophoric activity of eutypine, a toxin from Eutypa lata, in plant mitochondria. J Plant Physiol. 1996b;334:200–205. doi: 10.1006/abbi.1996.0447. [DOI] [PubMed] [Google Scholar]
- Durbin RD, ed (1981) Toxins in Plant Disease. Academic Press, New York
- Duthie JA, Munkvold GP, Marois JJ, Grant S, Chellemi DO. Relationship between the age of the vineyard and incidence of Eutypa lata dieback. Phytopathology. 1991;81:1183. [Google Scholar]
- Fallot J, Deswarte C, Dalmayrac C, Colrat S, Roustan JP. Eutypa dieback of grapevine: isolation of a molecule synthesized by Eutypa lata and toxic for grapevine. C R Acad Sci (Paris) 1997;320:149–158. [Google Scholar]
- Goffner D, Joffroy I, Grima-Pettenati J, Halpin C, Knight ME, Schuch W, Boudet AM. Purification and characterization of isoforms of cinnamyl alcohol dehydrogenase (CAD) from Eucalyptus xylem. Planta. 1992;188:48–53. doi: 10.1007/BF00198938. [DOI] [PubMed] [Google Scholar]
- Hayes JD, Judah DJ, Neal GE. Resistance to aflatoxin B1 is associated with the expression of a novel aldo-keto reductase which has a catalytic activity towards a cytotoxic aldehyde-containing metabolite of the toxin. Cancer Res. 1993;53:3887–3894. [PubMed] [Google Scholar]
- Heller W, Forkmann G, Britsch L, Grisebach H. Enzymatic reduction of (+)-dihydroflavonol to flavan -3, 4-cis-diols with flower extracts from Matthiola incana and its role in anthocyanin biosynthesis. Planta. 1985;165:284–287. doi: 10.1007/BF00395052. [DOI] [PubMed] [Google Scholar]
- Inoue S, Sharma RC, Schimke RT, Simoni RD. Cellular detoxification of tripeptidyl aldehyde by an aldo-keto reductase. J Biol Chem. 1993;268:5894–5898. [PubMed] [Google Scholar]
- Jez JM, Bennett MJ, Schlegel BP, Lewis M, Penning TM. Comparative anatomy of the aldo-keto reductase superfamily. Biochem J. 1997;326:625–636. doi: 10.1042/bj3260625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johal GS, Briggs SP. Reductase activity encoded by the HM1 disease resistance gene in maize. Science. 1992;258:985–987. doi: 10.1126/science.1359642. [DOI] [PubMed] [Google Scholar]
- Keen NT. The molecular biology of disease resistance. Plant Mol Biol. 1993;19:109–122. doi: 10.1007/BF00015609. [DOI] [PubMed] [Google Scholar]
- Kolb NA, Hunsaker LA, Vander Jagt DL. Aldose reductase catalyzed reduction of acrolein: implication in cyclophosphamide toxicity. Mol Pharmacol. 1994;45:797–801. [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the bacteriophage T4. Nature. 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- March J (1985) Advanced Organic Chemistry, 3rd Ed. John Wiley & Sons, New York, pp 242–249
- Mato M, Ishikura N. Flavonol changes in seedlings of Vigna mungo during growth. Plant Physiol. 1993;142:647–650. [Google Scholar]
- Meeley RB, Johal GS, Briggs SP, Walton JD. A biochemical phenotype for a disease resistance gene of maize. Plant Cell. 1992;4:71–77. doi: 10.1105/tpc.4.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeley RB, Walton JD. Enzymatic detoxification of HC-toxin, the host selective cyclic peptide from Cochliobolus carbonum. Plant Physiol. 1991;97:1080–1086. doi: 10.1104/pp.97.3.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller MJ, Kasimatis N. Dieback of grapevines caused by Eutypa armeniacae. Plant Dis Rep. 1978;62:254–258. [Google Scholar]
- Moller MJ, Kasimatis N. Further evidence that Eutypa armeniacae – not Phomopsis viticola – incites dead arm symptoms on grape. Plant Dis. 1981;65:429–431. [Google Scholar]
- Munkvold GP, Duthie JA, Marois JJ. Reduction in yield and vegetative growth of grapevines due to Eutypa dieback. Phytopathology. 1994;84:186–192. [Google Scholar]
- Robertson EF, Dannelly HK, Malloy PJ, Reeves HC. Rapid isoelectric focusing in a vertical polyacrylamide minigel system. Anal Biochem. 1987;167:290–294. doi: 10.1016/0003-2697(87)90166-7. [DOI] [PubMed] [Google Scholar]
- Schäfer W. Molecular mechanism of fungal pathogenicity to plant. Annu Rev Phytopathol. 1994;32:461–477. [Google Scholar]
- Shaw JP, Rekik M, Schwager F, Harayama S. Kinetic studies on benzyl alcohol dehydrogenase encoded by TOL plasmid pWWW0. J Biol Chem. 1993;268:10842–10850. [PubMed] [Google Scholar]
- Shaw JP, Schwager F, Harayama S. Substrate specificity of benzyl alcohol dehydrogenase and benzaldehyde dehydrogenase encoded by TOL plasmid pWWW0. Biochem J. 1992;283:789–794. doi: 10.1042/bj2830789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somssich IE, Wernert P, Kiedronwski S, Hahlbrock K. Arabidopsis thaliana defense-related protein ELI3 is an aromatic alcohol: NADP+ oxidoreductase. Proc Natl Acad Sci USA. 1996;93:14199–14203. doi: 10.1073/pnas.93.24.14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tey Rulh P, Philippe I, Renaud JM, Tsoupras G, De Angelis P, Roustan JP, Fallot J, Tabacchi R. Eutypine, a phytotoxin produced by Eutypa lata, the causal agent of dying-arm disease of grapevine. Phytochemistry. 1991;30:471–473. [Google Scholar]
- Utsumi R, Hadama T, Noda M, Toyoda H, Hashimoto H, Ohuchi S. Cloning of fusaric acid-detoxifying gene from Cladosporium werbeckii: a new strategy for prevention of plant diseases. J Biotechnol. 1988;8:311–316. [Google Scholar]
- Vander Jagt DL, Robinson B, Taylor KK, Hunsaker LA. Aldose reductase from human skeletal and heart muscle. J Biol Chem. 1990;265:20982–20987. [PubMed] [Google Scholar]
- Winters CJ, Molowa DT, Guzelian PS. Isolation and characterization of cloned cDNAs encoding human liver chlordecone reductase. Biochemistry. 1990;29:1080–1087. doi: 10.1021/bi00456a034. [DOI] [PubMed] [Google Scholar]
- Wyrambik D, Grisebach H. Purification and properties of isoenzymes of cinnamyl alcohol dehydrogenase from soybean cell suspension cultures. Eur J Biochem. 1975;59:9–15. doi: 10.1111/j.1432-1033.1975.tb02418.x. [DOI] [PubMed] [Google Scholar]
- Zhang L, Birch RG. The gene for albicidin detoxification from Pantoea dispersa encodes an esterase and attenuates pathogenicity of Xanthomonas albilineans to sugarcane. Proc Natl Acad Sci USA. 1997;94:9984–9989. doi: 10.1073/pnas.94.18.9984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweillemuller M, Antus S, Kovacs T, Sonnenbichler J. Biotransformation of the fungal toxin fomannoxin by conifer cell cultures. Biol Chem. 1997;378:915–921. doi: 10.1515/bchm.1997.378.8.915. [DOI] [PubMed] [Google Scholar]