Abstract
The movement protein (MP) of Cowpea mosaic virus forms tubules in plasmodesmata to enable the transport of mature virions. Here it is shown that the MP is capable of specifically binding riboguanosine triphosphate and that mutational analysis suggests that GTP binding plays a role in the targeted transport of the MP. Furthermore, the MP is capable of binding both single-stranded RNA and single-stranded DNA in a non-sequence-specific manner, and the GTP- and RNA-binding sites do not overlap.
Most plant virus genomes code for one or more movement proteins (MPs), which are required for viral cell-to-cell movement. Based on their primary structure, MPs can be divided into several superfamilies, one of which is the “30K” superfamily, related to the Tobacco mosaic virus (TMV) MP (20). Within this 30K superfamily, two basic mechanisms for cell-to-cell movement have been proposed (18). TMV MP typifies one mechanism whereby the MP modifies plasmodesmata, allowing viral RNA-MP complexes to move from cell to cell. The other type of movement, best known from Cowpea mosaic virus (CPMV) MP, is the tubule-guided movement of mature virus particles through drastically modified plasmodesmata. Secondary-structure comparisons of MPs belonging to the 30K superfamily predicted a common central core (20). In this core, one aspartic acid, referred to as the D motif, is almost absolutely conserved (16, 21), but its function has so far remained unresolved. MPs of como- and tobamoviruses have been suggested to contain a Walker B-like ribonucleoside triphosphate (rNTP)-binding motif (4, 27), and some MPs of the 30K superfamily (for example, TMV MP) have been shown to bind GTP (19). However, the GTP-binding site has not been identified, and a function for GTP binding has not been described yet, although it has been suggested that GTP hydrolysis provides the energy needed for cell-to-cell transport (19).
To investigate potential rNTP-binding properties of CPMV MP, GTP-coupled agarose beads (Sigma), prewashed three times with binding buffer (24), were incubated for 60 min at 4°C with 5 μg of purified wild-type MP (wtMP; prepared as described previously [3]). The beads were then pelleted by centrifugation (5 min at 18,000 × g) and washed three times with binding buffer to remove unbound proteins. Pellet (bound protein) and supernatant (unbound protein) fractions were analyzed by immunoblotting with anti-MP antibodies (15), which showed that MP binds to the GTP-coupled beads (Fig. 1A). When the MP was incubated for 60 min at 4°C with 2.5 mM GTP prior to incubation with the GTP-agarose beads, most of the MP remained in the supernatant fraction, showing that the binding of MP is to GTP and not directly to the agarose beads (Fig. 1B). Furthermore, preincubation with 2.5 mM ATP, CTP, or UTP did not prevent the binding of MP to the GTP-agarose beads (Fig. 1C), indicating that the CPMV MP specifically binds GTP, like the MPs of TMV and Cucumber mosaic virus (19).
FIG. 1.
GTP-binding capacity of MP and mutant AM5MP. (A through C) wtMP specifically binds GTP. Purified MP was incubated with GTP-agarose beads. Unbound proteins (sup) were separated from GTP-agarose beads (beads) by centrifugation, and both were analyzed by Western blotting with anti-CPMV MP. For the competition assays, wtMP was preincubated with 2.5 mM GTP (B), ATP, CTP, or UTP (C). (D) Mutant AM5MP does not bind GTP. The arrow indicates the position of the MP.
In CPMV MP mutant AM5 (1), V142 and D143 (the D motif), which are located in the putative rNTP-binding site (4), were replaced with alanines. To express His-tagged AM5MP in insect cells, the AM5 mutation was introduced into pFASTBAC-MP (3) by ligation of the NdeI/BamHI fragment from pTMAM5 (1) and the NcoI/NdeI fragment from pFASTBAC-MP into NcoI/BamHI-digested pFASTBAC-MP with a triple ligation. Mutant AM5MP, expressed and purified as described for wtMP (3), almost completely failed to bind to the GTP-agarose beads (Fig. 1D), indicating that the D motif is part of the GTP-binding site, as suggested by Chen and Bruening (4). Based on the resemblance of the D motif to the Walker B motif (9, 17), the aspartic acid (D143) might play a role in binding an Mg cation complexed with phosphate groups of the GTP (9, 17). The observation that the D motif is highly conserved among members of the 30K superfamily suggests that GTP binding may be a general feature of these MPs. Although the lack of GTP binding of this mutant could be due to misfolding, this is very unlikely, as the two amino acid changes are rather mild and the mutant still binds RNA, which, as we show below, is dependent on the conformation of the MP.
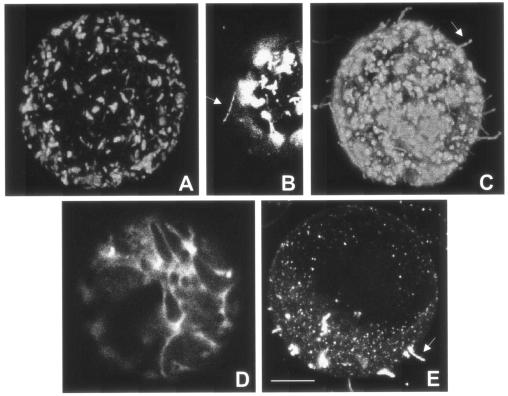
So far, a function for GTP binding by MPs has not been demonstrated, although it was suggested that rNTP binding and hydrolysis provide energy for cell-to-cell movement, which is supposed to be an energy-dependent process (2, 10). At least two steps during MP-mediated cell-to-cell movement might involve rNTP-dependent events: (i) intracellular targeting of MP or MP-RNA complexes to plasmodesmata and (ii) modification of plasmodesmata either by the trafficking of MP-RNA complexes or by the formation of tubules (10, 12, 18). To determine whether the mutation in the rNTP-binding site has an effect on the subcellular localization of the MP, insect cells (Spodoptera frugiperda; Sf21) expressing mutant AM5MP were fixed for immunofluorescent detection of the protein (3). In these cells, AM5MP (Fig. 2A and B), like wtMP (Fig. 2C), accumulated in aggregates in the cytoplasm which might be caused by the overexpression of MP in insect cells. Rarely, i.e., in approximately 1% of the AM5MP-expressing cells, relatively short tubules were found at the cell surface (Fig. 2B), while in approximately 80% of the cells expressing wtMP, tubules were present (Fig. 2C), indicating that GTP binding plays a role in tubule formation.
FIG. 2.
Intracellular localization of wt and AM5 mutant MPs in insect cells and cowpea protoplasts. (A through C) Immunofluorescent detection of MP in insect cells expressing AM5MP (A and B) and wtMP (C); (D and E) cowpea protoplasts transiently expressing AM5MP-YFP (D) and wtMP-YFP (E). Arrows indicate tubules formed at the cell surface. Bar, 5 μm.
Since wtMP formed aggregates in the cytoplasm of insect cells, a process that is not observed in plant cells, and insect cells are not hosts for CPMV, the localization of AM5MP in cowpea protoplasts was also studied. Therefore, the AM5 mutation was introduced into pMON-wtMP-YFP (25), which encodes wtMP fused C terminally to the yellow fluorescent protein (wtMP-YFP) under the control of a double 35S promoter by ligation of the BglII/XhoI fragment from pTMAM5 into BglII/XhoI-digested pMON-wtMP-YFP. Cowpea mesophyl protoplasts were isolated and inoculated as described previously (30), and images from the protoplasts were obtained with a Zeiss LSM510 confocal microscope with standard YFP filters. At 24 h posttransfection, AM5MP-YFP had accumulated uniformly in the cytoplasm of transfected protoplasts (Fig. 2D), in contrast to wtMP-YFP, which had accumulated in punctate structures at the plasma membrane and formed tubules protruding from the cell surface into the culture medium (Fig. 2E), indicating a role for GTP binding in targeting MP to the cell periphery. As the D motif is conserved throughout the 30K MP superfamily, GTP binding by other MPs may also be necessary for targeting to the cell periphery. The observations that the targeting deficiency of AM5 could not be complemented by wt protein (data not shown) and that targeting to the plasma membrane very likely requires di- or multimerization of MPs (25) suggest that GTP binding might play a role in the multimerization of MP. It is tempting to speculate that the mechanism by which CPMV MP forms tubules resembles microtubule formation, since β-tubulin requires GTP binding for incorporation into microtubules (13).
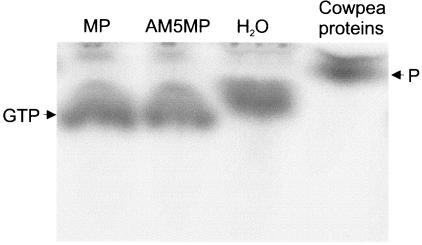
As GTP binding often coincides with the GTPase activity of a protein, a GTPase assay was performed with wtMP and AM5MP by incubating 1 μl of [γ-32P]GTP with 1 μg of wtMP or AM5MP in binding buffer for 2 h at 25°C. The soluble (S30) fraction of a cowpea leaf was used as a positive control, and water was used as a negative control. The mixtures were applied directly to thin-layer chromatography plates, and the GTP and P were separated by using 0.3 M sodium phosphate (pH 3.8) containing 1 M LiCl as the solvent. The results obtained demonstrate that neither wtMP nor AM5MP exhibits GTPase activity in this assay (Fig. 3) but that GTPases present in the cytoplasmic fraction from cowpea leaves were able to hydrolyze GTP. This experiment, however, does not rule out the possibility that CPMV MP, like tubulin (8, 22), needs to form multimers or requires a host factor or membrane binding for GTP hydrolysis.
FIG. 3.
GTPase activity assay of CPMV MP. wtMP or AM5MP was incubated with [γ-32P]GTP and applied to thin-layer chromatography plates. A cytoplasmic extract of cowpea leaves was used as a positive control, and H2O was used as a negative control. The GTP and P are indicated.
GTP binding by CPMV MP may be the result of an RNA-binding capacity. For many MPs of the 30K superfamily RNA binding has been confirmed (5, 6, 19, 23, 26, 28), but this information was not available for the CPMV MP. RNA binding by CPMV MP was studied in a gel shift assay by using a radiolabeled RNA fragment derived from CPMV. This RNA fragment was obtained by in vitro transcription of M19GFP7 (11) (linearized with PvuII) with T7 RNA polymerase (GIBCO BRL) in the presence of [α-32P]CTP, resulting in the transcription of the 3′-terminal 707 nucleotides (nt) of CPMV RNA2. Fifty nanograms of this radioactive RNA fragment was incubated with different amounts of purified wtMP in 20 μl of binding buffer for 60 min at room temperature and then used for electrophoresis in a 1% agarose gel. The results of this experiment showed that wtMP is able to bind single-stranded RNA (ssRNA) in a cooperative manner (Fig. 4A). Heating MP for 5 min at 95°C prior to binding drastically decreased the RNA binding capacity (data not shown), indicating that the native protein conformation is important for RNA binding.
FIG. 4.
Nucleic acid binding of CPMV MP, evaluated by gel shift assay. The protein-RNA complex and the free RNA were separated by electrophoresis in a 1% agarose gel. Fifty nanograms of 32P-labeled ssRNA corresponding to the C-terminal 707 nt of CPMV RNA2 was used in all experiments. For reasons yet unknown, two RNA molecules were produced during in vitro transcription. (A) Binding of different amounts of MP to ssRNA; (B) binding of CPMV MP (500 ng) to ssRNA under increasing NaCl concentrations; (C) various amounts of AM5MP incubated with 50 ng of 32P-labeled ssRNA. The wtMP was used as a control.
Competition experiments (data not shown) with 1 μg (a 20-fold excess) of ssRNA, double-stranded RNA (dsRNA) (both corresponding to the 3′-terminal 203 nt of brome mosaic virus RNA3 [14]), ssDNA (M13 DNA), or dsDNA (linearized pFASTBAC-HT; GIBCO BRL) revealed that CPMV MP binds ssRNA or ssDNA, but not dsRNA or dsDNA, without sequence specificity just like the related Broad bean wilt virus 2 MP and many other MPs (5, 6, 19, 23, 26, 28). The ability to bind single-stranded nucleic acids therefore seems to be a general property of plant viral MPs. However, the significance of the RNA-binding capacity for CPMV MP has yet to be determined, since for CPMV, virus particles rather than MP-RNA complexes are transported from cell to cell. The formation of an MP-RNA complex, however, might be required for systemic infection. More likely, the capacity to bind RNA is not important for MP but may be required for the function of the overlapping cofactor for RNA2 replication (CR), which is translated from a start codon in frame with that of the MP and plays a role in the replication of RNA2 (29). However, we have not studied whether CR is able to bind RNA. A third possibility is that RNA binding is not needed at all for MPs that form tubular structures but is just an evolutionary remnant, although it seems unlikely that a protein of an RNA virus, which generally has a high mutation rate (7), would retain such activity for no reason.
RNA binding also decreased with increasing concentrations of NaCl (Fig. 4B), suggesting that RNA binding involves ionic interactions. The CPMV MP-RNA complex was disrupted at NaCl concentrations of 300 mM and higher, which are similar to values reported for Alfalfa mosaic virus MP (28) and Cauliflower mosaic virus MP (6). The MP-RNA complexes made by TMV (5), Cucumber mosaic virus (19), Red clover necrotic mosaic virus (23), and Broad bean wilt virus 2 (26), however, dissociated at higher NaCl concentrations and therefore seem more stable. Whether the differences in the stabilities of the MP-RNA complex are biologically significant is unclear.
Similar RNA-binding experiments were carried out with mutants AM5MP (Fig. 4C) and Δ48CMP (data not shown), which lacks the C-terminal 48 amino acids and was previously shown to be unable to bind CPMV virions or coat proteins (3). Both mutant MPs retained their RNA-binding capacity, indicating that different domains in the MP are involved in RNA binding, GTP binding, and virion binding.
Acknowledgments
We thank Jan Verver and Gerard van der Krogt for providing M13 DNA and pMON-YFP, respectively.
C. M. Carvalho was financially supported by a fellowship from the CAPES Foundation (Brazil).
REFERENCES
- 1.Bertens, P., J. Wellink, R. Goldbach, and A. van Kammen. 2000. Mutational analysis of the cowpea mosaic virus movement protein. Virology 267:199-208. [DOI] [PubMed] [Google Scholar]
- 2.Carrington, J. C., K. D. Kasschau, S. K. Mahajan, and M. C. Schaad. 1996. Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8:1669-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carvalho, C. M., J. Wellink, S. G. Ribeiro, R. W. Goldbach, and J. W. M. van Lent. 2003. The C-terminal region of the movement protein of Cowpea mosaic virus is involved in binding to the large but not to the small coat protein. J. Gen. Virol. 84:2271-2277. [DOI] [PubMed] [Google Scholar]
- 4.Chen, X., and G. Bruening. 1992. Nucleotide sequence and genetic map of cowpea severe mosaic virus RNA 2 and comparisons with RNA 2 of other comoviruses. Virology 187:682-692. [DOI] [PubMed] [Google Scholar]
- 5.Citovsky, V., D. Knorr, G. Schuster, and P. Zambryski. 1990. The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell 60:637-647. [DOI] [PubMed] [Google Scholar]
- 6.Citovsky, V., D. Knorr, and P. Zambryski. 1991. Gene I, a potential cell-to-cell movement locus of cauliflower mosaic virus, encodes an RNA-binding protein. Proc. Natl. Acad. Sci. USA 88:2476-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Domingo, E., and J. J. Holland. 1997. RNA virus mutations and fitness for survival. Annu. Rev. Microbiol. 51:151-178. [DOI] [PubMed] [Google Scholar]
- 8.Erickson, H. P., and E. T. O'Brien. 1992. Microtubule dynamic instability and GTP hydrolysis. Annu. Rev. Biophys. Biomol. Struct. 21:145-166. [DOI] [PubMed] [Google Scholar]
- 9.Fry, D. C., S. A. Kuby, and A. S. Mildvan. 1986. ATP-binding site of adenylate kinase: mechanistic implications of its homology with ras-encoded p21, F1-ATPase, and other nucleotide-binding proteins. Proc. Natl. Acad. Sci. USA 83:907-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ghoshroy, S., R. Lartey, J. Sheng, and V. Citovsky. 1997. Transport of proteins and nucleic acids through plasmodesmata. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:27-49. [DOI] [PubMed] [Google Scholar]
- 11.Gopinath, K., J. Wellink, C. Porta, K. M. Taylor, G. P. Lomonossoff, and A. van Kammen. 2000. Engineering cowpea mosaic virus RNA-2 into a vector to express heterologous proteins in plants. Virology 267:159-173. [DOI] [PubMed] [Google Scholar]
- 12.Heinlein, M. 2002. Plasmodesmata: dynamic regulation and role in macromolecular cell-to-cell signaling. Curr. Opin. Plant Biol. 5:543-552. [DOI] [PubMed] [Google Scholar]
- 13.Howard, J., and A. A. Hyman. 2003. Dynamics and mechanics of the microtubule plus end. Nature 422:753-758. [DOI] [PubMed] [Google Scholar]
- 14.Jansen, K. A., C. J. Wolfs, H. Lohuis, R. W. Goldbach, and B. J. Verduin. 1998. Characterization of the brome mosaic virus movement protein expressed in E. coli. Virology 242:387-394. [DOI] [PubMed] [Google Scholar]
- 15.Kasteel, D. T., M. C. Perbal, J. C. Boyer, J. Wellink, R. W. Goldbach, A. J. Maule, and J. W. van Lent. 1996. The movement proteins of cowpea mosaic virus and cauliflower mosaic virus induce tubular structures in plant and insect cells. J. Gen. Virol. 77:2857-2864. [DOI] [PubMed] [Google Scholar]
- 16.Koonin, E. V., A. R. Mushegian, E. V. Ryabov, and V. V. Dolja. 1991. Diverse groups of plant RNA and DNA viruses share related movement proteins that may possess chaperone-like activity. J. Gen. Virol. 72:2895-2903. [DOI] [PubMed] [Google Scholar]
- 17.la Cour, T. F., J. Nyborg, S. Thirup, and B. F. Clark. 1985. Structural details of the binding of guanosine diphosphate to elongation factor Tu from E. coli as studied by X-ray crystallography. EMBO J. 4:2385-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lazarowitz, S. G., and R. N. Beachy. 1999. Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11:535-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, Q., and P. Palukaitis. 1996. Comparison of the nucleic acid- and NTP-binding properties of the movement protein of cucumber mosaic cucumovirus and tobacco mosaic tobamovirus. Virology 216:71-79. [DOI] [PubMed] [Google Scholar]
- 20.Melcher, U. 2000. The ′30K' superfamily of viral movement proteins. J. Gen. Virol. 81:257-266. [DOI] [PubMed] [Google Scholar]
- 21.Mushegian, A. R., and E. V. Koonin. 1993. Cell-to-cell movement of plant viruses. Insights from amino acid sequence comparisons of movement proteins and from analogies with cellular transport systems. Arch. Virol. 133:239-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nogales, E., M. Whittaker, R. A. Milligan, and K. H. Downing. 1999. High-resolution model of the microtubule. Cell 96:79-88. [DOI] [PubMed] [Google Scholar]
- 23.Osman, T. A., R. J. Hayes, and K. W. Buck. 1992. Cooperative binding of the red clover necrotic mosaic virus movement protein to single-stranded nucleic acids. J. Gen. Virol. 73:223-227. [DOI] [PubMed] [Google Scholar]
- 24.Peters, S. A., J. Verver, E. A. Nollen, J. W. van Lent, J. Wellink, and A. van Kammen. 1994. The NTP-binding motif in cowpea mosaic virus B polyprotein is essential for viral replication. J. Gen. Virol. 75:3167-3176. [DOI] [PubMed] [Google Scholar]
- 25.Pouwels, J., N. Kornet, N. van Bers, T. Guighelaar, J. van Lent, T. Bisseling, and J. Wellink. 2004. Identification of distinct steps during tubule formation by the movement protein of Cowpea mosaic virus. J. Gen. Virol. 84:3485−3494. [DOI] [PubMed]
- 26.Qi, Y. J., X. P. Zhou, X. Z. Huang, and G. X. Li. 2002. In vivo accumulation of Broad bean wilt virus 2 VP37 protein and its ability to bind single-stranded nucleic acid. Arch. Virol. 147:917-928. [DOI] [PubMed] [Google Scholar]
- 27.Saito, T., Y. Imai, T. Meshi, and Y. Okada. 1988. Interviral homologies of the 30K proteins of tobamoviruses. Virology 167:653-656. [PubMed] [Google Scholar]
- 28.Schoumacher, F., C. Erny, A. Berna, T. Godefroy-Colburn, and C. Stussi-Garaud. 1992. Nucleic acid-binding properties of the alfalfa mosaic virus movement protein produced in yeast. Virology 188:896-899. [DOI] [PubMed] [Google Scholar]
- 29.Van Bokhoven, H., O. Le Gall, D. Kasteel, J. Verver, J. Wellink, and A. B. Van Kammen. 1993. Cis- and trans-acting elements in cowpea mosaic virus RNA replication. Virology 195:377-386. [DOI] [PubMed] [Google Scholar]
- 30.van Bokhoven, H., J. Verver, J. Wellink, and A. van Kammen. 1993. Protoplasts transiently expressing the 200K coding sequence of cowpea mosaic virus B-RNA support replication of M-RNA. J. Gen. Virol. 74:2233-2241. [DOI] [PubMed] [Google Scholar]