Abstract
The physiology of small mammalian hibernators shifts profoundly over a year, from summer homeothermy to winter heterothermy. Torpor-arousal cycles define high-amplitude tissue activity fluctuations in winter, particularly for skeletal muscle, which contributes to the energetically demanding rewarming process via shivering. To better understand the biochemistry underlying summer-winter and torpor-arousal transitions, we applied two-dimensional gel electrophoresis coupled with liquid chromatography/mass spectrometry/mas spectrometry to the soluble proteins from hindlimb muscle of 13-lined ground squirrels (Ictidomys tridecemlineatus) in two summer and six winter states. Two hundred sixteen protein spots differed by sampled state. Significantly, intrawinter protein adjustment was a minor component of the dataset despite large discrepancies in muscle activity level among winter states; rather, the bulk of differences (127/138 unequivocally identified proteins spots) occurred between summer and winter. We did not detect any proteomic signatures of skeletal muscle atrophy in this hibernator nor any differential seasonal regulation of protein metabolism. Instead, adjustments to metabolic substrate preferences dominated the detected proteomic differences. Pathways of carbohydrate metabolism (glycolysis and gluconeogenesis) were summer enriched, whereas the winter proteome was enriched for fatty acid β-oxidation. Nevertheless, our data suggest that some reliance on carbohydrate reserves is maintained during winter. Phosphoglucomutase (PGM1), which reversibly prepares glucose subunits for either glycolysis or glycogenesis, showed apparent winter state-specific phosphorylation. PGM1 was phosphorylated during rewarming and dephosphorylated by interbout arousal, implying that glucose supplements lipid fuels during rewarming. This, along with winter elevation of TCA cycle enzymes, suggests that hindlimb muscles are primed for rapid energy production and that carbohydrates are an important fuel for shivering thermogenesis.
Keywords: DIGE, glycogen, PDK4, Ca2+ATPase, FABP, mitofilin
mammalian hibernators including ground squirrels undergo extreme annual changes in physiology and morphology. Among these changes are adjustments in body composition and a switch between summer homeothermy and winter heterothermy (reviewed in Refs. 4 and 14). Each year animals fatten during a summer active period and then fast and catabolize those stores through winter. Hibernating ground squirrels capitalize on energy savings associated with low body temperature (Tb) during multiday torpor bouts (reviewed in Ref. 38) to permit survival solely on endogenous reserves. Short (<1 day) arousal intervals punctuate winter torpor bouts (50). The switch between summer homeothermy and winter heterothermy therefore has two major components: 1) a transition from fat anabolism to catabolism; and 2) orchestration of reversible metabolic depression, i.e., torpor-arousal cycles. In fact, a model describing two regulatory switches has been advanced to explain the cyclical nature of hibernation (28, 57). Switch 1 identifies the transition from summer homeothermy to winter heterothermy, and is likely facilitated by reprogramming gene expression (57, 62). Switch 2 controls intrawinter cycling between torpor and arousal. Given that ground squirrel cells are not able to maintain transcription and translation at low Tb (67, 68), switch 2 is likely facilitated by small biomolecules and posttranslational regulation of proteins (19, 57, 58).
Torpor-arousal cycles reflect the temporal partitioning of depressed and active physiological states and biochemical processes, respectively, throughout winter. Metabolic, heart, and respiration rates decline precipitously during torpor bouts (reviewed in Refs. 4, 14, and 38). These rates, along with processes such as translation and transcription are restored during interbout arousals (14). Likewise, the functions of specific tissues are temporally partitioned in winter; the skeletal muscles of ground squirrels provide a striking example. During arousal periods, squirrels rewarm from near-ambient Tb to euthermic (i.e., summer) Tb using endogenous mechanisms of heat production. Rewarming episodes are therefore periods of intense metabolic activity where whole body metabolic rates can exceed basal euthermic rates by severalfold (see Table 2 in Ref. 28). A portion of the metabolic work devoted to rewarming occurs in the form of skeletal muscle shivering thermogenesis, which appears to be a necessary component of the arousal process in rodents (29, 36).
Table 2.
Functional annotation of gene products elevated in winter states
| Cluster | Enrichment Score | No. of Annotations | Mean Fold Enrichment |
|---|---|---|---|
| Mitochondrion | 14.2 | 3 | 13.8 |
| Mitochondrial matrix | 7.6 | 3 | 11.5 |
| Aerobic respiration | 5.5 | 21 | 52.0 |
| Monosaccharide metabolism | 4.8 | 3 | 12.8 |
| Monosaccharide catabolism | 4.1 | 7 | 23.2 |
| Mitochondrial envelope | 3.5 | 7 | 5.6 |
| Fatty acid catabolism | 3.4 | 9 | 25.7 |
| Membrane-enclosed lumen | 3.0 | 3 | 2.5 |
| Reactive O2 species metabolism | 2.6 | 15 | 29.0 |
| Negative regulation of cell death | 2.5 | 3 | 5.7 |
| Regulation of cell death | 2.1 | 3 | 3.3 |
| Nucleoside binding | 2.1 | 4 | 2.3 |
| Flavoprotein | 1.7 | 4 | 11.3 |
| Membrane-bounded vesicle | 1.6 | 5 | 4.1 |
| Peroxisome | 1.5 | 3 | 10.8 |
| Nucleotide binding | 1.4 | 6 | 2.1 |
Functional annotation clusters were generated using DAVID bioinformatics database with κ similarity = 0.85 (default settings, otherwise). All significantly enriched clusters (≥1.3) are listed. Clusters are described by the top 3 annotation with the widest biological relevance, and the mean pathway fold enrichment for all annotations is reported from each cluster.
The torpid state contrasts markedly with arousals. Shivering is suppressed as animals reenter torpor (15, 60) and tissue functions are rapidly diminished. During torpor, skeletal muscles are essentially quiescent, displaying no EMG activity (15). This disuse scenario of torpor has prompted considerable interest in the possibility of skeletal muscle atrophy during hibernation. In fact, an inactive period with the duration of a single torpor bout (∼2 wk in ground squirrels) would be sufficient to evoke disuse atrophy in a nonhibernator of similar size (e.g., 44). In the torpid state, skeletal muscle certainly represents an attractive fuel resource that can be tapped to meet organismal energetic requirements. Skeletal muscle depletes and replenishes a significant portion of its glycogen stores during each torpor/arousal cycle (22, 23). Carbohydrate catabolism is evidenced by the 100 times increase in circulating plasma lactate as arousal progresses (23), implying that anaerobic glycolysis is an essential aspect of shivering energetics. In light of this, it is surprising that only limited skeletal muscle atrophy has been documented in hibernating rodents (48, 63, 71). Moreover, there is evidence that this attenuation of muscle atrophy is due to an intrinsic, seasonally regulated feature of skeletal muscle physiology or biochemistry, rather than simply a temperature-reduction of protein degradation rates occurring alongside reduced activity during torpor (48).
Skeletal muscle faces two contradictions during the winter heterothermic period: the need to 1) maintain contractile functionality for arousals and spring emergence, yet inactivate contractility during both the entrance and maintenance phases of torpor; and 2) catabolize local fuels for shivering and locomotion, yet limit tissue atrophy. Hibernator skeletal muscle must therefore be capable of regulating energy savings during extended inactivity, the transient motor requirements of winter arousal periods, and sustained activity following spring emergence.
To better understand the observed partitioning of skeletal muscle function in terms of associated cellular biochemistry and two-switch regulatory control, we set out to define changes in the skeletal muscle proteome of 13-lined ground squirrels (Ictidomys tridecemlineatus) over several stages that represent key transitions across the hibernator's year. While a number of unbiased screens in hibernators (summarized in Refs. 17 and 20) have previously been conducted for gene expression (also see Ref. 21), proteins, and metabolites (19, 45, 46, 57), our proteomic analysis across a range of functional states (2 summer, 6 winter) makes this the largest scale screen published to date on a hibernating species. Our aim was to exploit this multistate sampling design to compare the biochemical markers and hibernation switch regulation in a mixed hindlimb sample. Summer homeothermy was examined during the spring emergence and summer fattening phases and winter heterothermy across the torpor-arousal cycle. We sampled early and late torpor states along with arousing and interbout aroused animals as well as those reentering torpor. As perfusion (37, 38) and shivering amplitude lag in posterior regions (38), we expected early- and late-arousing animals, defined by abdominal Tb, to represent markedly different physiology in hindlimb muscle.
MATERIALS AND METHODS
Animals
All animal care and procedures were approved by the University of Colorado School of Medicine.
Laboratory-bred 13-lined ground squirrels were purchased from Dana Vaughan at the University of Wisconsin OshKosh in July-August, 2006–2008. Upon arrival at the University of Colorado, squirrels were housed individually at 18°C (14:10-h light-dark cycle) and provided with food (dry cat food and sunflower seeds) and water ad libitum. Animals were classified by hibernation state at the time of tissue collection (Fig. 1). Summer active (SA, n = 6) animals were euthanized August 4–5, 1 wk after arrival. The remaining squirrels were each implanted with two abdominal Tb monitors, to provide both a telemetered (MiniMitter, Bend, OR) and logged (iButton; Embedded Data Systems, Dallas, TX) Tb record through the winter. After a minimum 2-wk surgical recovery period, squirrels were transferred to a cold room (4°C, 0:24-h light-dark cycle) in early October. Food and water were removed after each squirrel exhibited regular torpor in the cold room and was returned to spring dark (SD, n = 6) at least 10 days before tissue collection.
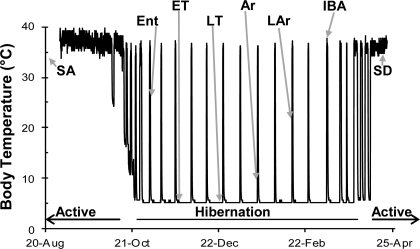
Fig. 1.
Body temperature (Tb) of a laboratory-housed 13-lined ground squirrel (Ictidomys tridecemlineatus) over ∼8 mo. The hibernation label identifies the winter heterothermic period and the annual cycle of this species is completed by a spring-fall active period. The physiological state at sampling (n = 8) over the year are indicated by the abbreviations. SA, summer active; Ent, entering torpor; ET, early torpor; LT, late torpor; Ar, early arousing; LAr, late arousing; IBA, interbout aroused, SD, spring dark.
Winter hibernation states (n = 6 per group) were defined as follows (Fig. 1): entering torpor (Ent) at 27°C > Tb > 23°C; early torpor (ET) within 5–10% of previous torpor bout duration; late torpor (LT) at 80–95% previous bout duration; early arousing (Ar) at 7°C < Tb < 12.8°C; late arousing (LAr) at 18°C < Tb < 25°C; and interbout aroused (IBA) animals at ∼3 h after the Tb inflection point between rapid rewarming and stable euthermic values of ∼37°C. SD animals remained in the cold room, and were euthanized 11–20 days after their last recorded torpor bout.
Protein Extracts
Squirrels were euthanized under isoflurane anesthesia by exsanguination via cardiac draw, and were then cold-saline perfused after which a mixed skeletal muscle sample comprised of biceps femoris, semitendinosus, and semimembranosus was collected; all muscles were composed predominantly of type II glycolytic fibers in rats (15a). Dissected muscle was frozen immediately in liquid N2 and stored at −80°C. Samples were pulverized in liquid N2 and then homogenized on ice, centrifuged to retain the soluble fraction, and aliquotted as previously described (17). Homogenization buffer differed from the previous description only in the addition of 2 times phosphatase inhibitor cocktail (HALT, cat. no. 78420; Thermo Scientific, Rockford, IL). Protein concentrations of the soluble fractions were determined using a BCA assay (Pierce, Rockford, IL).
DIGE Labeling and 2D Gels
The procedure for denaturing and labeling protein samples with Cy2, Cy3, or Cy5 (CyDye DIGE Fluors; GE Healthcare, Piscataway, NJ) has been described (17). Homogenate samples (with equal protein content) from each animal were combined to generate a pooled reference standard. A mixed sample consisting of one Cy2-labeled reference sample, and one each of Cy3- and Cy5-labeled samples were run on a single gel. Protein from the mixed homogenate sample was precipitated in methanol/chloroform (18) and resuspended as previously described (17). Samples were then absorbed onto Immobiline DryStrips (pH 3–10 NL, 18 cm; GE Healthcare) for 18- to 24-h, focused overnight (Multiphor II; GE Healthcare), and then incubated 15 min each in reducing and alkylating buffers before mounting them to 9–16% polyacrylamide gels (17, 18, 20). Gradient gel pouring, running, and scanning procedures for analytical and spot-picking gels are described fully in Epperson et al. (17). Gels used for spot picking were fixed overnight in 10% methanol/7.5% acetic acid solution, stained with SYPRO Ruby (Bio-Rad, Hercules, CA) for 24 h, and then destained (10% methanol/7.5% acetic acid) prior to scanning.
Protein spots were picked and digested in the University of Colorado School of Medicine Mass Spectrometry and Proteomics Core Facility by using an automated system (Ettan spot picker and Ettan DIGEster, version 1.10; GE Healthcare). Gel plugs were extracted with a 1.4-mm diameter, 1.5-mm depth picker head, and after overnight digestion with trypsin (10 μg/ml in 20 mM ammonium bicarbonate) were kept at −20°C until analysis by liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS). Tryptic peptides in 0.1% formic acid were separated using an HPLC-Chip Cube (chip no. G4240–62001 Zorbax 300SB-C18; Agilent Technologies, Santa Clara, CA) along a 3–50% acetonitrile hydrophobicity gradient (7 min) followed by a 3-min wash in 90% acetonitrile for detection of full and tandem mass spectra in an ESI ion trap (LC/MSD XCT Ultra; Agilent Technologies) in positive ion mode. Tandem MS was performed for the four highest peaks with a 30-s dynamic exclusion. Spectral data were collected with 6300 Series TrapControl software (version 6.1, Build 83; Agilent Technologies).
We searched raw mass spectral data against an in-house database including all National Center for Biotechnology Information (NCBI) mammal sequences as of April 2010 plus the actual and predicted protein sequences of 13-lined ground squirrel sequences available from Ensembl and Arctic ground squirrels (Urocitellus parryii) from a previously published database (58), using Spectrum Mill MS Proteomics Workbench (revision A.03.03.075). The database contained 792,655 entries. Settings, such as precursor and fragment ion mass tolerances, allowed missed cleavage sites and fixed amino acid modifications followed Epperson et al. (17). Additionally, oxidized methionine was included as a variable modification. If a picked spot returned a unique protein identification (ID), with ≥ 4 peptides recovered and a score ≥ 50, no further attempt was made toward identification. Otherwise, spots were picked from one to four additional gels, and their spectral data merged. An in-house program, ExtracTags (17), compiled the results from multiple MS runs and from multiple species hits within the database. The program identified the single best ID for each spot from all species in the database, and these results are presented in Table S1. (Supplemental data is available with the online publication of this article.) Spectral data that supported unequivocal protein IDs are available in the PRIDE database (69) (www.ebi.ac.uk/pride) under accession no. 18461. Data were converted using PRIDE Converter (7) (http://pride-converter.googlecode.com). IDs at minimum required two supporting peptides and a score of 30 to be retained; however, after applying these criteria (and after removing trypsin and keratin IDs), some gel spots still produced multiple IDs. These multiple results were filtered according to score, peptides recovered, and relative average intensity of each possible ID. When one of these criteria was at least fourfold greater for a possible ID compared with all others, this was retained as the single ID for that gel spot (42) (Table S1). Ten protein IDs were resolved outside the framework of the filters described above and warrant specific mention. These multiple IDs occurred in close proximity to a highly abundant protein, and the spot ID was contaminated with peptides from the more prevalent protein. Single IDs were retained when the contaminating protein had a molecular mass inconsistent with the spot's location in the gel. We applied this criterion to recover single IDs from protein spots 246, 534, 565, 607, 657, 771, 776, 806, 1532, and 2280. The complete file output, including weak IDs and multiple IDs, which could not be resolved, is reported in Table S2.
Data Analyses
Protein spots from DIGE analytical gels were matched and quantified by densitometry. The Cy2 images from each analytical gel were matched to a master gel using DeCyder software (version 7.0; GE Healthcare). The abundance of each analytical spot (Cy3- and Cy5-labeled) was normalized to the corresponding reference gel (Cy2) spot. DeCyder software was also used to match spots from SYPRO Ruby pick gels to the master analytical Cy2 gel image and to generate pick location coordinates.
We applied one-way ANOVA to identify protein spots whose abundance differed significantly among hibernation states. Protein spots bearing the same ID, i.e., different protein isoforms, were considered independently via ANOVA and post hoc tests to account for the possibility that only a portion of a protein's total pool was changing. ANOVA was initially conducted on spots matched in three out of six animals (gels) for every sampling group. However, the matching of all protein spots found significant by ANOVA were double checked and ultimately, all of these 216 reported protein spots were matched in 5/6 gels for every sampling group. ANOVA P values were subsequently corrected for multiple tests (resulting in ANOVA q values) by Benjamini-Hochberg criteria (8). Protein spots with ANOVA q < 0.05 were selected for ID by LC-MS/MS. Tukey's post hoc tests were employed for pairwise comparisons between all hibernation states when the global F-test was significant after correction for multiple tests. To examine possible large-scale gene expression changes associated with hibernation switch 1 (57; see introduction), this same data subset (spots present in 5/6 gels per sampling state) was also examined by t-test to determine which protein spots were significantly more abundant in grouped summer versus winter samples. We analyzed the data using Random Forests unsupervised classification (n = 50,000 trees), and hierarchical clustering was visualized by heat map. All statistical analyses were conducted in R. Random Forests is a classification technique based on an ensemble of decision trees (11), in which a randomly selected subset of data features is applied to generate classifications at each node when growing a tree. Random Forests takes advantage of two powerful machine learning techniques: bagging (10) and random feature selection.
We used the NCBI and Ensembl protein and gene databases to gather functional information about identified proteins, which are denoted by gene symbol and NCBI official gene names (human accession nos. are also reported; see Table S1). When a protein was not named in the best-matching species record (NCBI), the name of the human RefSeq homolog identified by BLink was reported. The DAVID bioinformatics database (version 6.7) was used to examine gene functional annotations (31), specifically, to determine which pathways were significantly enriched in summer compared with winter states. This enrichment analysis used human as the reference organism and a kappa similarity of 0.85 (default settings otherwise). We report all significantly enriched functional annotation clusters having a mean enrichment score ≥ 1.3 (Tables 1 and 2). Clusters are named here by the description of widest biological relevance appearing in the top three annotations.
Table 1.
Functional annotation of gene products elevated in summer states
| Cluster | Enrichment Score | No. of Annotations | Mean Fold Enrichment |
|---|---|---|---|
| Carbohydrate catabolism | 7.2 | 6 | 34.3 |
| Glycolysis | 5.7 | 6 | 54.6 |
| Gluconeogenesis | 2.7 | 7 | 44.7 |
| Nucleotide binding | 2.2 | 10 | 3.2 |
| Sarcomere | 1.9 | 4 | 14.1 |
| Transit peptide | 1.6 | 10 | 5.8 |
| Cytoskeleton | 1.3 | 3 | 4.7 |
| Magnesium ion binding | 1.3 | 3 | 10.4 |
Functional annotation clusters were generated using DAVID bioinformatics database with κ similarity = 0.85 (default settings, otherwise). All significantly enriched clusters (≥1.3) are listed. Clusters are described by the top 3 annotation with the widest biological relevance, and the mean pathway fold enrichment for all annotations is reported from each cluster.
Phosphoprotein Analysis
As an initial examination of posttranslational phosphorylation modifications to protein spots of interest, we applied a phosphoprotein stain (ProQ Diamond; Molecular Probes, Eugene, OR) to IBA and LAr samples (n = 3 of each). Then 120 μg of unlabeled protein from each sample was prepared and run on 2D gels as described for the DIGE experiment, and then stained for phosphoproteins by using a published protocol (3). After scanning to record phosphoprotein staining (excitation: 532 nm, emission: 560 nm), gels were rinsed in water and then stained for total protein with SYPRO ruby as described above. Images of the paired scans were analyzed using ImageJ software (version 1.43u; National Institutes of Health) and merged for viewing using Adobe Photoshop (version 10.1).
Western Blot Analysis
Protein abundance data generated with the DIGE method in several tissues have been previously validated in our laboratory using Western blot analysis (17, 20, 42). As this study represents the first application of the DIGE technique in our laboratory to skeletal muscle, we also conducted a confirmatory Western blot analysis. Relative protein content of the Ca2+-ATPase ion pump (ATP2A1) in the hibernation states SA, Ent, LT, LAr, and IBA was analyzed from the same homogenized samples used for DIGE (n = 3 per state). Homogenates containing 20 μg protein were denatured with heat and Laemmli buffer and separated using SDS-PAGE (10% polyacrylamide gels). Protein was transferred to PVDF membranes (Immobilon-FL; Millipore, Bedford, MA), and then membranes were cut at the 100-kDa mark. The lower membrane portion was stained (MemCode cat. no. 24585; Pierce) to assess the consistency of protein loading and transfer. The upper portion was blocked for 1 h (Odyssey blocking buffer; Li-Cor, Lincoln, NE), incubated with anti-ATP2A1 overnight at 4°C (1:1,000, mouse monoclonal CaF2–5D2; Developmental Studies Hybridoma Bank, Iowa City, IA), then for 1.5 h with a secondary antibody for anti-mouse IgG (1:20,000 IRDye 800CW-conjugated; Li-Cor). Blots were imaged with Odyssey near-infrared fluorescence detection (Li-Cor). Samples were normalized to the lowest intensity lane on each membrane and quantified with ImageJ.
RESULTS
Protein Identification
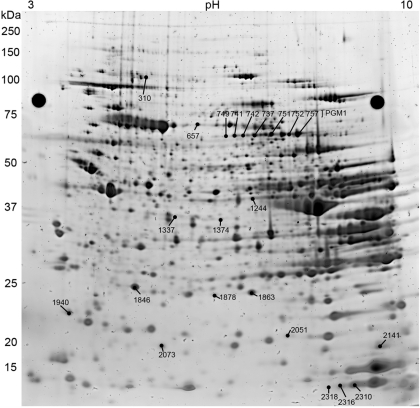
When separated and analyzed with the DIGE method, up to 2,426 skeletal muscle protein spots were recovered on an analytical gel (Fig. 2), resulting in 216 spot densities, which differed significantly among the eight sampled hibernation states of 13-lined ground squirrels (ANOVA q < 0.05). LC-MS/MS provided a single protein identity for 138 of these 216 spots (Table S1). Of the remainder, 53 gave multiple IDs and 25 could not be identified (Table S2).
Fig. 2.
Proteins from a hindlimb muscle sample fractionated by two-dimensional (2D) gel electrophoresis and stained with SYPRO ruby. Reference markers for spot picking are the black circles on the upper portion of the gel on either side. Left: molecular mass ruler (kDa). Top: isoelectric point (pH). All known protein strong contributors to the Random Forests clustering (listed in Table 3) are identified by spot number. Also note the elements of the phosphoglucomutase (PGM1) spot train, which differed significantly among sampling states.
Seasonal Proteomic Differences
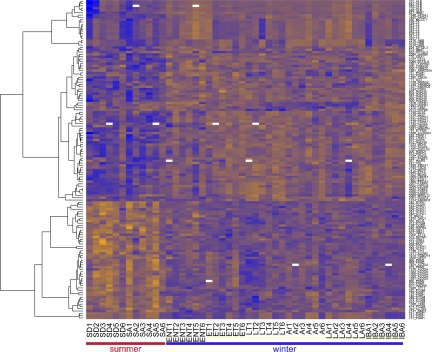
A notable feature in hindlimb muscle of this hibernator was a clear distinction between a summer and a winter proteome (Figs. 3, 4, and 5, A–C), as evidenced by the reciprocal abundance patterns of protein spots between the two seasons visible by heat map (Fig. 3). Indeed, of the 138 identified protein spots, 127 displayed an overall summer versus winter difference. When the hibernation cycle was examined as a state-by-state progression (Fig. 4), the greatest number of significant Tukey post hoc tests also occurred between the euthermic winter (IBA) and summer (SA or SD) states, rather than among intraseason comparisons. The majority of protein spot abundance increases occurred for winter (Fig. 3, orange) such that t-tests comparing pooled summer versus winter data identified 78 spots containing 45 unique winter-elevated proteins, but only 49 spots containing 23 unique proteins higher in the summer groups.
Fig. 3.
Proteins that differed significantly by ANOVA after multiple test correction and were confidently identified with liquid chromatography/mass spectrometry/mas spectrometry were clustered hierarchically and visualized by heat map. Orange represents higher relative protein abundance, while blue denotes lower. Shown on the x-axis: each sampled animal (see materials and methods for sampled state definitions); y-axis: identified proteins. This heat map reveals a marked switch from a summer to winter skeletal muscle proteome in 13-lined ground squirrels and that the majority of protein abundance increases occur in the winter season.
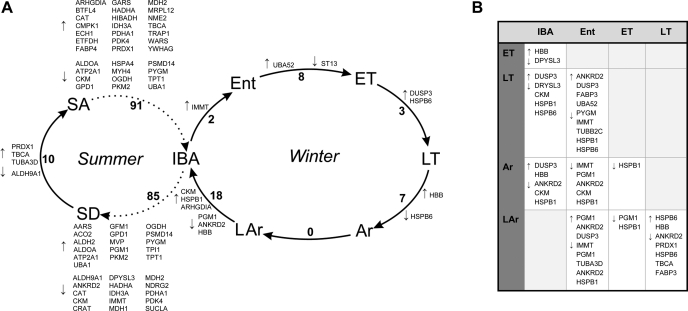
Fig. 4.
Significant (P < 0.05) state-by-state Tukey post hoc tests demonstrate that the majority of pairwise differences occurred between summer (SA or SD) and winter (IBA) samples from the skeletal muscle of 13-lined ground squirrels. A: consecutive states in which the number of significant pairwise comparisons are indicated along each connecting arrow, as are the unique protein identities, denoted by gene symbol, which increased (↑) or decreased (↓). A broken arrow connects SA and SD to IBA to indicate their similarity as euthermic states but that animals transition indirectly between them. The number of significant Tukey comparisons is greater than the number of listed proteins because pairwise comparisons could reflect multiple protein spots representing isoforms of the same protein as well as ambiguous or missing protein identifications. As adjustments to winter cellular protein content were often too gradual to elicit significant pairwise significance between consecutive states, proteins with significant abundance change between nonconsecutive states are displayed in B (state comparisons are top row vs. left column in order through the cycle as depicted in A). Sampling states are described fully in materials and methods.
Fig. 5.
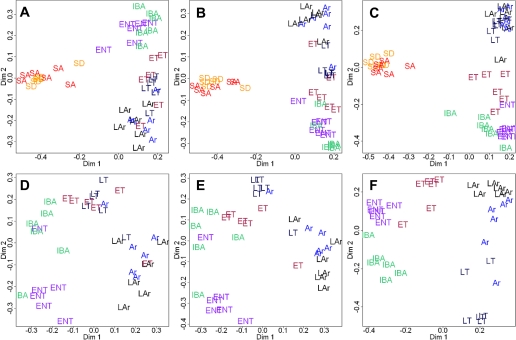
Unsupervised classification by Random Forests (50,000 trees) reveals a distinct summer (SA, SD) vs. winter (Ent, ET, LT, Ar, LAr, IBA) switch (A–C) as well as intrawinter cycling (D–F) in the skeletal muscle proteome of hibernating 13-lined ground squirrels (n = 6 per state, see materials and methods). Patterns of clustering were maintained or improved by limiting the data inputs (left to right), ultimately revealing the minimum number of proteins required to define the states. A (all states) and D (winter only) show Random Forests outputs from all completely matched spots (i.e., present on all analytical gels), while B (all) and E (winter) reflect all 133 of those that were identified. C and D: minimum number of known proteins required for equivalent or improved clustering compared with the expanded data inputs. This was accomplished with 10 protein spots for all states, and 12 proteins for winter states only (listed in Table 3).
Functional annotation analysis for those proteins that met seasonal t-test significance revealed pathways enriched during either summer or winter. Summer was dominated by carbohydrate metabolism, having upregulation of both glycolysis and gluconeogenesis pathway elements (Table 1). Conversely, the winter proteome of skeletal muscle supported lipid over carbohydrate catabolism and revealed a striking overrepresentation for energy-producing pathways (Table 2). Specifically, several structural and enzymatic components of mitochondria were elevated in winter, such as those that support fatty acid β-oxidation (25.7 times) and the tricarboxylic acid cycle (52.0 times), suggesting that oxidative capacity is enhanced in winter. While the summer proteome was enriched for cellular building blocks, such as cytoskeletal elements (e.g., actin binding protein 3, translationally controlled tumor protein, cofilin-2) and components of the sarcomere (e.g., Ca2+-ATPase, Fig. 6A and myosin heavy chain 4), management of cell death and oxidative stress were enriched during winter (Tables 1 and 2).
Fig. 6.
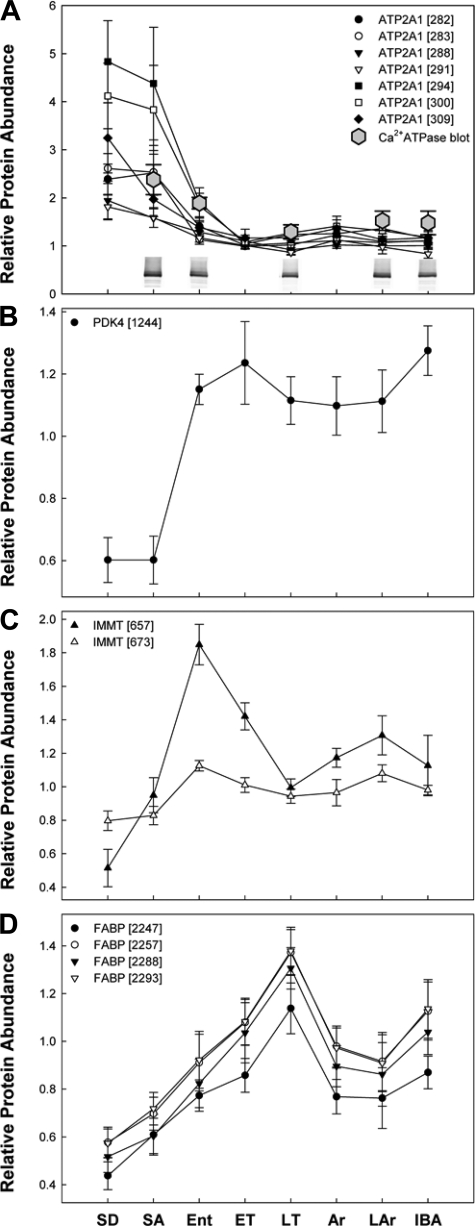
Several patterns of protein abundance among sampling states (x-axis: defined in materials and methods) were observed in the skeletal muscle proteome of 13-lined ground squirrels (n = 6 per state). Ca2+ATPase, (ATP2A1; A) and pyruvate dehydrogenase kinase 4, (PDK4; B) displayed summer to winter reprogramming, a pattern that was the major feature of the dataset. A more limited suite of proteins had altered abundance within the winter hibernation season, being significantly elevated in warm (C; IMMT, mitofilin) or cold-Tb states (D; FABP3, fatty acid binding protein). Relevant spot numbers appear with protein IDs. Western blot analysis quantification of Ca2+ATPase can be found in A (n = 3 per state). Inset: representative lanes from anti-Ca2+ATPase Western blot: lanes from a single blot have been reordered to align with the progression of physiological states and images have not otherwise been retouched. All data are presented means ± SE.
Protein spots that contribute to the separation of all sampled states via Random Forests clustering (Fig. 5, A–C, Table 3) may do so because they define some aspect of the annual cycle in 13-lined ground squirrel skeletal muscle. Those spots that contribute strongly to the clustering and separation of all states, but not the winter states alone, may be informative for the summer-winter transition in this tissue. Pyruvate dehydrogenase kinase 4 (1244; Fig. 6B) was one such protein, found to be 2.1 times elevated in winter (IBA vs. SA, Tukey P < 0.001; Tables S3 and S4), but displaying no significant intrawinter cycling.
Table 3.
Minimum biomarker proteins required to define the skeletal muscle proteome during hibernation using Random Forests clustering
| Gene Symbol | Protein name | Spot No. |
|---|---|---|
| All states | ||
| DUSP3 | Dual specificity phosphatase 3 | 2051(1) |
| PDK4 | Pyruvate dehydrogenase kinase isozyme 4 | 1244(2) |
| HSPB1 | Heat shock protein β1 | 1863(3) |
| HSPB6 | Heat shock protein β6 | 2073(4) |
| APOA1 | Apolipoprotein A1 | 1846(5) |
| TPT1 | Translationally controlled tumor protein 1 | 1940(6) |
| HBB | β-hemoglobin | 2316(7), 2310(9), 2318(10) |
| IMMT | Mitofilin | 657(8) |
| Winter states only | ||
| HSPB1 | Heat shock protein β1 | 1863(1), 1878(7) |
| DUSP3 | Dual specificity phosphatase 3 | 2051(2) |
| HSPB6 | Heat shock protein β6 | 2073(3) |
| ANKRD2 | Ankyrin repeat domain 2 | 1374(4), 1337(12) |
| NME2 | Nucleoside diphosphate kinase B | 2141(5) |
| HBB | β-hemoglobin | 2316(6), 2310(8), 2318(10) |
| UBA1 | Ubiquitin-activating enzyme E1 | 310(9) |
| IMMT | Mitofilin | 657(11) |
Known proteins required for equivalent or improved clustering compared with all matched spots are presented. Rankings of spot importance to the Random Forests clustering are noted in parentheses.
Intrawinter Protein Dynamics
Despite a clear summer-to-winter shift in the majority of identified skeletal muscle proteins, a modest amount of intrawinter cycling in protein spot abundance also occurred (Fig. 4) as evidenced by the discrete winter state clusters defined by Random Forests (Fig. 5, D–F). This clustering pattern became more obvious when only the top known Random Forests contributors were examined (Fig. 5F, Table 3), suggesting that these protein spots reflect some aspects of reversible metabolic depression and can serve as useful biomarkers for winter stages. For example, the regulatory protein dual-specificity phosphatase 3 (DUSP3, 2051) displayed similar abundance levels in summer and warm winter (IBA and Ent) states. However, DUSP3 abundance increased during torpor, reaching a maximum from LT to LAr (2.1 times increase from Ent to LT, P = 0.002; Tables S3 and S4). Two small heat shock proteins (HSPβ1 and -β6) were also strong contributors to the Random Forests clustering (Table 3); and also showed significant intrawinter abundance changes. HSPβ1 (1863, 1878) reached its minimum in LAr, whereas HSPβ6 (2073) was lowest in LT (1.8–2.9 times decline from Ent; Tables S3 and S4).
Examination of intrawinter protein spot cycles provides clues to the nature of protein regulation during heterothermy. Significant increases in protein spot abundance during high Tb states (IBA and Ent), relative to abundance in low-Tb winter states, likely reflect protein resynthesis during these intervals following loss to catabolism during torpor. For I. tridecemlineatus skeletal muscle, examples of this protein dynamic (significantly elevated by Tukey post hoc in either Ent or IBA compared with low-Tb states and having no abundance increase before the resumption of euthermia) included: a small heat shock protein (HSPβ1 i.e., HSP27, 1863, 1878); dihydropyrimidinase-related protein 3 (832); β-tubulin (900); 70 kDa heat shock binding protein (1002); and mitofilin (657; Fig. 6C).
On the other hand, a significant increase in protein spot abundance while Tb is low is unlikely to be the result of protein synthesis (67, 68). Rather, such increases may result from posttranslational modification or the release of that protein from subcellular sequestration that resulted in its segregation into the insoluble fraction of the homogenate. Protein spots that undergo significant abundance elevations while Tb is low (as revealed by Tukey pairwise significance between torpid or arousing states-ET, LT, Ar, or LAr, and relatively high-Tb states-Ent or IBA) were identified as: small HSPβ6 (2073, 2084); dual-specificity protein phosphatase 3 (2051); fatty acid binding protein (2257, 2293; Fig. 6D); peroxiredoxin 1 (1888); tubulin folding cofactor A (2194); and ubiquitin (2386).
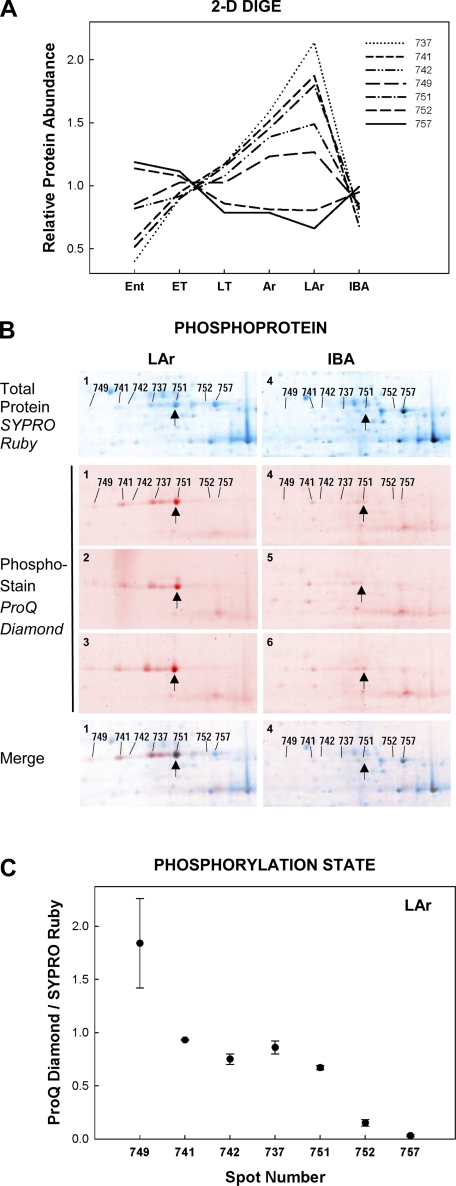
Posttranslational modifications such as phosphorylation or acetylation, for example, shift the isoelectric point for a given protein isoform, and should therefore be detectable on 2D gels by first-dimension separation as additional protein spots. Specifically, posttranslational modification resulting in isoforms of differing activity should present as a spot train consisting of a single protein (Fig. 2) whose distinct isoforms demonstrate reciprocal abundance. Several examples of reciprocal abundance patterns within trains of spots, possibly reflecting posttranslational protein modifications, were apparent in the data. Reciprocal patterns were observed for phosphoglucomutase (PGM1; 7 spots; Fig. 7A); ankyrin repeat domain protein 2 (3 spots), and creatine kinase (2 spots). For PGM1, it is the presumed dephosphorylated forms (752, 757) that bear a reverse pattern of abundance across winter hibernation states relative to the more acidic phosphorylated forms (737, 741, 742, 749, 751; Fig. 7A) based on their position within the spot train. Further evidence for posttranslational modification by phosphorylation to PGM1 was obtained via direct phosphostaining of 2D gels (Fig. 7B), which revealed a generally increasing phosphorylation state toward the more acidic portion of the PGM1 spot train (Fig. 7C). The phosphorylated forms were more abundant in LAr compared with IBA (Fig. 7B).
Fig. 7.
Patterns of relative protein abundance across winter hibernation states, defined in materials and methods, elucidates a mechanism of protein regulation during heterothermy. A: reciprocal Cy2-normalized abundance patterns of 7 protein spots all identified as PGM1 imply that this protein undergoes hibernation state-specific posttranslational modification. Phosphoprotein staining (ProQ Diamond) of 2D gels from n = 3 squirrels in each of 2 winter states [LAr vs. IBA (B)] supports a specific posttranslational modification, phosphorylation, of the more acidic spots (749, 741, 742, 737, 751) in the PGM1 train. B: false-color overlay to compare the SYPRO Ruby total protein stain (blue) with ProQ Diamond phosphoprotein stain (red) and merged images for LAr and IBA. A representative gel from each state poststained for total protein with SYPRO Ruby is labeled for each PGM1 spot. The PGM1 spot train region of each ProQ Diamond-stained gel (LAr 1–3, IBA 4–6) is also shown, along with 1 ProQ Diamond-SYPRO Ruby merged image from each state. Spot 751 is indicated (↑) in each image for comparison. The phospho-forms comprised a larger proportion of the overall PGM1 signature in LAr, while the dephosphorylated forms (752 and 757) were more prevalent in IBA. Quantification of phosphorylation state (ProQ Diamond/SYPRO Ruby staining intensity) is presented for each LAr gel (n = 3 gels stained in sequence, means ± SE; C). Staining intensity for both variables was normalized to the lowest-staining spot on each gel. The general increase in phosphorylation state moving left along the spot train (i.e., toward increasingly acidic protein spots) is expected if the posttranslational modification is indeed phosphorylation.
DISCUSSION
The summer physiology of small mammalian hibernators, such as ground squirrels, profoundly differs from that of winter. Given that our multistate proteomic analysis of I. tridecemlineatus skeletal muscle revealed that the majority (>80%) of detected protein spots (2,426) do not change significantly among physiologically distinct states across the hibernator's annual cycle, proteins that do fluctuate in correlation with hibernation state are likely informative for these physiology shifts as regulatory components or as participants in altered function. In the transition from summer homeothermy to winter heterothermy, skeletal muscle in particular faces the challenge of maintaining structural integrity while modifying contractile activity. The specifics of functional control during torpor-arousal cycles that define the periods of low versus high activity are likely to be uncovered by comparisons between the functionally distinct states directly (discussed below), yet in general, skeletal muscle contractile function must also be preserved across the entire winter season.
Seasonal Differences
The immediate locomotor requirements associated with foraging and breeding at spring emergence suggest that seasonal maintenance of skeletal muscle functionality is critical to individual fitness. Muscle size (total cross-sectional area) is the primary determinant of its contractile strength, implying that one method to preserve contractile ability is to protect skeletal muscle from remodeling and atrophy associated with winter disuse. Ground squirrels rely on the catabolism of stored reserves to survive the food-limited winter season. As skeletal muscle is the body's primary amino acid store and its largest tissue by mass, it represents an attractive energy source during long-term fasting. In line with previous morphological studies in rodents suggesting limited or modulated atrophy during hibernation (48, 63, 71) and a previous proteomic screen in bat (Murina leucogaster) skeletal muscle (35), our protein assessment did not reveal any clear enhancement of muscle degradation machinery such as a winter-elevation of caspase, calpain, lysosomal autophagy, or ubiquitin-proteasome systems. Our single finding suggesting skeletal muscle remodeling in winter by the decline of MYH4 (myosin heavy chain 2b, spot 25) was consistent with fiber-type measurements from another ground squirrel (Callospermophilus lateralis) that indicated a seasonal shift (48, 55, 56). A proportional loss of this fast myosin isoform is counter to expectations for mammalian disuse remodeling (e.g., 25, 66), further supporting the atypical disuse response observed in hibernating mammals.
Seasonal reprogramming of fuel preference.
Preserving skeletal muscle function through winter hibernation accompanies the retention of muscle mass. One aspect of this is to limit amino acid catabolism for which muscle contains the body's major store during the winter fast. Accordingly, a winter fuel shift in hibernators toward a reliance on stored fat reserves and preservation of amino acids (19, 20, 73, 74) has been well documented at whole body (e.g., respiratory quotients) (12, 33, 47, 60) through molecular levels (e.g., lipolytic flux; reviewed in Refs. 4 and 14). It is perhaps not surprising therefore that the largest theme identified from this dataset was a profound shift between a summer and a winter proteome and that a primary component of this seasonal adjustment supports a transition toward reliance on lipid metabolic substrates in winter. Indeed, when overall summer and winter patterns are compared in the skeletal muscles of 13-lined ground squirrels, there is a relative seasonal decrease in lipid fuel reliance during summer. Rather, carbohydrate metabolism pathways are significantly summer enriched (Table 1), including the glycolytic enzymes ALDOA (1376, 1384, 1391, 1396, 1401), GPD1 (1575, 1609), TPI1 (1786), and PKM2 (865, 868, 869, 870, 875). Pyruvate kinase (PKM2), in particular, is a key regulatory enzyme of glycolysis, catalyzing the final, irreversible step in pyruvate formation. We also noted a summer-elevation of glycogen phosphorylase (glycogen breakdown, PYGM 379, 388, 412, 414, 423, 520), which suggests that muscle glycogen stores support the glucose substrate pool.
By contrast, several key elements of the lipid β-oxidation pathway (including HADHA 587, 600, 604, 607, 608, 609, 615 and ACADL 1144, 1152) are elevated in winter (Table 2). In addition to this seemingly higher capacity for lipid metabolism in muscle, our analysis also identified a winter abundance increase in pyruvate dehydrogenase kinase (PDK4 1244; Fig. 6B), which is a regulatory enzyme in a global tissue switch between carbohydrate and fatty acid metabolism. PDK4 gene expression in mammals is enhanced by fasting (51, 53, 61) and high-fat diets (52), is inhibited by insulin (40), and has become a well-described element of the winter fuel switch in hibernator skeletal muscles (6, 13) and other tissues (reviewed in Refs. 4 and 14). As unbiased screens from natural or experimentally induced long-term fasting studies become available, it will be possible to ascertain whether this hibernation-induced fuel shift is a common element of a universal fasting response.
This kinase phosphorylates the pyruvate dehydrogenase (PDH) E1 subunit, thereby inactivating the PDH complex, inhibiting pyruvate conversion to acetyl-coA and flux into the TCA cycle, and ultimately controlling the final breakdown of carbohydrate stores. Given that the observed PDK4 winter elevation should increase the overall phosphorylation state of the PDH pool during hibernation, it is interesting that we also observed a similar winter increase in this subunit of the PDH complex (PDHA1 1232, 1237, 1238; Table S4). This seasonal adjustment to PDH could simply suggest a mechanism to combat the Q10 effect and to conserve reaction rates of the entire enzyme pool in the cold. However, it is also plausible that this increase augments the capacity of the PDH enzyme pool under opportune conditions. During hibernation, such conditions are likely restricted to transient rewarming/arousal episodes. If the PDK4 isozyme has similar allosteric regulation to the well-characterized PDK2 (54), it is likely that as a torpor bout concludes, substrate accumulation (pyruvate and increased ADP/ATP ratio) at the inactive PDH gateway could in turn inactivate PDK4 for arousal and subsequent rapid processing of pyruvate. Elevated PDHA1 and the corresponding winter increase in a number of TCA enzymes (Tables 1 and S4) support a high capacity to exploit carbohydrate substrates (specifically, products of glycolysis) during hibernation, transiently as needed during arousal. Taken together, these enzyme enrichments imply that in skeletal muscle, energy homeostasis, and metabolic substrate regulation cycle dynamically during winter.
Seasonal reduction in energy expenditure.
Another aspect of preserving muscle mass and ultimately contractile function through the winter hibernation season is to reduce energy expenditures. Decreased Tb during torpor bouts is a significant component of this reduction, which correlates to a global depression of reaction rates by Q10 effects (24). Beyond the preservation of local substrates by lowering ATP-consuming processes, regulating skeletal muscle metabolism will impact the whole animal energy budget. Under resting conditions, skeletal muscle is the primary determinant of whole body metabolic rates (76). Furthermore, active transport processes are a significant element of cellular energy expenditure under resting euthermic conditions. Our finding of a marked winter reduction (Fig. 6A) of the Ca2+ ion transporter ATP2A1 (282, 283, 288, 291, 294, 300, 309) in skeletal muscle therefore suggests a mechanism for reducing ATP requirements during hibernation, particularly in the euthermic periods. ATP2A1 is a Ca2+-ATPase ion pump that transports cytosolic Ca2+ into the sarcoplasmic reticulum (SR) where it is sequestered in skeletal muscle until needed for contraction. Ca2+-ATPase manages Ca2+ homeostasis under normal conditions by combating passive ion leak and restoring Ca2+ gradients between each contraction. This pump alone is estimated to account for ∼5% of basal euthermic metabolic rate (59). A reduced abundance of this pump during winter would therefore reduce the overall ATP consumption rate of Ca2+ uptake into SR, and the investment into maintenance and turnover of this protein. Considered alone, our data suggest the possibility that muscle contraction is impaired during arousal events. However, a limited number of previous examinations of skeletal muscle Ca2+ handling during hibernation reveals possible compensatory mechanisms. Specifically, there is increased efficiency of Ca2+ transport into SR during winter in the skeletal muscles of hibernating hamsters (1, 2), which could counter the effects of reduced pump abundance to some degree. Alterations in the fatty acyl composition of SR membranes of Richardson's ground squirrels (Urocitellus richardsonii), which could elevate the activity of Ca2+-ATPase during arousal and IBA states, have also been documented (49), supporting the findings of increased Ca2+ transport efficiency.
Ion pumps represent only one-half of ion homeostasis. Indeed, the maintenance of ion channel/pump flux ratios near unity over a wide temperature range is a well-described feature of cold acclimation (30). Since the two components of ion homeostasis do not share a similar Q10 coefficient [pumps ∼2–3, channels ∼1; (30)] cold-acclimation is often supported by reduced ion channel density coupled with increased pump density. While this is precisely the case for hibernator cardiac muscle (70), which functions in cold, our data suggest that the situation in skeletal muscle differs. This difference in response of two muscle types may reflect disparate regulation of a constitutively active tissue, such as heart, compared with a transiently contractile tissue, such as skeletal muscle. Heart muscle appears to rely on upregulated Ca2+ handling in winter to maintain contractility, specifically relaxation rates, at very low Tb (reviewed in Ref. 4), a feature that is less critical in skeletal muscle, which is inactive during torpor. Although we only assessed Ca2+ transport and not Ca2+ leak form SR, our data do suggest an energy conservation mechanism during hibernation, as well as underscore the question of whether contraction is at all compromised in winter skeletal muscle.
Winter Proteome
For skeletal muscle in particular, winter physiology is associated with dramatic fluctuations in contractile activity across torpor and arousal cycles. The rewarming transition from torpor to euthermia is particularly intriguing, as skeletal muscle resumes contraction to contribute to rewarming in conjunction with perfusion restoration (37, 38), intracellular pH changes (43), and apparent adjustments in fuel selection (23; but see also 33, 47, 60). Skeletal muscle must balance the retention of contractile functionality for arousals with energy conservation tactics required for the hibernation season. Direct proteomic comparisons of different winter functional states should therefore reveal biochemical regulators and the protein requirements needed to support this balance across repeated swings in activity level throughout winter. Because skeletal muscle undergoes such large amplitude shifts in activity level during the winter season, it is significant that intrawinter protein adjustments were only a minor component of the proteomic changes detected in 13-lined ground squirrels correlated to hibernation state; rather, the bulk of those changes occurred between summer and winter (Table S1). In fact, we could not document any significant change in protein spot abundance between early and late-arousing states (Ar vs. LAr; Fig. 4A), states that should reflect start and end points of the transitions in activity, perfusion, and fuel selection that occur in skeletal muscle during rewarming. Perhaps it is the requirement to preserve contractile function that results in the relative uniformity of the winter proteome. However, a further complication to understanding the winter biochemistry of hibernators is that protein synthesis mechanisms (transcription and translation) are temporally partitioned by Tb (67, 68), making it likely that adjustments to mRNA expression drive the bulk of the summer-winter transitions (hibernation switch 1; 62), but have only limited contributions to the torpor-arousal cycles of winter (hibernation switch 2; 57). If protein synthesis is limited in winter to euthermic states, then low-energy mechanisms necessarily provide critical support for hibernation switch 2 (torpor-arousal). For instance, posttranslational modifications, such as phosphorylation or acetylation, can affect protein activity (65, 75) and these could manifest as fluctuations within trains of 2D gel spots. Alternatively, significant protein abundance differences during torpor, due to intracellular sequestration or release from the insoluble component, would impact the percent of a given protein pool recovered in the soluble homogenate. Investigation into these low-energy mechanisms of protein regulation is therefore a necessary component of deciphering intrawinter proteomic changes that we detected in skeletal muscle.
State-dependent protein abundance changes.
One such intrawinter protein spot abundance change occurred in the mitochondrial membrane protein mitofilin (IMMT 657, 673; Fig. 6C). Mitofilin content was highest in animals reentering torpor (Ent) but declined sharply through the two measured torpid states, suggesting that this finding is an example of protein degradation during torpor. Resynthesis of lost or damaged mitofilin could represent general mitochondrial structural repair during euthermic periods, which is completed before reentry into torpor. Mitofilin is required for proper inner mitochondrial membrane (specifically, cristae) morphology, and thus for normal mitochondrial function (32). Indeed, ultrastructural examination of edible dormouse (Glis glis) skeletal muscle mitochondria revealed a winter increase of the mitochondrial inner membrane (41), suggesting the importance of mitofilin to hibernation. Maintenance and even augmentation of inner mitochondrial membrane components during hibernation underscores the idea that skeletal muscles are primed for rapid energy production to capitalize on favorable but transient conditions. Furthermore, IMMT protein responds within 24 h of environmental hypoxia exposure in a hypoxia-tolerant ectotherm (16), suggesting that transient cellular hypoxia during the arousal process (39) may be the mechanism that regulates IMMT synthesis.
In contrast to the pattern displayed by IMMT (a decline in torpor followed by increase during warm Tb states), the muscle form of fatty acid binding protein (FABP3 2247, 2257, 2288, and 2293) content increased during cold Tb states (Fig. 6D). FABPs play an important role in delivery of long-chain fatty acids and in maintaining intracellular fatty acid concentrations (reviewed in Ref. 64). The gradual increase in FABP3 content during torpor supports a continued reliance of skeletal muscle on lipid fuels during this period. In fact, FABP3 knockout mice are unable to retain lipid as the primary metabolic fuel in the heart (9). However, rather than reflecting protein synthesis during torpor, the increased protein abundance of FABP3 at low Tb is likely due to intracellular sequestration. The majority of skeletal muscle FABP3 exists in the soluble cytoplasmic fraction (26), but some may be bound to cellular lipids (i.e., a variety of specific membranes within the cell, either organellar or sarcolemmal, or possibly intracellular lipid droplets), binding which could alter FABP recovery in the soluble homogenate fraction in the present study. As fatty acids are consumed during torpor, FABPs are released into the cytosol to transport fatty acids from the donor location to areas of lipid metabolism, such as mitochondria or peroxisomes. The observation that FABP protein accumulation does not continue into the arousal period provides evidence that additional metabolic substrates (e.g., carbohydrates) are important in powering rewarming in 13-lined ground squirrel muscles. However, our data clearly demonstrate that a transition to lipid and ketone fuels is essential for achieving a stable winter starvation state in this hibernator.
State-dependent posttranslational protein modification.
Evidence for reduced reliance on lipids during arousals is supported by evidence for increased reliance on carbohydrate substrates in the predominantly fast glycolytic muscles sampled. PGM1 is a key trafficking point in carbohydrate metabolism-specifically for glycogenesis and glycolysis-catalyzing the bidirectional conversion of glucose-1-phosphate to glucose-6-phosphate. This enzyme appeared in skeletal muscle as a train of spots on the 2D gels, and showed reciprocal patterns of abundance within the train (Fig. 7A), suggesting regulation by posttranslational modification. Although the low activation energy of this reaction makes it reversible under normal conditions, PGM1 phosphorylation at threonine-466 (by Pak1; 27) is proposed to drive the reaction toward glucose-6-phosphate and ultimately favor glycolysis. The sharp elevation of the apparent phospho form observed in I. tridecemlineatus in Ar and LAr (Fig. 7B) may support a persisting increased preference of PGM1 for glucose-1-phosphate as substrate, thereby maintaining a high glucose-6-phosphate substrate pool for glycolysis. It is possible that the ultimate effect of this posttranslational modification to PGM1 is to elevate pyruvate concentration to the point of inactivating PDK4 and thereby funnel the glucose released from glycogen stores into the TCA cycle once perfusion and aerobic metabolism are restored. The striking decrease in phosphorylated PGM1 from LAr to IBA (Fig. 7B) would lead to a reversal of carbohydrate metabolism to glycogenesis during the euthermic period, supporting a replenishment of muscle glycogen stores (22, 23). This provides an excellent example of a low-temperature regulatory mechanism for a key energy transition in skeletal muscle during torpor-arousal cycles of hibernation.
Perspectives and Significance
Our eight-state proteomic examination of the 13-lined ground squirrel hindlimb muscle reveals critical adjustments both between seasons and within winter to facilitate the maintenance of tissue size and function during hibernation. It is clear that adjustments to metabolic substrate preferences comprise a large portion of the proteomic differences we detected in correlation with hibernation or summer state in this species. Such adjustments likely maximize the exploitation of whole body, rather than local, energy stores to fuel winter activities. Of particular note is the lack of detected seasonal differences that would suggest catabolism of local protein stores. Since hibernators such as ground squirrels are well-known to preserve protein and amino acid pools in winter (19, 20, 73, 74), this is likely not due to an inability to detect an upregulation of protein catabolic pathways, but rather due to their absence. Skeletal muscle tissue is the body's primary amino acid store, and protecting that store from catabolism is consistent with a general observation of limited atrophy (35, 48, 63, 71) among small mammalian hibernators, in turn preserving contractile function for arousals and ultimately for spring emergence. Rather than protein stores, hindlimb muscles in 13-lined ground squirrels predominantly rely on lipid fuels during hibernation, consistent with the bulk of previous work in hibernators highlighting a global reliance on fat and ketone substrates (e.g., 5, 19, 34, 72) and active limitation of carbohydrate use during most of winter (6, 13). Furthermore, our data suggest a mechanism, i.e., phosphorylation of PGM1 specifically during rewarming, to supplement lipid substrates with local carbohydrate stores during periods of high contractile activity. While this mechanism has been previously described in tumorigenesis (27), its occurrence in the torpor-arousal cycles of hibernation was previously undocumented. The same mechanism for transient and targeted winter use of glucose may be employed by more oxidative skeletal muscles and other tissues as well. The broad sampling in the present study, which includes, among others, LT, arousing, and IBA samples in a single experiment, combined with the 2D gel protein isoform separation, enables identification of protein expression and posttranslational modifications that play key regulatory roles in maintenance of hibernation physiology.
GRANTS
This work was supported by National Institutes of Health Grants 5T15-LM-009451 (to A. Karimpour-Fard), R01-LM-008111 and R01-LM-009254 (to L. E. Hunter) and R01-HL-89049 (to S. L. Martin). We acknowledge the Institutional Proteomics Mass Spectrometry Facility, which is supported in part by NIH Grants to the Colorado Clinical and Translational Science Institute (ULI-RR025780) and the University of Colorado Cancer Center (P30-CA-046934).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We thank Rae Russell and Katharine Grabek for technical assistance on this project and for helpful comments on the manuscript. The Ca2+-ATPase (ATP2A1) monoclonal antibody developed by D. M. Fambrough was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by The University of Iowa, Department of Biology, Iowa City, IA.
REFERENCES
- 1. Agostini B, Martino LD, Hasselbach W. On the problem of season and cold dependence of calcium transport by skeletal muscle sarcoplasmic reticulum. Z Naturforsch C 45: 671–675, 1990 [DOI] [PubMed] [Google Scholar]
- 2. Agostini B, Martino LD, Soltau B, Hasselbach W. The modulation of the calcium transport by skeletal muscle sarcoplasmic reticulum in the hibernating European hamster. Z Naturforsch C 46: 1109–1126, 1991 [DOI] [PubMed] [Google Scholar]
- 3. Agrawal GK, Thelen JJ. Development of a simplified, economical polyacrylamide gel staining protocol for phosphoproteins. Proteomics 5: 4684–4688, 2005 [DOI] [PubMed] [Google Scholar]
- 4. Andrews MT. Advances in molecular biology of hibernation in mammals. Bioessays 29: 431–440, 2007 [DOI] [PubMed] [Google Scholar]
- 5. Andrews MT, Russeth KP, Drewes LR, Henry PG. Adaptive mechanisms regulate preferred utilization of ketones in the heart and brain of a hibernating mammal during arousal from torpor. Am J Physiol Regul Integr Comp Physiol 296: R383–R393, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Andrews MT, Squire TL, Bowen CM, Rollins MB. Low-temperature carbon utilization is regulated by novel gene activity in the heart of a hibernating mammal. Proc Natl Acad Sci USA 95: 8392–8397, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barsnes H, Vizcaino JA, Eidhammer I, Martens L. PRIDE Converter: making proteomics data-sharing easy. Nat Biotech 27: 598–599, 2009 [DOI] [PubMed] [Google Scholar]
- 8. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57: 289–300, 1995 [Google Scholar]
- 9. Binas B, Danneberg H, McWhir J, Mullins L, Clark AJ. Requirement for the heart-type fatty acid binding protein in cardiac fatty acid utilization. FASEB J 13: 805–812, 1999 [DOI] [PubMed] [Google Scholar]
- 10. Breiman L. Bagging predictors. Mach Learn 24: 123–140, 1996 [Google Scholar]
- 11. Breiman L. Random Forests. Mach Learn 45: 5–32, 2001 [Google Scholar]
- 12. Buck CL, Barnes BM. Effects of ambient temperature on metabolic rate, respiratory quotient, and torpor in an arctic hibernator. Am J Physiol Regul Integr Comp Physiol 279: R255–R262, 2000 [DOI] [PubMed] [Google Scholar]
- 13. Buck MJ, Squire TL, Andrews MT. Coordinate expression of the PDK4 gene: a means of regulating fuel selection in a hibernating mammal. Physiol Genomics 8: 5–13, 2002 [DOI] [PubMed] [Google Scholar]
- 14. Carey HV, Andrews MT, Martin SL. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol Rev 83: 1153–1181, 2003 [DOI] [PubMed] [Google Scholar]
- 15. Daan S, Barnes BM, Strijkstra AM. Warming up for sleep? – Ground squirrels sleep during arousals from hibernation. Neurosci Lett 128: 265–268, 1991 [DOI] [PubMed] [Google Scholar]
- 15a. Delp MD, Duan C. Composition and size of type I, IIA, IID/X, and IIB fibers and citrate synthase activity of rat muscle. J Appl Physiol 80: 261–270, 1996 [DOI] [PubMed] [Google Scholar]
- 16. Dowd WW, Renshaw GMC, Cech JJ, Kültz D. Compensatory proteome adjustments imply tissue-specific structural and metabolic reorganization following episodic hypoxia or anoxia in the epaulette shark (Hemiscyllium ocellatum). Physiol Genomics 42: 93–114, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Epperson L, Rose J, Russell R, Nikrad M, Carey H, Martin S. Seasonal protein changes support rapid energy production in hibernator brainstem. J Comp Physiol B 180: 599–617, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Epperson LE, Dahl TA, Martin SL. Quantitative analysis of liver protein expression during hibernation in the golden-mantled ground squirrel. Mol Cell Proteomics 3: 920–933, 2004 [DOI] [PubMed] [Google Scholar]
- 19. Epperson LE, Karimpour-Fard A, Hunter LE, Martin SL. Metabolic cycles in a circannual hibernator. Physiol Genomics 43: (13) 799–807, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Epperson LE, Rose JC, Carey HV, Martin SL. Seasonal proteomic changes reveal molecular adaptations to preserve and replenish liver proteins during ground squirrel hibernation. Am J Physiol Regul Integr Comp Physiol 298: R329–R340, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fedorov V, Goropashnaya A, Toien O, Stewart N, Chang C, Wang H, Yan J, Showe L, Showe M, Barnes B. Modulation of gene expression in heart and liver of hibernating black bears (Ursus americanus). BMC Genomics 12: 171, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Galster WA, Morrison P. Cyclic changes in carbohydrate concentrations during hibernation in the arctic ground squirrel. Am J Physiol 218: 1228–1232, 1970 [DOI] [PubMed] [Google Scholar]
- 23. Galster WA, Morrison PR. Gluconeogenesis in arctic ground squirrels between periods of hibernation. Am J Physiol 228: 325–330, 1975 [DOI] [PubMed] [Google Scholar]
- 24. Geiser F. Metabolic rate and body temperature reduction during hibernation and daily torpor. Annu Rev Physiol 66: 239–274, 2004 [DOI] [PubMed] [Google Scholar]
- 25. Giger JM, Bodell PW, Zeng M, Baldwin KM, Haddad F. Rapid muscle atrophy response to unloading: pretranslational processes involving MHC and actin. J Appl Physiol 107: 1204–1212, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Glatz JFC, Schaap FG, Binas B, Bonen A, Van Der Vusse GJ, Luiken JJFP. Cytoplasmic fatty acid-binding protein facilitates fatty acid utilization by skeletal muscle. Acta Physiol Scand 178: 367–371, 2003 [DOI] [PubMed] [Google Scholar]
- 27. Gururaj A, Barnes CJ, Vadlamudi RK, Kumar R. Regulation of phosphoglucomutase 1 phosphorylation and activity by a signaling kinase. Oncogene 23: 8118–8127, 2004 [DOI] [PubMed] [Google Scholar]
- 28. Hampton M, Andrews MT. A simple molecular mathematical model of mammalian hibernation. J Theor Biol 247: 297–302, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hayward JS, Lyman CP, Taylor CR. The possible role of brown fat as a source of heat during arousal from hibernation. Ann NY Acad Sci 131: 441–446, 1965 [DOI] [PubMed] [Google Scholar]
- 30. Hochachka PW. Channels and pumps–determinants of metabolic cold adaptation strategies. Comp Biochem Physiol B 90: 515–519, 1988 [Google Scholar]
- 31. Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protocols 4: 44–57, 2009 [DOI] [PubMed] [Google Scholar]
- 32. John GB, Shang Y, Li L, Renken C, Mannella CA, Selker JML, Rangell L, Bennett MJ, Zha J. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol Biol Cell 16: 1543–1554, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Karpovich S, Tøien Ø, Buck C, Barnes B. Energetics of arousal episodes in hibernating arctic ground squirrels. J Comp Physiol B 179: 691–700, 2009 [DOI] [PubMed] [Google Scholar]
- 34. Krilowicz BL. Ketone body metabolism in a ground squirrel during hibernation and fasting. Am J Physiol Regul Integr Comp Physiol 249: R462–R470, 1985 [DOI] [PubMed] [Google Scholar]
- 35. Lee K, Park JY, Yoo W, Gwag T, Lee JW, Byun MW, Choi I. Overcoming muscle atrophy in a hibernating mammal despite prolonged disuse in dormancy: proteomic and molecular assessment. J Cell Biochem 104: 642–656, 2008 [DOI] [PubMed] [Google Scholar]
- 36. Lyman CP, Chatfield PO. Mechanisms of arousal in the hibernating hamster. J Exp Zool 114: 491–515, 1950 [DOI] [PubMed] [Google Scholar]
- 37. Lyman CP, O'Brien RC. Autonomic control of circulation during the hibernating cycle in ground squirrels. J Physiol 168: 477–499, 1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lyman CP, Willis JS, Malan A, Wang LCH. Hibernation and Torpor in Mammals and Birds. New York: Academic, 1982, p. 317 [Google Scholar]
- 39. Ma YL, Zhu X, Rivera PM, Toien O, Barnes BM, LaManna JC, Smith MA, Drew KL. Absence of cellular stress in brain after hypoxia induced by arousal from hibernation in Arctic ground squirrels. Am J Physiol Regul Integr Comp Physiol 289: R1297–R1306, 2005 [DOI] [PubMed] [Google Scholar]
- 40. Majer M, Popov KM, Harris RA, Bogardus C, Prochazka M. Insulin downregulates pyruvate dehydrogenase kinase (PDK) mRNA: potential mechanism contributing to increased lipid oxidation in insulin-resistant subjects. Mol Genet Metab 65: 181–186, 1998 [DOI] [PubMed] [Google Scholar]
- 41. Malatesta M, Perdoni F, Battistelli S, Muller S, Zancanaro C. The cell nuclei of skeletal muscle cells are transcriptionally active in hibernating edible dormice. BMC Cell Biol 10: 19, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Martin SL, Epperson LE, Rose JC, Kurtz CC, Ane C, Carey HV. Proteomic analysis of the winter-protected phenotype of hibernating ground squirrel intestine. Am J Physiol Regul Integr Comp Physiol 295: R316–R328, 2008 [DOI] [PubMed] [Google Scholar]
- 43. McArthur MD, Hanstock CC, Malan A, Wang LCH, Allen PS. Skeletal muscle pH dynamics during arousal from hibernation measured by 31P NMR spectroscopy. J Comp Physiol B 160: 339–347, 1990 [Google Scholar]
- 44. Musacchia XJ, Steffen JM, Fell RD, Dombrowski MJ. Skeletal muscle response to spaceflight, whole body suspension, and recovery in rats. J Appl Physiol 69: 2248–2253, 1990 [DOI] [PubMed] [Google Scholar]
- 45. Nelson CJ, Otis JP, Carey HV. Global analysis of circulating metabolites in hibernating ground squirrels. Comp Biochem Physiol D 5: 265–273, 2010 [DOI] [PubMed] [Google Scholar]
- 46. Nelson CJ, Otis JP, Martin SL, Carey HV. Analysis of the hibernation cycle using LC-MS-based metabolomics in ground squirrel liver. Physiol Genomics 37: 43–51, 2009 [DOI] [PubMed] [Google Scholar]
- 47. Nestler JR. Relationships between respiratory quotient and metabolic rate during entry to and arousal from daily torpor in deer mice (Peromyscus maniculatus). Physiol Zool 63: 504–515, 1990 [Google Scholar]
- 48. Nowell M, Choi H, Rourke B. Muscle plasticity in hibernating ground squirrels (Spermophilus lateralis) is induced by seasonal, but not low-temperature, mechanisms. J Comp Physiol B 181: 147–164, 2011 [DOI] [PubMed] [Google Scholar]
- 49. Pehowich DJ. Modification of skeletal muscle sarcoplasmic reticulum fatty acyl composition during arousal from hibernation. Comp Biochem Physiol B 109: 571–578, 1994 [Google Scholar]
- 50. Pengelley ET, Fisher KC. Rhythmical arousal from hibernation in the golden-mantled ground squirrel, Citellus lateralis tescorum. Can J Zool 39: 105–120, 1961 [Google Scholar]
- 51. Peters SJ, Harris RA, Heigenhauser GJF, Spriet LL. Muscle fiber type comparison of PDH kinase activity and isoform expression in fed and fasted rats. Am J Physiol Regul Integr Comp Physiol 280: R661–R668, 2001 [DOI] [PubMed] [Google Scholar]
- 52. Peters SJ, Harris RA, Wu P, Pehleman TL, Heigenhauser GJF, Spriet LL. Human skeletal muscle PDH kinase activity and isoform expression during a 3-day high-fat/low-carbohydrate diet. Am J Physiol Endocrinol Metab 281: E1151–E1158, 2001 [DOI] [PubMed] [Google Scholar]
- 53. Pilegaard H, Saltin B, Neufer PD. Effect of short-term fasting and refeeding on transcriptional regulation of metabolic genes in human skeletal muscle. Diabetes 52: 657–662, 2003 [DOI] [PubMed] [Google Scholar]
- 54. Roche T, Hiromasa Y. Pyruvate dehydrogenase kinase regulatory mechanisms and inhibition in treating diabetes, heart ischemia, and cancer. Cell Mol Life Sci 64: 830–849, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rourke B, Yokoyama Y, Milmsom W, Caiozzo V. Myosin isoform expression and MAFbx mRNA levels in hibernating golden-mantled ground squirrels (Spermophilus lateralis). Physiol Biochem Zool 77: 582–593, 2004 [DOI] [PubMed] [Google Scholar]
- 56. Rourke BC, Qin A, Haddad F, Baldwin KM, Caiozzo VJ. Cloning and sequencing of myosin heavy chain isoform cDNAs in golden-mantled ground squirrels: effects of hibernation on mRNA expression. J Appl Physiol 97: 1985–1991, 2004 [DOI] [PubMed] [Google Scholar]
- 57. Serkova NJ, Rose JC, Epperson LE, Carey HV, Martin SL. Quantitative analysis of liver metabolites in three stages of the circannual hibernation cycle in 13-lined ground squirrels by NMR. Physiol Genomics 31: 15–24, 2007 [DOI] [PubMed] [Google Scholar]
- 58. Shao C, Liu Y, Ruan H, Li Y, Wang H, Kohl F, Goropashnaya AV, Fedorov VB, Zeng R, Barnes BM, Yan J. Shotgun proteomics analysis of hibernating arctic ground squirrels. Mol Cell Proteomics 9: 313–326, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Simonides W, Thelen M, van der Linden CG, Muller A, van Hardeveld C. Mechanism of thyroid-hormone regulated expression of the SERCA genes in skeletal muscle: implications for thermogenesis. Biosci Rep 21: 139–154, 2001 [DOI] [PubMed] [Google Scholar]
- 60. Snapp BD, Heller HC. Suppression of metabolism during hibernation in ground squirrels (Citellus lateralis). Physiol Zool 54: 297–307 1981 [Google Scholar]
- 61. Spriet LL, Tunstall RJ, Watt MJ, Mehan KA, Hargreaves M, CameronSmith D. Pyruvate dehydrogenase activation and kinase expression in human skeletal muscle during fasting. J Appl Physiol 96: 2082–2087, 2004 [DOI] [PubMed] [Google Scholar]
- 62. Srere HK, Wang LC, Martin SL. Central role for differential gene expression in mammalian hibernation. Proc Natl Acad Sci USA 89: 7119–7123, 1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Steffen JM, Koebel DA, Musacchia XJ, Milsom WK. Morphometric and metabolic indices of disuse in muscles of hibernating ground squirrels. Comp Biochem Physiol B 99: 815–819, 1991 [DOI] [PubMed] [Google Scholar]
- 64. Storch J, Thumser AE. The fatty acid transport function of fatty acid-binding proteins. Biochim Biophys Acta 1486: 28–44, 2000 [DOI] [PubMed] [Google Scholar]
- 65. Storey KB, Storey JM. Metabolic rate depression in animals: transcriptional and translational controls. Biol Rev 79: 207–233, 2004 [DOI] [PubMed] [Google Scholar]
- 66. Templeton GH, Sweeney HL, Timson BF, Padalino M, Dudenhoeffer GA. Changes in fiber composition of soleus muscle during rat hindlimb suspension. J Appl Physiol 65: 1191–1195, 1988 [DOI] [PubMed] [Google Scholar]
- 67. van Breukelen F, Martin SL. Reversible depression of transcription during hibernation. J Comp Physiol B 172: 355–361, 2002 [DOI] [PubMed] [Google Scholar]
- 68. van Breukelen F, Martin SL. Translational initiation is uncoupled from elongation at 18C during mammalian hibernation. Am J Physiol Regul Integr Comp Physiol 281: R1374–R1379, 2001 [DOI] [PubMed] [Google Scholar]
- 69. Vizcaíno JA, Côté R, Reisinger F, Barsnes H, Foster JM, Rameseder J, Hermjakob H, Martens L. The Proteomics Identifications database: 2010 update. Nucleic Acids Res 38: D736–D742, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang SQ, Lakatta EG, Cheng H, Zhou ZQ. Adaptive mechanisms of intracellular calcium homeostasis in mammalian hibernators. J Exp Biol 205: 2957–2962, 2002 [DOI] [PubMed] [Google Scholar]
- 71. Wickler SJ, Horwitz BA, Kott K S. Muscle function in hibernating hamsters: a natural analog to bed rest? J Therm Biol 12: 163–166, 1987 [Google Scholar]
- 72. Williams CT, Goropashnaya AV, Buck CL, Fedorov VB, Kohl F, Lee TN, Barnes BM. Hibernating above the permafrost: effects of ambient temperature and season on expression of metabolic genes in liver and brown adipose tissue of arctic ground squirrels. J Exp Biol 214: 1300–1306, 2011 [DOI] [PubMed] [Google Scholar]
- 73. Williams DR, Epperson LE, Li W, Hughes MA, Taylor R, Rogers J, Martin SL, Cossins AR, Gracey AY. Seasonally hibernating phenotype assessed through transcript screening. Physiol Genomics 24: 13–22, 2005 [DOI] [PubMed] [Google Scholar]
- 74. Yan J, Barnes BM, Kohl F, Marr TG. Modulation of gene expression in hibernating arctic ground squirrels. Physiol Genomics 32: 170–181, 2008 [DOI] [PubMed] [Google Scholar]
- 75. Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science 327: 1000–1004, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zurlo F, Larson K, Bogardus C, Ravussin E. Skeletal muscle metabolism is a major determinant of resting energy expenditure. J Clin Invest 86: 1423–1427, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.