Abstract
To identify candidate genes involved in the aggressive behavior of worker honeybees, we used the differential display method to search for RNAs exclusively detected in the brains of aggressive workers that had attacked a hornet. We identified a novel, 10,152-nucleotide RNA, termed Kakugo RNA. Kakugo RNA encodes a protein of 2,893 amino acid residues that shares structural features and sequence similarities with various picorna-like virus polyproteins, especially those from sacbrood virus, which infects honeybees. The Kakugo protein contains several domains that correspond to the virion protein, helicase, protease, and RNA-dependent RNA polymerase domains of various picorna-like virus polyproteins. When the worker bee tissue lysate was subjected to sucrose density gradient centrifugation, Kakugo RNA, except for the material at the bottom, was separated into two major peaks. One of the peaks corresponded to the position of Kakugo mRNA, and the other corresponded to the position of the poliovirus virion. These results suggest that the Kakugo RNA exists as an mRNA-like free RNA and virion RNA in the honeybee. Furthermore, injection of the lysate supernatant from the attacker heads into the heads of noninfected bees resulted in a marked increase in Kakugo RNA. These results demonstrate that Kakugo RNA is a plus-strand RNA of a novel picorna-like virus and that the brains of aggressive workers are infected by this novel virus. Kakugo RNA was detected in aggressive workers but not in nurse bees or foragers. In aggressive workers, Kakugo RNA was detected in the brain but not in the thorax or abdomen, indicating a close relation between viral infection in the brain and aggressive worker behaviors.
The European honeybee Apis mellifera L. is a eusocial insect, and the workers perform diverse tasks to maintain colony activity, such as comb-building, nursing, guarding, and foraging, according to age after eclosion (age polyethism) (36). Guard bees gathering at the entrance of the hive are highly aggressive and often scramble to counterattack natural enemies, such as hornets, to protect the colony (4, 5). The worker honeybee stinger is part of a highly modified ovipositor that evolved for defensive functions. The stinger is hooked and the worker loses it after use, resulting in the death of the bee. The advantage of losing the stinger is that the venom sac is then activated to inject additional venom after the sac detaches from the abdomen of the workers. The attacking behavior of guard bees is self-sacrificing and is therefore considered to be a typical altruistic behavior exhibited by the workers (35). Thus, the honeybee is an attractive model for the study of altruistic aggressive behaviors. Quantitative trait locus analysis has been used to identify the loci related to aggressive worker behaviors (11, 12). The genes responsible for the aggressive behaviors, however, have not yet been identified.
Previously, members of our laboratories used the differential display method to identify genes expressed preferentially in the mushroom bodies (MBs), which are important for sensory integration, memory, and learning in the insect brain, with the aim of identifying candidate genes involved in the highly advanced behaviors of the honeybees (17, 30, 32, 33). We expected that this method would also be useful for identifying genes expressed in an aggressive behavior-selective manner in the honeybee brain.
The giant hornet Vespa mandarinia japonica is the most formidable natural enemy of honeybees in Japan (21, 25). For the present study, to identify a candidate gene(s) involved in the aggressive behavior of honeybee workers, we used the differential display method to search for genes expressed selectively in the brains of workers that exhibited attacking behavior against hornets. A novel RNA was identified that was a genome-sense RNA of a putative picorna-like virus.
MATERIALS AND METHODS
Collection of honeybees.
Honeybee A. mellifera L. Italian race colonies were maintained at the Experimental Station for Medical Plant Studies (Chiba, Japan) of the University of Tokyo. Hornets, V. mandarinia japonica, caught near Tamagawa University, were used throughout the experiments. For collection of aggressive workers, a worker hornet that had its stinger removed was hung by a string around its neck and presented as a decoy to the guard bees. Some of the bees (attackers) scrambled and grappled with the hornet obstinately, shaking their wings and bending their abdomens. They were collected with tweezers and immediately anesthetized on ice. To collect nonaggressive workers, a comb was taken from the hive and the decoy hornet was presented to the workers that were gathered on the comb. Some of the bees escaped from the hornet and they were collected as escapers (control bees for attackers). Attackers and escapers were collected four times (October and November 1999 and August and November 2000) from two different colonies. Nurse bees and foragers were collected according to their behaviors. Nurse bees were feeding the brood and foragers had returned to the colony after foraging for pollen (19).
RNA extraction and differential display.
The MBs were dissected from the heads of 100 attackers, escapers, nurse bees, and foragers. The heads, thoraxes, and abdomens of five attackers were collected. The samples were stored at −80°C before use. Each tissue was homogenized with a Polytron homogenizer (Central Scientific Commerce Inc., Tokyo, Japan), and total RNA was extracted with RNAgent (Promega, Madison, Wis.). RNA was treated with RNase-free DNase I and then reverse transcribed, with or without Superscript II (Invitrogen Corp., Carlsbad, Calif.), by use of an anchored oligo(dT) primer. Differential display was performed as described previously (14, 17, 20, 30, 32, 33) by use of a fluorescence differential display kit (Takara, Shiga, Japan) and arbitrary 10-mers with 178 primer combinations and LA Taq polymerase (Takara). Differential display was performed with three different RNA samples, each of which was extracted from 100 MBs.
Subcloning and sequencing.
Bands of interest were excised, and the DNA was reamplified by PCR with the same primer combinations used for the differential display method, except that BamHI sites were added at both the 5′- and 3′-flanking regions. The reamplified DNA was ligated into a pGEM-T or pGEM-T Easy vector (Promega) at the SmaI site and transfected into Escherichia coli JM109 or DH5α (Takara). The nucleotide sequences of both strands for multiple independent clones were determined by DNA sequencing reactions performed by the dideoxynucleotide cycle sequencing method, using a Dye Terminator cycle sequencing kit (Applied Biosystems, Tokyo, Japan) and an ABI Prism 3100 genetic analyzer (Applied Biosystems).
Semiquantitative RT-PCR.
For reverse transcription (RT)-PCR, RNA from the MBs of the attackers and the escapers was treated with DNase and reverse transcribed as described above. PCR was performed with gene-specific primers (+8388 to +8415 and +8745 to +8765 for Kakugo cDNA and +2 to +21 and +376 to +395 for honeybee cytoplasmic actin cDNA [24]) and Ex Taq (Takara) under the following conditions: 94°C for 30 s, 55°C for 30 s, and 72°C for 40 s. Experiments were repeated for three RNA samples obtained from two different colonies on October and November 1999 and August 2000.
5′- and 3′-RACE.
For rapid amplification of cDNA ends (RACE), RNA from the MBs of the attackers was treated with DNase and reverse transcribed as described above. Experiments were performed with the Marathon-Ready cDNA kit (Clontech, Palo Alto, Calif.) according to the manufacturer's protocol, using Ex Taq polymerase (Takara). The amplified DNA fragments were subcloned, and the nucleotide sequences of both strands were determined.
Southern blotting.
DNA (10 μg each) extracted from the whole bodies of the nurse bees was digested with 100 U of EcoRI (Takara), subjected to 0.8% agarose gel electrophoresis, and transferred to a nylon membrane (GeneScreen Plus; NEN Life Science Products, Inc., Boston, Mass.). A 378-bp fragment of Kakugo cDNA (+8388 to +8765) and a 366-bp fragment of Mblk-1 cDNA (+12151 to +12516) (33) were used as probes. The probes were labeled with 32P by use of a StripEZ DNA kit (Ambion, Austin, Tex.). Hybridization was performed with ExpressHyb hybridization solution (Clontech) according to the manufacturer's protocol. After hybridization, membranes were washed and exposed to Kodak X-ray film (Rochester, N.Y.).
Phylogenetic analysis.
The highly conserved fragments of RNA-dependent RNA polymerase (RdRp) amino acid sequences encompassing motifs 1 to 8 of RdRps of the picornaviruses (amino acid positions 2559 to 2829) were used for phylogenetic analysis by the neighbor-joining method (34). The statistical significance of the branch order was estimated by performing 1,000 replications of bootstrap resampling of the original aligned amino acid sequences.
Infection of the worker honeybees with head lysate containing Kakugo virus.
Whole heads of 70 attackers or foragers were homogenized with 10 volumes of phosphate-buffered saline (PBS; 8.1 mM phosphate buffer, pH 7.4, containing 137 mM NaCl, 2.68 mM KCl, and 1.47 mM KH2PO4) in a glass homogenizer. The homogenates were centrifuged at 1,500 × g for 15 min at 4°C, and the supernatants were used as the inoculum for infection. The presence of Kakugo virus in the attacker head supernatants was confirmed by RT-PCR analysis.
For inoculation of workers with Kakugo virus, 1 μl of the supernatant or PBS was injected into the heads of foragers ventrally from the neck. The inoculated foragers were kept in separate cages at 25°C and collected at different times after injection. Each whole head was homogenized with 300 μl of PBS in a glass homogenizer, and the resulting homogenates were centrifuged at 1,500 × g for 15 min at 4°C. The total RNA was extracted from 250 μl of the supernatants with 750 μl of TRIzol LS, according to the manufacturer's protocol (Invitrogen), and was subjected to quantitative RT-PCR to detect Kakugo RNA.
Sucrose density gradient centrifugation.
Whole thoraxes from seven foragers inoculated with Kakugo virus were homogenized with 10 volumes of PBS in a glass homogenizer, and the homogenate was centrifuged at 1,500 × g for 15 min at 4°C. The supernatant (100 μl) was homogenized with 10 μl of 20% N-lauroylsarcosine sodium salt, added to 20 μg of yeast tRNA and 40 U of RNase out (Invitrogen), and then loaded in a 15 to 30% sucrose density gradient column prepared in diethyl pirocarbonate-treated distilled water. Centrifugation was performed at 200,000 × g for 1 h at 4°C with a Beckman SW55Ti rotor (Beckman Coulter, Inc., Fullerton, Calif.). Next, 520-μl fractions were collected and 250 μl of each fraction was used to determine the Kakugo RNA content by quantitative RT-PCR. The total RNA (500 ng) extracted from the same lysate of forager thoraxes that were inoculated with Kakugo virus and the partially purified virion of the type 1 poliovirus (PV) Mahoney strain [PV1(M)OM] (31) were also loaded in the sucrose density gradient centrifugation column, separately, as controls.
Quantitative RT-PCR.
PCR was performed with Light Cycler and LightCycler-DNA master hybridization probes (Roche, Basel, Switzerland) according to the manufacturer's protocol, using gene-specific primers (+8388 to +8415 and +8745 to +8765 for Kakugo cDNA and +2 to +21 and +376 to +395 for honeybee cytoplasmic actin cDNA) and fluorescent probes (+8654 to +8679 and +8681 to +8716 for Kakugo cDNA and +297 to +313 and +315 to +354 for actin cDNA) in a total volume of 20 μl. The PCR conditions used were as follows: 95°C for 1 s, 55°C for 10 s, and 72°C for 10 s. Kakugo and actin cDNA clones of known concentrations were used as standards.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been submitted to the DDBJ/EMBL/GenBank data bank under the accession number AB070959.
RESULTS
Identification of Kakugo RNA, which was detected exclusively in the brains of aggressive workers.
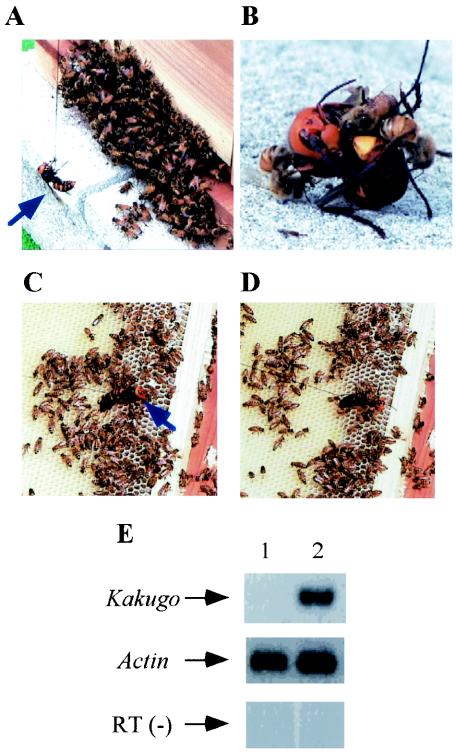
We selected workers based on their attacking behavior against the hornet V. mandarinia japonica. Guard bees that counterattacked a hornet presented as a decoy were collected as aggressive workers (attackers) (Fig. 1A and B) and workers that were in the hive and escaped from the hornet were collected as nonaggressive workers (escapers) (Fig. 1C and D). Total RNAs were extracted, and RNAs that were differentially detected in the brains of the attackers and the escapers were screened by the differential display method. One of the candidate bands for attacker-specific genes, termed Kakugo (which means “ready to attack” in Japanese), was subcloned and sequenced. To confirm the attacker-specific presence of Kakugo RNA, semiquantitative RT-PCR analysis was performed with the total RNAs from the MBs of the attackers and escapers. Kakugo RNA was detected specifically in the attackers, dependent on the RT reaction, although the control actin mRNA was detected almost equally between the attackers and escapers. Essentially the same results were obtained with three RNA samples obtained from two different colonies. These results indicate that Kakugo RNA was present in the MBs of the attackers but not in those of escapers.
FIG. 1.
Identification of Kakugo RNA in the brains of attacker honeybees. (A) A hornet (blue arrow) was hung by a thread and presented as a decoy to the guard bees. (B) Some guard bees scrambled and obstinately attacked the hornet. (C) The hornet was presented to the workers inside the hive. (D) Some of the workers escaped from it. (E) Semiquantitative RT-PCR using total RNA from the MBs of the escapers (lane 1) and attackers (lane 2). Gene-specific primer sets for Kakugo and actin were used. RT (−), control experiment with RT-negative template and Kakugo-specific primer set.
cDNA cloning of Kakugo RNA, which encodes a putative polyprotein for a novel picorna-like virus.
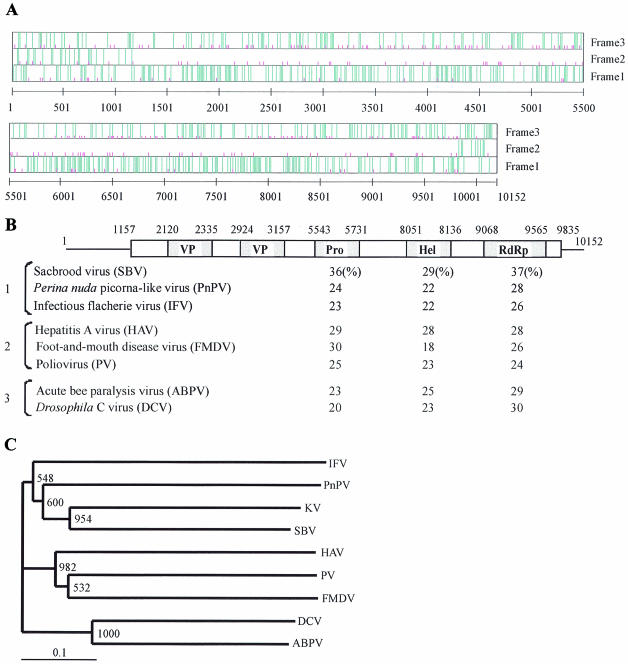
By repeating the 5′- and 3′-RACE methods, we identified a full-length Kakugo cDNA, of 10,152 bp without the poly(A) tail, that contained a large open reading frame (ORF) encoding 2,893 amino acid residues (Fig. 2A). The ORF started at nucleotide 1157 in a context (GAAAUGG) that matches Kozak's rule for invertebrate initiation sites (A/GNNAUGG/A) (6). A database search revealed significant sequence similarities between this protein and polyproteins of various picorna-like viruses, especially sacbrood virus (SBV), which is infectious to the honeybee (8) (Fig. 2B).
FIG. 2.
Kakugo RNA encodes a polyprotein of a putative picorna-like virus. (A) Graphic representation of the positions of the initiation codons and termination codons in the Kakugo cDNA. Termination codons (TAA, TAG, and TGA) and the initiation codon (ATG) are indicated by long blue and short pink vertical lines, respectively, in each of three reading frames. The numbers indicate base numbers from the 5′ end. (B) Structure of Kakugo cDNA and comparison of putative amino acid sequence of the Kakugo protein with those of other picorna-like virus polyproteins. Open and shaded boxes show the ORFs carried by the Kakugo cDNA and the domains of the Kakugo polyprotein, respectively. Numbers indicate the base positions corresponding to each domain. Sequence identities with the Kakugo protein in the helicase (Hel), protease (Pro), and RdRp domains are indicated below the corresponding domains. Bracket 1, the insect picorna-like viruses SBV (8), Perina nuda picorna-like virus (PnPV) (37), and infectious flacherie virus (IFV) (13); bracket 2, the mammalian picornaviruses hepatitis A virus (HAV) (22), foot-and-mouth disease virus (FMDV) (7), and PV (23); bracket 3, the cricket paralysis-like viruses acute bee paralysis virus (ABPV) (10) and Drosophila C virus (DCV) (16). (C) Phylogenetic tree constructed with the highly conserved amino acid sequences encompassing motifs 1 to 8 of the RdRp domains by the neighbor-joining method. KV, putative Kakugo virus. Numbers at each node represent bootstrap values as the results of 1,000 replicons.
Picornavirus is an RNA virus and its genome is replicated by RdRp, which is encoded by the viral genome. The viral RNA is translated into a single polyprotein followed by processing in the following four domains: virion protein (VP), helicase (Hel), protease (Pro), and RdRp domains (18). The order of the domains for the Kakugo protein is the same as that for other picornavirus polyproteins. There are significant sequence identities (18 to 37%) among the domains of the Kakugo protein and other picornavirus polyproteins (Fig. 2B). Phylogenetic tree analysis using amino acid sequences corresponding to the RdRp domains of various picorna-like viruses revealed that this putative virus belongs to the insect picorna-like virus family (Fig. 2C). Kakugo RNA has a total G+C content of 38% and resembles other insect picorna-like viruses in being A/U rich.
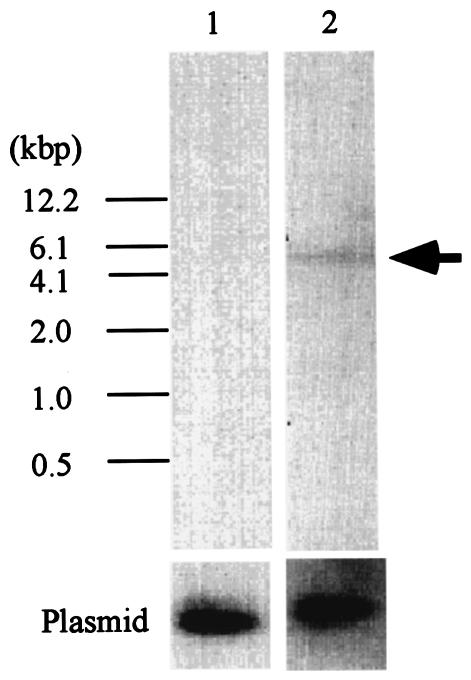
To further examine whether Kakugo RNA is encoded by the honeybee genome, we performed Southern blotting analysis, using Kakugo cDNA as a probe (Fig. 3). Kakugo RNA was not detected in the honeybee genome, whereas a single band was detected for the MB-preferential transcription factor Mblk-1 (26, 27, 33), indicating that Kakugo RNA is not encoded by the honeybee genome. Furthermore, the Kakugo RNA sequence is not included in the genomic sequences annotated and registered to date by the honeybee genome project (Human Genome Sequencing Center at Baylor College of Medicine [http://hgsc.bcm.tmc.edu/projects/honeybee/]). These results strongly suggest that Kakugo RNA is a foreign RNA derived from an RNA virus and that the attackers' brains were infected by this putative RNA virus, Kakugo virus.
FIG. 3.
Southern blotting analysis of Kakugo DNA. Southern blotting analysis was performed with honeybee genomic DNA (10 μg) digested with EcoRI, with a partial Kakugo cDNA as a probe (lane 1). The Mblk-1 gene, which encodes a transcription factor (26, 27, 32), was also analyzed as a loading control (lane 2). Plasmid cDNAs (5 pg each) for Kakugo and Mblk-1 were included as hybridization controls (bottom). The band positions are indicated by arrows.
Kakugo RNA has a 5′ untranslated region (UTR) of 1,156 bases, which is a similar size to those of the UTRs of other picornaviruses (610 to 1,200 nucleotides [nt]) (15) and is long enough to contain an internal ribosomal entry site, which is usually approximately 450 bases in length (15). There were many AUG sequences upstream of the putative translation initiation codon for the polyprotein, which is the 32nd AUG sequence from the 5′ end of the Kakugo RNA sequence (data not shown). The number of AUG sequences in the 5′ UTR is remarkably higher than that of other picornaviruses. This region might function as an internal ribosomal entry site.
Comparison of the domain structure of Kakugo polyprotein with those of other picornavirus polyproteins.
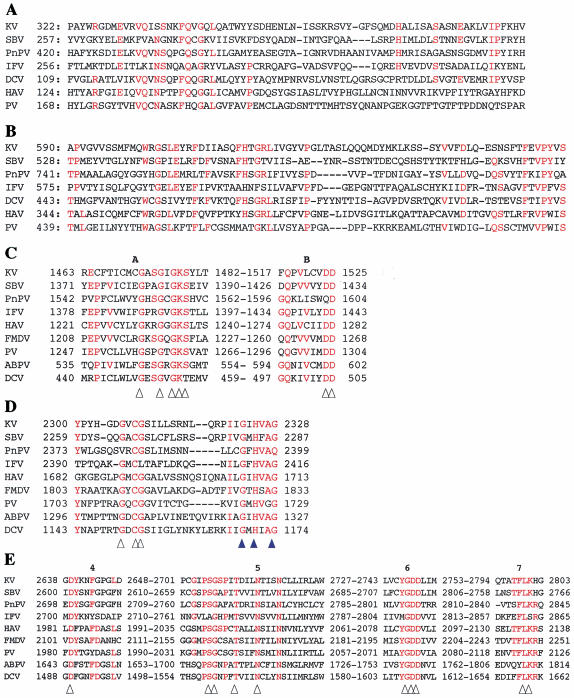
For further sequence analysis, the amino acid residues of the Kakugo protein were compared with those of various picornavirus polyproteins. First, most of the amino acid residues conserved among the VP domains of picornavirus polyproteins are also conserved in residues 322 to 392 and 590 to 667 of the Kakugo protein (Fig. 4A and B), whereas the other regions were not conserved. Many picornaviruses have three or four major structural proteins; however, not all of them are necessarily conserved among viruses. Usually two of them are conserved among insect picorna-like viruses. The Kakugo protein also had two domains similar to those of other insect picorna-like viruses, consistent with previous findings (8, 10, 29, 37).
FIG. 4.
Comparison of the amino acid sequences of functional domains of the Kakugo polyprotein and other picorna-like virus polyproteins. (A) One of the VP regions of Kakugo virus with the corresponding region of SBV, CP1 of PnPV, VP3 of IFV, VP1 of DCV, VP2 of HAV, and the corresponding region of PV. Residues that are identical for at least four of the seven viruses are shown in red (A and B). (B) The other VP region of the Kakugo virus with the corresponding region of SBV, CP3 of PnPV, VP1 of IFV, VP2 of DCV, VP3 of HAV, and the corresponding region of PV. (C) The putative motifs A and B in the helicase domain of the Kakugo virus with those of SBV, PnPV, IFV, HAV, FMDV, PV, ABPV, and DCV. Amino acid residues that are essential for enzymatic activity are indicated by arrowheads (C and E). (D) The C terminus of the putative protease domain of Kakugo virus with those of SBV, PnPV, IFV, HAV, FMDV, PV, ABPV, and DCV. The protease motif and amino acid residues involved in substrate binding are indicated with white and blue arrowheads, respectively. (E) The putative motifs 4 through 7 in the RdRp domains of Kakugo virus with those of SBV, PnPV, IFV, HAV, FMDV, PV, ABPV, and DCV. Residues that are identical in at least five of the nine viruses are shown in red (C, D, and E). Arrowheads indicate the consensus amino acids within RdRp motifs. Numbers show amino acid positions from the N terminus.
Second, most of the amino acid residues considered essential for enzymatic activity are also conserved in the Kakugo protein. The amino acid sequence for positions 1463 to 1525 was homologous to that of the helicase domain, and both helicase motifs A and B are included in this region (Fig. 4C). This region also contained two conserved motifs that are characteristic of the NTP binding sequence (Fig. 4C, arrowheads) (9). The amino acid sequence for positions 2300 to 2328 was similar to those of the protease domains of various picorna-like virus polyproteins. In particular, the cysteine protease motif (GXCG) (Fig. 4D, white arrowheads) and putative substrate binding residues corresponding to G2323, H 2325, and G2328 (Fig. 4D, blue arrowheads) were all conserved in this domain (18). Moreover, the C-terminal region of the Kakugo protein was homologous to the RdRp domain. The amino acid sequence for positions 2638 to 2803 was similar to motifs 4 to 7 of the RdRp domains of various picorna-like viruses (Fig. 4E), and most of the invariant amino acids of the RdRp domain (28) were also conserved (Fig. 4E, arrowheads). These results provide further support that Kakugo RNA encodes active viral proteins.
Kakugo RNA as the genomic RNA of an infectious virus.
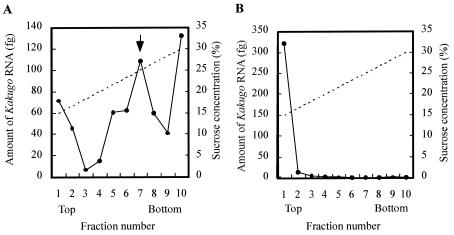
If the Kakugo RNA encodes a viral polyprotein, it could exist as mRNA as well as genomic RNA within a mature virion in honeybee tissues. For testing of this possibility, the whole worker abdomen lysate that contained Kakugo RNA was subjected to sucrose density gradient centrifugation, and the amount of Kakugo RNA in each fraction was determined by quantitative RT-PCR. When the fractions were analyzed, Kakugo RNAs, except for the material at the bottom, from the thorax lysate were detected in two major peaks, fractions 1 and 2 and fractions 6 to 8 (Fig. 5A), whereas Kakugo RNA from the total thorax RNA was detected as a single peak in fractions 1 and 2 (Fig. 5B). PV virion used as a control was detected mainly in fraction 7. These results suggest that Kakugo RNAs detected in these fractions correspond to free RNA and/or mRNA (fractions 1 and 2) and to genome-sense RNA constituting the virion (fractions 6 to 8). The material at the bottom might correspond to aggregates and/or cell debris that contains Kakugo RNA (fraction 10).
FIG. 5.
Sucrose density gradient centrifugation of the honeybee tissue lysate that contained Kakugo RNA. The lysate (A) and the total RNA (B) prepared from thoraxes of the foragers inoculated with Kakugo virus were each subjected to sucrose density gradient centrifugation, and the amount of the Kakugo RNA in each fraction was determined by quantitative RT-PCR. Arrow, the fraction in which the PV virion was mainly found. The solid and broken lines indicate the amount of Kakugo RNA and the putative sucrose concentration, respectively.
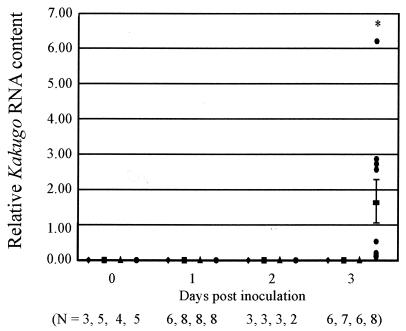
Next, to demonstrate that Kakugo RNA is also the virion RNA for an infectious virus, we examined the infectivity of the Kakugo virus. Head lysate (1 μl) from the attackers or the foragers or PBS was injected into the heads of noninfected foragers (see Fig. 7). Total RNA was extracted from the heads 0, 1, 2, and 3 days after the inoculation and was subjected to quantitative RT-PCR to detect Kakugo RNA. There was a marked increase in the amount of Kakugo RNA 3 days after inoculation with the attacker head lysate (Fig. 6), although no significant amount of Kakugo RNA was detected from the heads of foragers 1 to 3 days after inoculation with noninfected forager head lysate or PBS. These results indicate that Kakugo RNA constitutes an infectious virus (Kakugo virus) in the honeybee. The workers inoculated with attacker head lysate or PBS were viable for at least 3 days.
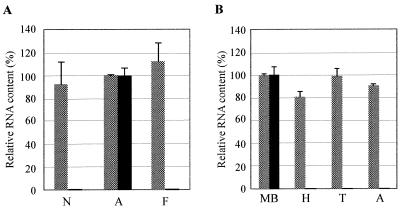
FIG. 7.
Kakugo RNA is detected almost exclusively in the brains of attackers. (A) Relative Kakugo RNA content in the MBs of nurse bees (N), attackers (A), and foragers (F). As many as 100 of each were examined by quantitative RT-PCR with Kakugo-specific primers and probes (black bar). actin mRNA was also examined to show that essentially the same amount of RNA was contained in each sample (dotted bars). Values are represented as percentages of Kakugo or actin mRNA relative to that in the attacker brains. (B) Relative RNA content of the MBs, heads (H), thoraxes (T), and abdomens (A) from five attackers, examined with Kakugo-specific (black bar) and actin-specific (dotted bars) primers and probes.
FIG. 6.
Kakugo RNA constitutes an infectious virus. Foragers inoculated either with head lysate from the attackers (circle), head lysate from the foragers (triangle), or PBS (square) were collected at different times after inoculation. Noninjected foragers (lozenge) were also collected as controls. Kakugo RNA and actin mRNA levels, which served as a control, were determined by quantitative RT-PCR. The mean value of relative Kakugo RNA content normalized to that of actin mRNA is shown with a bold bar showing standard errors. N, the number of samples used for each experiment; asterisk, differences were significant, with a P value of <0.05 by the unpaired t test.
Exclusive presence of Kakugo RNA in the brains of aggressive workers.
Honeybee workers shift their tasks according to age after eclosion, from nursing to guarding to foraging (36). To examine whether the presence of Kakugo RNA is closely related to aggressive honeybee behaviors, we performed quantitative RT-PCR, using RNAs from the brains of the attackers as well as from nurse bees and foragers. Kakugo RNA was detected specifically in the attackers' brains, whereas there were no significant signals in the brains of nurse bees or foragers (Fig. 7A). We also examined the body parts of the attackers in which Kakugo RNA was present by quantitative RT-PCR. Kakugo RNA was detected almost exclusively in the brains of the attackers and not in other body parts, including the thorax and abdomen (Fig. 7B). The whole head content of Kakugo RNA was also below the detection level, possibly due to the concentrated presence of Kakugo RNA in the brain. We could not eliminate the possibility that other organs in the thoraxes and abdomens might also be infected by Kakugo virus, but at levels below the detection limit. Taken together, our results indicated a close correlation between the presence of Kakugo RNA in the brain and aggressive honeybee behaviors.
DISCUSSION
In the present paper, we identified Kakugo RNA as the genome-sense RNA of a novel insect picorna-like virus which is infectious specifically in the brains of aggressive worker honeybees. The identification of Kakugo RNA as a viral genome-sense RNA was based on the following observations. (i) Kakugo RNA encodes a protein homologous to the picorna-like virus polyproteins. Particularly, the presence of the RdRp domain is characteristic of the RNA viruses. (ii) The amino acid residues conserved among the domains of the picorna-like virus polyproteins were also conserved in the Kakugo protein, further supporting the idea that Kakugo RNA encodes active viral proteins. (iii) Kakugo RNA is not encoded by the honeybee genome. (iv) Kakugo RNA exists as mRNA-type RNA as well as genome-sense RNA in infected worker tissues. (v) Kakugo RNA constitutes an infectious virus, as revealed by inoculation experiments. To our knowledge, this is the first study to demonstrate a close relationship between a viral infection and honeybee aggressive behaviors. Some questions then arise regarding the ecology of this novel virus. How and when are the attackers infected with this virus and what is the possible relationship between Kakugo virus infection and honeybee aggressive behaviors?
SBV infects honeybees. SBV infection during the larval stage is lethal, and infection in the adult shortens the worker life span (2). Honeybee adults exchange nutrients orally, and SBV infects honeybees orally (1). Similarly, Kakugo virus might be transmitted by oral infection between the colony members. Alternatively, Kakugo virus might be transmitted accidentally by bodily injury from infected colony members or by other organisms (e.g., a mite that is parasitic for honeybees or hornets) that carry the virus. Since Kakugo RNA is detected specifically in the attackers but not in the nurse bees or foragers in normal colonies, Kakugo virus infection might affect the honeybee behaviors so that the infected workers tend to become attackers. The relationship between Kakugo virus infection and honeybee aggressive behaviors, however, is not clear at present, as it was difficult to observe the aggressive behaviors of the workers that were removed from the hive, inoculated with Kakugo virus, and kept in a cage. To overcome this issue, it will be necessary to establish a method for inoculation of Kakugo virus into workers without damage to observe their behaviors in the normal colony. It is also unclear whether the virus has some pathogenic or lethal effects on honeybees, like most other animal viruses at present, as the inoculated workers were viable for at least 3 days in the cage. If the Kakugo virus has some pathogenic effect on the honeybees, however, the attacking behavior of the infected workers might act as a self-defense system at the colony level, as the attacking behavior might shorten the life span of the infected workers and help to eliminate the virus from the colony.
The amount of Kakugo RNA detected in attacker heads was much less than that detected in forager heads artificially inoculated with Kakugo virus. This might be explained by the fact that the amount of Kakugo virus inoculated artificially (1 μl of attacker head lysate) into the foragers was much higher than that transmitted from infected workers to the other noninfected workers in a normal colony. Alternatively, heavily infected attackers might be eliminated from the colony so that we could not detect any attackers with a high titer of Kakugo RNA in the head.
Very recently, sequences similar to Kakugo RNA (98 and 97% homology) were registered in GenBank (accession numbers AJ489744 and AY292384) as the genome-sense RNA of the deformed wing virus (DWV) of the honeybee (unpublished data). Kakugo RNA and the registered DWV genome-sense RNAs differ in the following two ways. (i) There are at least 201 nucleotide substitutions, deletions, or insertions which are unique to Kakugo RNA; 21 of them result in amino acid substitutions in the polyproteins and 7 of them are located in the conserved domain structures. (ii). The 5′ UTRs of the registered DWV genome-sense RNAs are 16 and 11 nt shorter than that of Kakugo RNA. DWV is transmitted from the bee mite, Varroa jacobsoni, into the honeybee, and the viral infection is suggested to cause morphological deformity of the adult wing when it infects honeybee larvae (3). Although we do not have any more information about the relationship between Kakugo virus and DWV, they might be closely related viruses carrying different pathogenicities, as no significant DWV symptoms are observed in our colonies. Further analyses are needed to clarify the relationship between these two virus species.
Acknowledgments
This work was partially supported by grants-in-aid from the Bio-Oriented Technology Research Advancement Institution (BRAIN), the Ministry of Education, Science, Sports, and Culture of Japan, Terumo Life Science Foundation, and The Naito Foundation. T.F. is the recipient of a grant-in-aid for JSPS Fellows.
REFERENCES
- 1.Bailey, L. 1969. The multiplication and spread of sacbrood virus of bees. Ann. Appl. Biol. 63:483-491. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, L., and E. F. W. Fernando. 1972. Effects of sacbrood virus on adult honey-bees. Ann. Appl. Biol. 72:27-35. [Google Scholar]
- 3.Bowen-Walker, P. L., S. J. Martin, and A. Gunn. 1999. The transmission of deformed wing virus between honeybees (Apis mellifera L.) by the ectoparasitic mite Varroa jacobsoni Oud. J. Invertebr. Pathol. 73:101-106. [DOI] [PubMed] [Google Scholar]
- 4.Breed, M. D., G. E. Robinson, and J. R. E. Page. 1990. Division of labor during honey bee colony defense. Behav. Ecol. Sociobiol. 27:395-401. [Google Scholar]
- 5.Breed, M. D., T. A. Smith, and A. Torres. 1992. Role of guard honey bees (Hymenoptera: Apidae) in nestmate discrimination and replacement of removed guards. Ann. Entomol. Soc. 85:633-637. [Google Scholar]
- 6.Cavener, D. R., and S. C. Ray. 1991. Eucaryotic start and stop translation sites. Nucleic Acids Res. 19:3185-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forss, S., K. Strebel, E. Beck, and H. Schaller. 1984. Nucleotide sequence and genome organization of foot-and-mouth disease virus. Nucleic Acids Res. 12:6587-6601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghosh, R. C., B. V. Ball, M. M. Willcocks, and M. J. Carter. 1999. The nucleotide sequence of sacbrood virus of the honey bee: an insect picorna-like virus. J. Gen. Virol. 80:1541-1549. [DOI] [PubMed] [Google Scholar]
- 9.Gorbalenya, A. E., and E. V. Koonin. 1989. Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res. 17:8413-8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Govan, V. A., N. Leat, M. Allsopp, and S. Davison. 2000. Analysis of the complete genome sequence of acute bee paralysis virus shows that it belongs to the novel group of insect-infecting RNA viruses. Virology 277:457-463. [DOI] [PubMed] [Google Scholar]
- 11.Hunt, G. J., A. M. Collins, R. Rivera, R. E. Page, Jr., and E. Guzman-Novoa. 1999. Quantitative trait loci influencing honeybee alarm pheromone levels. J. Hered. 90:585-589. [DOI] [PubMed] [Google Scholar]
- 12.Hunt, G. J., E. Guzman-Novoa, M. K. Fondrk, and J. R. E. Page. 1998. Quantitative trait loci for honey bee stinging behavior and body size. Genetics 148:1203-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isawa, H., S. Asano, K. Sahara, T. Iizuka, and H. Bando. 1998. Analysis of genetic information of an insect picorna-like virus, infectious flacherie virus of silkworm: evidence for evolutionary relationships among insect, mammalian and plant picorna(-like) viruses. Arch. Virol. 143:127-143. [DOI] [PubMed] [Google Scholar]
- 14.Ito, T., K. Kito, N. Adati, Y. Mitsui, H. Hagiwara, and Y. Sakaki. 1994. Fluorescent differential display: arbitrarily primed RT-PCR fingerprinting on an automated DNA sequencer. FEBS Lett. 351:231-236. [DOI] [PubMed] [Google Scholar]
- 15.Jackson, R. J., M. T. Howell, and A. Kaminski. 1990. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem. Sci. 15:477-483. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, K. N., and P. D. Christian. 1998. The novel genome organization of the insect picorna-like virus Drosophila C virus suggests this virus belongs to a previously undescribed virus family. J. Gen. Virol. 79:191-203. [DOI] [PubMed] [Google Scholar]
- 17.Kamikouchi, A., H. Takeuchi, M. Sawata, K. Ohashi, S. Natori, and T. Kubo. 1998. Preferential expression of the gene for a putative inositol 1,4,5-trisphosphate receptor homologue in the mushroom bodies of the brain of the worker honeybee Apis mellifera L. Biochem. Biophys. Res. Commun. 242:181-186. [DOI] [PubMed] [Google Scholar]
- 18.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 19.Kubo, T., M. Sasaki, J. Nakamura, H. Sasagawa, K. Ohashi, H. Takeuchi, and S. Natori. 1996. Change in the expression of hypopharyngeal-gland proteins of the worker honeybees (Apis mellifera L.) with age and/or role. J. Biochem. (Tokyo) 119:291-295. [DOI] [PubMed] [Google Scholar]
- 20.Liang, P., and A. B. Pardee. 1992. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257:967-971. [DOI] [PubMed] [Google Scholar]
- 21.Matsuura, M. 1988. Ecological study on vespine wasps (Hymenoptera: Vespidae) attacking honeybee colonies. 1. Seasonal changes in the frequency of visits to apiaries by vespine wasps and damage inflicted, especially in the absence of artificial protection. Appl. Entomol. Zool. 23:428-440. [Google Scholar]
- 22.Najarian, R., D. Caput, W. Gee, S. J. Potter, A. Renard, J. Merryweather, G. Van Nest, and D. Dina. 1985. Primary structure and gene organization of human hepatitis A virus. Proc. Natl. Acad. Sci. USA 82:2627-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nomoto, A., T. Omata, H. Toyoda, S. Kuge, H. Horie, Y. Kataoka, Y. Genba, Y. Nakano, and N. Imura. 1982. Complete nucleotide sequence of the attenuated poliovirus Sabin 1 strain genome. Proc. Natl. Acad. Sci. USA 79:5793-5797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohashi, K., M. Sawata, H. Takeuchi, S. Natori, and T. Kubo. 1996. Molecular cloning of cDNA and analysis of expression of the gene for α-glucosidase from the hypopharyngeal gland of the honeybee Apis nellifera L. Biochem. Biophys. Res. Commun. 221:380-385. [DOI] [PubMed] [Google Scholar]
- 25.Ono, M., T. Igarashi, E. Ohno, and M. Sasaki. 1995. Unusual thermal defence by a honeybee against mass attack by hornets. Nature 377:334-336. [Google Scholar]
- 26.Park, J. M., T. Kunieda, and T. Kubo. 2003. The activity of Mblk-1, a mushroom body-selective transcription factor from the honeybee, is modulated by the Ras/MAPK pathway. J. Biol. Chem. 278:18689-18694. [DOI] [PubMed] [Google Scholar]
- 27.Park, J. M., T. Kunieda, H. Takeuchi, and T. Kubo. 2002. DNA-binding properties of Mblk-1, a putative transcription factor from the honeybee. Biochem. Biophys. Res. Commun. 291:23-28. [DOI] [PubMed] [Google Scholar]
- 28.Sankar, S., and A. G. Porter. 1992. Point mutations which drastically affect the polymerization activity of encephalomyocarditis virus RNA-dependent RNA polymerase correspond to the active site of Escherichia coli DNA polymerase 1. J. Biol. Chem. 267:10168-10176. [PubMed] [Google Scholar]
- 29.Sasaki, J., N. Nakashima, H. Saito, and H. Noda. 1998. An insect picorna-like virus, Plautia stali intestine virus, has genes of capsid proteins in the 3′ part of the genome. Virology 244:50-58. [DOI] [PubMed] [Google Scholar]
- 30.Sawata, M., D. Yoshino, H. Takeuchi, A. Kamikouchi, K. Ohashi, and T. Kubo. 2002. Identification and punctate nuclear localization of a novel noncoding RNA, Ks-1, from the honeybee brain. RNA 8:772-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiroki, K., T. Ishii, T. Aoki, M. Kobashi, S. Ohka, and A. Nomoto. 1995. A new cis-acting element for RNA replication within the 5′ noncoding region of poliovirus type 1 RNA. J. Virol. 69:6825-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeuchi, H., T. Fujiyuki, K. Shirai, Y. Matsuo, A. Kamikouchi, Y. Fujinawa, A. Kato, A. Tsujimoto, and T. Kubo. 2002. Identification of genes expressed preferentially in the honeybee mushroom bodies by combination of differential display and cDNA microarray. FEBS Lett. 513:230-234. [DOI] [PubMed] [Google Scholar]
- 33.Takeuchi, H., E. Kage, M. Sawata, A. Kamikouchi, K. Ohashi, M. Ohara, T. Fujiyuki, T. Kunieda, K. Sekimizu, S. Natori, and T. Kubo. 2001. Identification of a novel gene, Mblk-1, that encodes a putative transcription factor expressed preferentially in the large-type Kenyon cells of the honey bee brain. Insect Mol. Biol. 10:487-494. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibsons. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilson, E. O. 1975. Sociobiology: the new synthesis. The Belknap Press of Harvard University Press, Cambridge, Mass.
- 36.Winston, M. L. 1987. The biology of the honeybee. Harvard University Press, Cambridge, Mass.
- 37.Wu, C. Y., C. F. Lo, C. J. Huang, H. T. Yu, and C. H. Wang. 2002. The complete genome sequence of Perina nuda picorna-like virus, an insect-infecting RNA virus with a genome organization similar to that of the mammalian picornaviruses. Virology 294:312-323. [DOI] [PubMed] [Google Scholar]