Abstract
The proton-pumping ATPase (H+-ATPase) of the plant plasma membrane is encoded by two major gene subfamilies. To characterize individual H+-ATPases, PMA2, an H+-ATPase isoform of tobacco (Nicotiana plumbaginifolia), was expressed in Saccharomyces cerevisiae and found to functionally replace the yeast H+-ATPase if the external pH was kept above 5.0 (A. de Kerchove d'Exaerde, P. Supply, J.P. Dufour, P. Bogaerts, D. Thinès, A. Goffeau, M. Boutry [1995] J Biol Chem 270: 23828–23837). In the present study we replaced the yeast H+-ATPase with PMA4, an H+-ATPase isoform from the second subfamily. Yeast expressing PMA4 grew at a pH as low as 4.0. This was correlated with a higher acidification of the external medium and an approximately 50% increase of ATPase activity compared with PMA2. Although both PMA2 and PMA4 had a similar pH optimum (6.6–6.8), the profile was different on the alkaline side. At pH 7.2 PMA2 kept more than 80% of the maximal activity, whereas that of PMA4 decreased to less than 40%. Both enzymes were stimulated up to 3-fold by 100 μg/mL lysophosphatidylcholine, but this stimulation vanished at a higher concentration in PMA4. These data demonstrate functional differences between two plant H+-ATPases expressed in the same heterologous host. Characterization of two PMA4 mutants selected to allow yeast growth at pH 3.0 revealed that mutations within the carboxy-terminal region of PMA4 could still improve the enzyme, resulting in better growth of yeast cells.
The plasma membrane H+-ATPase plays a pivotal role in plant physiology. By extruding protons from the cell, an electrochemical gradient is created across the plasma membrane, providing the driving force for ion and nutrient uptake, intracellular pH regulation, maintenance of cell turgidity, and related phenomena such as cell growth, organ and stomata movement, and salinity tolerance (for review, see Serrano, 1989; Sussman, 1994; Michelet and Boutry, 1995; Palmgren, 1998).
The plant H+-ATPase is encoded by a multigene family of approximately 10 genes (Harper et al., 1990, 1994; Perez et al., 1992; Ewing and Bennett, 1994). However, all of the H+-ATPase genes that have been identified at the cDNA level in several species can be classified into two subfamilies according to their sequence identity. Divergence between these subfamilies that represent the most highly expressed genes seems to predate the evolution of current monocotyledonous and dicotyledonous species (Moriau et al., 1993).
Biochemical characterization of individual H+-ATPase isoforms and the unraveling of their possible differences in structure, kinetics, and regulatory features will help us understand the respective roles of the two major H+-ATPase subfamilies. Unfortunately, this kind of study is hindered in plants because of the simultaneous presence of several isoforms within a single organ. However, the heterologous expression of individual H+-ATPases in the yeast Saccharomyces cerevisiae has provided a way of overcoming this difficulty. Three Arabidopsis H+-ATPase genes, AHA1, AHA2, and AHA3, all belonging to the second subfamily, have been expressed in yeast (Villalba et al., 1992; Palmgren and Christensen, 1994). AHA1 and AHA3 did not allow growth and AHA2 permitted slow growth when the yeast H+-ATPase gene, under the control of a Gal-induced promoter, was turned off. Characterization of the three gene products that accumulated in ER-derived internal membranes revealed quantitative differences in enzymatic properties (Palmgren and Christensen, 1994).
On the other hand, pma2, a Nicotiana plumbaginifolia H+-ATPase gene from the first subfamily, complemented a strain of S. cerevisiae deprived of its H+-ATPase genes if the external pH was kept above 5.0 (de Kerchove d'Exaerde et al., 1995). Therefore, this expression system made it possible to characterize biochemically a plant H+-ATPase in a purified plasma membrane fraction devoid of contaminant yeast H+-ATPase. It was thus important to test a N. plumbaginifolia isoform of the second subfamily in the same expression system.
In the present study we show that the plant pma4 also successfully replaced the yeast H+-ATPase gene and supported yeast growth. However, in contrast to the strain expressing pma2, which is unable to grow at an external pH lower than 5.0, the yeast strain containing pma4 can grow at a pH as low as 4.0. Biochemical characterization of PMA4 and PMA2 revealed differences in enzymatic properties, pH sensitivity, and LPC stimulation. In addition, we selected two pma4 mutants with improved ATPase, which allow yeast growth at an external pH of 3.0.
MATERIALS AND METHODS
Chemicals
5-Fluoroorotic acid was purchased from PCR, Inc. (Gainesville, FL), Na2ATP from Sigma, protease inhibitors from Boehringer Mannheim, yeast extract from KAT (Ohly, Hamburg, Germany), and yeast nitrogen base amino acids from Difco (Detroit, MI). All other reagents were of analytical grade.
Plasmid Construction
The pma4 cDNA from Nicotiana plumbaginifolia, cpma4 described by Moriau et al. (1993), was cloned as a 3363-bp EcoRI restriction fragment in the plasmid vector PTZ19U starting 105 bp upstream from the initiation ATG of the PMA open reading frame and extending 353 bp beyond the stop codon. Within the 5′ untranslated region is a small, 5-codon upstream open reading frame located 83 nucleotides upstream from the start of the PMA open reading frame. To prevent any potential interference of the 5′ and 3′ untranslated regions in the expression of the pma4 gene in yeast, these were eliminated by PCR.
In the 5′ region we made use of a BamHI restriction site in the plasmid and of a PstI site 371 nucleotides downstream of the PMA4 initiation codon. Two oligonucleotides, one corresponding to nucleotides −14 to +5 of the PMA4 translation initiation codon (CGCGGATCCGTGGAGCAGAGATGGC; the introduced BamHI site is underlined), and the other complementary to nucleotides 445 to 427 downstream of the PMA4 translation initiation codon (CGGCTTCCTGTTCACTCC), were synthesized to amplify the corresponding region in between. This amplified fragment, digested with BamHI and PstI, was then used to replace the corresponding original BamHI-PstI fragment, producing a new 5′ region without the upstream open reading frame.
The same strategy was applied for the removal of the 3′ untranslated region using a HindIII site in the plasmid and an AflII site 417 nucleotides upstream of the stop codon. Two oligonucleotides, one corresponding to a sequence upstream of the AflII site (−369 to −351 upstream of the stop codon; AGCATTATTTTCTACCTC) and the other complementary to nucleotides +18 to +36 downstream of the stop codon (CGCAAGCTTCCTCTCCCTTTCTTGTGT; the introduced HindIII site is underlined), were synthesized for PCR amplification. The amplified fragment was then introduced to replace the original AflII-HindIII region. The correct sequence of the modified pma4 cDNA was verified by sequencing.
This modified pma4 was then released as a BamHI-HindIII fragment and introduced into the yeast Yeplac181 plasmid (bearing the 2μ origin of replication and the LEU2 marker) containing the promoter region of the yeast PMA1 gene (de Kerchove d'Exaerde et al., 1995). The transcription-terminator region of the yeast PMA1 gene was then released as a 1.2-kb XbaI fragment, blunted by Klenow DNA polymerase, and inserted into the HindIII site (also blunted by the Klenow polymerase) of Yeplac181, yielding 2μp(PMA1)pma4.
Culture Conditions
Yeast was grown at 30°C in a rich medium containing 2% (w/v) yeast extract and 2% Glu (YGlu medium) or Gal (YGal medium), or in a synthetic medium containing 0.7% yeast nitrogen base without amino acids, 0.11% drop mix (Treco, 1989), all of the amino acids except those used for selection (His, Leu, Ura, and Trp), and 2% Glu (MGlu-His,Leu,Ura,Trp) or 2% Gal (MGal-His,Leu,Ura,Trp). Solid media also contained 2% agar (Difco). These media were supplemented with 20 mm K2HPO4 and buffered to the pH indicated using HCl or KOH.
Yeast Strains
Several yeast strains were used in this study. YPS14-4 (Supply et al., 1993) was the wild-type S. cerevisiae. YAK2 (de Kerchove d'Exaerde et al., 1995) was deleted from its own H+-ATPase genes, PMA1 and PMA2, and survived with the yeast PMA1 under the control of the GAL1 promoter in a centromeric plasmid PRS-316 (Sikorski and Hieter, 1989). YAKpma2 (de Kerchove d'Exaerde et al., 1995) corresponds to YAK2, in which the centromeric plasmid bearing the yeast PMA1 was replaced by a multicopy plasmid bearing the plant pma2 cDNA under the control of the yeast PMA1 promoter. The strain YAKpma4 was obtained as follows: YAK2 was transformed with 2μp(PMA1)pma4 (see above) and was plated on MGal-His,Leu,Trp,Ura. Transformants were replicated on MGlu-His,Leu,Trp medium containing 0.1% 5-fluoroorotic acid to delete the strain of the URA3 plasmid pRS-316 containing the yeast PMA1 gene under the control of the GAL1 promoter. Loss of this plasmid was verified by Southern-blot analysis. Yeast cells were transformed after treatment with lithium acetate and PEG according to the method of Ito et al. (1983).
Selection of Mutants
Several single colonies of the YAKpma4 strain were inoculated individually into 5 mL of YGlu medium, pH 5.5, and grown until they reached the stationary phase. After centrifugation and resuspension in 200 μL of water, the yeast cells from each culture were plated onto solid YGlu medium, pH 3.0. Spontaneous mutants growing under these nonpermissive conditions appeared after 3 d at 30°C.
Sequencing of pma4 and Its Mutants
The 2μp(PMA1)pma4 plasmid from both wild-type and mutant yeast strains was retrieved and transferred to Escherichia coli. The plasmid DNA was sequenced using a series of synthetic primers scattered throughout the pma4 gene. The wild-type and mutated 2μp(PMA1)pma4 were finally reintroduced into the YAK2 strain to verify the phenotype they conferred.
Plasma Membrane Preparation and Protein Determination
Plasma membranes were prepared according to the method of Goffeau and Dufour (1988). Yeast cells were grown in a 1.5-L culture (YGlu medium) and harvested at a density of 80 × 106 to 100 × 106 cells/mL. The cell pellet was then washed three times with ice-cold water and resuspended (1.5 mL/g fresh weight) in 250 mm sorbitol, 1 mm MgCl2, 50 mm imidazole, pH 7.5 (NaOH), 1 mm PMSF, and 5 mm DTT. After disruption, subcellular fractionation, and plasma membrane enrichment, the plasma membranes were finally resuspended in 10 mm imidazole, pH 7.5 (NaOH), 1 mm MgCl2, and 1 mm PMSF, and then divided into aliquots, frozen in liquid nitrogen, and stored at −80°C. The protein concentration was determined using the Bradford method (1976) with BSA as the standard.
ATPase Assays
ATPase activity was assayed at 30°C in 50 μL of a medium containing 6 mm MgATP, 1 mm free Mg2+ (MgCl2), 50 mm Mes-NaOH, pH 7.0, 10 mm sodium azide (a mitochondrial ATPase inhibitor), 0.2 mm Na molybdate (a phosphatase inhibitor), 20 mm KNO3 (a vacuolar ATPase inhibitor), and 4 μg of proteins. The reaction was stopped after 12 min by the addition of 60 μL of 5% TCA, 30 μL of 50% NH4 molybdate (in 4 n H2SO4), 30 μL of 1% aminonaphthosulfonic acid, and 3% NaHSO3. A700 was read after 15 min. Km and Vmax were determined using an ATP-regenerating system (100 μg/mL pyruvate kinase and 5 mm PEP). The concentration of MgATP varied between 40 μm and 10 mm. Km and Vmax were determined using Eadie-Hofstee plots. The optimal pH was determined as described above except that three buffers were mixed (50 mm Mes, 50 mm Mops, and 50 mm Tris) and adjusted to the pH indicated with HCl or NaOH. The amounts of ATP and Mg2+ to be added to the reaction mixture at the pH indicated were calculated as described by Wach et al. (1990) to obtain the desired concentration of MgATP and Mg2+. All data presented in the figures represent the means ± sd of at least three independent experiments using two separate membrane preparations.
Measurement of Glu-Induced Acid Efflux from Yeast Cells
Yeast cells were grown in a 100-mL culture (YGlu medium at pH 5.5 [YAKpma4] or 6.5 [YAKpma2]), harvested at the late-exponential phase, washed four times with ice-cold water, and stored on ice. For each assay 109 cells were incubated in 10 mL of 250 mm sorbitol at 30°C in a vial with a magnetic barrel and a pH electrode of a microprocessor pH meter (Wissenschaftlich-Technische Werkstätten, Weiheim, Germany). Once a stable pH baseline was established (after about 5 min), 250 mm Glu was added as an energy source and the external pH was recorded over time.
SDS-PAGE
Proteins (30 μg) were suspended in a buffer containing 60 mm Tris-HCl, pH 6.8, 5% (w/v) glycerol, 1% (w/v) SDS, 10 mm DTT, 1 mm PMSF, 2 μg/mL each of leupeptin, aprotinin, antipain, pepstatin, and chymotrypsin, and 0.005% bromphenol blue. The samples were kept on ice for 15 min and centrifuged at 12,000g (15,000 rpm) for 1 min (Biofuge 15, Heraleus Sepatech GmbH, Hanau, Germany). The supernatant was loaded onto a 10% SDS-polyacrylamide gel using the Bio-Rad Mini-Protean II system (Laemmli, 1970).
RESULTS
pma4 Supports Yeast Growth at Low pH
A cDNA clone for the N. plumbaginifolia pma4 gene was placed on a yeast 2μ-derived multicopy plasmid under the control of the strong and constitutive transcriptional promoter of PMA1, the major S. cerevisiae H+-ATPase gene. The plasmid was introduced into the yeast strain YAK2 deleted from its own two H+-ATPase genes, PMA1 and PMA2, and surviving on Gal medium with a centromeric plasmid carrying the yeast PMA1 controlled by the GAL1 promoter. Independent transformants obtained in a Gal medium were still able to grow when shifted to a Glu medium, indicating that the plant H+-ATPase allowed yeast growth. To eliminate the potential residual expression of yeast PMA1 under control of the GAL1 promotor and any recombination between the yeast PMA1 and the plant pma4, we induced the loss of the plasmid bearing the yeast PMA1 by spreading the transformants on a Glu medium containing 5-fluoroorotic acid, which becomes toxic in the presence of the URA3 gene present in the latter plasmid. Colonies (YAKpma4) growing in the presence of the drug lost the URA3 plasmid containing the yeast PMA1 gene, as confirmed by Southern-blot analysis (data not shown).
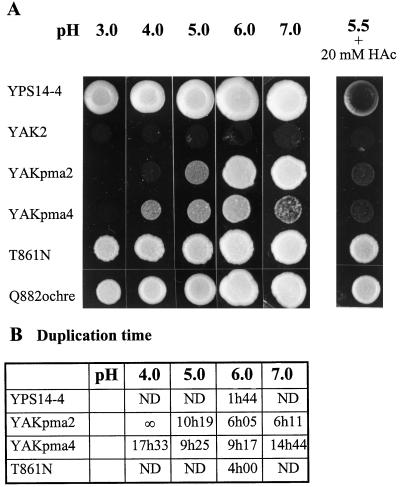
The plant H+-ATPase PMA4 isoform was thus able to sustain growth of cells devoid of yeast H+-ATPase. Moreover, it allowed yeast growth at a pH as low as 4.0 (YAKpma4; Fig. 1A), whereas PMA2, a N. plumbaginifolia isoform belonging to the first subfamily, could replace the yeast H+-ATPase only if the pH medium was kept above 5.0 (YAKpma2; Fig. 1A). However, at a more alkaline pH (7.0) PMA2 still allowed yeast growth, whereas PMA4 barely allowed growth. These observations were confirmed by measurement of the duplication time in liquid cultures (Fig. 1B). At the optimal pH (6.0), YAKpma2 and YAKpma4 grew 3.4- or 5.3-fold slower, respectively, than a wild-type yeast. At a lower pH, however, YAKpma4 grew better than YAKpma2, confirming its capacity to better sustain a low pH.
Figure 1.
Growth at different pH levels of yeast cells expressing wild-type and mutant plant plasma membrane H+-ATPases. A, The wild-type YPS14-4 strain (yeast PMA1 under its own promoter), the recipient strain YAK2 (yeast PMA1 under the GAL1 promoter), YAKpma2, YAKpma4, and the mutants T861N and Q882ochre were grown in YGlu medium at pH 6.5 and spotted onto solid YGlu medium at pH 3.0, 4.0, 5.0, 6.0, and 7.0, and at pH 5.5 in the presence of 20 mm acetic acid (HAc). B, The indicated strains were inoculated in liquid YGlu medium at pH 4.0, 5.0, 6.0, and 7.0, and the cell number was scored periodically. The duplication time was calculated during the exponential phase. ND, Not determined; ∞, no division.
Single Mutations of pma4 Conferred Growth at Low pH
We recently showed that point mutations in various domains of N. plumbaginifolia pma2 expressed in yeast improved the H+-pumping activity and conferred growth at pH 4.0 (Morsomme et al., 1996, 1998). Because the wild-type N. plumbaginifolia pma4 already allowed yeast growth at pH 4.0, we searched for mutants able to grow at pH 3.0. When independent colonies were cultured at a permissive pH (5.5) and streaked on selective plates at pH 3.0, five spontaneous mutants were obtained. The plasmid containing the pma4 gene from YAKpma4 and each of the selected mutants was retrieved and reintroduced into E. coli. The coding sequence of pma4 in the wild type and each of the mutants was then determined using a series of synthetic primers. In three cases, no modification of the pma4 gene was found, indicating that genetic modification occurred elsewhere in the plasmid or in the host genome (extragenic mutation). However, a single nucleotide change within the pma4 gene did occur in another two mutants (intragenic mutation). In the mutant T861N, a point mutation (C to A) transformed the amino acid Thr-861 (ACT) into Asn (AAT), whereas in the mutant Q882ochre, a point mutation (C to T) created a stop codon at codon 882 (CAG to TAG), resulting in an H+-ATPase shortened of the last 71 amino acids.
To determine whether the two mutations detected in the plant pma4 were indeed responsible for improved growth, the retrieved plasmids containing wild-type or mutant plant pma4 genes were reintroduced into the YAK2 strain, which was then depleted of the centromeric-URA3 plasmid containing the yeast H+-ATPase gene, as described above. The yeast strains containing wild-type and mutant PMA4 were tested for growth at various pH levels. Unlike YAKpma4, the two mutants grew at pH 3.0 and 7.0 (Fig. 1A). A preliminary interpretation of these data suggested that the mutations that had occurred in T861N and Q882ochre improved H+-ATPase functioning. To test this hypothesis, we grew the different strains at pH 5.5 in the presence of 20 mm acetate. This weak acid is thought to acidify the cytosol and prevent the growth of cells containing partially defective H+-ATPase that is unable to create a strong electrochemical H+ gradient across the plasma membrane (McCusker et al., 1987). Only YPS14-4, a wild-type yeast strain expressing its own H+-ATPase genes, and the two mutants T861N and Q882ochre were able to grow in the presence of acetate (Fig. 1A), suggesting that their H+-ATPases function better.
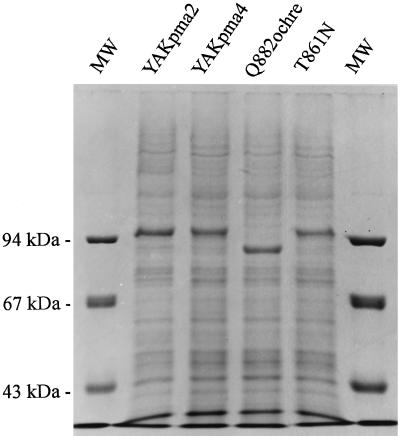
We have seen so far that the plant pma4 allows yeast growth at a lower pH than pma2 and that two pma4 mutants allow growth at an even lower pH. These different growth properties could be attributed to distinct amounts of H+-ATPase assembled in the plasma membrane or to distinct intrinsic enzyme properties. To clarify this point, plasma membranes were purified from the various transformants and the proteins were analyzed by SDS-PAGE (Fig. 2). Plant H+-ATPases appeared as the major band in the plasma membrane fraction. Their identity was confirmed by western-blot analysis (data not shown). The Q882ochre H+-ATPase had a lower apparent Mr (approximately 90,000), in agreement with the nonsense mutation identified. The pattern shown by this mutant also indicated that the major band observed in the other strains integrally represents the H+-ATPase. Considering the H+-ATPase amount, no large difference could be identified between the strains, except for a slightly larger quantity for YAKpma2 and Q882ochre. Therefore, better growth performances could not be explained by a larger amount of H+-ATPase in the plasma membrane. Therefore, we compared the kinetic properties of the H+-ATPase isoforms.
Figure 2.
Electrophoretic analysis of plasma membrane proteins of yeast cells expressing wild-type and mutant plant plasma membrane H+-ATPases. Purified plasma membranes (30 μg) prepared from YAKpma2, YAKpma4, and the two mutants Q882ochre and T861N were analyzed by electrophoresis on a 10% polyacrylamide gel and stained with Coomassie brilliant blue. Molecular mass markers (MW) are shown on the left in kilodaltons.
PMA2 and PMA4 H+-ATPases Have Distinct Enzymatic Properties
The apparent Km for MgATP and Vmax for ATP hydrolysis were determined on a purified plasma membrane fraction (Table I). We observed a higher Vmax for PMA4 compared with PMA2, whereas their Km values were not significantly different. Because the amount of PMA4 was slightly lower in the plasma membrane fraction (Fig. 2), we conclude that the molecular activity for the PMA4 H+-ATPase was clearly increased. A decrease of Km and a further increase of Vmax was observed for the two mutants derived from YAKpma4 (Table I).
Table I.
Kinetics of the wild-type and mutant plant H+-ATPases expressed in S. cerevisiae
| Strain | Km | Vmax |
|---|---|---|
| μm MgATP | μmol Pi min−1 mg−1 | |
| YAKpma2 | 312 ± 101 | 0.43 ± 0.14 |
| YAKpma4 | 246 ± 57 | 0.75 ± 0.15 |
| T861N | 189 ± 41 | 0.94 ± 0.05 |
| Q882ochre | 118 ± 21 | 1.47 ± 0.30 |
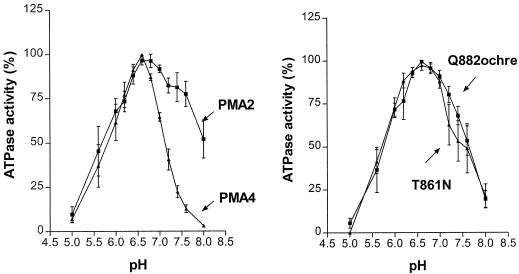
The pH dependence of H+-ATPase activity provided another means of distinguishing the isoforms. Both PMA2 and PMA4 had a similar pH optimum (around 6.6–6.8; Fig. 3). However, sensitivity to alkaline pH was much more acute in PMA4. For example, at pH 7.4 the activity of PMA2 was still around 80% of the optimum, whereas that of PMA4 decreased to less than 25%. The pH optima for both PMA4 mutants were not modified but their activity did not decrease as sharply as that of PMA4 at alkaline pH.
Figure 3.
pH dependence of ATP hydrolysis by purified plasma membrane fractions prepared from YAKpma2, YAKpma4, Q882ochre, and T861N measured as described in Methods at the pH values indicated. ATPase activity is expressed as the percentage of maximal activity observed for each strain. The mean specific activities at the optimal pH were: 0.46 (PMA2), 0.60 (PMA4), 0.69 (T861N), and 1.01 (Q882ochre) μmol Pi min−1 mg−1.
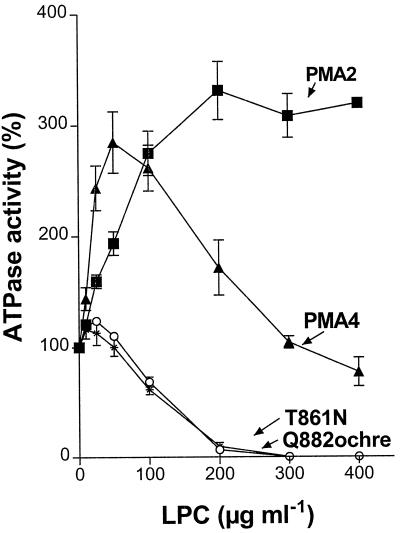
LPC is a phospholipid that specifically stimulates plant H+-ATPase (Palmgren et al., 1988). Maximal stimulation for PMA2 (about 3-fold) was obtained at 200 μg/mL LPC (Fig. 4). Stimulation of PMA4 occurred at a lower LPC concentration but decreased above 50 μg/mL LPC. A more dramatic effect was observed for both mutants; their H+-ATPase was only slightly increased at low LPC concentrations and became almost completely inhibited at 200 μg/mL LPC.
Figure 4.
Effect of LPC on H+-ATPase activity. ATP hydrolysis was measured as described in Methods on purified plasma membrane fractions prepared from YAKpma2, YAKpma4, Q882ochre, and T861N in the presence of the LPC concentrations indicated, and is expressed as the percentage of activity observed in the absence of LPC. Mean specific activities in the absence of LPC were: 0.44 (PMA2), 0.61 (PMA4), 0.65 (T861N), and 1.14 (Q882ochre) μmol Pi min−1 mg−1.
PMA2 and PMA4 H+-ATPases Confer Different Rates of External Acidification
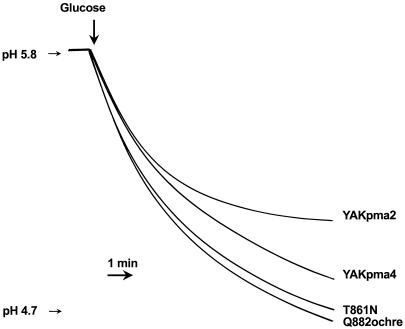
Acidification of the external medium of a yeast suspension has usually been interpreted as a reflection of H+-ATPase activity (Foury et al., 1977; Serrano et al., 1986). Therefore, we recorded the rate of acidification of the external medium induced by the addition of an energy source (Glu). Although Glu might also operate as a regulator of the yeast H+-ATPase (Eraso and Portillo, 1994), this is unlikely for the plant enzymes expressed in yeast, because sequences involved in Glu regulation in the yeast pump are not present in the plant enzymes. Despite the similarity in their initial rate, the steady-state level of acidification by the YAKpma4 strain exceeded that of YAKpma2 (Fig. 5). The mutants T861N and Q882ochre performed even better. These in vivo observations are in agreement with the phenotypes observed on solid media at different pH values (Fig. 1).
Figure 5.
Acidification of the external medium by yeast cells expressing plant H+-ATPase. Cells of YAKpma2, YAKpma4, T861N, and Q882ochre were grown in YGlu medium, washed, and incubated at 30°C for 5 min, and then Glu (250 mm) was added and the pH was recorded.
DISCUSSION
All of the cDNA encoding for plasma membrane H+-ATPases cloned so far from various organs of plant species fall into two subfamilies according to their predicted amino acid sequence identity. Therefore, we can assume that throughout the whole plant these subfamilies account for the major part of the expression of the H+-ATPase. It was not possible to determine directly by means of plasma membranes prepared from plant material whether the H+-ATPases encoded by these two subfamilies had similar enzymatic properties. Indeed, in N. plumbaginifolia (Perez et al., 1992; Moriau et al., 1993), as in tomato (Ewing and Bennett, 1994), transcripts for members of the two subfamilies were found in all of the organs analyzed. However, within an organ, expression might have been restricted to particular cell types, as has already been shown for several genes in Arabidopsis (De Witt et al., 1991; Harper et al., 1994) and N. plumbaginifolia (Michelet et al., 1994; L. Moriau, B. Michelet, P. Bogaerts, and M. Boutry, unpublished data). Heterologous expression in the yeast S. cerevisiae appeared to be an alternative method for comparing the basic properties of plant H+-ATPases.
The pma2 gene, as the most highly expressed from the first N. plumbaginifolia subfamily (Perez et al., 1992), had already been characterized in this way (de Kerchove d'Exaerde et al., 1995). Here we have shown that PMA4, the unique member of the second subfamily in N. plumbaginifolia, like PMA2, is able to replace the yeast H+-ATPase.
The present study provides several lines of evidence demonstrating that PMA2 and PMA4 possess distinct enzymatic properties. Kinetic differences were previously reported for three Arabidopsis H+-ATPases, AHA1, AHA2, and AHA3, which were also expressed in yeast (Palmgren and Christensen, 1994). The Km for ATP varied between 0.15 and 1.5 mm, and the Vmax varied between 1.25 and 2.45 μmol Pi min−1 mg−1. Their pH optimum was similar (6.4–6.5), but AHA3 was more sensitive to inactivation at lower pH values, whereas AHA2 showed a slightly more rapid decrease of activity on the alkaline side of the peak. AHA2 was stimulated by LPC to a larger extent (160%) than AHA1 (60%) or AHA3 (50%) (Palmgren and Christensen, 1994). The differences observed between N. plumbaginifolia PMA2 and PMA4 are also related to kinetics, pH profile, and LPC activation. An important observation was that PMA4, like AHA1, AHA2, and AHA3 (all four belong to the same subfamily), showed a sharp decline in activity above pH 7.0, whereas PMA2 had a much broader profile.
At the cellular level, the capacity to allow yeast growth seems to be a major difference between the Arabidopsis and N. plumbaginifolia H+-ATPases. Among the three AHA genes, only AHA2 was able to allow yeast growth (to a very low degree) when the yeast H+-ATPase controlled by a Gal-induced promoter was silenced by shifting the cells from a Gal to a Glu medium (Palmgren and Christensen, 1993, 1994). On the contrary, both N. plumbaginifolia pma2 and pma4 complemented well enough to allow definitive removal of the yeast H+-ATPase gene. The origin of this functional difference is not clear. It does not seem to be explained by differences in ATPase activity, because the Arabidopsis H+-ATPases displayed a 2- to 3-fold higher specific ATPase activity than the N. plumbaginifolia PMA2 or PMA4. However, a direct comparison has to be made with care because the Arabidopsis H+-ATPases were characterized in the ER-derived membranes where they accumulate, whereas PMA2 and PMA4 were analyzed in a plasma membrane fraction. A more appropriate comparison would thus require a quantitative analysis of H+-ATPase in the same membrane fraction. A different potential in H+-ATPase isoforms to be directed to the plasma membrane might have a direct consequence on their ability to allow yeast growth. Finally, we cannot exclude the possibility that more trivial factors such as minor differences in the expression system or growth media are responsible for the differences in behavior between the Arabidopsis and N. plumbaginifolia H+-ATPases.
The data clearly suggest that PMA4 performs better as an enzyme than PMA2: the Vmax at the optimum pH is higher and this is probably reflected at the cell level by the faster and higher acidification of the external medium. The physiological consequence of this is that pma4 still allowed yeast growth at pH 4.0, whereas pma2 did not. This begs the question of whether differences between PMA2 and PMA4 are linked to kinetic or thermodynamic properties. To answer this, H+ pumping and the H+:ATP ratio should be measured. For example, we might consider the possibility that PMA2 and PMA4 differ in their H+:ATP ratios. A ratio of 1.09 H+ pumped per ATP hydrolyzed was found for the red beet plasma membrane H+-ATPase (Briskin et al., 1995). However, this ratio might have been an average of data provided by several isoforms expressed in the same organ.
The cells expressing PMA4 grew more slowly at pH 7.0 than at pH 6.0, which was not the case for PMA2. If we assume that in the yeast transformant the pH of the growth medium exerted some influence on the internal pH (Smith and Raven, 1979), the difficulty that YAKpma4 had in growing at pH 7.0 might be explained by the sharp decrease in ATPase activity above pH 6.6 observed for PMA4, whereas the activity-versus-pH profile was much broader for PMA2.
A detailed examination of pma2 and pma4 expression at the cell level is being developed by means of a reporter gene. Our current observations are that both are expressed in various plant cell types, some of them common to both genes (L. Moriau, B. Michelet, P. Bogaerts, and M. Boutry, unpublished data). If this was confirmed at the protein level, it would indicate that two different isoforms might be found in the same cell. Such a situation might be interesting if a large H+-ATPase activity is required for activating intense transport, and also if different H+-ATPase isoforms must respond to separate regulatory systems. It has been proposed, and in some cases shown, that H+-ATPases are involved in many physiological functions, such as internal pH regulation, nutrient uptake, turgor control, and cell elongation, and are regulated by various endogenous or external factors (Serrano, 1989; Sussman, 1994; Michelet and Boutry, 1995; Palmgren, 1998).
An enzyme such as PMA4, which has a sharp alkaline pH profile, might be well adapted to the regulation of the cytosolic pH (Smith and Raven, 1979). The difference in sensitivity to LPC observed for PMA2 and PMA4 might be another sign of differential regulation. LPC is considered to be a natural regulator because its presence within the plasma membrane may result from the activity of phospholipase A2 (Palmgren et al., 1988). Phospholipases act within regulatory pathways such as that resulting in the wound response (Lee et al., 1997), and thus might also intervene in the regulatory signaling leading to H+-ATPase activation. The reduced stimulation (PMA4) or even the strong inhibition (T861N and Q882ochre) found after increasing the LPC concentration has not been observed for Arabidopsis wild-type or mutant H+-ATPase (Palmgren and Christensen, 1993, 1994). The physiological implications of this are not clear, although it might be an indication of differential regulation of ATPase isoforms. For example, although a low LPC concentration might be involved in displacing the C-terminal region (Palmgren and Christensen, 1993), a high LPC concentration might disrupt some interactions in the hydrophobic region of PMA4 and consequently reduce its activity.
We previously characterized PMA2 mutants that allow yeast to grow at pH 4.0 (Morsomme et al., 1996). Although the YAKpma4 strain could already grow at this pH, the discovery of two pma4 mutants that grew at pH 3.0 showed that there was still room for improvement. Like the majority of the mutants derived from YAKpma2, the two YAKpma4 mutants were modified in their C-terminal regions. We previously showed that the mutations in pma2 effectively changed the H+-ATPase into an activated conformation, possibly moving away from the C-terminal domain (Morsomme et al., 1998). This presumably also applies to the PMA4 mutants, because the mutation T861N in PMA4 is localized in one residue adjacent to the mutation R865T identified in PMA2 (Morsomme et al., 1996). Moreover, mutants of both enzymes lead to similar changes (e.g. increased enzyme activity, reduced LPC stimulation, and faster acidification of the external medium).
The improved activity of wild-type PMA4 compared with PMA2 could be explained by a change in conformation similar to that suggested for the mutants. Thus, compared with PMA2, PMA4 would be at a more active stage, but would still be subject to improvement by point mutations. The activity difference between PMA2 and PMA4 might also reflect differences in intrinsic properties between these enzymes that are not necessarily relevant to the regulatory C-terminal region.
ACKNOWLEDGMENTS
We are indebted to J. Nader and P. Gosselin for their excellent technical assistance, and to Dr A. de Kerchove d'Exaerde for helpful discussions during this study.
Abbreviation:
- LPC
lysophosphatidylcholine
Footnotes
This work was supported by the Interuniversity Poles of Attraction Program (Belgian State Prime Minister's Office, Federal Office for Scientific, Technical and Cultural Affairs), by the European Community's BIOTECH Program, and by the Belgian Fund for Scientific Research.
LITERATURE CITED
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Briskin DP, Basu S, Assman SM. Characterization of the red beet plasma membrane H+-ATPase reconstituted in a planar bilayer system. Plant Physiol. 1995;108:393–398. doi: 10.1104/pp.108.1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kerchove d'Exaerde A, Supply P, Dufour JP, Bogaerts P, Thinès D, Goffeau A, Boutry M. Functional complementation of a null mutation of the yeast Saccharomyces cerevisiae plasma membrane H+-ATPase by a plant H+-ATPase gene. J Biol Chem. 1995;270:23828–23837. doi: 10.1074/jbc.270.40.23828. [DOI] [PubMed] [Google Scholar]
- De Witt ND, Harper J, Sussman MR. Evidence for a plasma membrane proton pump in phloem cells of higher plants. Plant J. 1991;1:121–128. doi: 10.1111/j.1365-313x.1991.00121.x. [DOI] [PubMed] [Google Scholar]
- Eraso PE, Portillo F. Molecular mechanism of regulation of yeast plasma membrane H+-ATPase by glucose. J Biol Chem. 1994;14:10393–10399. [PubMed] [Google Scholar]
- Ewing NN, Bennett AB. Assessment of the number and expression of P-type H+-ATPase genes in tomato. Plant Physiol. 1994;106:547–557. doi: 10.1104/pp.106.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foury F, Boutry M, Goffeau A. Efflux of potassium induced by DIO-9, a plasma membrane ATPase inhibitor in the yeast Schizosaccharomyces pombe. J Biol Chem. 1977;252:4577–4583. [PubMed] [Google Scholar]
- Goffeau A, Dufour JP. Plasma membrane preparation from the yeast Saccharomyces cerevisiae. Methods Enzymol. 1988;157:528–533. doi: 10.1016/0076-6879(88)57101-x. [DOI] [PubMed] [Google Scholar]
- Harper JF, Manney L, Sussman MR. The Arabidopsis thaliana plasma membrane H+-ATPase multigene family. J Biol Chem. 1990;265:13601–13608. [PubMed] [Google Scholar]
- Harper JF, Manney L, Sussman MR. The plasma membrane H+-ATPase gene family in Arabidopsis: genomic sequence of AHA10 which is expressed primarily in developing seeds. Mol Gen Genet. 1994;244:572–587. doi: 10.1007/BF00282747. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee S, Suh S, Kim S, Crain RC, Kwak JM, Nam HG, Lee Y. Systemic elevation of phosphatidic acid and lysophospholipid levels in wounded plants. Plant J. 1997;12:547–556. [Google Scholar]
- McCusker JH, Perlin DS, Haber JE. Pleiotropic plasma membrane ATPase mutations of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:4082–4088. doi: 10.1128/mcb.7.11.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelet B, Boutry M. The plasma membrane H+-ATPase, a highly regulated enzyme with multiple physiological functions. Plant Physiol. 1995;108:1–6. doi: 10.1104/pp.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelet B, Lukaszewicz M, Dupriez V, Boutry M. A plant plasma membrane proton-ATPase gene is regulated by development and environment and shows signs of a translational regulation. Plant Cell. 1994;6:1375–1389. doi: 10.1105/tpc.6.10.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriau L, Bogaerts P, Jonniaux JL, Boutry M. Identification and characterization of a second plasma membrane H+-ATPase gene subfamily in Nicotiana plumbaginifolia. Plant Mol Biol. 1993;21:955–963. doi: 10.1007/BF00023594. [DOI] [PubMed] [Google Scholar]
- Morsomme P, Dambly S, Maudoux O, Boutry M. Single point mutations distributed in ten soluble and membrane regions of the Nicotiana plumbaginifolia plasma membrane PMA2 H+-ATPase activate the enzyme and modify the structure of the C-terminal region. J Biol Chem. 1998;273:34837–34842. doi: 10.1074/jbc.273.52.34837. [DOI] [PubMed] [Google Scholar]
- Morsomme P, de Kerchove d'Exaerde A, De Meester S, Thinès D, Goffeau A, Boutry M. Single point mutations in various domains of a plant plasma membrane H+-ATPase expressed in Saccharomyces cerevisiae increase H+-pumping and permit yeast growth at low pH. EMBO J. 1996;15:5513–5526. [PMC free article] [PubMed] [Google Scholar]
- Palmgren MG. Proton gradients and plant growth: role of the plasma membrane H+-ATPase. Adv Bot Res. 1998;28:1–70. [Google Scholar]
- Palmgren MG, Christensen G. Complementation in situ of the yeast plasma membrane H+-ATPase gene pma1 by an H+-ATPase gene from a heterologous species. FEBS Lett. 1993;317:216–222. doi: 10.1016/0014-5793(93)81279-9. [DOI] [PubMed] [Google Scholar]
- Palmgren MG, Christensen G. Functional comparisons between plant plasma membrane H+-ATPase isoforms expressed in yeast. J Biol Chem. 1994;269:3027–3033. [PubMed] [Google Scholar]
- Palmgren MG, Sommarin M, Ulvskov P, Jorgensen PL. Modulation of plant plasma membrane H+-ATPase from oat roots by lysophosphatidylcholine, free fatty acids and phospholipase A2. Physiol Plant. 1988;74:11–19. [Google Scholar]
- Perez C, Michelet B, Ferrant V, Bogaerts P, Boutry M. Differential expression within a three-gene subfamily encoding a plasma membrane H+-ATPase in Nicotiana plumbaginifolia. J Biol Chem. 1992;267:1204–1211. [PubMed] [Google Scholar]
- Serrano R. Structure and function of plasma membrane ATPase. Annu Rev Plant Physiol Plant Mol Biol. 1989;40:61–91. [Google Scholar]
- Serrano R, Montesinos C, Cid A. A temperature-sensitive mutant of the yeast plasma membrane ATPase obtained by in vitro mutagenesis. FEBS Lett. 1986;208:143–146. [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FA, Raven JA. Intracellular pH and its regulation. Annu Rev Plant Physiol. 1979;30:289–311. [Google Scholar]
- Supply P, Wach A, Thinès-Sempoux D, Goffeau A. Proliferation of intracellular structure upon overexpression of the PMA2 ATPase in Saccharomyces cerevisiae. J Biol Chem. 1993;268:19744–19752. [PubMed] [Google Scholar]
- Sussman MR. Molecular analysis of proteins in the plasma membrane. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:211–234. [Google Scholar]
- Treco DA. Basic techniques of yeast genetics. In: Ausubel FA, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley-Interscience; 1989. pp. 13.1.1–13.1.7. [Google Scholar]
- Villalba JM, Palmgren MG, Berberian GE, Ferguson C, Serrano R. Functional expression of plant plasma membrane H+-ATPase in yeast endoplasmic reticulum. J Biol Chem. 1992;267:12341–12349. [PubMed] [Google Scholar]
- Wach A, Ahlen J, Gräber P. The H+-ATPase of the plasma membrane of yeast. Eur J Biochem. 1990;189:675–682. doi: 10.1111/j.1432-1033.1990.tb15536.x. [DOI] [PubMed] [Google Scholar]