Abstract
Although many eukaryotic proteins are N-terminally acetylated, structural mechanisms by which N-terminal acetylation mediates protein interactions are largely unknown. Here we found that N-terminal acetylation of the E2 enzyme, Ubc12, dictates distinctive E3-dependent ligation of the ubiquitin-like protein, Nedd8, to Cul1. Structural, biochemical, biophysical, and genetic analyses revealed how complete burial of Ubc12’s N-acetyl-methionine in a hydrophobic pocket in the E3, Dcn1, promotes cullin neddylation. The results suggest that the N-terminal acetyl both directs Ubc12’s interactions with Dcn1, and prevents repulsion of a charged N-terminus. Our data provide a link between acetylation and ubiquitin-like protein conjugation, and define a mechanism for N-terminal acetylation-dependent recognition.
Many eukaryotic proteins are N-terminally acetylated (1–4). Genetic data underscore the importance of N-terminal methionine acetylation (1, 5–10), although specific interactions mediated by N-acetyl-methionine are largely unknown. We examined how N-acetyl-methionine can direct protein interactions by studying an E2 enzyme. E2s play central roles in E1>E2>E3 ubiquitin-like protein (UBL) conjugation cascades. First, an E2 transiently binds E1 for generation of a thioester-linked E2~UBL intermediate, which then interacts with an E3. For RING E3s, the UBL is transferred from E2 to an E3-associated target’s lysine, producing an isopeptide-bonded target~UBL complex. E2 core domains are sufficient for binding E1s and RING E3s (11). Contacts beyond E2 cores often mediate pathway-specific interactions. A unique N-terminal extension on Nedd8’s E2, Ubc12, binds both E1 and E3 (12–16). Nedd8 transfer from Ubc12 to cullins involves a “dual E3” mechanism (16): a RING E3, Rbx1, is essential for cullin neddylation; a co-E3, Dcn1, contains a “potentiating neddylation” domain (Dcn1P) thought to bind different Ubc12 and cullin surfaces (15–18). Notably, human Dcn1 acts as an oncogene (19).
Because Dcn1’s E3 activity was elusive with bacterially-expressed Ubc12 (14, 17), we asked whether in eukaryotes Ubc12 might be modified. Tandem mass spectrometry identified exogenous yeast (y) and human (h), and endogenous human Ubc12 as being N-terminally acetylated on Met1 (Fig 1A, Fig S1). Although mammalian N-terminal acetyltransferases (Nats) appear partially redundant (3), yNat specificities are well-defined (20). yUbc12’s N-terminal Met-Leu sequence was predicted to retain Met1 and be acetylated by the Mak3p-Mak10p-Mak31p complex comprising yNatC (21). Indeed, Mak3 gene deletion prevented yUbc12 N-terminal acetylation in yeast, and bacterially-expressed yNatC catalyzed yUbc12 N-terminal acetylation (Fig 1). Thus, yNatC performs yUbc12 N-terminal acetylation.
Figure 1. Ubc12 is N-terminally acetylated in eukaryotic cells.
A, LC-MS/MS spectrum from endogenous hUbc12’s N-terminal peptide after Glu-C digestion/desalting, indicating XCorr and ΔCN values, y (red) and b (blue) ions used to match peptide sequence. B, In vitro bacterially-expressed yNatC reactions with [14C]-Acetyl-CoA and Ubc12Met (free N-terminus) or Ubc12AcMet (pre-acetylated negative control). C, MaxEnt LC-TOF spectra of yUbc12-His6 purified from WT orΔmak3 yeast, or from coexpression with yNatC in E. coli. D, Immunoblot of yCul1 (Cdc53p) from indicated yeast strains. E, Genetic interactions between NatC subunit mak10 and cdc34-2.
To address whether Ubc12 N-terminal acetylation influences Nedd8 ligation, we examined yNedd8~yCul1 steady-state levels in yeast with Nat gene deletions. Only yeast lacking NatC components displayed decreased yNedd8~yCul1 (Fig 1D). Furthermore, loss of NatC activity was synthetically lethal in combination with the cdc34-2 temperature-sensitive allele (Fig 1E) - a hallmark for Nedd8 pathway components due to roles in yCul1/SCF-regulated cell division (14, 22). Thus, yUbc12 N-terminal acetylation is important for yCul1 neddylation and function in vivo.
In vitro, Ubc12 N-terminal acetylation dictated Dcn1P-mediated Nedd8 transfer to Cul1 in pulse-chase assays comparing N-terminally acetylated Ubc12AcMet, Ubc12Met (identical sequence but not N-terminally acetylated), and Ubc12GSMet (unacetylated with Gly-Ser-Met at the N-terminus) (16). yDcn1P E3 activity was substantially magnified for yUbc12AcMet, with lower yDcn1P-enhancement of yNedd8 transfer to yCul1 from yUbc12Met or yUbc12GSMet consistent with the residual neddylation in NatC null yeast (Fig 1D, 2A). hUbc12 N-terminal acetylation was absolutely required for hDcn1P-mediated potentiation of neddylation (Fig 2B, Fig S2). In all cases, Ubc12 N-terminal acetylation was specific for Dcn1P E3 activity, because Dcn1P-independent Rbx1-mediated transfer of Nedd8 to Cul1 was independent of the state of Ubc12’s N-terminus (Fig 2, Fig S2). Nonetheless, even in the presence of Dcn1P and Ubc12 N-terminal acetylation, Cul1 neddylation required Rbx1’s RING E3 activity and was blocked by the CAND1 inhibitor (Fig S2). Thus, in addition to roles of the acetylated Ubc12-Dcn1 E2–E3 complex, in vivo steady-state Cul1~Nedd8 levels may also reflect Dcn1- and acetylation-independent regulation.
Figure 2. Dependence of Dcn1 co-E3 activity on Ubc12 N-acetylation.
A, Pulse-chase [32P]~yNedd8 transfer from yUbc12 variants to yCul1 C-terminal domain (yCul1C) complexed with Rbx1 −/+ yDcn1P. B, Same as A, but with human proteins. C, Thermodynamic parameters for Ubc12 or its peptides binding to Dcn1P and E1 by ITC. NB = no binding.
To obtain mechanistic insights, Ubc12-Dcn1P interactions were quantified by isothermal titration calorimetry (ITC). N-terminal acetylation increased Ubc12’s affinity for Dcn1P by ~2 orders-of-magnitude, and this was recapitulated by synthetic Ubc12 N-terminal peptides, which also inhibited neddylation reactions (Fig. 2C, Fig S3, S4). In contrast, N-terminal acetylation had little effect on Ubc12 peptide binding to E1 (Fig. 2C, Fig S5).
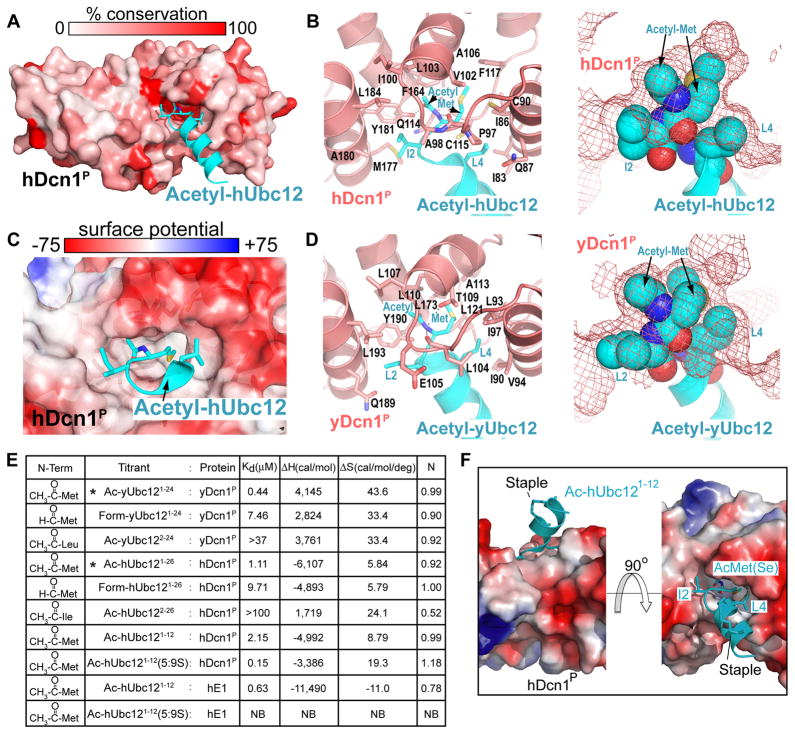
To understand how N-acetyl-methionine mediates interactions, we determined crystal structures of yeast and human Dcn1P bound to acetylated Ubc12 peptides (Table S1, Fig S6). As in prior structures, Dcn1P forms a helical domain containing two EF-hand-like folds (14, 16, 23). The Ubc12 N-terminal peptides are α-helical, as in full-length yUbc12GSMet (16). A Dcn1P groove at the junction between the two EF-hand-like subdomains cradles Ubc12’s helix, culminating in Ubc12’s N-acetyl-methionine burial in a conserved, deep, hydrophobic pocket in Dcn1P (Fig 3, Fig S7, S8).
Figure 3. Dcn1P recognition of Ubc12’s N-acetyl-methionine.
A, hCul1WHB (not shown)-hDcn1P-AcetylhUbc121–15 structure, with hDcn1P surface colored by conservation among human and yeast orthologs and Acetyl–hUbc121–15 peptide in cyan. B, Close-up of hUbc12’s acetylated N-terminus (cyan) binding hDcn1P (salmon) in cartoon (left) or hUbc12’s N-acetyl-Met1 and residues 2 and 4 as spheres in a mesh view of hDcn1P (right). C, Close-up of hUbc12’s acetylated N-terminus (cyan) binding hDcn1P surface colored by electrostatic potential. D, Same as B, but with yeast proteins. E, Thermodynamic parameters for Ubc12 peptide binding to Dcn1P by ITC. 5:9S refers to helical staple. *reference from 2C. NB = no binding. F, Solvent-exposure of helical staple in hUbc12 peptide (cyan) bound to hDcn1P (surface, colored by electrostatic potential).
N-acetyl-methionine recognition consists of three major components (Fig 3). First, the methyl portion of the acetyl group fits snuggly in a hydrophobic pocket. Second, the amide makes a hydrogen bond with a structurally conserved carbonyl oxygen from Dcn1. Third, the Met1 side-chain is also fully enwrapped by the hydrophobic pocket.
The structures suggest two mechanisms by which Ubc12’s N-terminal acetylation dictates binding to Dcn1P. First, the acetyl group interacts directly with Dcn1P. Second, acetylation eliminates an N-terminal positive charge, which would impede burial in the hydrophobic pocket. These concepts were substantiated by assaying Dcn1P binding to N-terminally formylated Ubc12 peptides, which lack the acetyl methyl but retain the amide and are uncharged. N-terminal capping via formylation did improve binding ~10-fold in comparison to unacetylated peptides, although the Kds were decreased ~9- and ~17-fold compared with the acetylated human and yeast peptides, respectively, highlighting the importance of the acetyl methyl (Fig 3E). Further agreeing with the structures, Ubc12’s N-terminal Met1 was also required to bind Dcn1P (Fig 3E).
Ubc12’s N-acetyl-methionine is sealed into Dcn1P’s hydrophobic pocket by Ubc12’s N-terminal helix positioning hydrophobic residues 2 and 4 (Fig 3). On one side, yLeu2/hIle2 buries the acetyl. On the other side, Leu4 seals the Met1 side-chain into place. Downstream residues from Ubc12’s helix also interact with Dcn1P (Fig S9).
Although yUbc12GSMet’s N-terminal extension is helical (16), we wished to test the structurally-observed role for the helix with human proteins because hUbc12Met’s N-terminal region forms an extended structure in complex with E1 (12, 13). A 2.0 Å resolution structure with a stapled-helix (24) peptide superimposed with unstapled hUbc12AcMet-hDcn1P, confirming solvent exposure of the staple (Fig 3F). Helical stapling improved binding to hDcn1P ~14-fold, largely due to decreasing the entropic cost (Fig 3E). Moreover, helical stapling eliminated E1-binding. Thus, locking the flexible hUbc12 N-terminal region into a helix contributes to the hDcn1P interaction.
Additional Dcn1P elements secure Ubc12’s N-acetyl-methionine in place. First, yDcn1’s Tyr190/hDcn1-Tyr181 clamps between Ubc12’s N-acetyl-Met1 and yLeu2/hIle2, pressing Ubc12’s N-acetyl-methionine into Dcn1P’s hydrophobic pocket. Second, the loop between Dcn1P’s E- and F-α-helices closes down on Ubc12’s N-acetyl-methionine. In prior yDcn1P structures lacking Ubc12(14, 16, 23) these elements are repositioned to occlude the hydrophobic pocket (Fig S10). yDcn1P apparently initially engages Ubc12’s acetylated N-terminus, and subsequently clamps it down. Conformational flexibility may account for yDcn1P’s low-level activity toward yUbc12 even without N-terminal acetylation.
Given yDcn1P’s structural malleability, we reasoned that mutations alleviating repulsion of an N-terminal charge might enhance yDcn1P’s low-level E3 activity toward yUbc12Met. The structure indicated that Asp substitutions for yDcn1P Leu110 or Leu173 would approach yUbc12Met’s N-terminus to balance the positive charge. Also, an Ala replacement for the Tyr190 “clamp” would not force a charged yUbc12Met’s N-terminus directly into the hydrophobic pocket. Indeed, the three Dcn1P mutants showed enhanced E3 activity specifically toward unacetylated yUbc12 (Fig 4A).
Figure 4. Structure-based Dcn1 mutant compensation for lack of Ubc12 N-terminal acetylation.
A, Pulse-chase [32P]-yNedd8 transfer from yUbc12Met (top) or yUbc12AcMet (bottom) to yCul1C-yRbx1 with structure-based yDcn1P mutants (note different time-courses). B, Thermodynamic parameters for binding between yDcn1P or the Tyr190Ala mutant to unacetylated and acetylated yUbc12. *reference from 2C. C, Immunoblots for yCul1 (Cdc53p, top) or HA-tag (bottom) from mid-log whole cell extracts from Δdcn1 or Δdcn1/Δmak10 yeast harboring empty, WT Dcn1-HA, or Y190A Dcn1-HA expression vectors.
We asked whether a structure-based mutation could compensate for in vivo defects in cullin neddylation resulting from loss of NatC-mediated yUbc12 acetylation. Thus, we expressed HA-tagged Dcn1 or the Tyr190Ala mutant (we could not express comparable levels of the other mutants in yeast), in strains deleted for Dcn1 alone, or both Dcn1 and the NatC subunit Mak10. As with the in vitro enzymology and improved binding, the Tyr190Ala mutant rescued the defect in yCul1~yNedd8 conjugate formation that resulted from lack of NatC activity (Fig 4).
We showed N-terminal acetylation of Ubc12 to be an avidity enhancer, contributing a critical interaction within a highly interconnected neddylation complex. As only part of molecular recognition within large multicomponent complexes, many interactions depending on N-terminal acetylation likely remain unknown, and may be auxiliary (2, 20, 25). Our study raises the question of whether rules dictating N-terminal acetylation determined evolution of interactions controlling functions of N-terminally acetylated proteins. Specificity may also involve proximal elements, such as Ubc12’s N-terminal helix. Because N-acetyl-methionine can be completely enwrapped in a hydrophobic environment where it would be unfavorable to bury the positive charge masked by acetylation, we propose that N-acetyl-methionine can serve as a distinctive residue type allowing burial of protein N-termini into hydrophobic pockets of interacting proteins. Such N-acetyl-methionine binding sites may serve as targets for small molecules disrupting these critical interactions.
Supplementary Material
Acknowledgments
This was supported by ALSAC/St. Jude, NIH, and HHMI (BAS), grants from NIH and Millennium Pharmaceuticals (JWH), and Damon Runyon Cancer Research Foundation (EJB). We thank S Gygi, I Kurinov, C Ralston, R Cassell, P Rodrigues, K Kodali, V Pagala, R Schekman, DW Miller, S Bozeman, DJ Miller, J Bollinger and C Rock for assistance/reagents/discussions. DCS, JKM, BAS, SJCRH have applied for a patent on uses of Ubc12 N-terminal acetylation for inhibiting neddylation.
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
RCSB structural accession codes: 3TDI, 3TDU, 3TDZ.
Author contributions: DCS, JKM, EJB designed, performed, analyzed experiments; DCS and BAS wrote the manuscript with all authors contributing; JWH and BAS advised and assisted on all aspects.
www.sciencemag.org, Materials and Methods, Figs. S1–S10, Table S1, References (26–44)
References and Notes
- 1.Arnesen T. PLoS Biol. 2011;9:e1001074. doi: 10.1371/journal.pbio.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnesen T, et al. Proc Natl Acad Sci U S A. 2009;106:8157–8162. doi: 10.1073/pnas.0901931106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Damme P, et al. PLoS Genet. 2011;7:e1002169. doi: 10.1371/journal.pgen.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yi CH, et al. Cell. 146:607–620. [Google Scholar]
- 5.Askree SH, et al. Proc Natl Acad Sci U S A. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanki T, et al. Mol Biol Cell. 2009;20:4730–4738. doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon SJ, et al. Proc Natl Acad Sci U S A. 2008;105:16653–16658. doi: 10.1073/pnas.0806261105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behnia R, Panic B, Whyte JR, Munro S. Nat Cell Biol. 2004;6:405–413. doi: 10.1038/ncb1120. [DOI] [PubMed] [Google Scholar]
- 9.Setty SR, Strochlic TI, Tong AH, Boone C, Burd CG. Nat Cell Biol. 2004;6:414–419. doi: 10.1038/ncb1121. [DOI] [PubMed] [Google Scholar]
- 10.Hwang CS, Shemorry A, Varshavsky A. Science. 2010;327:973–977. doi: 10.1126/science.1183147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye Y, Rape M. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang DT, et al. Nat Struct Mol Biol. 2004;11:927–935. doi: 10.1038/nsmb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang DT, et al. Nature. 2007;445:394–398. doi: 10.1038/nature05490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurz T, et al. Mol Cell. 2008;29:23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Kim AY, et al. J Biol Chem. 2008;283:33211–33220. doi: 10.1074/jbc.M804440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scott DC, et al. Mol Cell. 2010;39:784–796. doi: 10.1016/j.molcel.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurz T, et al. Nature. 2005;435:1257–1261. doi: 10.1038/nature03662. [DOI] [PubMed] [Google Scholar]
- 18.Huang G, Kaufman AJ, Ramanathan Y, Singh B. J Biol Chem. 2011 doi: 10.1074/jbc.M110.203729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarkaria I, et al. Cancer Res. 2006;66:9437–9444. doi: 10.1158/0008-5472.CAN-06-2074. [DOI] [PubMed] [Google Scholar]
- 20.Polevoda B, Sherman F. J Mol Biol. 2003;325:595–622. doi: 10.1016/s0022-2836(02)01269-x. [DOI] [PubMed] [Google Scholar]
- 21.Polevoda B, Sherman F. J Biol Chem. 2001;276:20154–20159. doi: 10.1074/jbc.M011440200. [DOI] [PubMed] [Google Scholar]
- 22.Lammer D, et al. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, et al. J Biol Chem. 2007;282:24490–24494. doi: 10.1074/jbc.C700038200. [DOI] [PubMed] [Google Scholar]
- 24.Bird GH, Bernal F, Pitter K, Walensky LD. Methods Enzymol. 2008;446:369–386. doi: 10.1016/S0076-6879(08)01622-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polevoda B, Sherman F. Biochem Biophys Res Commun. 2003;308:1–11. doi: 10.1016/s0006-291x(03)01316-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.