Abstract
CD81 has been described as a putative receptor for hepatitis C virus (HCV); however, its role in HCV cell entry has not been characterized due to the lack of an efficient cell culture system. We have examined the role of CD81 in HCV glycoprotein-dependent entry by using a recently developed retroviral pseudotyping system. Human immunodeficiency virus (HIV) pseudotypes bearing HCV E1E2 glycoproteins show a restricted tropism for human liver cell lines. Although all of the permissive cell lines express CD81, CD81 expression alone is not sufficient to allow viral entry. CD81 is required for HIV-HCV pseudotype infection since (i) a monoclonal antibody specific for CD81 inhibited infection of susceptible target cells and (ii) silencing of CD81 expression in Huh-7.5 hepatoma cells by small interfering RNAs inhibited HIV-HCV pseudotype infection. Furthermore, expression of CD81 in human liver cells that were previously resistant to infection, HepG2 and HH29, conferred permissivity of HCV pseudotype infection. The characterization of chimeric CD9/CD81 molecules confirmed that the large extracellular loop of CD81 is a determinant for viral entry. These data suggest a functional role for CD81 as a coreceptor for HCV glycoprotein-dependent viral cell entry.
Hepatitis C virus (HCV) is an enveloped, positive-stranded RNA virus classified in the family Flaviviridae. An estimated 170 million individuals worldwide are infected with HCV. Infection is associated with the development of chronic hepatitis, cirrhosis, and hepatocellular carcinoma. The principal site of virus replication is thought to be the liver; however, the specific cell types targeted by HCV remain unclear. Recent reports of HCV antigen detection in the chronically infected liver (43) and serum-derived virus infection of primary liver cell cultures (7) suggest that hepatocytes are the primary target cells in vivo. However, authors from several laboratories have suggested that HCV may infect a wider range of cell types, including B cells and cells of the monocyte/macrophage lineage within the central nervous system (14, 18, 32, 40). Since attachment of a virus to a target cell is determined by specific interactions between the viral glycoproteins (gp) and cell surface receptors, this suggests that liver-specific molecules may act as receptors for HCV.
HCV encodes two putative envelope gp's, E1 and E2, which are believed to be type I integral transmembrane proteins. Our understanding of gp maturation and virus assembly is limited by the lack of a tissue culture system supporting virus assembly and release. Hence, the mechanism(s) by which HCV enters target cells is currently unknown. In the absence of a cell culture system, surrogate assays have been developed to study HCV entry, including the expression of a truncated version(s) of the E2 gp (10, 29), E1E2-liposomes (15), vesicular stomatitis virus (VSV)-HCV pseudotypes (6, 22, 24), and viruslike particles expressed in insect cell systems (4, 42, 44). Truncated versions of E2 bind specifically to human cells and were used to identify interactions with a number of cell surface molecules. These include CD81 (10, 29), scavenger receptor class B type 1 (SR-BI) (35), and dendritic cell-specific intracellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN) (11, 20, 30). In addition, HCV purified from human plasma is associated with low-density lipoprotein (LDL), suggesting that the virus may use the LDL receptor to enter cells (1, 45). While these studies characterized gp-receptor binding, the significance of these receptor candidates for HCV entry has not been tested.
CD81 is a member of the tetraspanin membrane protein superfamily, whose members function to organize signaling complexes at the cell surface by association with other tetraspanins, integrins, and signaling proteins in a cell-type-dependent manner (19). Human CD81 was identified to interact with soluble HCV E2 and virus in serum and was proposed to play a role in HCV entry (10, 29). However, several reports suggested that soluble E2 cloned from diverse genotypes fails to interact with CD81 (34, 35, 38). Despite numerous studies on the CD81-E2 interaction, definitive data showing its role in HCV infection were missing due to the lack of a functional assay. Indeed, the role of CD81 as a putative receptor was questioned due to the ubiquitous expression of CD81 in vivo and the ability of CD81 in tamarins (2, 23), a monkey species known to be refractory to HCV infection, to bind E2, suggesting that CD81 expression is not the sole determinant of HCV species and/or tissue tropism.
Recent studies (3, 13) have reported on the generation of retroviral pseudotypes harboring HCV envelope gp's, allowing both HCV gp-dependent binding and entry to be studied. HCV pseudotypes infect human liver cell lines, and entry is pH dependent and can be neutralized by monoclonal antibodies (MAbs) specific for E2 (13). Pseudotype infection of both primary human hepatocytes and Huh-7 hepatoma cells was blocked by a recombinant soluble form of CD81 and a MAb specific for CD81, suggesting that CD81 is required for HCV pseudotype infection (3, 13). In this report, we show that HCV pseudotype infection of several target cells is CD81 dependent. This conclusion is supported by (i) the inhibition of HCV pseudotype infection by a MAb specific for CD81 and CD81-specific small interfering RNAs (siRNAs) and (ii) the conferment of permissivity of HCV pseudotype infection by expression of CD81. The large extracellular loop (LEL) of CD81 was found to be a critical determinant for viral entry.
MATERIALS AND METHODS
Cells.
Hos.CD4.R5 cells were obtained through the National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, and propagated in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) and 1 μg of puromycin/ml. HeLa, SW13, and 293-T cells (obtained from the American Type Culture Collection [ATCC]) and Huh-7 (gift of R. Lanford, Southwestern Foundation for Biomedical Research), Huh-7.5 (5), PLC/PR5 (gift of J. Garson, University College London, London, United Kingdom), HepH (ATCC), HH29 (gift of A. Schwartz, Washington University, Saint Louis, Mo.), Hep3B (ATCC), and HepG2 (gift of Y. Matsuura, Osaka University, Osaka, Japan) cells were propagated in DMEM-10% FBS. HepG2 cells were cultured on collagen type 1-coated tissue culture plastic. The mouse liver cell lines Hepa1-6, H2-35, and AML 12 were obtained from the ATCC and propagated according to ATCC instructions. SK Hep1 and FT0-2B (gifts of L. Reid, University of North Carolina School of Medicine, Chapel Hill, N.C.) were propagated in RPMI medium-10% FBS. Fet Hep1.3 cells (gift of H. Hsu, Washington University) were propagated in Williams E medium-10% FBS-5 μg of insulin/ml-5 μg of transferrin/ml. Human lymphoid cells (MT-2, Molt 4, Hut-78, and Daudi) and peripheral blood mononuclear cells (PBMC) (gift of C. Cheng-Mayer, Aaron Diamond AIDS Research Center, New York, N.Y.) were propagated in RPMI medium-10% FBS. Caco-2 cells and HT-29 cells (gifts of M. Poles, Aaron Diamond AIDS Research Center) were propagated in Eagle's minimal essential medium-10% FBS and McCoys 5A medium-10% FBS, respectively. All cells were grown at 37°C in 5% CO2.
Plasmids and antibodies.
The plasmids carrying HCV strain H and Con1 E1E2 gp's (polyprotein residues 171 to 746), SF162 gp160, and VSV G protein (VSV G) were described previously (13). Wild-type CD81 and CD81 mutants (T163A, F186L, E188K, and D196E) were described previously and were cloned into the BamHI/XhoI site of the lentiviral vector TRIP (12, 46). CD81 and CD9 LEL chimeras (CD81-9LEL and CD9-81LEL) were generated by four-primer PCR by switching the reciprocal nucleotide sequences encoding the CD81 (amino acids [aa] 116 to 201) and CD9 (aa 114 to 192) LELs. Both chimeric constructs were cloned into the lentiviral vector TRIP, and sequences were verified.
Murine MAbs used in this study included 5A6 (a gift of S. Levy, Stanford University, Stanford, Calif.), 1D6 (Serotec Ltd., Oxford, United Kingdom), and 1.3.3.22 (Santa Cruz, Santa Cruz, Calif.), specific for CD81; C3-3A2 (Ancell Immmunology Research Products), specific for CD9; and CLA1 (BD Biosciences), specific for SR-BI. Rat MAbs specific for HCV E2 were previously described (10).
Flow cytometric analysis.
Expression of CD81 was quantified as previously described (13). All cells were incubated with an irrelevant isotype-matched immunoglobulin G (IgG) or the antibody of interest, and the fluorescence signal(s) was used to establish threshold values of detection for the test MAbs. Analyses were performed using a FACScalibur flow cytometer (Becton Dickinson) and FlowJo software (Tree Star, San Carlos, Calif.).
Pseudotype production and infection.
Human immunodeficiency virus (HIV) pseudotypes were generated by cotransfection of 293-T cells with equal amounts of expression plasmids expressing the viral gp's or an empty vector and the pNL4-3.Luc.R−E− plasmid containing the env-defective proviral genome (9, 13). The supernatants were collected 48 h posttransfection, and HIV p24 antigen contents were assessed using a commercially available enzyme immunoassay (Coulter Beckman). Target cells were seeded onto 96-well plates (8 × 103 cells/well) 24 h before infection. Equal volumes of p24 antigen-normalized viral supernatants were diluted in 3% FBS-DMEM plus 4 μg of Polybrene/ml, and 100 μl was added per well for 4 h. Virus was removed, and the cells were incubated at 37°C for 72 h and lysed with 40 μl of cell lysis buffer (Promega)/well. Thirty-five microliters of lysate was tested for luciferase activity by the addition of 50 μl of luciferase substrate and measured for 10 s in a luminometer (Lumat LB 9507).
In antibody blocking experiments, target cells were incubated with anti-CD81 at 5 μg/ml (100 μl/well) for 30 min on ice and 100 μl of pseudotyped virus, diluted in 3% FBS-DMEM plus 4 μg of Polybrene/ml, and incubated for 4 h. Cultures were incubated at 37°C for 72 h, and luciferase activity was measured. MAb 11/20, specific for E2 (final concentration, 5 μg/ml), was incubated with pseudotype virus for 30 min at 37°C, and virus-ligand mixtures were tested for infectivity of target cells.
RNA interference assay.
Silencing RNAs (siRNAs) were designed with the following sense strand sequences (a complementary oligonucleotide was synthesized for each): CD81 siRNA (siCD81), 5′-ugauguucguuggcuuccuTT, and irrelevant siRNA (siIRR), 5′-ggcgcuuguggacauucugTT. Chemically synthesized RNA oligonucleotides were annealed, deprotected, and desalted as recommended by the manufacturer (Dharmacon) (33). Four nanomoles of RNA duplexes in annealing buffer (100 mM potassium acetate, 30 mM HEPES-KOH [pH 7.4], 2 mM magnesium acetate) was electroporated into 2.5 × 106 Huh-7.5 cells (5 pulses of 900 V with 1-s intervals on a BTX electroporator). Cells were propagated and tested for CD81 cell surface expression and for their ability to support pseudotype infection.
Transduction of cells to express CD81.
HepG2, HH29, and Huh-7.5 cells were plated at 8 × 105 cells per well on a 6-well dish and infected 24 h later with a packaged lentivirus expressing human CD81 (TRIP-CD81) at an approximate multiplicity of infection of 1 to 3 in 3% FBS-DMEM plus 4 μg of Polybrene/ml for 12 h. Cells were washed, trypsinized, and seeded at 8 × 103 cells/well on a 96-well plate, infected 48 h later with pseudotypes, and stained for cell surface CD81 expression. HepG2 cells were transduced with lentivirus-expressing wild-type and mutant human CD81 proteins and stained with the CD81-specific MAb 1D6 and phycoerythrin-conjugated secondary antibody, and positively stained cells were sorted using a FACSVantage sorter (Becton Dickinson). Expression-positive cell populations were infected with pseudotypes.
Soluble E2 binding to CD81-transduced HepG2 cells.
Binding of soluble E2661 to HepG2 cells expressing wild-type or mutant CD81 proteins was as previously described (10). Briefly, 2 × 105 cells were incubated with a saturating amount of E2-containing tissue culture supernatant at room temperature for 1 h, and unbound soluble protein was removed by washing. Cell-bound E2 was detected with rat anti-E2 MAb 6/1a and phycoerythrin-conjugated anti-rat IgG. Analyses were performed using a FACScalibur flow cytometer (Becton Dickinson) and FlowJo software (Tree Star).
RESULTS
Identification of cell types able to support HCV pseudotype infection.
The principal site of HCV replication is thought to be the liver, though authors from several laboratories have suggested that the virus may infect a wider range of cell types, including monocytes/macrophages and B cells (17, 28, 40). We were interested to use the HCV pseudotype system to determine which cell types could support HCV gp-mediated infection. A range of human and rodent liver, epithelial, and lymphoid cells were tested for their ability to support infection with HIV pseudotypes bearing no envelope gp (no Env), HCV E1E2 gp's (strains H and Con1), HIV SF162 gp160, and VSV G (Table 1). As expected, pseudotypes bearing SF162 gp160 infected only Hos.CD4.R5 cells and activated PBMC. HIV-VSV G infected most cell types, with the exception of PBMC and some lymphoid cell lines. HCV pseudotypes infected only four human liver cell lines (Huh-7, Huh-7.5, PLC/PR5, and Hep3B) among the cell lines studied, which included nine of human liver origin (Table 1). HIV strain H E1E2 demonstrated 50% tissue culture infective dose titers of 2 × 104, 2.2 × 103, and 1.2 × 103/ml for Hep3B, Huh-7, and PLC/PR5 cells, respectively. Consistent with a previous observation, the pseudotypes harboring strain H or Con1 HCV gp's showed similar infectivities in the permissive cell lines (13). HCV pseudotype infection was gp dependent since particles lacking gp's (no Env) failed to infect any cell type tested and HIV strain H E1E2 infection was neutralized by the anti-E2 MAb 11/20 (data not shown). While all of the human cell lines, with the exception of HepG2 and HH29, expressed CD81 to various levels, only four were permissive to HCV pseudotype infection (Table 1). In conclusion, HCV pseudotypes infect a subset of human liver cells that express CD81; however, CD81 expression alone is not sufficient to confer susceptibility to infection.
TABLE 1.
Susceptibility of cells to HCV pseudotype infection
| Cell line | Cell type | Human CD81 expressiona | Infectivity (RLU, 103) of pseudotypes expressing:
|
||||
|---|---|---|---|---|---|---|---|
| No Env | VSV G | SF162 | H E1E2 | Con1 E1E2 | |||
| Hos.CD4.R5 | Human osteosarcoma | 24.5 | 0.1 | 950.4 | 120.0 | 0.1 | 0.1 |
| Huh-7 | Human hepatoma | 48.0 | 0.1 | 4,986.9 | 0.8 | 68.6 | 58.3 |
| Huh-7.5 | Human hepatoma | 81.3 | 0.1 | 5,469.3 | 0.1 | 89.3 | 63.2 |
| HepG2 | Human hepatoma | 1.9 | 0.1 | 890.0 | 0.9 | 0.1 | 0.1 |
| PLC/PR5 | Human hepatoma | 108.0 | 0.1 | 3,553.3 | 0.1 | 9.4 | 7.5 |
| HepH | Human liver | 67.5 | 0.1 | 218.7 | 0.1 | 0.1 | 0.1 |
| HH29 | Human liver | 2.2 | 0.1 | 3,031.7 | 0.1 | 0.1 | 0.1 |
| Hep3B | Human liver | 49.4 | 0.7 | 5,123.6 | 0.1 | 120.3 | 89.5 |
| SK Hep1 | Human liver | 67.9 | 0.2 | 3,812.3 | 0.1 | 0.1 | 0.2 |
| Fet Hep1.3 | Human liver | 99.4 | 0.2 | 2,869.5 | 0.1 | 0.1 | 0.2 |
| Hepa1-6 | Mouse liver | NA | 0.1 | 410.7 | 0.2 | 0.1 | 0.1 |
| H2-35 | Mouse liver | NA | 0.1 | 232.1 | 0.2 | 0.2 | 0.1 |
| AML 12 | Mouse liver | NA | 0.1 | 332.6 | 0.1 | 0.1 | 0.1 |
| FT0-2B | Rat liver | NA | 0.1 | 109.1 | 0.1 | 0.1 | 0.1 |
| PBMC | Human peripheral blood, resting | NT | 0.1 | 0.1 | 0.1 | 0.1 | 0.1 |
| PBMC | Human peripheral blood, phytohemagglutinin activated | NT | 0.1 | 0.9 | 66.3 | 0.1 | 0.1 |
| MT-2 | Human T lymphoid | 95.6 | 0.1 | 913.1 | 0.1 | 0.1 | 0.1 |
| Molt 4 | Human T lymphoid | 67.3 | 0.2 | 3.9 | 0.1 | 0.1 | 0.1 |
| Hut-78 | Human T lymphoid | 171.5 | 0.1 | 2.6 | 0.1 | 0.1 | 0.1 |
| Daudi | Human B cell | 257.3 | 0.1 | 589.3 | 0.1 | 0.1 | 0.1 |
| HeLa | Human epithelial | 114.4 | 0.1 | 61.3 | 0.1 | 0.1 | 0.1 |
| 293-T | Human embryonal kidney | 189.4 | 0.1 | 1,300.1 | 0.1 | 0.1 | 0.1 |
| SW13 | Human colorectal adenocarcinoma | 76.4 | 0.1 | 559.2 | 0.2 | 0.1 | 0.1 |
| Caco-2 | Human colon adenocarcinoma | 45.9 | 0.1 | 250.3 | 0.1 | 0.1 | 0.1 |
| HT-29 | Human fibrosarcoma | NT | 0.1 | 1,156.8 | 0.1 | 0.1 | 0.1 |
Median fluorescence intensity, in relative fluorescence units. NA, not applicable; NT, not tested.
CD81 is required for HCV gp-dependent entry into human liver cells.
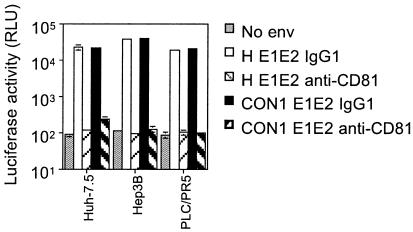
We next examined the requirement of CD81 for HIV-HCV infection of human cells by two independent approaches: (i) blocking infection with a CD81-specific MAb and (ii) silencing CD81 expression in Huh-7.5 cells with siRNAs. It has been previously reported that HIV-HCV pseudotype infection of Huh-7.5 cells can be inhibited by MAbs specific for CD81 and a recombinant soluble form of human CD81, suggesting that CD81 may be involved in mediating infection (13). We tested whether HCV pseudotype infection of the other permissive hepatoma cell lines was CD81 dependent. Target cells were incubated with either an irrelevant isotype control antibody or anti-CD81 MAb 5A6 and infected with pseudotypes bearing HCV H and Con1 gp's. The CD81 MAb inhibited HIV-HCV H and Con1 infection of Huh-7.5, Hep3B, and PLC/PR5 cells, suggesting a CD81-dependent route of infection for the HCV pseudotypes (Fig. 1).
FIG. 1.
Anti-CD81 MAb inhibits HCV pseudotype infection of Huh-7.5, Hep3B, and PLC/PR5 cells. Huh-7.5, Hep3B, and PLC/PR5 cells were incubated with anti-CD81 MAb (5A6) or an irrelevant isotype-matched IgG at 5 μg/ml and infected with pseudotypes bearing HCV H or Con1 gp's. Viral stocks were normalized for p24 HIV core antigen and used for infection at 1 ng/well. All infections were performed in triplicate, and the mean luciferase activity (in relative light units [RLU]) is shown. In this and subsequent figures, some standard deviation bars for RLU values are not visible due to compression from the log scale used, but all are within 10%.
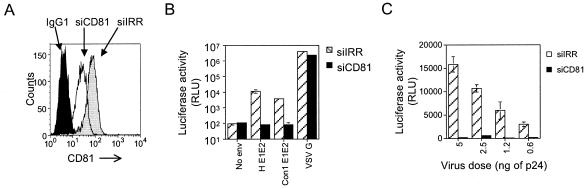
To further investigate whether CD81 expression in Huh-7.5 cells is required for HCV pseudotype infection, we silenced CD81 expression by the use of siRNAs. It was previously reported that siRNAs targeted to either HCV or lamin A/C effectively silence their targets in Huh-7.5 cells (33). Using this strategy, we designed siRNAs to specifically target CD81 (siCD81) or an irrelevant sequence (siIRR). siRNAs were transfected into Huh-7.5 cells, and 54 h later, the cells were harvested and examined for cell surface CD81 expression. Introduction of siCD81 resulted in a threefold reduction (median fluorescence intensity, 25 versus 71) in CD81 cell surface expression compared to that in cells transfected with siIRR (Fig. 2A). The levels of expression of an untargeted cell surface receptor, SR-BI, were equivalent in both transfected cell populations (data not shown). HIV-HCV H and Con1 infection was greatly reduced in siCD81-treated Huh-7.5 cells, whereas HIV-VSV G infection was unaffected (Fig. 2B). We tested the effect of silencing CD81 expression on infection with a range of concentrations of HIV-HCV H virus. siIRR-treated cells showed a linear increase in luciferase activity with increasing virus dose (Fig. 2C). Conversely, siCD81-treated cells showed minimal luciferase activity over background with the same range of virus inocula (Fig. 2C). These data, in combination with the CD81 MAb blocking data in Fig. 1, confirm that CD81 is required for HIV-HCV pseudotype infection of Huh-7.5 cells.
FIG. 2.
siRNA silencing of CD81 expression in Huh-7.5 cells inhibits HCV pseudotype infection. (A) Huh-7.5 cells transfected with either siIRR or siCD81 were stained with IgG1 or anti-CD81 MAb (1.3.3.22) at 54 h posttransfection and analyzed by flow cytometry. (B) siRNA-transfected Huh-7.5 cells were infected with the indicated pseudotypes and analyzed for luciferase activity. (C) siRNA-transfected Huh-7.5 cells were infected similarly with the indicated range of HIV-HCV H pseudotype inocula and analyzed for luciferase activity. All infections were performed in triplicate, and the mean luciferase activity is shown.
CD81 expression in HepG2 and HH29 liver cells confers susceptibility to HIV-HCV pseudotype infection.
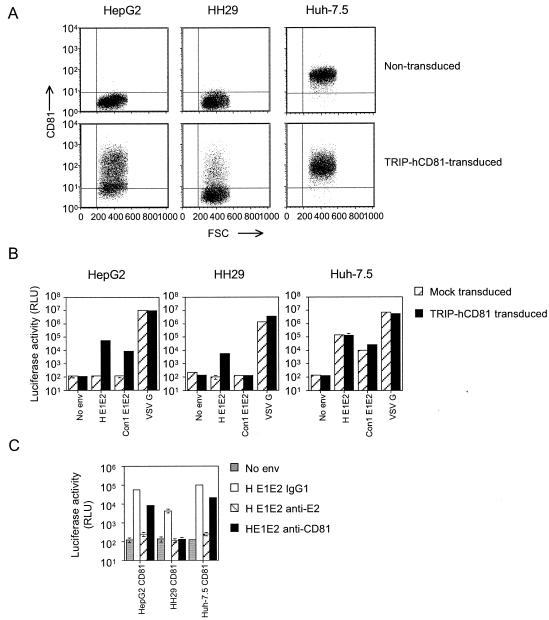
Our initial screening of cell lines identified HepG2 and HH29 as human liver cell lines that were both resistant to HIV-HCV infection and negative for cell surface CD81 expression. To confirm a direct role of CD81 in HCV pseudotype infection, we tested the ability of CD81 to confer susceptibility to infection of HepG2 and HH29 cells. Both cell lines were infected with a lentiviral vector encoding human CD81 and tested for their ability to support HCV pseudotype infection. Transduced HepG2 and HH29 cells expressed CD81 at the cell surface in 74.7 and 19.1% of the cells, respectively (Fig. 3A). CD81-transduced HepG2 cells were susceptible to infection with HIV-HCV H or Con1 (Fig. 3B). HIV-HCV H infected the CD81-expressing HH29 cells, albeit much less efficiently, while HIV-HCV Con1 failed to produce luciferase activity significantly above background (Fig. 3B). It is likely that the differences in susceptibilities to infection of HepG2 and HH29 cells reflects the different transduction efficiencies of the cell types. HIV-VSV G infection of both cell types was not affected by CD81 expression. To confirm that HCV pseudotype infection of the transduced cells was CD81 and HCV gp dependent, all infections were performed in the presence of the anti-CD81 MAb 5A6 and the neutralizing anti-E2 MAb 11/20 (13). HIV-HCV H infection of CD81-expressing cells was reduced by both MAbs, confirming the specificity of infection (Fig. 3C). The relative efficiency of the anti-CD81 blocking of pseudotype infectivity was lower on the transduced HepG2 and Huh-7.5 cells than on the HH29 cells, and this is most likely due to the high levels of CD81 expression after transduction.
FIG. 3.
Expression of CD81 in HepG2 and HH29 cells confers susceptibility to HCV pseudotype infection. (A) HepG2, HH29, and Huh-7.5 cells alone (upper panels) or after transduction with TRIP-CD81 (lower panels) were stained with an irrelevant IgG and anti-CD81 MAb (1.3.3.22) and analyzed for CD81 expression by flow cytometry. Quadrants were established based on irrelevant isotype-matched IgG staining of the cells. FSC, forward scatter. (B) Transduced (shaded bars) and nontransduced (hatched bars) HepG2, HH29, and Huh-7.5 cells were infected with the indicated pseudotypes 48 h posttransduction and analyzed for luciferase activity. (C) Transduced cells were incubated with either MAbs specific for CD81 (5A6) or an irrelevant isotype-matched IgG at 5 μg/ml and infected with HIV-HCV H E1E2 or with HIV-HCV H E1E2 preincubated with anti-E2 MAb 11/20. All infections were performed in triplicate, and the mean luciferase activity is shown.
To ascertain whether CD81 expression levels were rate limiting for HCV pseudotype infection of Huh-7.5, the cells were transduced with TRIP-CD81 and tested for their ability to support pseudotype infection. Transduced Huh-7.5 cells demonstrated a twofold increase in surface CD81 expression levels but no detectable change in their ability to support HIV-HCV H infection (Fig. 3B). However, HIV-HCV Con1 infection was increased more than twofold in the transduced Huh-7.5 cells (Fig. 3B). These data show that cell surface expression levels of CD81 in Huh-7.5 cells limit HIV-HCV Con1 infection but not HIV-HCV H infection, suggesting that these gp's may have different affinities for CD81. This conclusion is consistent with the inability of HIV-HCV Con1 to infect the TRIP-CD81-transduced HH29 cells (Fig. 3B).
Mapping the CD81 determinants for HCV pseudotype infection of HepG2 cells.
The LEL of CD81 has been shown to interact with HCV E2 (10, 29). The ability of CD81 to confer susceptibility to HIV-HCV infection on HepG2 cells provides an ideal system to test the determinants of CD81 required for viral infection. To determine whether the LEL domain of CD81 is critical for HCV pseudotype infection, we engineered a set of chimeras with CD81 and the related tetraspanin CD9 in which the LEL domains of CD81 and CD9 were exchanged (Fig. 4A). HepG2 cells were transduced to express CD9, CD81, CD9-81LEL, or CD81-9LEL and infected with pseudotypes bearing no gp (no Env), VSV G, or HCV H or Con1 gp's. Levels of cell surface expression of CD9, CD81, and the chimeras were comparable, and expression was detected in more than 95% of the cells (data not shown). Expression of CD9, CD81, or the chimeras did not affect the infectivity of HIV-VSV G. HCV pseudotypes failed to infect cells transduced with CD9 or CD81-9LEL, both of which express the CD9 LEL. Conversely, HCV pseudotypes infected cells expressing the CD81 LEL, both in the context of the native protein and as a chimeric CD9 molecule (CD9-81LEL) (Fig. 4B). We conclude that the CD81 LEL is critical for receptor activity.
FIG. 4.
The LEL of CD81 is the essential structural component for conferring CD81-dependent susceptibility to HCV pseudotype infection. (A) Schematic representation of CD81 (solid line), CD9 (dotted line), the chimeric molecules CD81-9LEL and CD9-81LEL, and the CD81 mutants with point mutations. (B) HepG2 cells expressing wild-type CD81 (CD81wt) and CD81 variants were infected with the indicated pseudotypes and analyzed for luciferase activity. All infections were performed in triplicate, and the mean luciferase activity is shown. CD9wt, wild-type CD9. (C) Binding of strain H and Con1 soluble E2 to HepG2 cells transduced to express wild-type CD81 and CD81 variants. Results are expressed as median fluorescence intensities. RFU, relative fluorescence units.
It was previously reported that soluble E2 failed to interact with African green monkey CD81, which differed from the human molecule at four amino acid residues within the LEL (12). Mutation of the human CD81 sequence at each of the four residues corresponding to the African green monkey sequence identified aa 186 to be critical for maintaining an interaction with soluble E2. The CD81 variants T163A, F186L, E188K, and D196E were expressed in HepG2 cells and tested for cell surface expression and their abilities to interact with soluble E2 and to support HIV-HCV pseudotype infection. All CD81 variants were expressed at comparable levels on the cell surface and in more than 95% of cells (data not shown). Cells expressing all of the CD81 variants, with the exception of F186L, interacted with HCV strain H E2, consistent with previous reports that the F186L mutation abolishes the interaction with E2 (Fig. 4C). CD81 E188K and CD81 D196E showed reduced binding to soluble E2. The Con1 strain of E2 showed minimal binding to HepG2 cells expressing wild-type and mutant CD81 proteins (Fig. 4C). In contrast to the soluble E2 binding, pseudotypes bearing H and Con1 gp's infected cells expressing wild-type and mutant CD81 proteins equivalently (Fig. 4B). All four mutants conferred susceptibility to infection of HepG2 cells by the HCV pseudotypes. This result, while surprising, highlights the limitations of studying soluble E2-CD81 interactions and suggests that the regions of CD81 which are critical for HCV pseudotype infection and soluble E2 interaction differ and that the former may extend beyond aa residues 186 and 188 and their immediately adjacent residues.
DISCUSSION
In this study, we show that CD81 is required for HCV gp-dependent infection with retroviral pseudotypes. HIV-HCV strain H and Con1 pseudotype infection of human liver hepatoma cell lines is blocked by a CD81-specific MAb, confirming that CD81 is involved in the infection of all susceptible cell types. Silencing CD81 expression in Huh-7.5 cells with siRNAs reduced HCV pseudotype infection, suggesting that CD81 is required for infectivity. The observation that CD81 expression in two previously nonpermissive human liver cell lines renders them susceptible to infection demonstrates that expression of CD81 in the appropriate target cell allows HCV gp-dependent pseudotype entry.
The traditional experiments for virus receptor characterization, such as antibody and soluble receptor blocking of virus infection, were supplemented with siRNA-mediated gene silencing. RNA interference has proven to be a valuable tool in studying the function of host genes, including viral receptors. HIV infection is precluded by the silencing of either of its coreceptors, CD4 or CCR5 (26, 31). Silencing of CD81 in Huh-7.5 cells produced a threefold reduction in total CD81 cell surface expression that significantly reduced HCV pseudotype infection. It is unclear why the residual CD81 is insufficient to promote viral entry. It is possible that a threshold of CD81 expression is required for viral entry or, alternatively, that distinct subpopulations of CD81 exist on the cell surface that are able to function as receptors. CD81 and other members of the tetraspanin family have been reported to reside within lipid rafts (8). One possibility is that CD81 within a receptor complex may be present in lipid rafts that are turned over with more-rapid kinetics and thus preferentially silenced. In support of this model, methyl-beta-cyclodextrin treatment of Huh-7 and Hep3B cells, which depletes cholesterol from the plasma membrane and thereby disrupts lipid rafts, specifically inhibited HCV pseudotype infection without affecting HIV-VSV G infection (data not shown).
The soluble form of HCV E2 has been used widely as a tool to identify interacting molecules and was used to clone CD81 and identify it as a receptor candidate (29). The truncated soluble form of strain H E2 has been reported to bind CD81 with 10−9 M affinity (27), whereas E2 proteins cloned from other genotypes show minimal interaction with CD81 (34, 38). In contrast, we demonstrate that pseudotypes bearing HCV gp's of strain H or Con1 infect cells in a CD81-dependent manner, despite the differing affinities of their soluble E2 for CD81 (Fig. 4). These data demonstrate that the interaction between soluble E2 and CD81 does not predict the CD81 dependence of HCV gp-mediated infection. This interpretation is reinforced by the observation that the CD81 F186L mutant, which was previously reported to disrupt the soluble E2-CD81 interaction, supported HCV pseudotype infection of HepG2 cells (12). In summary, these data suggest that pseudotype virus interaction with CD81 and other cellular molecules is more complex than the interaction of monomeric soluble E2 with CD81.
The specific role of CD81 in HCV pseudotype infection is yet to be defined. Tetraspanin molecules such as CD9 and CD81 have been implicated in the fusion between gametes, myoblasts, and virus-infected cells (16, 25, 36, 41). However, the mechanism(s) is unclear, and these molecules may play a role in facilitating the fusion reaction rather than a direct role of interacting with ligands on the cell surface. Silvie and colleagues recently demonstrated that CD81 expression on hepatocytes is essential for Plasmodium falciparum sporozoite infection, demonstrating that although ubiquitously expressed, CD81 can contribute to tissue-specific tropism (39).
The observation that retroviral pseudotypes bearing HCV gp's display a restricted tropism for cells of human liver origin is consistent with the liver being the primary reservoir for HCV replication in vivo and supports a model in which a liver-specific coreceptor(s) may contribute to the tissue specificity of HCV infection. The inability of HCV pseudotypes to infect lymphoid cells may reflect the phenotypes of the HCV strains being tested (H and Con1, genotype 1b), and future experiments will study the tropism of pseudotypes harboring gp's cloned directly from the PBMC of HCV-infected individuals. Although CD81 is required for HCV gp-mediated virus entry, CD81 expression alone is not sufficient to confer susceptibility to infection. Indeed, transgenic mice expressing human CD81 failed to support HCV infection, suggesting that CD81 is not the sole determinant of HCV tissue and species specificity (21). It was previously reported that several human cell lines (SW13, Hos, and U937) expressing CD81 and the other candidate HCV receptors, LDL receptor and SR-BI, were refractory to HIV-HCV pseudotype infection, suggesting that CD81 together with the other putative receptors is not sufficient for HCV gp-mediated infection. Since the only cell lines able to support HCV pseudotype infection are of liver origin, we propose that one or more liver-specific cell surface proteins function with CD81 as a receptor for HCV. Recent studies show that several virus families utilize receptors comprising more than one cellular protein to infect their host cells (37). Efforts to identify the liver cell-specific coreceptor molecule(s) and to further analyze the CD81-HCV pseudotype interaction will provide insights into the role of these molecules in the initial steps of HCV infection.
Acknowledgments
We are grateful to Hernan Jaramillo, Jack Hietpas, and James Fan for excellent technical support and to Pat Holst for obtaining many of the liver cell lines used in this study. We thank Mike Flint and Peter Balfe for reading the manuscript and for their helpful comments. We thank Shoshana Levy for antibody reagents.
J.Z., C.M.R., and J.A.M. are supported by the Greenberg Medical Research Institute and PHS grants CA57973 and AI40034. G.R. is supported by postdoctoral fellowship American Cancer Society Grant PF-02-016-01-MBC.
REFERENCES
- 1.Agnello, V., G. Abel, M. Elfahal, G. B. Knight, and Q. X. Zhang. 1999. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96:12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allander, T., X. Forns, S. U. Emerson, R. H. Purcell, and J. Bukh. 2000. Hepatitis C virus envelope protein E2 binds to CD81 of tamarins. Virology 277:358-367. [DOI] [PubMed] [Google Scholar]
- 3.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumert, T. F., S. Ito, D. T. Wong, and T. J. Liang. 1998. Hepatitis C virus structural proteins assemble into viruslike particles in insect cells. J. Virol. 72:3827-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buonocore, L., K. J. Blight, C. M. Rice, and J. K. Rose. 2002. Characterization of vesicular stomatitis virus recombinants that express and incorporate high levels of hepatitis C virus glycoproteins. J. Virol. 76:6865-6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castet, V., C. Fournier, A. Soulier, R. Brillet, J. Coste, D. Larrey, D. Dhumeaux, P. Maurel, and J. M. Pawlotsky. 2002. Alpha interferon inhibits hepatitis C virus replication in primary human hepatocytes infected in vitro. J. Virol. 76:8189-8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claas, C., C. S. Stipp, and M. E. Hemler. 2001. Evaluation of prototype transmembrane 4 superfamily protein complexes and their relation to lipid rafts. J. Biol. Chem. 276:7974-7984. [DOI] [PubMed] [Google Scholar]
- 9.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 10.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardner, J. P., R. J. Durso, R. R. Arrigale, G. P. Donovan, P. J. Maddon, T. Dragic, and W. C. Olson. 2003. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA 100:4498-4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higginbottom, A., E. R. Quinn, C. C. Kuo, M. Flint, L. H. Wilson, E. Bianchi, A. Nicosia, P. N. Monk, J. A. McKeating, and S. Levy. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 74:3642-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kato, N., T. Nakazawa, T. Mizutani, and K. Shimotohno. 1995. Susceptibility of human T-lymphotropic virus type I infected cell line MT-2 to hepatitis C virus infection. Biochem. Biophys. Res. Commun. 206:863-869. [DOI] [PubMed] [Google Scholar]
- 15.Lambot, M., S. Fretier, A. Op De Beeck, B. Quatannens, S. Lestavel, V. Clavey, and J. Dubuisson. 2002. Reconstitution of hepatitis C virus envelope glycoproteins into liposomes as a surrogate model to study virus attachment. J. Biol. Chem. 277:20625-20630. [DOI] [PubMed] [Google Scholar]
- 16.Le Naour, F., E. Rubinstein, C. Jasmin, M. Prenant, and C. Boucheix. 2000. Severely reduced female fertility in CD9-deficient mice. Science 287:319-321. [DOI] [PubMed] [Google Scholar]
- 17.Lerat, H., F. Berby, M. A. Trabaud, O. Vidalin, M. Major, C. Trepo, and G. Inchauspe. 1996. Specific detection of hepatitis C virus minus strand RNA in hematopoietic cells. J. Clin. Investig. 97:845-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lerat, H., S. Rumin, F. Habersetzer, F. Berby, M. A. Trabaud, C. Trepo, and G. Inchauspe. 1998. In vivo tropism of hepatitis C virus genomic sequences in hematopoietic cells: influence of viral load, viral genotype, and cell phenotype. Blood 91:3841-3849. [PubMed] [Google Scholar]
- 19.Levy, S., S. C. Todd, and H. T. Maecker. 1998. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol. 16:89-109. [DOI] [PubMed] [Google Scholar]
- 20.Lozach, P. Y., H. Lortat-Jacob, A. De Lacroix De Lavalette, I. Staropoli, S. Foung, A. Amara, C. Houles, F. Fieschi, O. Schwartz, J. L. Virelizier, F. Arenzana-Seisdedos, and R. Altmeyer. 2003. DC-SIGN and L-SIGN are high-affinity binding receptors for hepatitis C virus glycoprotein E2. J. Biol. Chem. 278:20358-20366. [DOI] [PubMed] [Google Scholar]
- 21.Masciopinto, F., G. Freer, V. L. Burgio, S. Levy, L. Galli-Stampino, M. Bendinelli, M. Houghton, S. Abrignani, and Y. Uematsu. 2002. Expression of human CD81 in transgenic mice does not confer susceptibility to hepatitis C virus infection. Virology 304:187-196. [DOI] [PubMed] [Google Scholar]
- 22.Matsuura, Y., H. Tani, K. Suzuki, T. Kimura-Someya, R. Suzuki, H. Aizaki, K. Ishii, K. Moriishi, C. S. Robison, M. A. Whitt, and T. Miyamura. 2001. Characterization of pseudotype VSV possessing HCV envelope proteins. Virology 286:263-275. [DOI] [PubMed] [Google Scholar]
- 23.Meola, A., A. Sbardellati, B. Bruni Ercole, M. Cerretani, M. Pezzanera, A. Ceccacci, A. Vitelli, S. Levy, A. Nicosia, C. Traboni, J. McKeating, and E. Scarselli. 2000. Binding of hepatitis C virus E2 glycoprotein to CD81 does not correlate with species permissiveness to infection. J. Virol. 74:5933-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meyer, K., A. Basu, and R. Ray. 2000. Functional features of hepatitis C virus glycoproteins for pseudotype virus entry into mammalian cells. Virology 276:214-226. [DOI] [PubMed] [Google Scholar]
- 25.Miller, B. J., E. Georges-Labouesse, P. Primakoff, and D. G. Myles. 2000. Normal fertilization occurs with eggs lacking the integrin alpha6beta1 and is CD9-dependent. J. Cell Biol. 149:1289-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novina, C. D., M. F. Murray, D. M. Dykxhoorn, P. J. Beresford, J. Riess, S. K. Lee, R. G. Collman, J. Lieberman, P. Shankar, and P. A. Sharp. 2002. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 8:681-686. [DOI] [PubMed] [Google Scholar]
- 27.Petracca, R., F. Falugi, G. Galli, N. Norais, D. Rosa, S. Campagnoli, V. Burgio, E. Di Stasio, B. Giardina, M. Houghton, S. Abrignani, and G. Grandi. 2000. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J. Virol. 74:4824-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pietschmann, T., and R. Bartenschlager. 2003. Tissue culture and animal models for hepatitis C virus. Clin. Liver Dis. 7:23-43. [DOI] [PubMed] [Google Scholar]
- 29.Pileri, P., Y. Uematsu, S. Compagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 30.Pohlmann, S., J. Zhang, F. Baribaud, Z. Chen, G. J. Leslie, G. Lin, A. Granelli-Piperno, R. W. Doms, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J. Virol. 77:4070-4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin, X. F., D. S. An, I. S. Chen, and D. Baltimore. 2003. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. USA 100:183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radkowski, M., J. Wilkinson, M. Nowicki, D. Adair, H. Vargas, C. Ingui, J. Rakela, and T. Laskus. 2002. Search for hepatitis C virus negative-strand RNA sequences and analysis of viral sequences in the central nervous system: evidence of replication. J. Virol. 76:600-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Randall, G., A. Grakoui, and C. M. Rice. 2003. Clearance of replicating hepatitis C virus replicon RNAs in cell culture by small interfering RNAs. Proc. Natl. Acad. Sci. USA 100:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roccasecca, R., H. Ansuini, A. Vitelli, A. Meola, E. Scarselli, S. Acali, M. Pezzanera, B. B. Ercole, J. McKeating, A. Yagnik, A. Lahm, A. Tramontano, R. Cortese, and A. Nicosia. 2002. Binding of the hepatitis C virus E2 glycoprotein to CD81 is strain specific and is modulated by a complex interplay between the hypervariable regions 1 and 2. J. Virol. 77:1856-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scarselli, E., H. Ansuini, R. Cerino, R. M. Roccasecca, S. Acali, G. Filocamo, C. Traboni, A. Nicosia, R. Cortese, and A. Vitelli. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 21:5017-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmid, E., A. Zurbriggen, U. Gassen, B. Rima, V. ter Meulen, and J. Schneider-Schaulies. 2000. Antibodies to CD9, a tetraspan transmembrane protein, inhibit canine distemper virus-induced cell-cell fusion but not virus-cell fusion. J. Virol. 74:7554-7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider-Schaulies, J. 2000. Cellular receptors for viruses: links to tropism and pathogenesis. J. Gen. Virol. 81:1413-1429. [DOI] [PubMed] [Google Scholar]
- 38.Shaw, M. L., J. McLauchlan, P. R. Mills, A. H. Patel, and E. A. McCruden. 2003. Characterisation of the differences between hepatitis C virus genotype 3 and 1 glycoproteins. J. Med. Virol. 70:361-372. [DOI] [PubMed] [Google Scholar]
- 39.Silvie, O., E. Rubinstein, J. F. Franetich, M. Prenant, E. Belnoue, L. Renia, L. Hannoun, W. Eling, S. Levy, C. Boucheix, and D. Mazier. 2003. Hepatocyte CD81 is required for Plasmodium falciparum and Plasmodium yoelii sporozoite infectivity. Nat. Med. 9:93-96. [DOI] [PubMed] [Google Scholar]
- 40.Sung, V. M., S. Shimodaira, A. L. Doughty, G. R. Picchio, H. Can, T. S. Yen, K. L. Lindsay, A. M. Levine, and M. M. Lai. 2003. Establishment of B-cell lymphoma cell lines persistently infected with hepatitis C virus in vivo and in vitro: the apoptotic effects of virus infection. J. Virol. 77:2134-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tachibana, I., and M. E. Hemler. 1999. Role of transmembrane 4 superfamily (TM4SF) proteins CD9 and CD81 in muscle cell fusion and myotube maintenance. J. Cell Biol. 146:893-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Triyatni, M., J. Vergalla, A. R. Davis, K. G. Hadlock, S. K. Foung, and T. J. Liang. 2002. Structural features of envelope proteins on hepatitis C virus-like particles as determined by anti-envelope monoclonal antibodies and CD81 binding. Virology 298:124-132. [DOI] [PubMed] [Google Scholar]
- 43.Verslype, C., F. Nevens, N. Sinelli, C. Clarysse, J. Pirenne, E. Depla, G. Maertens, J. van Pelt, V. Desmet, J. Fevery, and T. Roskams. 2003. Hepatic immunohistochemical staining with a monoclonal antibody against HCV-E2 to evaluate antiviral therapy and reinfection of liver grafts in hepatitis C viral infection. J. Hepatol. 38:208-214. [DOI] [PubMed] [Google Scholar]
- 44.Wellnitz, S., B. Klumpp, H. Barth, S. Ito, E. Depla, J. Dubuisson, H. E. Blum, and T. F. Baumert. 2002. Binding of hepatitis C virus-like particles derived from infectious clone H77C to defined human cell lines. J. Virol. 76:1181-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wunschmann, S., J. D. Medh, D. Klinzmann, W. N. Schmidt, and J. T. Stapleton. 2000. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J. Virol. 74:10055-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]