Abstract
The HET-s prion-forming domain from the filamentous fungus Podospora anserina is gaining considerable interest since it yielded the first well-defined atomic structure of a functional amyloid fibril. This structure has been identified as a left-handed beta solenoid with a triangular hydrophobic core. To delineate the origins of the HET-s prion-forming protein and to discover other amyloid-forming proteins, we searched for all homologs of the HET-s protein in a database of protein domains and fungal genomes, using a combined application of HMM, psi-blast and pGenThreader techniques, and performed a comparative evolutionary analysis of the N-terminal alpha-helical domain and the C-terminal prion-forming domain of HET-s. By assessing the tandem evolution of both domains, we observed that the prion-forming domain is restricted to Sordariomycetes, with a marginal additional sequence homolog in Arthroderma otae as a likely case of horizontal transfer. This suggests innovation and rapid evolution of the solenoid fold in the Sordariomycetes clade. In contrast, the N-terminal domain evolves at a slower rate (in Sordariomycetes) and spans many diverse clades of fungi. We performed a full three-dimensional protein threading analysis on all identified HET-s homologs against the HET-s solenoid fold, and present detailed structural annotations for identified structural homologs to the prion-forming domain. An analysis of the physicochemical characteristics in our set of structural models indicates that the HET-s solenoid shape can be readily adopted in these homologs, but that they are all less optimized for fibril formation than the P. anserina HET-s sequence itself, due chiefly to the presence of fewer asparagine ladders and salt bridges. Our combined structural and evolutionary analysis suggests that the HET-s shape has “limited scope” for amyloidosis across the wider protein universe, compared to the ‘generic’ left-handed beta helix. We discuss the implications of our findings on future identification of amyloid-forming proteins sharing the solenoid fold.
Introduction
The exact atomic structure adopted by amyloid fibrils is a topic of intense debate, as high molecular weights and the polymeric character and insolubility of amyloid fibrils remain obstacles for high resolution structure determination methods such as nuclear magnetic resonance (NMR) spectroscopy [1], [2], [3]. Several structural studies of peptide amyloid fibrils have shown that the fibrils are arranged in a “cross-beta” sheet, a pattern characterized by repetitive arrays of beta-sheets that are parallel to the fibril axis, with their strands perpendicular to the axis [1], [2], [3], [4], [5]. While atomic-resolution structures of the infectious fibrils for many prions and amyloid-forming proteins are still lacking, recent studies have presented the first well-defined atomic structure of a functional amyloid, based on amyloid fibrils of the HET-s yeast prion [6], [7].
The het-s gene locus has two antagonistic alleles, het-s and het-S, which encode for HET-s and HET-S, respectively, and which give rise to the compatibility phenotypes [Het-s] and [Het-S] [8], [9], [10]. In comparison to its polymorphic variant, HET-S, only HET-s undergoes a transition to an infectious prion state. The HET-s prion of the filamentous fungus Podospora anserina is involved in heterokaryon incompatibility, a programmed cell death reaction that regulates the fusion between genetically distinct individuals [8], [9], [10], [11]. HET-s is a 289 residue protein with an N-terminal domain (residues 1–227) and a prion-forming C-terminal domain (residues 218–289). The crystal structure of the HET-s N-terminal domain comprises an alpha-helical fold of 8–9 helices and a short two-stranded beta sheet [8]. The HET-s prion forming domain (PFD) is necessary and sufficient for amyloid formation in vitro, as well as prion propagation in vivo [8], [11], [12]. Fibrils formed from this PFD are described as a left-handed β-solenoid composed of four parallel, stacked pseudo-repeated β-helices; the pseudo-repeats are a result of one molecule forming two turns of the solenoid [6], [7]. The first three β-strands of each pseudo-repeat enclose a dense triangular hydrophobic core [6], [7]. In addition to intra- and inter-molecular hydrogen bonds between the pseudo-repeats, the solenoid structure is also stabilized by favourable side-chain contacts, such as salt bridges, between oppositely charged residues facing outside of the triangular core [6], [7].
Since its discovery, the HET-s solenoid, both in its native and fibrillar forms, has been well characterized [6], [7], [10], [11]. However, studies on the evolutionary analysis of this fold, and identification of possible homologs to HET-s, remain largely lacking, despite the observation that a structural homolog of HET-s contributes to efficient cross-seeding of the amyloid form [10]. Accordingly, analysis of the evolution of the complete HET-s protein may allow for the identification of newer, potential amyloid-forming proteins that can adopt the HET-s solenoid shape. To this end, we perform an exhaustive search for all homologs of the prion-forming solenoid, as well as the homologs to the HET-s N-terminal domain. Based on our findings, we perform an evolutionary analysis of both domains to determine when the solenoid fold arose in evolution, and its point of attachment to the HET-s N-terminal domain. Additionally, we identify and model structural homologs to the C-terminal solenoid fold, and we present an analysis of the conserved physicochemical properties we have observed in these generated solenoids, and how they compare to the current understanding of the β-solenoid structure. Our data sheds light on the relationship between the HET-s solenoid fold and understanding the amyloid disease state.
Methods
Datasets
We downloaded the NCBI NR (non-redundant database: 14,261,927 protein sequences, database assembly dated 5/31/2011) from ftp://ftp.ncbi.nih.gov/blast/db/FASTA/. The Podospora anserina proteome (21,408 sequences) was downloaded from the NCBI Taxonomy Browser [13] (Taxonomy ID 5145). An additional 99 fungal proteomes (including mitochondrial proteomes, where available) from finished and ongoing projects were downloaded from the Broad Fungal Genome Initiative [14]. The 100 proteomes (Supplementary Text S1) were grouped together into one in-house database (total of 715,255 protein sequences), and will be collectively referred to as BROAD throughout the manuscript.
Identification of HET-s homologs using sequence analysis
Using the genomes from NR and BROAD, we searched for homologs to the HET-s protein using (i) the N-terminal domain (residues 1–227), and (ii) the C-terminal prion-forming domain (PFD) (residues 218–289). For each query, sequence similarity searches were performed using Psi-blast [version 2.2.23] [15] with default parameters and masking for low complexity regions. Searches were performed until convergence was reached or up to a maximum of 20 iterations, whichever was earlier. Significant hits were considered with E value<0.0001.
HMMs (Hidden Markov Models) for each of the queried regions were generated using HMMER [version 3.0, March 2010] [16], based on blastp [version 2.2.23] [15], [17] hits of each query against the NR database. For the N-terminal domain, 86 hits were identified from which only significant hits (E<0.0001) were used to create the HMM (n = 52). For the PFD, separate HMMs were generated for significant hits (E<0.0001) to the PFD from blastp (n = 7) as well as psiblast (n = 12). HMMs were also generated using the entire sequences of all members that shared a conserved prion domain (n = 12), as indicated by CDART (Conserved Domain Architecture Retrieval Tool) [18]. The CDART sequences were also refined and an HMM was generated only from the subsequences that match the prion-forming domain itself. A final HMM for the prion domain was generated based on sequences of the HET-s_218–289 family from Pfam (PF11558) (n = 2) [19]. While such small number of sequences may raise concern about the quality of the resulting PFD HMMs, for HMMs generated from blastp, psiblast, or pfam multiple sequence alignments, we opted to generate these domain-specific HMMs to reduce the number false positive homologs to the solenoid fold when querying the HMM against NR, as opposed to relying on an HMM based on a multidomain (Nterm and Cterm PFD) sequence alignment. The pfam-based HMM is an extreme case of a “restricted” HMM, but which reflects on the highly restricted nature of the HET-s solenoid. Conserved protein domains were identified by querying the HMMs against the NR database to increase chances of detecting remote homologs to the Nterm and C-term PFD.
Identification of structural homologs based on protein fold recognition
All significant hits from Psiblast runs against NR and BROAD, as well as significant hits from HMMER searches were threaded against the HET-s solenoid [PDB: 2RNM] chains A–E, using pGenThreader [20]. Corresponding alignments of the significant hits were used to generate 3D models with MODELLER [21]. If needed, these alignments were modified based on sequence-alignments of the C-terminal region of HET-s and its homologs [10]. 500 models for each protein were generated and the best model was selected with the lowest Discrete Optimized Protein Energy (DOPE) score. Stereochemistry of the models was assessed using the PROCHECK summary [22] of EBI PDBsum [23]. Selected models were viewed and rendered in PyMOL [24]. The RMSD calculation between the generated model and 2RNM template was calculated based on a structural alignment using the ‘super’ function in PyMOL [24]. Where applicable, the presence of salt bridges at specific positions within the models was determined using the ESBRI Server [25].
Functional analysis of homologs
We downloaded a non-redundant set of ‘genetic’ single-chain domain protein sequences (n = 10,569) from ASTRALSCOP, based on PDB SEQRES records (release 1.75). This was the non-redundant set made such that all sequences in it have pairwise similarity ≤40%. Entire protein sequences of all the identified homologs to the prion-forming and N-terminal domains were searched against this dataset using Blastp [version 2.2.23] [15], [17]. Significant hits from ASTRALSCOP (E≤0.0001) were submitted to the SUPERFAMILY HMM search engine for further classification of protein domains and protein domain families [26], [27]. To search for HET-s/LopB (HeLo) domains specifically, an HMM was constructed based on a previously identified loss-of-pathogenicity (LopB) protein and HeLo domains (n = 24 sequences) [8], [28], and queried against the entire sequence of the N-terminal homologs identified from this study. Significant hits were selected based on a cutoff E≤0.0001. Protein sequences of identified structural homologs to the HET-s PFD were also searched against the Conserved Domain Database (38,392 PSSMs) using the NCBI CD-Search and Batch Web CD-Search Tools [29], [30], [31].
Phylogenetic analysis
The NCBI taxonomy browser [13] and the taxonomy common tree generation tool (http://www.ncbi.nlm.nih.gov/Taxonomy/CommonTree/wwwcmt.cgi) were used to determine the taxonomic lineage for identified homologs. Additional taxonomic trees were generated using the Interactive Tree of Life (iTOL) server [32]. PHYLIP v3.69 [33] was used to make neighbor-joining majority-rule consensus trees based on MUSCLE [34] multiple alignments. These trees were produced based on 100 replicates using the PHYLIP seqboot, protdist, neighbor, and consense programs. Briefly, 100 bootstrapped datasets were generated using seqboot. Bootstrapped datasets were then used as input into protdist, and distance matrices were generated for all sets using the Janet-Taylor-Thornton (JTT) matrix, with default parameters. Neighbor joining trees were generated based on these distance matrices using neighbor. Lastly, the consense tool was used to pick the final neighbor-joining bootstrapped tree. Selected trees were viewed using TreeDyn [35] within the Phylogeny.fr server [36]. Similarity matrices for N- and C-terminal domains of PFD homologs were generated based on the BLOSUM matrix using the EBI ClustalW [37] program, at default settings.
To make the neighbor-joining tree for phylogenetic analysis of horizontal transfer, we used the CLUSTALW [37] phylogenetic option, with 1000 bootstrap iterations. The tree was visualized using ProWeb tree server (www.proweb.org/treeviewer/).
Results
Identification of homologs to the HET-s domains
Homologs of the HET-s N-terminal and prion-forming domain (PFD) have been searched against the non-redundant database (NR) and genomes from the Broad Fungal Genome Initiative (here, termed ‘BROAD’), using Psiblast and HMMER as described in Methods . A total of 408 significant hits against both domains were observed, 217 hits were from NR and an additional 191 hits were from BROAD. In the initial comprehensive homology search, 29 hits were observed to match the prion-forming domain (PFD), and 400 hits matched against the HET-s N-terminal domain. Using Blastclust to remove identical sequences (100% identity cutoff), 16 hits to the PFD and 338 hits to the N-terminal domain are observed.
Evolution of the Prion-Forming Domain
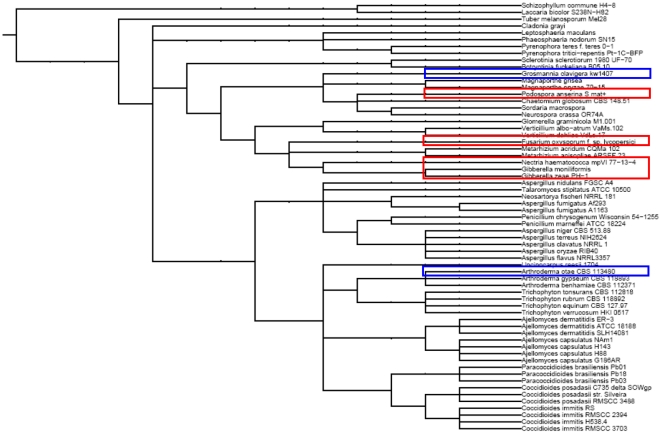
Despite the inclusion of the NR database, which represents all kingdoms of life, all the identified homologs of the prion-forming domain are restricted to the fungal kingdom, and they all belong to Saccharomyceta, more specifically, the Sordariomyceta ( Figure 1 ). Twenty-nine homologs to the PFD were identified using Psiblast and HMMer, in the initial comprehensive homology search. Manual curation to remove different genbank entries for the same gene (including provisional genbank entries), as well as removal of allelic variants with very high sequence similarity (>80% sequence identity) yielded 10 homologs to the PFD that were used in further evolutionary study (Supplementary Data S1). In addition to Podospora anserina, these 10 homologs were from 4 other fungal species, including Nectria haematococca mpVI 17-13-4, Fusarium oxysporum, Fusarium graminearum (Gibberella zeae), and Fusarium verticilliodes ( Figure 1 ). Almost all of these hits from our initial homology search have been previously identified as homologs to HET-s [37], with the exception of a newly identified homolog, EEU39630.1 [GI: 256726268] from Nectria haematococca mpVI 17-13-4.
Figure 1. Taxonomic lineage of homologs to the HET-s PFD.
The expanded taxonomic lineage of all species is presented. Actual species of HET-s PFD homologs are highlighted in bold and underlined. The non-significant PFD homologs from Arthroderma otae and Grosmannia clavigera (red boxes) are also included for comparison.
Interestingly, searching through non-significant hits to the HET-s PFD revealed the presence of newly-identified remote HET-s homologs that lend a more complete picture about the evolution of the HET-s PFD within fungi. We identified a HET-s homolog with a PFD domain in Grosmannia clavigera kw1407 [Genbank: EFX05012.1, GI: 320592582], which is a species that also belongs to the Sordariomyceta ( Figure 1 ). This protein was identified in the NR database with marginal significance levels (E< = 0.010 in psiblast iterations). Performing a reverse PSI-BLAST of this homologous PFD domain in the NR database yields a significant match to Podospora anserina HET-s residues 218–282 (E-value<0.005). We have also observed the presence of another small s protein annotation in Arthroderma otae CBS 113480 (anamorph: Microsporum canis CBS 113480), which is a more divergent Saccharomyceta species ( Figure 1 ). This protein was identified in both the NR [Genbank: XP_002843091,GI: 296804478] and BROAD (MCYG_08174) datasets with marginal significance levels in BROAD (E< = 0.030 in psiblast iterations). Unlike the PFD homolog identified in G. clavigera, which spans almost the entire length of the PFD (68 residues in G. clavigera compared to 72 residues in HET-s), the subsequence of A.otae matching against the PFD is much shorter (49 residues). By taking the segment in A.otae that matches only the PFD of HET-s, and performing a reverse PSI-BLAST with default parameters for short sequences, we find a significant match to Podospora anserina HET-s residues 271–289 (E-value<0.005). Interestingly, the N-term of the A.otae small s protein exhibits significant homology to the N-term of HET-s (E-value 2e-35 in a web-based search). Given that the remote homology of the A. otae segment to HET-s PFD is unlikely to occur beside a homology to the N-terminal HET-s domain, simply by chance, this marginally detectable homology likely indicates a horizontal transfer from the Sordariomycetes to Arthroderma otae (a Eurotiomycetes species). Indeed, the most similar sequences to the N-terminal domain of the A. otae protein come from the Sordariomycetes species P. anserina and Fusarium oxysporum (43% and 42% respectively, over 215 residues). Also, 6/10 of the most similar N-terminal domain sequences come from Sordariomycetes species, and not Eurotiomycetes). To investigate further this likely horizontal transfer, neighbor-joining phylogenetic analysis was performed on the N-terminal domains of HET-s orthologs that significantly align to the A. otae N-terminal domain protein sequence (Supplementary Figure S1). Regardless of the parameters used, the A. otae sequence always clusters with high bootstrap support (>80%) with the sequence from Fusarium oxysporum, within a larger grouping of Sordariomycetes sequences (green box in Supplementary Figure S1). Indeed, this is the only well-supported clustering between sequences from different phylogenetic fungal classes.
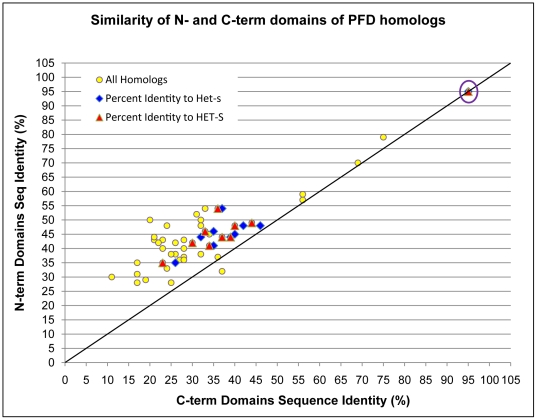
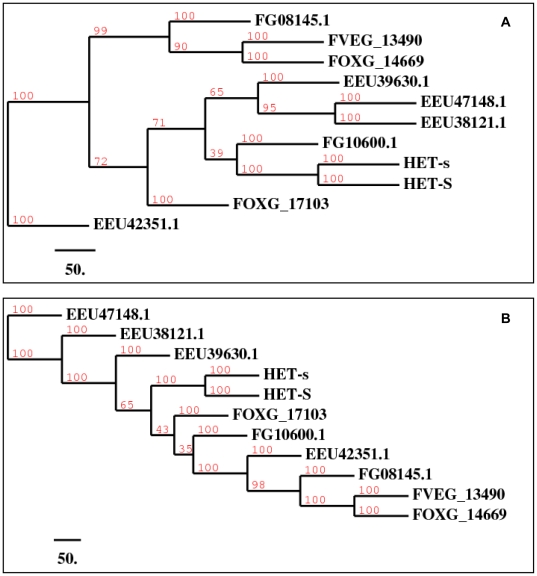
To compare the evolution of the N-terminal and C-terminal (prion-forming) domains that occur in the HET-s protein, we generated a similarity matrix for all proteins containing significant homologs of both HET-s domains (n = 11) ( Figure 2 , Table S1). We compared all pairwise similarities for the N-terminal domains to the corresponding pairwise similarities for the C-terminal PFD ( Figure 2 , Table S1). The plot clearly shows that the C-terminal PFD is evolving more rapidly that the N-terminal domain, with higher percentages of sequence identity between the N-terminal domains as opposed to the C-terminal domains, and only one pairwise comparison in disagreement amongst HET-s sequences from species other than Podospora anserina. Despite this, the majority-rule consensus neighbor-joining trees have similar clusterings of sequences (ignoring the tree branchings with <60% support) ( Figure 3 ). Taken collectively, the rapid evolution of the HET-s PFD we have demonstrated, coupled with the limited phyletic distribution of PFD homologs we have observed, suggests innovation of the PFD in Sordariomyceta, followed by rapid evolution in this domain, relative to the N-terminal domain. The additional marginal homolog in A. otae most likely arose by horizontal transfer, after innovation of the domain in Sordariomycetes.
Figure 2. Graphical representation of the similarity matrix between N- and C-terminal homologs of the PFD.
Each point on the graph represents the percent similarity of the C-terminal domains and the N-terminal domains for a pair of PFD homologs. In addition to HET-s, ten PFD homologs are represented. Pairs of homologs that include the HET-s or HET-S proteins have been colored differently for comparison. Comparison of Podospora anserina sequences to each other are circled (purple).
Figure 3. Phylogenetic trees of homologs to the HET-s prion-forming and N-terminal domains.
A) Phylogenetic tree of homologs to the HET-s prion-forming domain. B) Phylogenetic tree of homologs to the HET-s N-terminal domain. The generated trees are neighbor-joining majority-rule consensus trees composed of 11 sequences. Sequences starting with EEU represent Nectria haematococca mpVI 77-13-4, FOXG represent Fusarium oxysporum f. sp. lycopersici, FVEG represent Fusarium verticillioides (Gibberella moniliformis), and FG represent Fusarium graminearum (Gibberella zeae). Branch numbers indicate the number of times the partition of the species into two sets which are separated by that branch occurs among the trees, out of 100 trees, as described by Phylip consense program [33].
Distribution of the HET-s solenoid fold in HET-s homologs
Threading of all identified homologs to the HET-s N-terminal and PFD against the prion-forming solenoid [PDB: 2RNM] using pGenThreader [20] , identified 11 structural homologs from 5 species, almost all of which had already been previously identified in the sequential analysis ( Table 1 ). One of these homologs (FG10600.1) has been addressed in a previous publication and a model similar to HET-s has been proposed based on experimental analysis [10]. Two of the identified homologs (FOXG17103 and FOXG17314) are 100% identical and were considered henceforth as one model ( Table 1 ). Interestingly, in addition to these homologs that have been identified both by sequential and structural analysis, we also identified one further potential structural homolog through threading alone, i.e., TSTA_087480, in Talaromyces stipitatus ( Table 1 ). However, for this case, absence of other known homologs to TSTA_087480 precludes further bioinformatic analysis.
Table 1. HET-s homologs showing significant structural homology to the 2RNM solenoid.
| Threading Score | Accession Numbera | Protein | DBb | Structural Model | |||
| Template Chain | % Identityc | RMSDd | Prochecke | ||||
| LOW | [GI: 242774612] | Hypothetical protein, Talaromyces stipitatus, TSTA_087480 | NR | C | 17.7 | 0.816 | 88.1 |
| LOW | EEU47148.1 | Hypothetical protein, Nectria haematococca mpVI 77-13-4 | BROAD | A | 36.7 | 0.616 | 84.7 |
| LOW | EEU42351.1 | Hypothetical protein, Nectria haematococca mpVI 77-13-4 | BROAD | C | 31.6 | 0.736 | 82.8 |
| LOW | EEU39630 | Hypothetical protein, Nectria haematococca mpVI 77-13-4 | BROAD | C | 24.1 | 1.048 | 82.3 |
| MEDIUM | EEU38121.1 | Hypothetical protein, Nectria haematococca mpVI 77-13-4 | BROAD | A | 35.4 | 0.487 | 77.6 |
| LOW | FOXG14669 | Conserved hypothetical protein, Fusarium oxysporum | BROAD | C | 34.2 | 1.460 | 81.8 |
| LOW | FOXG17103 or FOXG17314 | Conserved hypothetical protein, Fusarium oxysporum | BROAD | C | 29.1 | 1.073 | 80 |
| LOW | FVEG13490 | Fusarium verticilliodes, hypothetical protein | BROAD | C | 26.6 | 1.172 | 80.7 |
| LOW | FG 08145.1[GI: 46127535] | Hypothetical protein, Fusarium graminearum | NR | D | 31.6 | 0.667 | 75 |
| MEDIUM | FG 10600.1[GI: 46138171] | Hypothetical protein, Fusarium graminearum | NR | A structure based on experimental analysis is proposed by Wasmer et al, 2010 [10] | |||
| LOW | [GI: 320592582] | Small s protein, Grosmannia clavigera kw1407 | NR | A | 26.6 | 0.492 | 83.3 |
a : The Genbank (GI) identification number from NR and BROAD accession numbers are provided, where available.
b : NR: non-redundant database, BROAD: Broad Fungal Genomes Initiative.
c : Percentage identity based on comparison with template in pGenThreader.
d : RMSD calculations are performed against the NMR model 9 of the [PDB: 2RNM] template.
e : Represents percentage of residues in the most favored region.
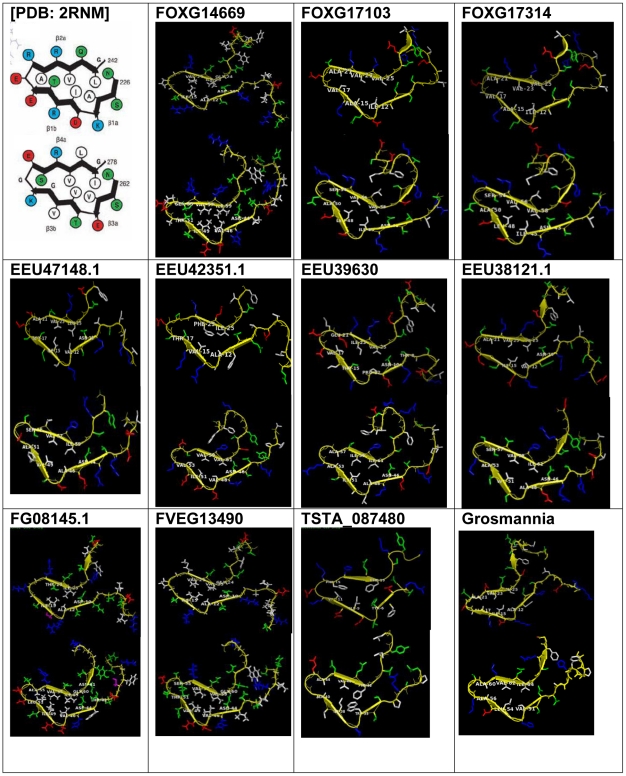
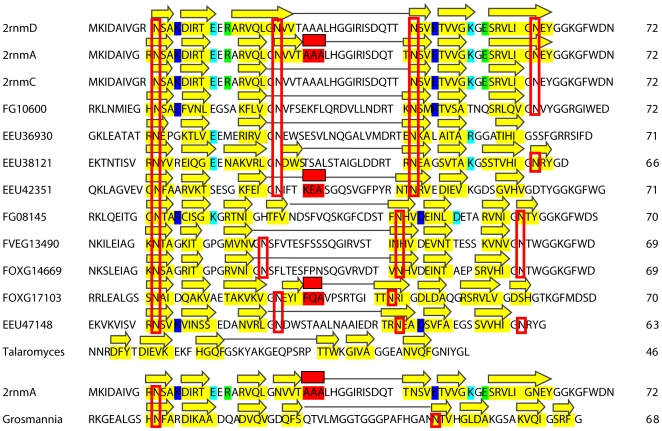
We were able to successfully generate solenoid structural models for all identified structural threadings of the C-terminal PFD using the MODELLER tool [21] and pGenThreader-generated sequence alignments ( Figure 4 ). The RMSD and PROCHECK [22] calculations of our generated models compare favorably against the template solenoid fold [PDB: 2RNM] ( Table 1 ). Similar to the HET-s PFD, the modeled proteins adopt a pseudorepetitive structure, where one chain is composed of two turns of the solenoid, in addition to a conserved triangular hydrophobic core with similar compositions of alanine (A) and the bulky hydrophobic residues of valine (V), isoleucine (I), and phenyalanine (F) ( Figure 4 , Figure 5 ). The asparagine ladder, as previously noted by Wasmer et al. [10] also remains largely conserved throughout the homologs ( Figure 5 ), although in some sequences, asparagines ladder residues are missing at the appropriate positions. Few of the models retain the ability for formation of a salt bridge pair at positions comparable to that of the 3 salt bridges of the PFD structure. Additionally, we have observed changes in the length of the pseudorepeats which may hinder the formation of a stable, repetitive fibril. For example, we have observed that the first pseudorepeat “rung” is shorter by 2 residues than the second rung in the homologs FVEG13490, FG08145, and FOXG14669. This length difference would yield an irregular fibrillar stacking of the solenoid.
Figure 4. Models of HET-s homologs with structural homology to the HET-s PFD.
The original solenoid [PDB: 2RNM] [6], [7] is shown in the top left corner. Ten structural homologs are represented, including the small s protein homolog from G. calivigera. The structure of FG10600.1 has been shown by Wasmer et al [10] and is not included here. For each structure, two rungs for each solenoid are represented, with the first rung at the top. Amino acids are color-coded as follows: acidic (Asp, Glu) in red, basic (Arg, Lys, His) in blue, nonpolor (Met, Phe, Pro, Trp, Val, Leu, Ile, Ala) in white, polar (Ser, Thr, Asn, Gln, Tyr) in green, and the protein backbone in yellow.
Figure 5. Conserved physicochemical properties of the HET-s structure in homologous solenoid models.
The three chains of 2RNM (A, C, D), used as templates in MODELLER, are represented (top). Beta sheet positions (as rendered in PyMOL) are highlighted in yellow (unless colored to represent other physicochemical properties), and helices are highlighted in red. The three salt bridge pairs (K229-E265, E234-K270, R236-E272) of HET-s are highlighted in dark blue, light blue, and light green, respectively. Asparagine ladders are represented in red boxes. The same coloring scheme has been adapted to the 11 generated models against the HET-s solenoid structure. The small s protein from G. clavigera (here, represented as Grosmannia), is also included for comparison against its 2rnm:A template. For all models, gapped positions have been removed for clarity, and the number of amino acids spanning the HET-s PFD are indicated (out of 72 residues). The secondary structure of each model has been placed above each sequence. Beta-strands are represented by yellow arrows and alpha-helices by red boxes. Blank spaces between the yellow arrows represent β arcs within each solenoid rung, and the long connecting loop between the two solenoid rungs in each model has been represented by a grey flat line.
We attempted to model structurally the small s proteins of the more divergent PFD sequence homologs from Grosmannia clavigera and Arthroderma otae, to determine if the conserved physicochemical properties of the HET-s structure could be observed in these marginal remote homologs. The small s protein from G.clavigera could easily be modeled against the solenoid structure, and similar to the other homologs, retains pseudorepeats, a conserved hydrophobic core, and asparagines ladders. Contrastingly, for the A.otae small s protein, all threading attempts using the entire sequence were ranked as “GUESS” in pGenThreader [20], with the exception of chain A of the solenoid structure [PDB: 2RNM], which ranked as “LOW” at 19% sequence identity. Interestingly, an unambiguous sequence alignment in the A. otae sequence could be generated for only one rung of the PFD solenoid (not shown), indicating perhaps that it comprises an obligate oligomer with a single solenoid rung.
Evolution of the HET-s N-terminal Domain across fungal clades
As opposed to the prion domain, which was likely innovated in Sordariomycetes, homologs to the HET-s N-terminal domain are more widespread within fungi ( Figure 6 ); however, the domain was not discovered outside of the fungal kingdom. As noted above, analysis of the N-terminal domains of the PFD homologs indicates that, while almost all of the domains share <50% identity with the HET-s or HET-S N-terminal domains, the sequence similarity between these domains still exceeds that of the PFDs ( Figure 2 ). Comparing the N-terminal domains of the homologs to one another also indicated that 8 pairs of homologous sequences (aside from those involving HET-s or HET-S) share >50% sequence identity, twice the number observed for the C-terminal PFDs (Table S1).
Figure 6. Taxonomic lineage of homologs to the N-Term domain.
Species with proteins homologous to the prion domain are highlighted in the red box. The marginal additional homologs observed in Grosmannia clavigera and Arthroderma otae are highlighted in the navy boxes.
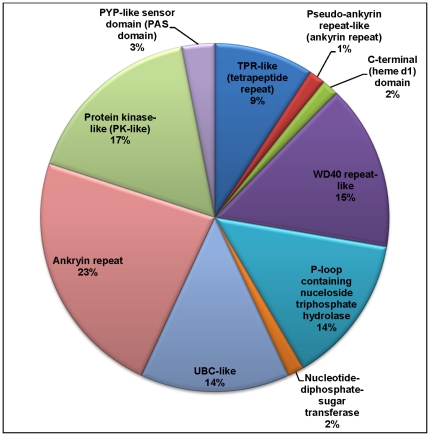
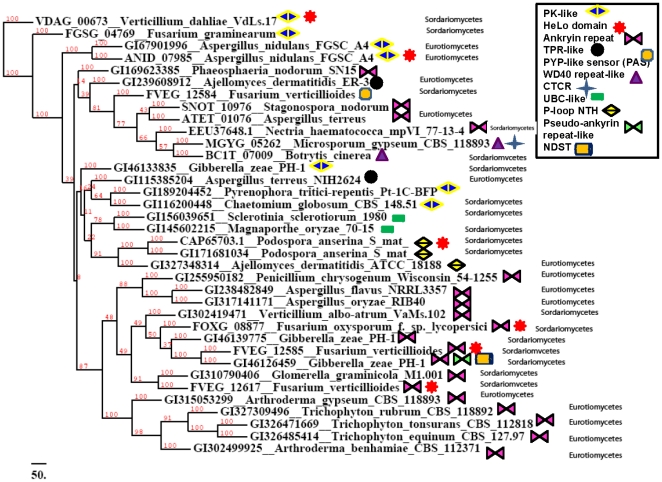
While an initial screen of the homologous sequences that contain the N-terminal HET-s domain indicates that many are labeled as hypothetical or predicted proteins, protein domain assignments reveal a wide diversity of domain architectures in HET-s homologs ( Figures 7 & 8 ). Forty HET-s homologs were mapped to 65 SCOP domains (Table S2, Table S3). Using the SUPERFAMILY HMM search engine [26], [27], these domains could be categorized into 10 superfamilies, with ankyrin being the most prevalent, followed by the WD40 repeat-like and the UBC-like domains ( Figure 7 ). A phylogenetic analysis of these 40 homologs indicates that the ankyrin repeat is largely predominant in Sordariomycetes ( Figure 8 ). Using HMMs, we also checked for the presence of HeLo (HET-s/LopB) domains in the entire sequences of identified homologs to the HET-s N-terminal domain, and we identified 212 HeLo domains in that set (Table S4). The HeLo domain had been previously identified based on >30% sequence similarity between the HET-s N-terminal domain and a fungal loss-of-pathogenicity (LopB) protein from Leptosphaeria maculans [8], [28]. In this study, we identified a second LopB protein [GI: 189205459] from Pyrenophora tritici-repentis Pt-1C-BFP with 30% similarity and 14% identity to the N-terminal domain. Searching for the conserved HeLo domains using the HMM also yielded a significant match to a HET-s/LopB domain from Metarhizium anisopliae ARSEF 23 [GI: 322703231, E-value 1.6e-10], as well as marginally significant matches [GI: 310797955, GI: 317157340, GI: 317033349] in several proteins from Glomerella graminicola, Aspergillus oryzae RIB40, and Aspergillus niger CBS 513.88, respectively [corresponding E-values 0.0042, 0.00082, 0.00083]. We visually inspected the remaining homologs of the N-terminal for any other HeLo domain-containing proteins and identified 3 more hits that are classified as containing a HeLo domain but which are not detected using the HMM ([GI:212532807] from Penicillium marneffei ATCC 18224, [GI:242776556] from Talaromyces stipitatus ATCC 10500, and [GI: 327353076] from Ajellomyces dermatitidis ATCC 1818).
Figure 7. Classification of 65 SCOP domains into superfamilies.
These are the SCOP domain superfamilies that co-occur with the HET-s protein domains. Ten superfamilies are represented.
Figure 8. SUPERFAMILY associations with the N-terminal homologs (n = 36).
A majority-rule consensus tree is generated for N-terminal homologs which are significantly associated with families identified using SUPERFAMILY. The clades of the different proteins are also annotated, especially for proteins which belong to ‘Sordariomycetes’ or ‘Eurotiomycetes’. Superfamilies associated with each protein are indicated, and are abbreviated as follows: CTCR: C-term (heme d1) of cytochrome reductase. P-loop NTH: P-loop containing nucleoside triphosphate hydrolase. NDST: Nucleoside-diphospho-sugar transferase.
Discussion
The HET-s solenoid remains the only atomic resolution of a fibril known to date, which raises an intriguing question of whether other amyloid-forming proteins that adopt the HET-s solenoid shape exist, and whether they can be identified. To probe this question, we have performed an exhaustive study for homologs of the HET-s prion-forming solenoid domain to identify potential amyloid-forming proteins that adopt such a shape in their native form or fibril states. Additionally, we investigated the evolutionary relationship between the prion-forming solenoid, and the HET-s N-terminal domain.
Our evolutionary analysis of the prion-forming domain reveals that the PFD, compared to the N-terminal domain, has limited phyletic distribution and has evolved rapidly. Despite the use of the NR database and multiple queries based on psi-blast and HMMs of the PFD, all results converge to the same set of homolog hits (n = 11). This indicated that a “restricted” profile HMM based on a small number of blast sequences has not influenced the results. Remote homologs to the P. anserina PFD were identified (in G. clavigera and A. otae), but with the exception of the remote homolog from A.otae, all the PFD homologs remain restricted to one fungal clade, Sordariomycetes. In several species, the HET-s homologs exist as paralogous gene families, as we observed a single HET-s protein in Podospora anserina, two in F. graminearum and four in N. haematococca. A comparison of the sequence similarities for the PFD and N-terminal domain of these homologs indicates a rapid divergence of the PFD compared to their companion N-terminal alpha-helical domains, as indicated by their sequence similarity matrix ( Figure 2 , Table S1). In stark contrast to the limited phyletic distribution of the PFD, we have identified a set of N-terminal homologs almost 14 times larger than the PFD homolog set, and not surprisingly, with a larger evolutionary spread within fungi ( Figure 6 ). Based on the phyletic distribution of these domains, the evolutionary point of attachment of the HET-s N-terminal domain and prion-forming domain can be attributed to Sordariomyceta, with a marginal homolog in A.otae that probably arose by horizontal transfer. Parsimoniously, horizontal transfer is a more likely event compared to multiple parallel gene loss events of the PFD in several fungal clades associated with the N-terminal domain.
The striking abundance and widespread phyletic distribution of homologs to the N-terminal domain implies that it may serve several functions beyond heterokaryon incompatibility and amyloidogenicity in many fungal species. Our protein domain assignment analysis of the homologous sequences that contain the N-terminal domain identified a wide diversity of protein domain partners. While many of the homologs to the N-terminal domain are hypothetical proteins, we have successfully identified 10 proteins superfamilies, based on SCOP and SUPERFAMILY, in 10% of our homolog dataset ( Figure 7 ). The most common superfamily is the ankyrin repeat, followed by the protein kinase-like (PK-like) domain, WD40 repeat-like, and UBC-like domains, among others. Interestingly, all of the above-mentioned families are involved in protein-protein interactions. The ankyrin repeat is of particular interest, as this repeat is predominant in the HET-s homologs in Sordariomycetes ( Figure 8 ). This repeat is a common protein-protein interaction motif found in a variety of functionally diverse proteins such as enzymes, toxins, and transcription factors [38]. Similarly, proteins containing WD40 or tetratricopeptide (TPR) repeats serve as platforms for protein complexes [39], [40], [41]; WD40 repeats are found in G proteins that participate in transmembrane signaling machinery, as well as proteins involved in RNA-processing complexes [39], [40].
In addition to protein-protein interactions, another underlying functionality we have observed, both in the HET-s N-terminal and prion-forming domains, is that of ‘pathogenicity’. While previous studies of the N-terminal homologs did not identify any homologs with a known function, a new HET-s/LopB (HeLo) domain had been identified based on a 31% similarity of the HET-s N-terminal domain to the loss-of-pathogenicity (LopB) protein from the Dothideomycete fungus Leptosphaeria maculans, a fungus that causes blackleg disease of Brassica napus [8], [28]. In current literature, 23 representative HeLo domains have been identified to date [8], [28]. We searched for these proteins in our list of homologs, and in addition to these representative proteins, we identified a second loss-of-pathogenicity protein (LopB) in the Dothideomycete fungus Pyrenophora tritici-repentis, and 212 HeLo domains in more than 40 species (Table S4). Notably, we observed that the species of many of the PFD structural homologs we have identified, such as Nectria haematococca mpVI 17-13-4, Fusarium oxsyporum, and Fusarium graminearum, are all plant pathogens, causing diseases such as wheat headblight disease and Fusarium wilt disease [42], [43].
Our evolutionary search for sequential homologs to the HET-s PFD, and subsequent analysis on structural homologs to the HET-s solenoid structure, sheds light on the contribution of the HET-s solenoid fold to fibril formation and stability in amyloid-forming proteins. As the HET-s solenoid shape remains the only atomic structure for a fibril to date, to what extent do other proteins share this fold? From an evolutionary perspective, our analysis of the PFD solenoid, and the limited phyletic distribution of PFD structural homologs we have observed, suggest that the HET-s solenoid shape has ‘limited scope’ for amyloidosis. The restriction of this particular left-handed β-solenoid to filamentous ascomycotes strikingly contrasts against that of a ‘generic’ left-handed beta-helix found in almost all phyla [44], and which is the current proposed model for fibrils of prions and other amyloid-forming proteins that are not necessarily fungal [45], [46], [47], [48], [49], [50], [51]. Interestingly, at face value, the HET-s solenoid is an attractive candidate for the formation of stable fibrils in the structural homologs we have identified: this shape is easily modelled in the homologs we have identified (despite poor sequence identity), and could even be modelled in remote homologs to the PFD, such as the small s protein of G. clavigera ( Figure 4 and Figure 5 ), and even in A.otae. Several characteristic physicochemical properties of HET-s remained conserved within these models, such as a conserved triangular hydrophobic core with enrichment for hydrophobic bulky residues, and conserved asparagine ladders at comparable positions to the HET-s PFD ( Figure 5 ). Such characteristics are amenable for fibril formation in some structural homologs such as FG10600.1, whereby the structural conservation in this solenoid allowed for HET-s and FG10600.1 amyloid cross-seeding experiments [10]. However, a closer inspection of structural homologs to the PFD indicates that the potential for salt-bridge formation is largely lacking, with several homologs only partaking in one possible salt-bridge pair compared to the 3 salt bridges in HET-s ( Figure 5 ). Additionally, in at least three of the structural homologs we have analyzed, we observe a discrepancy in the length of the rungs composing the pseudorepetitive solenoid, such that the first rung is shorter than the second rung in the solenoid monomer. If these homologs do indeed form fibrils, they would be built on the stacking of structurally different units, and as such, there would a noticeable “shift” in the hydrophobic core, asparagine ladders, and salt bridges between different units of the solenoid. These shifts in the inter- and intra-molecular bonds of the solenoid monomers may hinder stability of the resultant fibril; this remains to be determined by experimental analysis. Based on our analysis however, the contribution of the HET-s shape to future amyloid forming proteins is quite limited, and for many of the structural homologs that can adopt that shape, structural and energetic hindrances would need to be overcome before formation of a stable fibril.
We have performed an evolutionary, functional, and structural bioinformatics analysis of homologs to the HET-s prion-forming domain, and we compare our findings against the identified homologs of the HET-s N-terminal domain. Based on phylogenetic analysis, we conclude that the HET-s PFD has a limited phyletic distribution in the kingdom of life, especially within fungi, but is also highly evolving compared to the N-terminal domain. Using fold recognition techniques, we have predicted a set of PFD homologous structures which are amenable to adopting a β-solenoid fold, but which lack many of the characteristics of the HET-s solenoid that promote the formation of stable fibrils. Accordingly, we conclude that the HET-s shape has ‘limited scope’ for amyloidosis across the wider protein universe. Additionally, we assessed the tandem evolution of the HET-s N-terminal and prion-forming domains and identified functional linkages of the N-terminal homologs. Our research suggests that the HET-s N-terminal domain has a widespread phyletic distribution and may contribute to several protein-protein interactions besides heterokaryon incompatability.
Supporting Information
Neighbor-joining phylogentic tree of the N-terminal domains of Het-s orthologs that significantly align to the A. otae N-terminal domain protein sequence. This is a phylogenetic tree made with the neighbor-joining algorithm. The % bootstrap values are labeled at each node. The green box shows the clustering of A. otae with F. oxysporum. The phylogenetic class of each sequence is labeled after the species name (i.e., Sordariomycetes, Eurotiomycetes, etc.).
(TIF)
Blosum similarity matrix for the N-terminal domains and C-terminal domains of the homologs to the PFD. A percent similarity matrix is provided for each of the N-terminal and C-terminal domains based on 10 homologs to the PFD. Naming of the homologs matches the naming scheme of ( Figure 3 ).
(DOC)
List of N-terminal homologs with significant hits to SCOP domains (n = 40). For each homolog, the number of hits against the SCOP domains is also shown.
(XLS)
SCOP domains that are significant (E<0.0001) to the HET-s N-terminal homolog proteins (n = 65). 65 SCOP domains are represented, with a description of each domain, and the number of hits against the homologous proteins of the N-terminal domain.
(XLS)
212 HeLo domains identified in N-terminal homologs using HMMER. The HMMER output is shown. Detailed explanation of the output is provided with the file.
(XLS)
List of Proteomes constituting the BROAD database.
(TXT)
FASTA sequences of 10 homologs to the HET-s prion-forming domain (PFD).
(FASTA)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by a grant from the PrioNet Canada Network of Centres of Excellence to PMH, and by a CIHR McGill University Systems Biology Training Program to DMAG. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kajava AV, Baxa U, Wickner RB, Steven AC. A model for Ure2p prion filaments and other amyloids: The parallel superpleated β-structure. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7885–7890. doi: 10.1073/pnas.0402427101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kajava AV, Squire JM, Parry DAD, Andrey Kajava JMS, David ADP. [beta][hyphen (true graphic)]Structures in Fibrous Proteins. 2006. pp. 1–15. Advances in Protein Chemistry: Academic Press. [DOI] [PubMed]
- 3.Kajava AV, Steven AC, Andrey Kajava JMS, David ADP. Advances in Protein Chemistry: Academic Press; 2006. [beta][hyphen (true graphic)]Rolls, [beta][hyphen (true graphic)]Helices, and Other [beta][hyphen (true graphic)]Solenoid Proteins. pp. 55–96. [DOI] [PubMed] [Google Scholar]
- 4.Dobson CM. Structural biology: Prying into prions. Nature. 2005;435:747–749. doi: 10.1038/435747a. [DOI] [PubMed] [Google Scholar]
- 5.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, et al. Structure of the cross-[beta] spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Melckebeke Hln, Wasmer C, Lange A, Ab E, Loquet A, et al. Atomic-Resolution Three-Dimensional Structure of HET-s(218–289) Amyloid Fibrils by Solid-State NMR Spectroscopy. Journal of the American Chemical Society. 2010;132:13765–13775. doi: 10.1021/ja104213j. [DOI] [PubMed] [Google Scholar]
- 7.Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, et al. Amyloid Fibrils of the HET-s(218–289) Prion Form a {beta} Solenoid with a Triangular Hydrophobic Core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 8.Greenwald J, Buhtz C, Ritter C, Kwiatkowski W, Choe S, et al. The Mechanism of Prion Inhibition by HET-S. Molecular Cell. 2010;38:889–899. doi: 10.1016/j.molcel.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saupe SJ. The [Het-s] prion of Podospora anserina and its role in heterokaryon incompatibility. 2011. Seminars in Cell & Developmental Biology In Press, Corrected Proof. [DOI] [PubMed]
- 10.Wasmer C, Zimmer A, Sabaté R, Soragni A, Saupe SJ, et al. Structural Similarity between the Prion Domain of HET-s and a Homologue Can Explain Amyloid Cross-Seeding in Spite of Limited Sequence Identity. Journal of Molecular Biology. 2010;402:311–325. doi: 10.1016/j.jmb.2010.06.053. [DOI] [PubMed] [Google Scholar]
- 11.Balguerie A, Reis SD, Ritter C, Chaignepain S, Coulary-Salin B, et al. Domain organization and structure-function relationship of the HET-s prion protein of Podospora anserina. EMBO J. 2003;22:2071–2081. doi: 10.1093/emboj/cdg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coustou V, Deleu C, Saupe S, Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proceedings of the National Academy of Sciences. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cuomo CA, Birren BW, Jonathan W, Christine G, Gerald RF. Methods in Enzymology: Academic Press; 2010. The Fungal Genome Initiative and Lessons Learned from Genome Sequencing. pp. 833–855. [DOI] [PubMed] [Google Scholar]
- 15.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 17.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 18.Geer LYDM, Lipman DJ, Bryant SH. CDART: protein homology by domain architecture. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finn RD, Mistry J, Tate J, Coggill P, Heger A, et al. The Pfam protein families database. Nucleic Acids Research. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lobley A, Sadowski MI, Jones DT. pGenTHREADER and pDomTHREADER: new methods for improved protein fold recognition and superfamily discrimination. Bioinformatics. 2009;25:1761–1767. doi: 10.1093/bioinformatics/btp302. [DOI] [PubMed] [Google Scholar]
- 21.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, et al. Current Protocols in Bioinformatics: John Wiley & Sons, Inc; 2002. Comparative Protein Structure Modeling Using Modeller. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laskowski RAMMW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 23.Laskowski RA, Chistyakov VV, Thornton JM. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Research. 2005;33:D266–D268. doi: 10.1093/nar/gki001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schrodinger LLC. 2010. The PyMOL Molecular Graphics System, Version 1.3r1.
- 25.Costantini S, Colonna G, Facchiano AM. ESBRI: a web server for evaluating salt bridges in proteins. Bioinformation. 2008;3:137–138. doi: 10.6026/97320630003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gough J, Chothia C. SUPERFAMILY: HMMs representing all proteins of known structure. SCOP sequence searches, alignments and genome assignments. Nucl Acids Res. 2002;30:268–272. doi: 10.1093/nar/30.1.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. Journal of Molecular Biology. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]
- 28.Fedorova N, Badger J, Robson G, Wortman J, Nierman W. Comparative analysis of programmed cell death pathways in filamentous fungi. BMC Genomics. 2005;6:177. doi: 10.1186/1471-2164-6-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Research. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Research. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Research. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 33.Felsenstein J. PHYLIP - Phylogeny Inference Package (Version 3.2). Cladistics. 1989;5:164–166. [Google Scholar]
- 34.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chevenet F, Brun C, Banuls A-L, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Research. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bork P. Hundreds of ankyrin-like repeats in functionally diverse proteins: Mobile modules that cross phyla horizontally? Proteins: Structure, Function, and Bioinformatics. 1993;17:363–374. doi: 10.1002/prot.340170405. [DOI] [PubMed] [Google Scholar]
- 39.Li D, Roberts R. Human Genome and Diseases: WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cellular and Molecular Life Sciences. 2001;58:2085–2097. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith TF, Gaitatzes C, Saxena K, Neer EJ. The WD repeat: a common architecture for diverse functions. Trends in Biochemical Sciences. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 41.Blatch GL, Lässle M. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. BioEssays. 1999;21:932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 42.Bai G, Shaner G. Management and resistance in wheat and barley to Fusarium head blight. Annual Review of Phytopathology. 2004;42:135–161. doi: 10.1146/annurev.phyto.42.040803.140340. [DOI] [PubMed] [Google Scholar]
- 43.Takken F, Rep M. The arms race between tomato and Fusarium oxysporum. Molecular Plant Pathology. 2010;11:309–314. doi: 10.1111/j.1364-3703.2009.00605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi JH, Govaerts C, May BCH, Cohen FE. Analysis of the sequence and structural features of the left-handed β-helical fold. Proteins: Structure, Function, and Bioinformatics. 2008;73:150–160. doi: 10.1002/prot.22051. [DOI] [PubMed] [Google Scholar]
- 45.Govaerts C, Wille H, Prusiner SB, Cohen FE. Evidence for assembly of prions with left-handed beta-helices into trimers. Proc Natl Acad Sci USA. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi JH, May BCH, Wille H, Cohen FE. Molecular Modeling of the Misfolded Insulin Subunit and Amyloid Fibril. Biophysical Journal. 2009;97:3187–3195. doi: 10.1016/j.bpj.2009.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo J-t, Wetzel R, Xu Y. Molecular modeling of the core of Aβ amyloid fibrils. Proteins: Structure, Function, and Bioinformatics. 2004;57:357–364. doi: 10.1002/prot.20222. [DOI] [PubMed] [Google Scholar]
- 48.Langedijk JPM, Fuentes G, Boshuizen R, Bonvin AMJJ. Two-rung Model of a Left-handed [beta]-Helix for Prions Explains Species Barrier and Strain Variation in Transmissible Spongiform Encephalopathies. Journal of Molecular Biology. 2006;360:907–920. doi: 10.1016/j.jmb.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 49.Stork M, Giese A, Kretzschmar HA, Tavan P. Molecular Dynamics Simulations Indicate a Possible Role of Parallel [beta]-Helices in Seeded Aggregation of Poly-Gln. Biophysical Journal. 2005;88:2442–2451. doi: 10.1529/biophysj.104.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zanuy D, Gunasekaran K, Lesk AM, Nussinov R. Computational Study of the Fibril Organization of Polyglutamine Repeats Reveals a Common Motif Identified in [beta]-Helices. Journal of Molecular Biology. 2006;358:330–345. doi: 10.1016/j.jmb.2006.01.070. [DOI] [PubMed] [Google Scholar]
- 51.Iconomidou VA, Vriend G, Hamodrakas SJ. Amyloids protect the silkmoth oocyte and embryo. FEBS Letters. 2000;479:141–145. doi: 10.1016/s0014-5793(00)01888-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Neighbor-joining phylogentic tree of the N-terminal domains of Het-s orthologs that significantly align to the A. otae N-terminal domain protein sequence. This is a phylogenetic tree made with the neighbor-joining algorithm. The % bootstrap values are labeled at each node. The green box shows the clustering of A. otae with F. oxysporum. The phylogenetic class of each sequence is labeled after the species name (i.e., Sordariomycetes, Eurotiomycetes, etc.).
(TIF)
Blosum similarity matrix for the N-terminal domains and C-terminal domains of the homologs to the PFD. A percent similarity matrix is provided for each of the N-terminal and C-terminal domains based on 10 homologs to the PFD. Naming of the homologs matches the naming scheme of ( Figure 3 ).
(DOC)
List of N-terminal homologs with significant hits to SCOP domains (n = 40). For each homolog, the number of hits against the SCOP domains is also shown.
(XLS)
SCOP domains that are significant (E<0.0001) to the HET-s N-terminal homolog proteins (n = 65). 65 SCOP domains are represented, with a description of each domain, and the number of hits against the homologous proteins of the N-terminal domain.
(XLS)
212 HeLo domains identified in N-terminal homologs using HMMER. The HMMER output is shown. Detailed explanation of the output is provided with the file.
(XLS)
List of Proteomes constituting the BROAD database.
(TXT)
FASTA sequences of 10 homologs to the HET-s prion-forming domain (PFD).
(FASTA)