Abstract
The immediate-early 63-kDa (IE63) protein of varicella-zoster virus (VZV) is a phosphoprotein encoded by open reading frame (ORF) ORF63/ORF70. To identify functional domains, 22 ORF63 mutations were evaluated for effects on IE63 binding to the major VZV transactivator, IE62, and on IE63 phosphorylation and nuclear localization in transient transfections, and after insertion into the viral genome with VZV cosmids. The IE62 binding site was mapped to IE63 amino acids 55 to 67, with R59/L60 being critical residues. Alanine substitutions within the IE63 center region showed that S165, S173, and S185 were phosphorylated by cellular kinases. Four mutations that changed two putative nuclear localization signal (NLS) sequences altered IE63 distribution to a cytoplasmic/nuclear pattern. Only three of 22 mutations in ORF63 were compatible with recovery of infectious VZV from our cosmids, but infectivity was restored by inserting intact ORF63 into each mutated cosmid. The viable IE63 mutants had a single alanine substitution, altering T171, S181, or S185. These mutants, rOKA/ORF63rev[T171], rOKA/ORF63rev[S181], and rOKA/ORF63rev[S185], produced less infectious virus and had a decreased plaque phenotype in vitro. ORF47 kinase protein and glycoprotein E (gE) synthesis was reduced, indicating that IE63 contributed to optimal expression of early and late gene products. The three IE63 mutants replicated in skin xenografts in the SCIDhu mouse model, but virulence was markedly attenuated. In contrast, infectivity in T-cell xenografts was not altered. Comparative analysis suggested that IE63 resembled the herpes simplex virus type 1 US1.5 protein, which is expressed colinearly with ICP22 (US1). In summary, most mutations of ORF63 made with our VZV cosmid system were lethal for infectivity. The few IE63 changes that were tolerated resulted in VZV mutants with an impaired capacity to replicate in vitro. However, the IE63 mutants were attenuated in skin but not T cells in vivo, indicating that the contribution of the IE63 tegument/regulatory protein to VZV pathogenesis depends upon the differentiated human cell type which is targeted for infection within the intact tissue microenvironment.
Varicella-zoster virus (VZV), a member of the alphaherpesvirus subfamily of the Herpesviridae, causes varicella during primary infection and herpes zoster when it reactivates from latency in sensory ganglia. The VZV genome is a double-stranded DNA molecule of approximately 125 kb with open reading frames (ORFs) that are known or predicted to encode at least 70 distinct gene products (1). The genome consists of two main coding regions, the unique long (UL) and unique short (US) regions, each of which is flanked by internal repeat (IR) and terminal repeat (TR) sequences. Three ORFs, including ORF63/70 as well as ORF62/71 and ORF64/69, are located within the repeat sequences and are therefore duplicated within the VZV genome (7).
The development of VZV cosmid systems has permitted the functional analysis of VZV gene products by deleting ORFs or introducing targeted mutations into the viral genome (6, 15, 29). In previous experiments, we showed that IE63 protein was essential for generating infectious virus from our VZV cosmids (42). A single copy of ORF63 or ORF70 at the native site or inserted at a nonnative site was sufficient to permit typical VZV replication in cell culture in vitro and in skin xenografts in the SCIDhu mouse model of VZV pathogenesis in vivo (42). Experiments with another set of VZV cosmids showed recovery of infectious VZV despite ORF63/70 deletion, although growth of the ORF63/70 deletion mutant was diminished (J. Cohen, personal communication). These observations may reflect a differential requirement for IE63 coregulation or interaction with other viral proteins depending on the VZV genome from which cosmids were generated, or the addition of an ORF62-expressing plasmid to enhance VZV recovery in this cosmid system, which may compensate for the absence of IE63 during early stages of replication. The objective of our current studies was to use our single-copy ORF63 recombinant, rOka/ORF63rev, to examine the consequences of targeted mutations in ORF63.
The ORF63/70 gene product is a small, 278-amino-acid protein expressed at immediate-early times after infection, which localizes to the nuclei of infected cells, and is a component of the virion tegument (8, 11, 21). IE63 is heavily phosphorylated and is a substrate for phosphorylation by VZV ORF47 kinase and cellular kinases (5, 18, 34, 46). The contributions of IE63 to VZV replication are poorly understood, although it does not seem to have direct transactivating properties (5, 23, 46). IE63 binds to IE62, the major VZV transactivating protein, suggesting that IE63 might cooperate with IE62 to regulate viral gene expression, as indicated by the enhancement of IE62-induced transcription of the glycoprotein I (gI) promoter in the presence of IE63 (27, 43). Similar interactions have been demonstrated for the related herpes simplex virus type 1 proteins ICP22 and ICP4 (24, 36) and the equine herpesvirus 1 proteins IECP22 and IEP (10, 19, 25, 37, 39, 45). Understanding IE63 functions is of particular interest because of the evidence that it is expressed during latency in human ganglia and in the rat model (16, 26, 28, 43).
In order to assess IE63 function, we generated 22 mutations in ORF63 for evaluation in transient transfection experiments, and each ORF63 mutation was also introduced into the VZV cosmid used to generate the single-copy ORF63 virus, rOka/ORF63rev, in order to determine whether the change was lethal for replication (42). The ORF63 mutations were designed to map the site of IE62 binding within IE63, to determine the phosphorylation targets of serine/threonine (S/T) kinases in IE63, and to examine IE63 nuclear localization. Lynch et al. reported that the region of IE63 required for IE62 binding was located in the N-terminal half of IE63, amino acids 1 to 142 (27). The N terminus of IE63 includes region 2, amino acids 26 to 157, which is highly conserved in related proteins of other members of the alphaherpesvirus subfamily (9). This region mediates the binding of herpes simplex virus type 1 ICP22 and ICP4 (24) and equine herpesvirus 1 EICP22 to IEP (9).
Since protein alignments revealed eight highly conserved amino acid pairs within region 2 of IE63 and its homologues, eight paired amino acid deletions were made in this putative IE62 binding region within IE63 (2, 9). IE63 is heavily phosphorylated in infected cells, and Kenyon et al. showed that IE63 expressed in bacteria was a substrate for recombinant ORF47 kinase (18). IE63 has 19 residues predicted to be phosphorylated by cellular S/T kinases (4), or by the VZV kinases, the products of ORF47 and ORF66 (41). Changes were made, in combination or singly, that altered the putative phosphorylation targets within the N terminus, center, or C terminus of IE63.
To investigate IE63 nuclear localization, mutations were introduced to disrupt a potential nuclear localization signal (NLS), based upon previous mapping to the C terminus, amino acids 210 to 278 (46), and evidence that deletion of a KRRR sequence (amino acids 260 to 263) resulted in cytoplasmic localization of IE63 (5). When transfer of the ORF63 mutations into cosmids yielded infectious VZV, IE63 mutant viruses were evaluated for effects on VZV replication in vitro and virulence in skin and T-cell xenografts in vivo in the SCIDhu model of VZV pathogenesis (32, 33, 42).
These experiments showed that most modifications of the ORF63 coding sequence made with our cosmid system were lethal. The few changes in ORF63 that were tolerated resulted in VZV mutants that had substantially reduced infectivity in vitro and were markedly attenuated in their virulence for differentiated human skin cells, but not T cells, in vivo.
MATERIALS AND METHODS
Construction of ORF63 mutants in pET-ORF63.
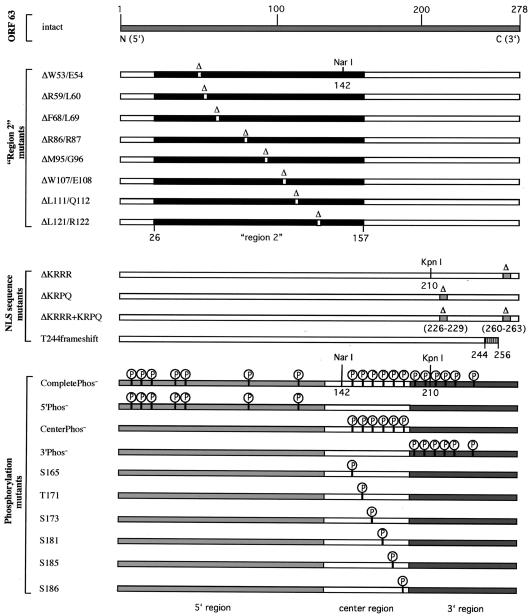
The AvrII fragment of pvSpe21ΔORF63/70 + 63@Avr (1.1 kb), which contains ORF63 (42), was blunt ended with T4 DNA polymerase and subcloned into the KpnI site of pETBlue-2 (Novagen, Madison, Wis.), yielding pET-ORF63, from which intact ORF63 could be isolated by AvrII digestion. Twenty-two mutations of ORF63 that were predicted to affect three characteristic regions or properties of IE63 protein were created by PCR mutagenesis (Fig. 1). The first set of ORF63 mutations was designed to alter eight highly conserved amino acid pairs within region 2 (amino acids 26 to 157). ORF63 mutations were made that deleted each of these amino acid pairs, designated ΔW53/E54, ΔR59/L60, ΔF68/L69, ΔR86/R87, ΔM95/G96, ΔW107/E108, ΔL111/Q112, and ΔL121/R122.
FIG. 1.
Overview of the mutational analysis of IE63 protein. This figure depicts all of the amino acid changes that were introduced into IE63 protein. Mutations in the IE63 region 2 deleted the conserved amino acid pairs ΔW53/E54, ΔR59/L60, ΔF68/L69, ΔR86/R87, ΔM95/G96, ΔW107/E108, ΔL111/Q112, and ΔL121/R122. Nuclear localization sequence mutations deleted the four amino acids 226 to 229 (ΔKRPQ) or 260 to 263 (ΔKRRR), by removing both putative NLS sequences (ΔKRRR+KRPQ) and by creating a frameshift, replacing the full-length 278-amino-acid protein with an IE63 truncation at amino acid 256 (T244 frameshift). Mutations of putative phosphorylation sites were made by alanine substitution of 19 serine (S) or threonine (T) residues that were predicted targets of cellular S/T kinases, including S12, S13, S15, T41, S42, S82, S129, S165, T171, S173, S181, S185, S186, S197, T201, S203, T222, S224, and T244 (CompletePhos− mutant). The mutant 5′Phos− contains alanine substitutions of the first seven putative phosphorylation targets (S12 to S129) in the IE63 N terminus, the mutant CenterPhos− has substitutions of the six putative phosphorylation sites in the center region (S165 to S186), and the mutant 3′Phos− has substitutions of the six putative phosphorylation sites (S197 to T244) in the IE63 C terminus. Single alanine substitutions were made in the S and T residues S165, T171, S173, S181, S185, and S186 in the center segment of the IE63 protein.
A panel of ORF63 mutations was generated to alter predicted phosphorylation sites of cellular S/T protein kinases, cellular kinase I, cellular kinase II, and protein kinase C, in IE63, based on NetPhos analysis (4). The program detected 19 possible serine (S) and threonine (T) residues: S12, S13, S15, T41, S42, S82, S129, S165, T171, S173, S181, S185, S186, S197, T201, S203, T222, S224, and T244. To map the phosphorylation sites of ORF63 in more detail, four multiple phosphorylation site mutations were created in which groups of S and T residues in the N terminus, center, or C terminus of IE63 were alanines. The mutant 5′Phos− has the first seven putative phosphorylation sites replaced with alanine (S12, S13, S15, T41, S42, S82, and S129), the CenterPhos− mutant has substitutions of the six putative phosphorylation targets in the center (S165, T171, S173, S181, S185, and S186), the mutant 3′Phos− has the last six putative residues substituted (S197, T201, S203, T222, S224, and T244), and the mutant CompletePhos− has alanine substitutions of all 19 S and T residues. Since initial kinase assay experiments showed that IE63 phosphorylation was localized predominantly within the center region, six single amino acid substitution mutants were constructed by replacing S165, T171, S173, S181, S185, or S186 with alanine.
Four mutations were made in ORF63 to disrupt the putative NLS in the IE63 C terminus (5, 46). The mutations were ΔKRRR (deletion of amino acids 260 to 263), ΔKRPQ (deletion of amino acids 226 to 229), ΔKRRR+KRPQ (deletion of amino acids 260 to 263 and amino acids 226 to 229), and a T244 frameshift that alters the C terminus of IE63 at amino acid 244 by deleting an adenosine at nucleotide 732. This change results in P(245)RKRSPRWARGV(256) and an earlier stop codon at amino acid 257 instead of amino acid 279. The T244 frameshift removes the putative NLS sequence KRRR (amino acids 260 to 263).
PCR mutagenesis was performed with two rounds of PCR per mutation. In the first PCR, a 5′-outside vector primer (pET-5′: 5′-TTCTGTACAGGCGCGCCTGCAGGAC-3′) of the construct pET-ORF63 was used in combination with the first mutagenic primer (mutation-A; see example below). In parallel, another PCR was performed with a 3′-ouside vector primer (pET-3′: 5′-GCTTCGAACGCGTATCGATGGTACC-3′) in combination with the second mutagenic primer (mutation-B; see example below). The two resulting PCR products divide the 1.1-kb ORF63 fragment into two smaller fragments. The mutagenic primers, mutation-A and mutation-B, were designed according to the following principle: 5′ — 10-bp complete sequence alignment — mutation — 20-bp complete sequence alignment — 3′. For example, the two mutagenic primers to create the mutation ΔKRRR (deletion of amino acids 260 to 263) read as follows: ΔKRRR-A: 5′-GGGCTTCGTG-deletion-CCAATCTACACCCCCCTCGC-3′; and ΔKRRR-B: 5′-TGTAGATTGG-deletion-CACGAAGCCCCGCGCCGGCA-3′.
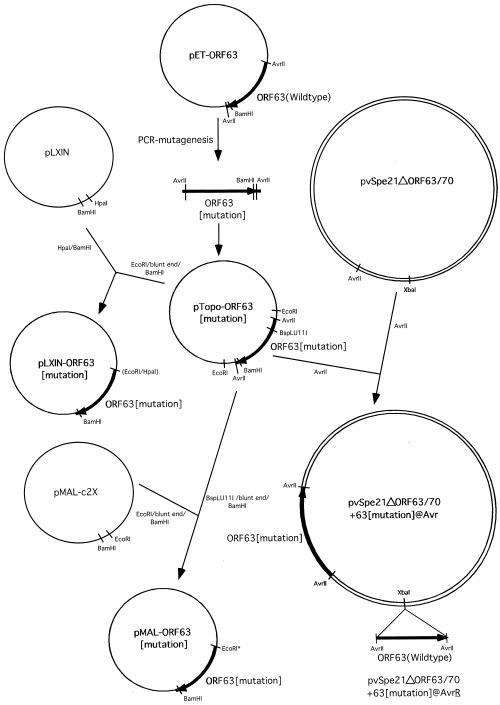
For the second PCR round, the two PCR products resulting from the first PCRs were gel purified and combined. The second PCR was performed with the outside vector primers pET-5′ and pET-3 only. The mutagenic primers yield a 20-bp overlap in the two PCR fragments resulting from the first PCR round at the site of the mutation, and therefore possess the ability to self-prime. The second PCR round resulted in a 1.1-kb fragment, which was Topo-cloned (Invitrogen, Carlsbad, Calif.) and sequenced to verify the mutation. Sequence analysis was done at the Stanford University PAN Facility. The resulting Topo vectors were named pTOPO-ORF63-[mutation] (Fig. 2).
FIG. 2.
Construction of ORF63 mutations. This figure is a schematic representation of the cloning steps involved in the construction of all plasmids and cosmids that were used to make ORF63 mutations, as described in Materials and Methods.
Construction of cosmids with ORF63 mutations in the ΔORF63/70 backbone.
The cosmid pvSpe21ΔORF63/70, from which ORF63 and ORF70 were deleted (42), was linearized by AvrII digest and gel purified with the QIAEX II kit (Qiagen, Inc., Chatsworth, Calif.). Each of the ORF63 mutations was isolated from the Topo-vector by AvrII digest, gel purified, and inserted into the AvrII site of pvSpe21ΔORF63/70, yielding a cosmid that had one copy of ORF63 containing the altered ORF63 sequence. The resulting constructs were designated pvSpe21ΔORF63/70 + 63[mutation]@Avr (Fig. 2). In order to create ORF63 rescued viruses, the intact ORF63 gene was isolated as an AvrII fragment from pET-ORF63 and was inserted into the XbaI site in the respective pvSpe21ΔORF63/70 + 63[mutation]@Avr construct. The resulting rescue cosmids were designated pvSpe21ΔORF63/70 + 63[mutation]@AvrR (Fig. 2).
Construction of pLXIN-based OR63 expression vectors.
The retroviral plasmid pLXIN (Clontech, Palo Alto, Calif.) was digested with HpaI and BamHI, and gel purified. The mutated ORF63 sequences were isolated from the Topo vectors by an EcoRI digest, followed by a blunt end reaction with T4 DNA polymerase, and a BamHI digest. The resulting fragments were gel purified and inserted into pLXIN. These constructs were named pLXIN-ORF63-[mutation].
Construction of pMAL-c2X-based IE63-MBP fusion proteins.
The vector pMAL-c2X (New England Biolabs, Beverly, Mass.) was digested with EcoRI, blunt ended with T4 DNA polymerase, and digested with BamHI. The coding region of each mutant ORF63 was isolated by BspLU11I digest, which cuts exactly at the site of the ORF63 start codon, followed by a blunt end reaction with T4 DNA polymerase and BamHI digest. The fragments were gel purified and inserted “in frame” into pMAL-c2X. The resulting plasmids were named pMAL-ORF63-[mutation].
Cosmid transfections.
The pvSpe21 mutated cosmids and the other three VZV cosmids, pvFsp4, pvSpe5, and pvPme19, were electroporated into Top 10F′ competent cells (Invitrogen Inc., Carlsbad, Calif.), grown in LB containing kanamycin and ampicillin, and purified with a Qiagen plasmid maxi prep kit (Qiagen, Inc., Chatsworth, Calif.). Cosmids were digested with AscI and mixed in water to a final concentration of 100 ng of pvFsp4, pvSpe5, and pvPme19 and 50 ng of pvSpe21 (42). Transfections were done in human melanoma cells with 20 or 30 μl of the cosmid mix in 31.5 μl of 2 M CaCl2 in water and Hanks' balanced salt solution. Human melanoma cells were grown in tissue culture medium (Dulbecco's modified Eagle's medium; Gibco, Gaithersburg, Md.) supplemented with heat-inactivated fetal calf serum. After transfection, melanoma cells were kept at 37°C for 3 to 4 days, trypsinized and transferred to a 75-cm2 flask; plaques appeared 5 to 10 days after transfection with intact cosmids. Cells transfected with mutant cosmids were passed at a 1:3 ratio every 3 to 4 days. Infectious virus was propagated in melanoma cells (42).
Preparation and PCR analysis of viral DNA.
DNA was recovered from infected cells with DNAzol (Gibco BRL, Inc., Grand Island, N.Y.). PCR was performed with Elongase enzyme mix (Gibco BRL, Grand Island, N.Y.). The primers used to assess the ORF63 mutations inserted at the AvrII site were 5′-CCACACAAACATCACCTG-3′ and 5′-TTACCACCGCTTCCATCA-3′ (42). Following PCR with infected cell DNA as a template, the products were cloned into the pCR-Topo cloning vector (Invitrogen, Carlsbad, Calif.). Sequencing reactions were primed with the M13 forward and reverse primers contained in the pCR-Topo vector. Sequence analysis was done at the Stanford University PAN Facility.
Kinase assays to assess ORF63 phosphorylation.
To measure the endogenous kinase activity associated with the immunoprecipitated IE63 proteins, 293T cells were transiently transfected with the pLXIN plasmids expressing the putative IE63 phosphorylation mutants and controls. Transient transfections were performed in 15-cm dishes with the Superfect transfection reagent (Qiagen, Hilden, Germany). Cells were harvested 36 h after transfection with radioimmunoprecipitation assay (RIPA) buffer; lysates were precleared with protein A-Sepharose beads (Sigma, St. Louis, Mo.) and rabbit preimmune serum and were incubated overnight at 4°C with protein A-Sepharose beads and ORF63 rabbit antiserum (a generous gift from Paul Kinchington); the beads were washed three times with HNTG buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.2 mM EDTA, 0.5% Nonidet P40). The HNTG supernatant was removed, and 1 ml of kinase buffer (20 mM HEPES, pH 7.5, 10 mM MgCl2, 100 μM orthovanadate, 2 mM dithiothreitol) was added to the beads. Beads were collected and kinase buffer was replaced with 30 μl of kinase reaction buffer (998 μl of kinase buffer, 2 μl of 10 mM cold ATP, and 24 μl of [γ-32P]ATP) per sample.
Following 20 min of incubation at 30°C, samples were boiled after addition of 6× sodium dodecyl sulfate sample buffer. Samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and gels were blotted on Immobilon transfer membranes (Millipore, Bedford, Mass.). Photographic film (Kodak) was used to detect the radioactive signals localized on the filter and the level of ORF63 phosphorylation was evaluated with a GS-710 imaging densitometer (Bio-Rad, Hercules, Calif.). Amounts of transiently expressed protein in the cell lysates were equivalent as assessed by immunoblot (data not shown). In order to perform kinase assays with VZV recombinant viruses carrying IE63 mutations, infected melanoma cells were grown to an equivalent cytopathic effect and harvested with RIPA buffer; kinase assays were done as described for transient transfections.
Immunofluorescence microscopy.
HeLa cells were grown on poly-d-lysine-coated cell culture slides (Becton Dickinson, Bedford, Mass.) and transiently transfected with the various pLXIN-ORF63[mutation] constructs, including pLXIN-ORF63[intact] and pLXIN as controls, with Superfect transfection reagent (Qiagen, Hilden, Germany). After 48 h, the cells were fixed in ice-cold 4% paraformaldehyde for 10 min at 4°C and washed once with ice-cold phosphate-buffered saline (PBS). Cells were blocked for 30 min at room temperature in PBS (10% donkey serum/0.3% Triton X-100). After blocking, cells were incubated in PBS (3% donkey serum/0.3% Triton X-100) with ORF63 rabbit antiserum (1:1,000) and mouse monoclonal antibody anti-human nuclei (1:250) (Chemicon, Temecula, Calif.) at 4°C overnight on a shaking device. Cells were washed three times for 30 min with PBS at room temperature and were incubated for 6 h in PBS (3% donkey serum/0.3% Triton X-100) with the secondary antibodies (1:1,000), including donkey anti-rabbit-Cy3 and donkey anti-mouse-fluorescein isothiocyanate (Jackson ImmunoResearch, West Grove, Pa.). Cells were washed in PBS, and slides were mounted with Vectashield (Vector Laboratories, Burlingame, Calif.) and stored in the dark. Immunofluorescence microscopy was performed with a Zeiss HAL-100 microscope. To examine infected cells, melanoma cells were grown on cell culture slides (Becton Dickinson, Bedford, Mass.) and infected with ORF63 mutant viruses or the rOKA control. Four days later, infected cells were fixed in ice-cold 4% paraformaldehyde for 10 min at 4°C and processed for immunofluorescence microscopy.
Immunoprecipitation of IE63 bound to IE62 in infected cells.
Suspensions of 50 μl of slurry protein G-Sepharose beads (Amersham, Uppsala, Sweden) were blocked with 500 μl of a 4% milk-PBS solution for 1 h at 4°C. The blocked protein G-Sepharose was resuspended in 500 μl of PBS and incubated with 10 μl of anti-IE62 monoclonal antibody (H6), or with 10 μl of a nonrelevant monoclonal antibody as a control, for 1 h at room temperature, followed by three washes in immunoprecipitation buffer (1% Triton X-100/50 mM Tris-Cl [pH 7.4]/150 mM NaCl/5 mM EDTA/0.02% NaN3).
Infected melanoma cells were harvested, lysed in immunoprecipitation buffer, and precleared with protein G-Sepharose beads. The precleared cellular lysates were added to Sepharose G beads to the H6 anti-IE62 monoclonal antibody preloaded and incubated for 2 h at 4°C, followed by four washing steps in washing buffer (0.1% Triton X-100/50 mM Tris-Cl [pH 7.4]/150 mM NaCl/5 mM EDTA/0.02% NaN3). Beads were collected, resuspended in 40 μl of 1× SDS sample buffer, and separated by SDS-PAGE, and gels were blotted on Immobilon transfer membranes (Millipore, Bedford, Mass.). Polyclonal anti-ORF63 antibody was used in Western blots to detect IE63 (21). Bands were visualized with goat anti-rabbit IgG conjugated with horseradish peroxidase in conjunction with ECL plus chemiluminescence substrate (Amersham Biosciences, Piscataway, N.J.).
IE62 pull-down assay with IE63-MBP fusion proteins.
ORF63 and mutants resulting in deletions of each of the eight paired N-terminal amino acids were generated in pMAL-c2X and expressed in Escherichia coli (BL21-AI) as fusion proteins at the maltose-binding protein (MBP) C terminus. The recombinant proteins were affinity purified with amylose resin (New England Biolabs, Beverly, Mass.). All steps were performed at 4°C. Bacterial cells were harvested from 250-ml cultures and resuspended in 25 ml of lysis buffer (10 mM NaPO, 30 mM NaCl, 0.25% Tween 20, 10 mM β-mercaptoethanol, 10 mM EDTA, 10 mM EGTA). The suspensions were sonicated and NaCl was added to a final concentration of 500 mM. Supernatants were collected after centrifugation of the lysates at 9,000 × g for 30 min.
Approximately 100 μl of amylose resin (New England Biolabs, Beverly, Mass.) was washed with 500 μl of PBST (1% Triton X-100 in PBS) for 15 min, blocked with 5% milk in PBST for 1 h, and washed with 500 μl of PBST for 10 min. The amylose resin was incubated with 400 μl of the MBP fusion protein supernatants. Then 40 μl of the MBP supernatant was used because MBP was expressed at levels approximately 10-fold higher than the IE63-MBP fusion proteins. These reactions were incubated for 1 h and the beads were washed with 500 μl of PBST four times for 10 min each. The MBP fusion conjugated beads were incubated with 40 μg of recombinant IE62 (44) and 200 μg of bovine serum albumin in 300 μl of PBST for 3 h. The beads were washed with 500 μl of PBST four times for 15 min each, and samples were boiled after adding 6× SDS sample buffer. Samples were separated by SDS-PAGE and gels were blotted on Immobilon transfer membranes (Millipore, Bedford, Mass.). A polyclonal anti-ORF62 antibody (a generous gift from Paul Kinchington) was used to detect bound IE62 in Western blots. Bands were visualized with goat anti-rabbit IgG conjugated with horseradish peroxidase in conjunction with ECL plus chemiluminescence substrate (Amersham Biosciences, Piscataway, N.J.).
Analysis of VZV protein expression.
Lysates of melanoma cells infected with IE63 mutant viruses were prepared as described above. Equal loading of viral proteins was adjusted with a polyclonal antibody against IE4 (a generous gift from Paul Kinchington). Viral proteins were loaded on SDS gels, blotted on membranes, and probed with antibodies against IE62 (rabbit polyclonal), IE63 (rabbit polyclonal) (21), ORF47 (rabbit polyclonal) (3), and glycoprotein E (mouse monoclonal antibody).
Infection of skin and T-cell xenografts in SCIDhu mice.
Skin and T-cell implants were made in homozygous CB-17scid/scid mice, with human fetal tissues obtained with informed consent according to federal and state regulations (32). Animal use was in accordance with the Animal Welfare Act and approved by the Stanford University Administrative Panel on Laboratory Animal Care. VZV recombinants, passed three times in primary human lung (HEL) cells, were used to inoculate xenografts; infectious virus titers were determined for each inoculum at the time the implants were injected. Skin xenografts were harvested after 14 and 21 days and analyzed by infectious focus assay; T-cell xenografts were evaluated at 10 and 20 days. Viruses recovered from implants were tested to show stability of the ORF63 mutation by PCR and sequencing.
RESULTS
Analysis of the IE62 binding site in IE63.
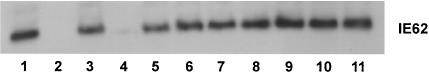
In order to map the IE62 binding region within IE63, which was localized to the N-terminal half (amino acids 1 to 142) by Lynch et al. (27), we performed IE62 pull-down assays with the panel of IE63-MBP fusion proteins that had deletions of each of the eight conserved amino acid pairs in IE63 region 2. IE62 binding to IE63 was detected with anti-IE63 rabbit polyclonal antiserum. IE62 bound to intact IE63, to the IE63 mutants ΔW53/E54, ΔF68/L69, ΔR86/R87, ΔM95/G96, ΔW107/E108, ΔL111/Q112, and ΔL121/R122, and to 5′Phos−, which was included as a control having no changes in amino acids 1 to 142 (Fig. 3). IE62 was not recovered in the pull-down assay done with MBP alone or with the mutant IE63-MBP fusion protein ΔR59/L60. This result indicated that amino acids R59 and L60 played a key role in the interaction of IE63 with IE62 and that IE63 residues required for IE62 binding were located between amino acids 55 and 67, since the capacity of IE63 mutants ΔW53/E54 and ΔF68/L69 to bind recombinant IE62 was not altered. By secondary structure analysis, R59/L60 was located at the beginning of a predicted α-helix.
FIG. 3.
Mapping of the IE62 binding site in IE63 protein. The IE62 pull-down assay was performed with IE63-MBP fusion proteins that had deletions of the conserved amino acid pairs in region 2. The ORF63 mutations were made in pMAL-c2X and expressed in E. coli (BL21-AI) as fusion proteins at the MBP C terminus. The recombinant proteins were affinity purified with amylose resin (New England Biolabs, Beverly, Mass.) and incubated with recombinant IE62. IE62 that bound to the IE63 recombinant proteins was detected with polyclonal rabbit anti-ORF62 antibody. IE62 binding is shown to intact IE63 (lane 1), the IE63 mutants ΔW53/E54 (lane 3), ΔF68/L69 (lane 5), ΔR86/R87 (lane 6), ΔM95/G96 (lane 7), ΔW107/E108 (lane 8), ΔL111/Q112 (lane 9), ΔL121/R122 (lane 10), and 5′Phos− (lane 11). No IE62 was recovered in the pull-down assay done with MBP alone (lane 2) or with the IE63-MBP fusion protein ΔR59/L60 (lane 4).
Phosphorylation targets of cellular S/T kinases in IE63.
The panel of IE63 mutations in putative phosphorylation sites, expressed in pLXIN vectors, was used to define targets of cellular S/T kinases by in vitro kinase assay. IE63 was immunoprecipitated from 293T cells transfected with pLXIN-based mutants or controls with polyclonal rabbit antiserum against IE63. IE63 phosphorylation resulted from bound cellular S/T kinases and was monitored by incubating the immunoprecipitate with radiolabeled ATP, followed by SDS gel electrophoresis and phospho-imager detection.
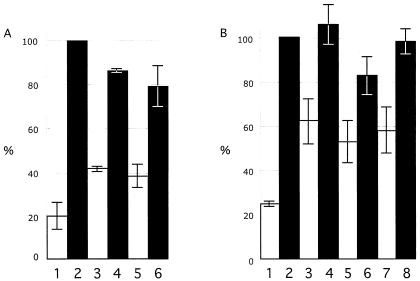
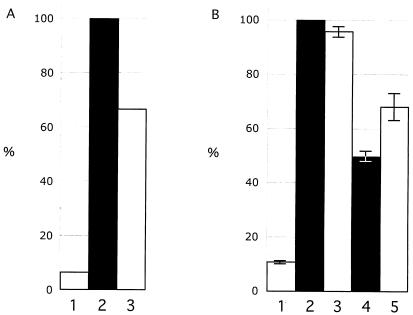
Kinase assays were done first with the four multiple site mutants, including the CompletePhos− mutant, in which all 19 putative phosphorylation targets were replaced with alanine residues, the 5′Phos− mutant, with substitutions of the first seven S and T residues in the N terminus, the CenterPhos− mutant, in which the next six S and T residues were replaced, and the 3′Phos− mutant, with substitutions of the last six S and T residues located in the IE63 C terminus. Three independent kinase assays were performed with the four multiple IE63 phosphorylation site mutants. The CompletePhos− and the CenterPhos− IE63 mutants exhibited almost no evidence of phosphorylation relative to intact IE63, whereas phosphorylation was observed despite the alanine substitutions that had been made in the N-terminal and C-terminal segments of IE63 (Fig. 4A). These experiments indicated that the IE63 center region, comprised of amino acids 165 to 186, contained the major targets for phosphorylation by cellular S/T kinases. These residues were within the region, amino acids 142 to 210, which was identified as containing targets for IE63 phosphorylation with bacterial IE63-MBP fusion proteins and recombinant cellular kinase II (46).
FIG. 4.
Kinase assay to evaluate phosphorylation of IE63 mutants expressed in 293T cells. 293T cells were transfected with the pLXIN-based ORF63 mutants designed to substitute alanine for S or T residues that were putative phosphorylation targets, or with control plasmids, and harvested in RIPA buffer after 36 h. IE63 was immunoprecipitated with polyclonal rabbit antiserum, and IE63 phosphorylation by cellular S/T kinases was assessed with radiolabeled ATP, followed by SDS gel electrophoresis and phosphorimager detection. The bar graph in panel A shows the results of three independent kinase assays performed with the multiple putative phosphorylation site mutants and reported as a mean percentage relative to intact IE63 (bar 2). The other bars are 1, negative control (pLXIN), 3, CompletePhos− mutant, 4, 5′Phos− mutant, 5, CenterPhos− mutant; and 6, 3′Phos− mutant. Panel B shows the combined results of three independent kinase assays performed with the mutants disrupting single putative phosphorylation targets relative to intact IE63 (no. 2); the other bars are 1, negative control (pLXIN), 3, S165, 4, T171, 5, S173, 6, S181, 7, S185; and 8, S186. The lines indicate standard errors.
In order to define the phosphorylation sites for cellular S/T kinases within the IE63 center region more precisely, kinase assays were done with the six constructs in which S165, T171, S173, S181, S185, or S186 was replaced with alanine. Three independent kinase assays showed that alanine substitution of S165, S173, or S185 resulted in significantly less phosphorylation compared to intact IE63, indicating that these three amino acids were the primary targets for IE63 phosphorylation by cellular S/T kinases in vitro (Fig. 4B). By NetPhos analysis, S165 is a potential cellular kinase II site and residues S173 and S185 are potential cellular kinase I sites.
Nuclear localization signal sequences in IE63.
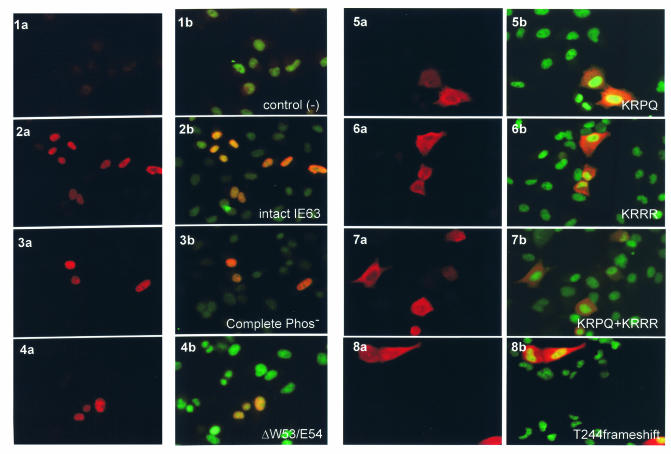
Intact IE63 shows a distinct nuclear localization in transiently transfected cells and at immediate-early times in VZV-infected cells (5, 27, 46). To assess whether mutations designed to disrupt the putative NLS resulted in altered cellular localization of IE63, HeLa cells were transfected with the pLXIN-IE63 mutant constructs and stained with polyclonal rabbit antiserum against IE63 (indocarbocyanine) and a murine monoclonal antibody against human nuclei (fluorescein isothiocyanate). The negative control, which was pLXIN alone, showed no IE63-indocarbocyanine signal (Fig. 5, 1a, b). Distinct nuclear localization of IE63 was evident with intact IE63 (Fig. 5: 2a, b) and all mutants of putative phosphorylation sites, as illustrated with the CompletePhos− construct (Fig. 5: 3a, b), as well as all region 2 mutants, as illustrated with ΔW53/E54 (Fig. 5, 4a, b). None of the mutations in the IE63 putative phosphorylation sites or in region 2 changed the predominant nuclear localization of IE63. In contrast, the four predicted NLS sequence mutants, ΔKRPQ (Fig. 5, 5a, b), ΔKRRR (Fig. 5, 6a, b), ΔKRPQ+KRRR (Fig. 5, 7a, b), and T244 frameshift (Fig. 5, 8a, b), showed an altered phenotype. The altered phenotype was the same in all four NLS sequence mutants and in most cells transfected with these mutants, the IE63-indocarbocyanine signal was detected in the cytoplasm as well as within nuclei, indicating that both predicted NLS sequences were involved in IE63 nuclear retention.
FIG. 5.
Effects of IE63 mutations in putative nuclear localization signal sequences. Intracellular distribution of the IE63 mutant proteins expressed as pLXIN constructs in transiently transfected HeLa cells was examined by immunofluorescence microscopy 48 h after transfection, with polyclonal rabbit antiserum against IE63 (indocarbocyanine label: red; panels 1a to 8a) and dual stain with anti-ORF63 and anti-human nuclei, a murine monoclonal antibody (fluorescein isothiocyanate label: green; panels 1b to 8b). The constructs tested were plasmid alone (1a, 1b), intact IE63 (2a, 2b), CompletePhos- (3a, 3b), ΔW53/W54 (4a, 4b), ΔKRPQ (5a, 5b), ΔKRRR (6a, 6b), ΔKRPQ+ΔKRRR (7a, 7b), and T244 frameshift (8a, 8b). Magnification: 40×.
FIG. 6.
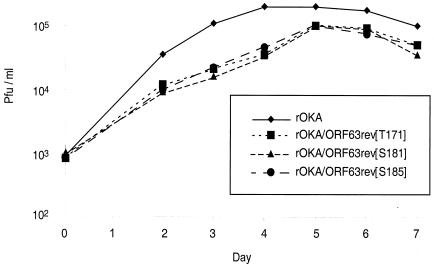
Replication of IE63 mutant viruses in melanoma cells. Melanoma cells were inoculated on day 0 with 103 PFU of rOka or rOka mutants rOKA/ORF63rev[T171], rOKA/ORF63rev[S181], and rOKA/ORF63rev[S185]. Aliquots were harvested for 7 days, and infectious foci were determined by titration on melanoma cell monolayers. Each point represents the mean of three wells.
FIG. 7.
Kinase assay to evaluate phosphorylation of IE63 as expressed in IE63 mutant viruses. IE63 was immunoprecipitated with polyclonal rabbit antiserum and phosphorylation was monitored by incubation of the immunoprecipitate with radiolabeled ATP, followed by SDS gel electrophoresis and phosphorimager detection. Panel A shows phosphorylation of IE63 in cells infected with rOka (bar 2) or rOKAORF47ΔC, an ORF47 kinase null mutant (bar 3), and uninfected melanoma cell control (bar 1). The bar graph in panel B shows the results of two independent kinase assays performed with IE63 mutant viruses and reported as a mean percentage relative to cells infected with rOKA/ORF63rev (bar 2); the other bars are uninfected melanoma cell control (bar 1), rOKA/ORF63rev[T171] (bar 3), rOKA/ORF63rev[S181] (bar 4), and rOKA/ORF63rev[S185] (bar 5). The lines indicate standard errors.
FIG. 8.
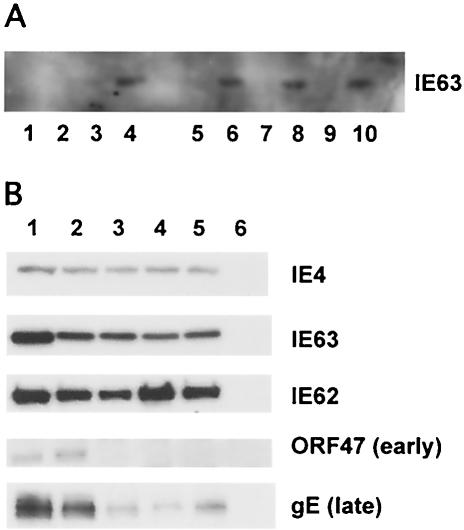
Immunoprecipitation of IE62 protein from melanoma cells infected with IE63 mutant viruses with anti-IE63 antiserum and expression of VZV immediate-early, early, and late proteins by IE63 mutant viruses. As shown in panel A, IE62 in lysates of infected cells was immunoprecipitated with monoclonal antibody against IE62. IE63 that coimmunoprecipitated with IE62 was detected with polyclonal rabbit antiserum against IE63 (even-numbered lanes) or nonspecific murine IgG (odd-numbered lanes). Samples tested included lanes 1 and 2, uninfected melanoma cells; lanes 3 and 4; rOKA/ORF63rev-infected cells; lanes 5 and 6, rOKA/ORF63rev[T171]-infected cells; lanes 7 and 8, rOKA/ORF63rev[S181]-infected cells; and lanes 9 and 10, rOKA/ORF63rev[S185]-infected cells. As shown in panel B, melanoma cells were infected with 5 × 104 PFU of the test virus and harvested at 4 days postinfection. Cell lysates were evaluated for expression of immediate-early proteins IE62 and IE63 and ORF4 proteins, the ORF47 viral kinase, which is an early protein, and the late protein gE, with rabbit polyclonal antiserum specific for IE62, IE63, ORF4, or ORF47, and a murine monoclonal antibody to gE. The specimens tested were lane 1, rOka (contains ORFs 63 and 70); lane 2, rOKA/ORF63rev (one copy of ORF63 at a nonnative site); lane 3, rOKA/ORF63rev[T171]; lane 4, rOKA/ORF63rev[S181]; and lane 5, rOKA/ORF63rev[S185].
Since some IE63 was also detected in the nuclei of HeLa cells transfected with each of these mutants, the alternative NLS may remain functional; in experiments with the ΔKRPQ+KRRR mutant, IE63 may bind to an abundant cellular protein that localizes to the nucleus, facilitating its nuclear transport. For example, cellular RNA polymerase II has been found to bind to IE63 in VZV-infected cells (27).
Effect of ORF63 mutations on VZV replication.
To assess the effects of the ORF63 mutations on VZV replication, all ORF63 mutations were transferred into the cosmid pvSpe21Δ63/70 (42). No infectious virus was obtained in 12 independent transfection experiments done when the ORF63 mutation resulted in deletions of one of the highly conserved amino acid pairs in region 2, including ΔW53/E54, ΔR59/L60, ΔF68/L69, ΔR86/R87, ΔM95/G96, ΔW107/E108, ΔL111/Q112, and ΔL121/R122. No recombinant virus was generated in three independent transfections following the introduction of ORF63 mutations that disrupted multiple phosphorylation sites, which included CompletePhos−, 5′Phos−, CenterPhos− and 3′Phos, or in five independent transfections done when the ORF63 mutation deleted one or both of the putative NLS sequences, KRPQ or KRRR, or with the T244 frameshift. In all cases, infectious VZV was recovered when intact ORF63 was inserted into the cosmid that contained the mutant ORF63. These experiments indicated that the failure to recover infectious virus was due to the ORF63 mutation and not to an unidentified change that had been introduced elsewhere in the VZV genome. Persistence of the mutant ORF63 and insertion of the intact ORF were confirmed by PCR and sequencing.
When cosmids were made that had ORF63 mutations altering each of the single putative phosphorylation sites in the IE63 center region, three of the six alanine substitutions were compatible with recovery of infectious VZV. T171, S181, and S185 substitutions were tolerated, but changes in S165, S173, or S186, were not. The recombinant IE63 mutant viruses with substitutions in the putative single phosphorylation sites were designated rOKA/ORF63rev[T171], rOKA/ORF63rev[S181], and rOKA/ORF63rev[S185].
Characterization of the IE63 mutant viruses for growth in cell culture and IE63 phosphorylation.
All three IE63 mutant viruses showed reduced growth in melanoma cells compared to rOKA (Fig. 6), and all had a small-plaque phenotype (data not shown). Since these mutants were designed to have altered S/T residues, kinase assays were done with IE63 immunoprecipitated from melanoma cells infected with rOKA/ORF63rev[T171], rOKA/ORF63rev[S181], and rOKA/ORF63rev[S185] in order to examine whether IE63 phosphorylation was altered. Under these conditions, IE63 phosphorylation may be due to the kinase activity of the bound cellular kinases or to the viral S/T kinases, the ORF47 and ORF66 proteins.
In an initial control experiment to prove that IE63 was phosphorylated by ORF47 kinase in infected cells, a kinase assay was performed with rOKA and rOKAORF47ΔC, a mutant that expresses a truncated, kinase-defective ORF47 protein (3) (Fig. 7A). IE63 that was immunoprecipitated from cells infected with rOKAORF47ΔC showed less phosphorylation than IE63 immunoprecipitated from rOKA-infected cells, confirming that IE63 was a substrate for active ORF47 kinase in infected melanoma cells. When kinase assays were performed with the IE63 mutant viruses, both rOKA/ORF63rev[S181] and rOKA/ORF63rev [S185] showed less IE63 phosphorylation than rOKA or rOKA/ORF63rev[T171] (Fig. 7B). These experiments indicated that S181 and S185 were targets of IE63 phosphorylation in infected melanoma cells.
Both S181 and S185 have flanking residues that are similar to the ORF47 consensus sequence described by Kenyon et al. (17). However, we cannot exclude some differential phosphorylation related to less ORF47 expression in cells infected with the ORF63 mutants. In contrast, under transient expression conditions, which tested IE63 phosphorylation by cellular kinases only, phosphorylation of the IE63 protein was reduced by alanine substitutions of S165, S173, and S185 but not S181 (Fig. 4B). Thus, the residues of IE63 that were phosphorylated differed when viral as well as cellular kinases were present, with S185 being the only common target.
Analysis of IE62/IE63 binding in the IE63 mutant viruses.
To examine whether the interaction between IE62 and IE63 was affected by the three mutations of IE63 that were compatible with viral replication, IE62 was immunoprecipitated from infected cell lysates with anti-IE62 monoclonal antibody. IE63 protein that coimmunoprecipitated with IE62 was detected with a polyclonal rabbit antiserum against IE63 (Fig. 8A). IE63 coimmunoprecipitation was equivalent when cells were infected with rOKA/ORF63rev (positive control), rOKA/ORF63rev[T171], rOKA/ORF63rev[S181], or rOKA/ORF63rev[S185]. This result showed, as expected, that binding of IE63 to IE62 was not affected by alanine substitutions in the IE63 center region in these IE63 mutant viruses.
Viral protein expression by IE63 mutant viruses.
To compare the pattern of viral protein expression by rOKA/ORF63rev[T171], rOKA/ORF63rev[S181], and rOKA/ORF63rev[S185], infected cell lysates were prepared at 2 days (data not shown) and 4 days (Fig. 8B) after infection; viral protein loading was adjusted based on detection of equivalent amounts of IE4 (40). The viral protein expression profiles of the IE63 mutants were compared to rOKA, which has two intact copies of ORF63, and to rOKA/ORF63rev, which has one intact copy of ORF63 (Fig. 8B). The expression of IE62 and IE63 proteins was not affected in cells infected with the IE63 mutant viruses compared to rOKA/ORF63rev. VZV rOka-infected cells had a relative increase in expression of IE63, which may be explained by the presence of the two intact ORF63 and ORF70 gene copies. However, the expression of ORF47 kinase, which is an early protein, and gE, which is a late protein, was reduced significantly in cells infected with each of the three IE63 mutant viruses compared to rOKA/ORF63rev; the results were the same at day 2 (data not shown). This finding indicated that IE63 did not affect the expression of immediate-early genes. In the case of rOKA/ORF63rev[S181] and rOKA/ORF63rev[S185], the effects on ORF47 and gE expression may reflect altered IE63 function due to altered phosphorylation.
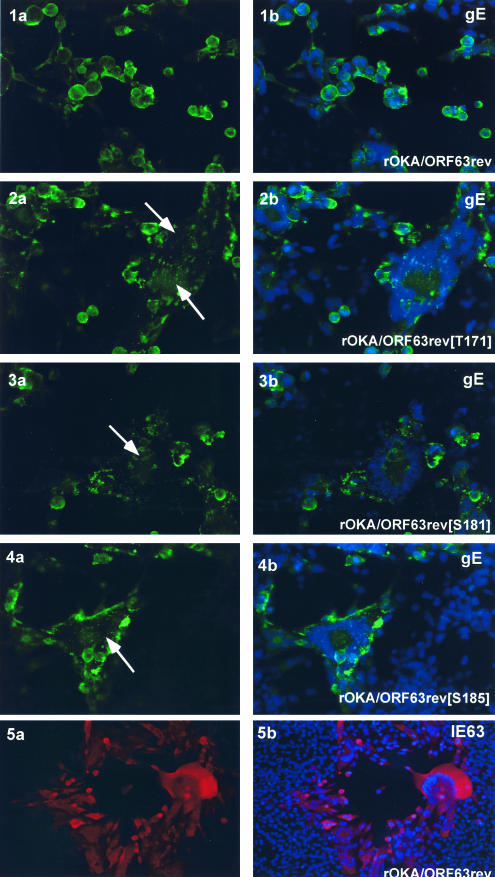
Reduced expression of gE was confirmed and an altered cellular distribution of gE was observed by immunofluorescence microscopy of cells infected with the IE63 mutant viruses and stained with anti-gE monoclonal antibody (Fig. 9). In rOKA-infected cells, gE protein was localized almost exclusively to the plasma membranes, and the nuclei within polykaryocytes were arranged in the typical symmetrical ring (29) (Fig. 9, 1a, b); the same pattern was detected in rOKA/ORF63rev-infected cells (data not shown). In contrast, monolayers of melanoma cells infected with the IE63 mutant viruses showed small plaques consisting of disorganized polykaryocytes. In these infected cells, gE protein was localized in a punctate pattern in the cytoplasm, as observed with rOKA/ORF63rev[T171] (Fig. 9, 2a, b), rOKA/ORF63rev[S181] (Fig. 9, 3a, b), and rOKA/ORF63rev[S185] (Fig. 9, 4a, b). These results suggested that less gE protein was made by the three IE63 mutant viruses than was necessary to preserve the characteristic distribution of gE within infected cells, or the regular organization of nuclei within fused infected cells that make up VZV syncytia. Altered gE localization could also be due to reduced expression of ORF47 protein (17). The localization of IE63 in melanoma cells infected with the three IE63 mutant viruses (data not shown) did not differ from IE63 distribution in cells infected with rOka (Fig. 9, 5a, b).
FIG. 9.
Intracellular localization of VZV IE63 and gE in cells infected with IE63 mutant viruses. Melanoma cells were infected with rOKA/ORF63rev (1 a,b; 5 a,b), rOKA/ORF63rev[T171] (2 a,b), rOKA/ORF63rev[T181] (3 a.b), or rOKA/ORF63rev[S185] (4 a, b). Monolayers were stained with anti-rabbit IE63 antiserum (red) and counterstained with the nuclear marker, 4′,6′-diamidino-2-phenylindole (DAPI) (blue) (panel 5 a, b) or with anti-gE monoclonal antibody (fluorescein isothiocyanate: green) (panels 1 to 4) and counterstained with the nuclear marker DAPI (blue). Panels 2a to 4a, arrows indicate abnormal, irregular polykaryocytes with punctate distribution of gE. Immunofluorescence microscopy was performed at 4 days after infection (late). Magnification, 40×.
Growth of IE63 mutants in skin and T-cell xenografts in vivo.
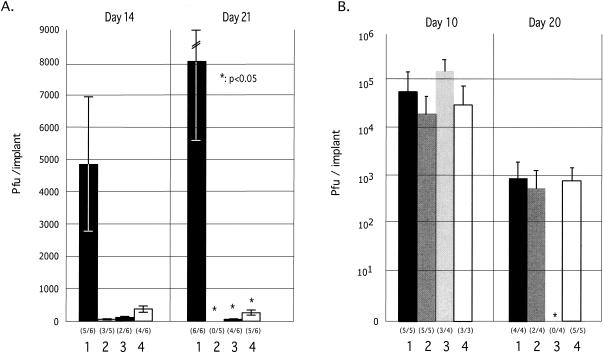
Analysis of the growth kinetics of the three IE63 mutant viruses in skin xenografts in SCIDhu mice demonstrated a significant reduction in growth in vivo when evaluated at 14 and 21 days after inoculation (Fig. 10A). The peak titers in skin xenografts were markedly reduced for all three IE63 mutant viruses at 21 days (P < 0.05). Eleven of 12 rOka-infected xenografts yielded infectious virus at either or both time points compared to 3 of 10 implants infected with rOKA/ORF63rev[T171], 6 of 12 with rOKA/ORF63rev[S181], and 9 of 12 with rOKA/ORF63rev[S185]. The T171 substitution had the most substantial effect on VZV virulence in vivo, based on relatively fewer infected implants (30%) and the lowest mean viral titers at days 14 and 21. Sequencing showed that each of the ORF63 mutations was retained after replication in skin xenografts for 21 days. In contrast, replication of the IE63 mutants in T-cell xenografts was not reduced compared to rOKA/ORF63rev (Fig. 10B). Infectious rOKA/ORF63rev[S185] was not recovered at 20 days, but this pattern was due to extensive infection and depletion of T cells, by immunohistochemistry analysis (data not shown); declining titers at days 21 to 28 days after T-cell infection with low passage clinical isolates or intact recombinants made from cosmids is characteristic of VZV replication in T-cell xenografts (3, 13, 32, 40).
FIG. 10.
Replication of IE63 mutant viruses in skin and T-cell xenografts in SCID-hu mice. Skin xenografts in SCID mice were injected with (left to right) rOKA, rOKA/ORF63rev[T171], rOKA/ORF63rev[S181], or rOKA/ORF63rev[S185] having equivalent inoculum titers. Virus titers in skin xenografts were assessed after harvest at day 14 (A, left panel) and day 21 (A, right panel) after inoculation and were graphed as mean titers for xenografts that yielded infectious virus, with lines indicating standard errors. The number of xenografts from which infectious virus was recovered per number that were inoculated is given in parentheses below the horizontal axis. The P values were <0.05 when titers of rOka and each of the IE63 mutant viruses were compared at day 21. Replication of VZV recombinants in T cells was assessed at days 10 and 20 (B). Lines indicate the standard errors.
DISCUSSION
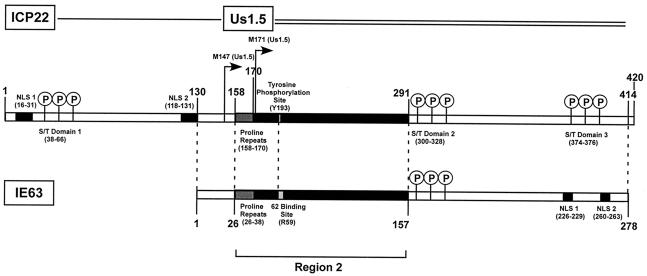
In this first mutational analysis of ORF63 in the context of the VZV genome, 19 of 22 changes in the coding sequence were lethal for VZV replication when each mutant ORF63 was introduced as a single ORF63 copy into our VZV cosmid from which we had deleted ORF63/70 (42). Three mutations of ORF63 that were tolerated resulted in IE63 mutant viruses that had substantially impaired infectivity in cell culture and a dramatic reduction of virulence in differentiated human skin cells, but not T cells, in the SCIDhu model of VZV pathogenesis in vivo. Comparative analysis of functional domains indicated that IE63 most closely resembles herpes simplex virus type 1 US1.5, which is a 274-amino-acid protein encoded by a gene that is colinear with the herpes simplex virus type 1 ICP22 (US1) C terminus (24, 36) (Fig. 11).
FIG. 11.
Comparative analysis of IE63 and herpes simplex virus type 1 ICP22 and US1.5 gene products. This diagram illustrates the similarities and differences between IE63 and the herpes simplex virus type 1 homologue ICP22 and the colinear US1.5 gene product. The N-terminal sequence of ICP22 is not present in IE63. IE63 is similar in size to the US1.5 gene product, and both proteins have similar regions arranged in the same linear pattern, e.g., initial proline repeats, the conserved region 2, and an S/T-rich center domain. Nuclear localization signal (NLS) sequences located in the C terminus of IE63 are not present in US1.5; US1.5 has another S/T domain where the NLS domains appear in IE63, and US1.5 has six amino acid residues at the C terminus that are not found in IE63. Amino acid identity between IE63 and US1.5 is 24.9%, and homology is 46%.
Previous experiments done with highly purified recombinant proteins showed that IE63 interacts directly with IE62 and that the binding site for IE62 was between amino acids 1 and 142 in IE63 (27, 42, 44). IE62 binding was located within IE63 region 2, comprised of amino acids 26 to 157. This region is highly conserved among alphaherpesviruses and is involved in the binding of herpes simplex virus type 1 ICP4 to ICP22, and of equine herpesvirus 1 IEP to EICP22 (2, 9, 10, 24, 36). Region 2 comprises the N-terminal half of VZV IE63, whereas it is located between amino acids 158 to 297 in herpes simplex virus type 1 ICP22. However, in herpes simplex virus type 1 US1.5, region 2 has the same N-terminal location as it does in IE63, and both IE63 and US1.5 have a proline-rich initial sequence, which has been suggested to promote protein-protein interactions (36) (Fig. 11).
Our experiments indicated that the specific binding site of IE62 in IE63 maps between amino acids 55 and 67, with R59/L60 being critical for this interaction. The R59/L60 pair is predicted to be near the N terminus of a long α-helix. IE63 interactions with IE62 may be essential for virion structure, since IE62 and IE63 are both tegument proteins, and their binding may be required for virion assembly (20, 21). Of interest, the R59/L60 pair corresponds to residues Y193/M194 within the herpes simplex virus type 1 ICP22 (Fig. 11). The tyrosine (Y193) is a target of tyrosine kinase phosphorylation, and alanine substitution results in decreased herpes simplex virus replication in fibroblasts (35), although whether this amino acid pair has a role in the interaction between herpes simplex virus ICP22 and ICP4 has not been determined. We observed that deletion of each of the other seven conserved amino acid pairs in IE63 region 2 was also lethal for viral replication. These residues may be required for IE63 protein folding that is necessary for IE63-IE62 binding, disrupt binding to other VZV proteins, e.g., ORF47, yield dominant negative mutants that block IE62 transactivation, or interfere with other as yet undefined IE63 functions.
IE63 binding to IE62 may also be required for VZV gene regulation during replication. IE63 has enhancing effects on IE62-mediated transactivation of VZV genes, as demonstrated for the gI promoter and IE63 also binds to cellular RNA polymerases (5, 27). Whereas IE63 alone does not function as a transactivator and does not bind directly to promoter DNA, IE63 binding to IE62 was associated with increased transcription from the gI promoter (27). Changes in IE63 in the three viable IE63 mutant viruses, rOKA/ORF63rev[T171], rOKA/ORF63rev[S181], and rOKA/ORF63rev[S185], were associated with altered expression of VZV early (ORF47 protein) and late (gE) genes, even though the mutations involved the IE63 center region, adjacent to region 2, rather than the IE63/IE62 binding site. In fact, the decreased synthesis of ORF47 protein and gE occurred despite the preservation of IE62 binding to IE63, and was not associated with decreased IE62 or IE63 expression. These experiments suggested that other regions of this small 278-amino-acid protein may be involved in its coregulatory functions, perhaps through altered intracellular localization at critical times during VZV replication, or other mechanisms that are independent of binding to IE62, such as effects on its binding to cellular RNA polymerase II (27).
IE63 is phosphorylated by the VZV ORF47 protein kinase and by cellular kinases (5, 17, 18, 46). Kinase assays done with IE63 mutants in which alanine was substituted for S and T residues showed that the center region, amino acids 165 to 186, contained major sites for IE63 phosphorylation. These observations further defined the predominant targets of S/T kinases in IE63 and were consistent with the previous mapping of key phosphorylation sites to amino acids 142 to 210 (46). Bontems et al. described decreased IE63 phosphorylation when all 10 IE63 residues, S150, S165, T171, S173, S181, S185, S186, S224, T201, and T244, were replaced with alanine in glutathione S-transferase fusion constructs (5).
Our kinase assays showed that S165, S173, and S185 were key targets of cellular S/T kinases. When kinase assays were performed with IE63 mutant viruses, rOKA/ORF63rev[T171], rOKA/ORF63rev[S181], and rOKA/ORF63rev[S185], S181 and S185 were phosphorylated in infected cells. These results suggested that S181 was phosphorylated predominantly by the viral S/T kinases, ORF47 and ORF66, and that S185 was a shared target for viral and cellular kinases. Although T171 was identified as being phosphorylated by cellular kinases in expression constructs, this finding was not predictive of its phosphorylation when IE63 was expressed as a viral gene product during replication in cell culture.
Phosphorylation of S181 by ORF47 kinase may block T171 phosphorylation by cellular kinases. The herpes simplex virus type 1 US1.5 protein has a similar S/T domain in the same region as the IE63 residues, S165 to 185 (Fig. 11) and US1.5 is phosphorylated by herpes simplex virus type 1 UL13, which is the ORF47 homologue (36). The observation that alanine substitutions of S165 and S173 were lethal mutations suggested that IE63 phosphorylation of these residues by cellular S/T kinases may be critical for viral replication. Alternatively, these mutations may have introduced conformational changes in IE63 or blocked other functions that do not depend on phosphorylation. In addition to ORF47-mediated phosphorylation of IE63, Kenyon et al. demonstrated that these two proteins form a tight complex, which differs from the usual transient binding required for kinase function (18). Since ORF47 and IE63 are both tegument proteins, IE63 binding to ORF47 may be essential for virion assembly. If so, the lethality of the S186 mutation may be explained by conformational changes in IE63 that block these interactions.
IE63 exhibits a distinct pattern of nuclear localization with transient expression as a single protein and at immediate-early times in VZV-infected cells (27, 46). Previous reports mapped NLS function to the IE63 C terminus (amino acids 210 to 278) (46) and showed that deletion of residues KRRR (amino acids 260 to 263) resulted in a cytoplasmic translocation of IE63 protein in transfected Vero and ND7 cells (5). We found that all 18 mutant constructs altering IE63 region 2 or putative phosphorylation targets retained the prominent nuclear localization phenotype. However, the four mutations that affected the two putative NLS sequences, which included ΔKRPQ (amino acids 226 to 229), ΔKRRR (amino acids 260 to 263), ΔKRPQ+KRRR, and T244 frameshift, resulted in a cytoplasmic as well as nuclear distribution of IE63. Disruption of both KRPQ and KRRR was not sufficient for complete cytoplasmic translocation of IE63. One explanation for this phenotype is that IE63 binds to a cellular protein that facilitates its nuclear import, such as cellular RNA polymerase II, which binds IE63 in infected cells (27). Herpes simplex virus type 1 ICP22 also has two NLSs (45); however, these NLSs are in the first 131 N-terminal residues of ICP22, which are not present in IE63, rather than in the US1.5 gene product (Fig. 11). The nuclear localization of ICP22 was also characterized by expression in small, dense nuclear structures (14, 30), whereas this pattern was observed in only a few VZV-infected cells during replication in melanoma cells.
The analysis of the three IE63 mutant viruses, rOKA/ORF63rev[T171], rOKA/ORF63rev[S181], and rOKA/ORF63rev[S185], in cell culture showed that these alanine substitutions resulted in decreased infectious virus yields and a small plaque phenotype, when compared to rOKA/ORF63rev, which has a single copy of ORF63 in the US region of the VZV genome (42), and to rOKA, which has both copies of ORF63 and ORF70 in their native sites. In addition to the reduced expression of a representative early gene product, ORF47 kinase, and of gE, which is a late protein, these changes were associated with disrupted intracellular localization of gE and disorganized polykaryocyte formation. ORF47 null and kinase defective VZV mutants exhibit no difference from parent viruses in growth kinetics and plaque size in vitro (3, 12, 33), whereas our mutational analyses of gE, gI, and the gI promoter indicate that the normal VZV replication and plaque phenotype requires adequate gE expression and formation of gE-gI complexes (13, 29, 31). Thus, the deleterious effect of these IE63 mutations on replication in melanoma cells probably reflects diminished gE synthesis, since ORF47 null mutants replicate normally in vitro.
The most important observation from our evaluation of the rOKA/ORF63rev[T171], rOKA/ORF63rev[S181], and rOKA/ORF63rev[S185] viruses was that the IE63 mutations had dramatic consequences for viral replication in differentiated human skin cells within their characteristic tissue microenvironment in vivo. In contrast, T-cell tropism was not affected by the IE63 mutations.
ORF47 protein as well as gE-gI interactions are required for VZV virulence in skin (3, 13, 31). Therefore, the reduced expression of either of these proteins as a result of IE63 mutation, is sufficient to account for altered virulence in skin in vivo, whether or not expression of other viral gene products was also diminished by interference with IE63 coregulatory functions (3, 31, 33). In contrast to skin infection, VZV T-cell tropism was not affected by IE63 mutations, suggesting that replication in T cells requires less gE and ORF47 protein, and possibly other viral proteins. VZV skin infection may depend on higher concentrations of gE for cell-cell spread, whereas infection of T-cell xenografts is not characterized by cell fusion. Since ORF47 protein is required for VZV replication in T cells in vivo, the lower concentrations of this protein made by the IE63 mutants must be sufficient to permit adequate kinase activity, virion assembly and release in T-cell xenografts. Herpes simplex virus type 1 mutants that do not express US1.5 make reduced amounts of herpes simplex virus immediate-early and γ-2 late genes (36, 38). In contrast, we observed no effect of IE63 mutations on IE gene expression. Herpes simplex virus mutants that do not express US1.5 were avirulent in mice, indicating that IE63 and US1.5 share a fundamental importance for viral pathogenesis in vivo (36). It would be of interest to determine whether US1.5 can complement IE63.
In summary, these experiments demonstrated that IE63 is a highly conserved protein, with almost all ORF63 disruptions in our single copy ORF63 mutant being lethal for VZV replication. The few changes in IE63 that were tolerated resulted in VZV mutant viruses that had significantly impaired replication in differentiated human skin cells in vivo, which is a necessary event in VZV pathogenesis, allowing viral transmission to other susceptible individuals in the population.
Acknowledgments
This work was supported by grants from the National Institute of Allergy and Infectious Diseases, AI053846 (A.M.A.), AI36884 (J.H., W.R., and A.M.A.), AI18449 (W.R., J.H.), and by a fellowship to A. Baiker from the Deutsche Forschungsgemeinschaft (DFG), BA 2035/1-1. H.I. received fellowship support from the Jikei University School of Medicine.
REFERENCES
- 1.Arvin, A. M. 2001. Varicella-zoster virus, p. 2731-2767. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
- 2.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, K. L. Howe, and E. L. Sonnhammer. 2000. The Pfam protein families database. Nucleic Acids Res. 28:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Besser, J., M. H. Sommer, L. Zerboni, C. Bagowski, H. Ito, J. Moffat, Ku, C.-C. and A. M. Arvin. 2003. Differentiation of varicella-zoster virus ORF47 protein kinase and IE62 protein binding domains and their contributions to replication in human skin xenografts in the SCIDhu mouse. J. Virol. 77:5964-5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blom, N., S. Gammeltoft, und S. Brunak. 1999. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294:1351-1362. [DOI] [PubMed] [Google Scholar]
- 5.Bontems, S., Valentin. D., E L. Baudox, B. Rentier, Sadzot-Delvaux, C., and J. Piette. 2002. Phosphorylation of varicella-zoster virus IE63 protein by casein kinases influences its cellular localization and gene regulation activity. J. Biol. Chem. 277:21050-21060. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, J. I., and K. E. Seidel. 1993. Generation of varicella-zoster virus (VZV) and viral mutants from cosmid DNAs: VZV thymidylate synthetase is not essential for viral replication in vitro. Proc. Natl. Acad. Sci. USA 90:7376-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davison. A. J., and J. Scott. 1986. The complete sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 8.Debrus, S., Sadzot-Delvaux, C., A. F. Nikkels, J. Piette, and B. Rentier. 1995. Varicella-zoster virus gene 63 encodes an immediate-early protein that is abundantly expressed during latency. J. Virol. 69:3240-3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derbigny, W. A., S. K. Kim, G. B. Caugham, and D. J. O'Callaghan. 2000. The EICP22 protein of equine herpesvirus 1 physically interacts with the immediate-early protein and with itself to form dimers and higher-order complexes. J. Virol. 74:1425-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Derbigny, W. A., S. K. Kim, H. K. Jang, and D. J. O'Callaghan. 2002. Equine herpesvirus 1 EICP22 protein sequence that mediate its physical interaction with the immediate-early protein are not sufficient to enhance the trans-activation activity of the IE protein. Virus Res. 84:1-15. [DOI] [PubMed] [Google Scholar]
- 11.Gomi, Y., H. Sunamachi, Y. Mori, K. Nagaike, M. Takahashi, and K. Yamanishi. 2002. Comparison of the complete DNA sequences of the varicella vaccine and its parental virus. J. Virol. 76:11447-11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heineman, T., and J. I. Cohen. 1995. The varicella-zoster virus (VZV) open reading frame 47 (ORF47) protein kinase is dispensable for viral replication and is not required for phosphorylation of ORF63 protein, the VZV homolog of herpes simplex virus ICP22. J. Virol. 69:7376-7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito, H., M. H. Sommer, L. Zerboni, H. He, D. Boucaud, J. Hay, W. Ruyechan, and A. M. Arvin. 2003. Promoter sequences of varicella-zoster virus glycoprotein I targeted by cellular transactivating factors Sp1 and USF determine virulence in skin and T cells in SCIDhu mice in vivo. J. Virol. 77:489-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jahedi, S., N. S. Markovitz, F. Filatov, and B. Roizman. 1999. Colocalization of the herpes simplex virus 1 UL4 protein with infected cell protein 22 in small, dense nuclear structures formed prior to onset of DNA synthesis. J. Virol. 73:5132-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kemble, G. W., P. Annunziato, O. Lungu, R. E. Winter, T. A. Cha, S. J. Silverstein, and R. R. Spaete. 2000. Open reading frame S/L of varicella-zoster virus encodes a cytoplasmic protein expressed in infected cells. J. Virol. 74:11311-11321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kennedy, P. G., E. Grinfeld, S. Bontemps, and Sadzot-Delvaux. 2001. Varicella-zoster virus gene expression in latently infected rat dorsal root ganglia. Virology 289:218-223. [DOI] [PubMed] [Google Scholar]
- 17.Kenyon, T. K., E. Homan, J. Storlie, M. Ikoma, and C. Grose. 2003. Comparison of a varicella-zoster virus ORF47 protein kinase and casein kinase II and their substrates. J. Med. Virol. 70:S95-S102. [DOI] [PubMed] [Google Scholar]
- 18.Kenyon, T. K., J. Lynch, J. Hay, W. Ruyechan, and C. Grose. 2001. Varicella-zoster virus ORF47 protein serine kinase: characterization of a cloned, biologically active phosphotransferase and two viral substrates, ORF62 and ORF63. J. Virol. 75:8854-8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, S. K., V. R. Holden, and D. J. O'Callaghan. 1997. The ICP22 protein of equine herpesvirus 1 cooperates with the IE protein to regulate viral gene expression. J. Virol. 71:1004-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinchington, P. R., J. K. Hougland, A. M. Arvin, W. T. Ruyechan, and J. Hay. 1992. The varicella-zoster virus immediate-early protein IE62 is a major component of virus particles. J. Virol. 66:359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinchington, P. R., D. Bookey, and S. E. Turse. 1995. The transcriptional regulatory proteins encoded by varicella-zoster virus open reading frames (ORFs) 4 and 63, but not 61, are associated with purified virus particles. J. Virol. 69:4274-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kinchington, P. R. and J. I. Cohen. 2000. Viral proteins, p. 74-104. In A. M. Arvin and A. A. Gershon (ed.), Varicella Zoster Virus: basic virology and clinical management. Cambridge University Press, Cambridge, United Kingdom.
- 23.Kost, R. G., H. Kupinsky, and S. E. Strauss. 1995. Varicella-zoster virus gene 63: transcript mapping and regulatory activity. Virology 209:218-224. [DOI] [PubMed] [Google Scholar]
- 24.Leopardi, R., P. L. Ward, W. O. Ogle, and B. Roizman. 1997. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes containing EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the UL13 protein kinase. J. Virol. 71:1133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long, M. C., V. Leong, P. A. Schaffer, C. A. Spencer, and S. A. Rice. 1999. ICP22 and the UL13 protein kinase are both required for herpes simplex virus-induced modification of the large subunit of RNA polymerase II. J. Virol. 73:5593-5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lungu, O., C. A. Panagiotidis, P. W. Annuziato, A. A. Gershon, and S. J. Silverstein. 1998. Aberrant intracellular localization of varicella-zoster virus regulatory proteins during latency. Proc. Natl. Acad. Sci. USA 95:7080-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch, J. M., T. K. Kenyon, C. Grose, J. Hay, and W. T. Ruyechan. 2002. Physical and functional interaction between the varicella zoster virus IE63 and IE62 proteins. Virology 302:71-82. [DOI] [PubMed] [Google Scholar]
- 28.Mahalingam, R., M. Wellish, R. Cohrs, S. Debrus, J. Piette, B. Renier, and D. H. Gilden. 1996. Expression of protein encoded by varicella-zoster virus open reading frame 63 in latently infected human ganglionic neurons. Proc. Natl. Acad. Sci. USA 93:2122-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mallory, S., M. Sommer, and A. M. Arvin. 1997. Mutational analysis of the role of glycoprotein I in varicella-zoster virus replication and its effects on glycoprotein conformation and trafficking. J. Virol. 71:8279-8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markovitz, N. S., and B. Roizman. 2000. Small dense nuclear bodies are the site of localization of herpes simplex virus 1 UL3 and UL4 proteins and of ICP22 only when the latter protein is present. J. Virol. 74:523-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mo, C., J. Lee, M. Sommer, C. Grose, and A. M. Arvin. 2002. The requirement of varicella zoster virus glycoprotein E (gE) for viral replication and effects of glycoprotein I on gE in melanoma cells. Virology 304:176-186. [DOI] [PubMed] [Google Scholar]
- 32.Moffat, J. F., M. D. Stein, H. Kaneshima, and A. M. Arvin. 1995. Tropism of varicella-zoster virus for human CD4+ and CD8+ T lymphocytes and epidermal cells in SCID-hu mice. J. Virol. 69:5236-5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moffat, J. F., L. Zerboni, M. H. Sommer, T. C. Heineman, J. I. Cohen, H. Kaneshima, and A. M. Arvin. 1998. The ORF47 and ORF66 putative kinases of varicella-zoster virus determine tropism for human T cells and skin in the SCID-hu mouse. Proc. Natl. Acad. Sci. USA 95:11969-11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ng, T. I., L. Keenan, P. R. Kinchington, and C. Grose. 1994. Phosphorylation of varicella-zoster virus open reding frame (ORF) 62 regulatory product by viral ORF47 associated protein kinase. J. Virol. 68:1350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ogle, W., O., and B. Roizman. 1999. Functional anatomy of herpes simplex virus 1 overlapping genes encoding infected-cell protein 22 and US1.5 protein. J. Virol. 73:4305-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Toole, J. M., M. Aubert, A. Kotsakis, and J. Blaho. 2003. Mutation of the protein tyrosin kinase consensus site in the herpes simplex virus 1 alpha22 gene alters ICP22 posttranslational modification. Virology 305:153-167. [DOI] [PubMed] [Google Scholar]
- 37.Poffenberger, K. L., A. D. Idowu, E. B. Smith, P. E. Raichlen, and R. C. Herman. 1994. A herpes simplex virus type 1 ICP22 deletion mutant is altered for virulence and latency in vivo. Arch. Virol. 139:111-119. [DOI] [PubMed] [Google Scholar]
- 38.Poon, A. P., W. O. Ogle, and B. Roizman. 2000. Posttranslational processing of infected cell protein 22 mediated by viral protein kinases is sensitive to amino acid substitutions at distant sites and can be cell-type specific. J. Virol. 74:11210-11214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice, S. A., M. C. Lomg, V. Lam, P. A. Schaffler, and C. A. Spencer. 1995. Herpes simplex virus immediate-early protein ICP22 is required for viral modification of host RNA polymerase II and establishment of the normal viral transcription program. J. Virol. 69:5550-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato, B., H. Ito, S. Hinchliffe, M. H. Sommer, L. Zerboni, and A. M. Arvin. 2003. Mutational analysis of open reading frames 62 and 71, encoding the varicella-zoster virus immediate-early transactivating protein, IE62, and effects on replication in vitro and in skin xenografts in the SCID-hu mouse in vivo. J. Virol. 77:5607-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, R. F., and T. F. Smith. 1989. Identification of new protein kinase-related genes in three herpesviruses, herpes simplex virus, varicella-zoster virus, and Epstein-Barr virus. J. Virol. 63:450-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sommer, M. H., E. Zagha, O. K. Serrano, et al. 2001. Mutational analysis of the repeated open reading frames, ORFs 63 and 70 and ORFs 64 and 69, of varicella-zoster virus. J. Virol. 75:8224-8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spengler, M. L., N. Niesen, C. Grose, W. T. Ruyechan, and J. Hay. 2001. Interactions among structural proteins of varicella-zoster virus. Arch. Virol. 17:71-79. [DOI] [PubMed] [Google Scholar]
- 44.Spengler, M. L., W. T. Ruyechan, and J. Hay. 2000. Physical interaction between two varicella-zoster virus gene regulatory proteins, IE4 and IE62. Virology. 272:375-381. [DOI] [PubMed] [Google Scholar]
- 45.Stelz, G., E. Rucker, O. Rossorius, G. Meyer, R. H. Stauber, M. Spatz, M. M. Eibl, and J. Hauber. 2002. Identification of two nuclear import signals in the alpha-gene product ICP22 of herpes simplex virus 1. Virology 295:360-370. [DOI] [PubMed] [Google Scholar]
- 46.Stevenson, D., M. Xue, J. Hay, and W. T. Ruyechan. 1996. Phosphorylation and nuclear localization of the varicella zoster virus gene 63 protein. J. Virol. 70:658-662. [DOI] [PMC free article] [PubMed] [Google Scholar]