Abstract
Identification of T-cell subsets that are infected in vivo is essential to understanding the pathogenesis of human immunodeficiency virus (HIV) disease; however, this goal has been beset with technical challenges. Here, we used polychromatic flow cytometry to sort multiple T-cell subsets to 99.8% purity, followed by quantitative PCR to quantify HIV gag DNA directly ex vivo. We show that resting memory CD4+ T cells are the predominantly infected cells but that terminally differentiated memory CD4+ T cells contain 10-fold fewer copies of HIV DNA. Memory CD8+ T cells can also be infected upon upregulation of CD4; however, this is infrequent and HIV-specific CD8+ T cells are not infected preferentially. Naïve CD4+ T-cell infection is rare and principally confined to those peripheral T cells that have proliferated. Furthermore, the virus is essentially absent from naïve CD8+ T cells, suggesting that the thymus is not a major source of HIV-infected T cells in the periphery. These data illuminate the underlying mechanisms that distort T-cell homeostasis in HIV infection.
Although human immunodeficiency virus type 1 (HIV-1) infection is characterized by the progressive depletion of CD4+ T cells, the mechanisms underlying this remain controversial (18, 22). Furthermore, the profound qualitative changes in both CD4+ and CD8+ T cells that are observed in HIV-infected individuals are still not well understood (1, 6, 24, 32, 43, 46). It has been estimated that the frequency of CD4+ T-cell infection by HIV in vivo is probably too low to account alone for the death and dysfunction of T cells throughout the disease (5, 17). CD4+ T-cell loss is likely effected by factors including increased memory T-cell turnover, damage to the thymus and other lymphoid tissues, limitations in CD4+ T-cell renewal, and the direct destructive effects of the virus on susceptible T-cell pools.
Developmental and homeostatic relationships between various T-cell compartments—thymocytes, naïve T cells, and resting memory, effector, and terminally differentiated T cells—are affected by HIV. A fuller appreciation of infection within these compartments may lead to a better understanding of HIV disease pathogenesis and events that result in latent infection (13, 20). For example, it has been shown that memory CD4+ T cells specific for HIV are preferentially infected by the virus, which may contribute to the loss of HIV-specific CD4+ T-cell responses (14, 17, 24). Furthermore, it has been shown that developing thymocytes at the CD4+ CD8+ double-positive stage can become infected by HIV and thus might contribute to infection of the peripheral naïve CD4+ and CD8+ T-cell pools (7, 9, 25, 28, 33, 34, 38, 44). Such infection of naïve T cells has important consequences, as they might serve as a quiescent reservoir for virus (19), supply the larger pool of infected memory CD4+ T cells, contribute to infection of CD8+ T-cell pools (30, 34, 44), and attenuate homeostatic maintenance of the diminishing memory CD4+ T-cell pool (18, 23). Additionally, the evidence that dividing CD4+ T cells are infected more efficiently than resting T cells (55) implies that HIV infection, through inhibition of proliferation or cytotoxicity, would affect the in vivo generation of terminally differentiated CD4+ T cells (8).
There are limited data on HIV infection of memory and naïve CD4+ T cells in vivo (10, 42) and CD8+ T cells (25, 30, 34, 44). While studies have demonstrated the presence of HIV in these T-cell subsets, the purity of T-cell populations analyzed ex vivo limits interpretations. However, recent advances in flow cytometry have allowed us to accurately define T-cell subsets with many more parameters than previously and to isolate them at high purity (16).
In this study we used polychromatic flow cytometry to sort multiple CD4+ and CD8+ T-cell subsets, defined by 11 phenotypic parameters, directly from the peripheral blood mononuclear cells (PBMC) of HIV-infected individuals. We compared the frequency of HIV infection by quantitative PCR within each of these subsets. Importantly, subjects with viremia may have increased unintegrated viral DNA compared to individuals with low viral load. Our quantitative PCR assay cannot distinguish between provirus and unintegrated viral DNA. Unintegrated viral DNA would have different physiological consequences compared to viral DNA; however, our assay offers a measure of infection history nonetheless. Our data provide detailed information of the degree to which different T-cell compartments serve as substrates and reservoirs for HIV in vivo and relationships between these compartments. Such ex vivo analysis of the differential infection of T-cell subsets provides a mechanistic framework to comprehend HIV pathogenesis in vivo.
MATERIALS AND METHODS
Study subjects.
Twenty-two HIV-1-infected subjects were recruited for this study. These subjects included men and women with CD4 counts varying from 101 to 1,746 and viral load varying from <50 to 170,004; 11 of the individuals were being treated with highly active antiretroviral therapy, and 11 of the individuals were highly active antiretroviral therapy naïve. Clinical details are shown in Table 1. Viral loads were determined with either the Roche Amplicor Monitor assay or the Roche Ultradirect assay. The subjects all gave informed consent in compliance with the appropriate institutional review board.
TABLE 1.
Subject cohort
| Subject | Age (yr) | Approx. time infected (yr) | CD4 count (cells/μl) | Viral load (copies/ml) | Antiretroviral treatment |
|---|---|---|---|---|---|
| 1 | 47 | 12 | 131 | 47,760 | No |
| 2 | 46 | 5 | 403 | 12,000 | Yes |
| 3 | 45 | 10 | 896 | 18,049 | Yes |
| 4 | 24 | 8 | 412 | 84,093 | No |
| 5 | 62 | 13 | 327 | 125 | Yes |
| 6 | 50 | 8 | 190 | 22,015 | No |
| 7 | 26 | 3 | 644 | <50 | Yes |
| 8 | 46 | 1 | 843 | 11,612 | Yes |
| 9 | 33 | 1 | 120 | 106,000 | No |
| 10 | 29 | 1 | 706 | 13,930 | No |
| 11 | 25 | 1 | 742 | 170,044 | No |
| 12 | 47 | 1 | 231 | 57,430 | No |
| 13 | 30 | 4 | 101 | 60,000 | No |
| 14 | 46 | 6 | 523 | 21,700 | Yes |
| 15 | 46 | 6 | 565 | 548 | Yes |
| 16 | 31 | 3 | 642 | 6,532 | Yes |
| 17 | 62 | 12 | 653 | 3,968 | No |
| 18 | 53 | 11 | 530 | 26,740 | Yes |
| 19 | 47 | 15 | 1,746 | <50 | No |
| 20 | 43 | 13 | 457 | <50 | No |
| 21 | 44 | 16 | 697 | 46,516 | Yes |
| 22 | 50 | 17 | 460 | 449 | No |
Peptides.
Fifteen-mer peptides overlapping by 11 amino acids corresponding to sequences of the chimeric HXBc2/Bal R5 HIV strain were synthesized as free acids, and lyophilized peptides were resuspended and grouped together in corresponding antigen mixtures as previously described (8).
HIV-specific stimulation assay.
Stimulation was performed on fresh or frozen PBMC as described elsewhere (40). Freshly isolated or freshly thawed PBMC were resuspended at 106/ml in RPMI medium supplemented with 10% heat-inactivated fetal calf serum and 1 μg of anti-CD28 and anti-CD49d antibodies per ml. Overlapping peptides were used to stimulate HIV-specific T cells in the presence of brefeldin A (1 μg/ml) (Sigma) for 5 h at 37°C. All cells were surface stained for phenotypic markers of interest and intracellularly stained for cytokines or surface stained for tetrameric major histocompatibility complexes.
Monoclonal antibodies.
Monoclonal antibodies used for phenotypic characterization of T-cell subsets were anti-CD19 conjugated to Cy5-phycoerythrin (Cy5PE), anti-CD14 conjugated to Cy5PE, anti-CD56 conjugated to Cy5PE, anti-CD57 conjugated to fluorescein isothiocyanate, anti-CD27 conjugated to phycoerythrin, anti-gamma interferon (anti-IFN-γ) conjugated to allophycocyanin (APC; Becton Dickinson Pharmingen, San Diego, Calif.), anti-CD45RO conjugated to energy-coupled dye (Coulter), CD3 conjugated to Cascade blue, CD8 conjugated to Cy7PE CD4 conjugated to Alexa 594, and CD11a conjugated to Cy7APC. Unconjugated antibodies against CD3, CD8, CD4, and CD11a were obtained from BD Pharmingen and were then conjugated with the appropriate fluorochrome (Molecular Probes, Eugene, Oreg.; Amersham, Piscataway, N.J.; Prozyme San Leandro, Calif.) with standard protocols (http://drmr.com/abcon). In some experiments, anti-CD31 conjugated to phycoerythrin (BD Pharmingen) and anti-CD27 conjugated to allophycocyanin (BD Pharmingen) were used in lieu of anti-IFN-γ conjugated to allophycocyanin and anti-CD27 conjugated to phycoerythrin.
Flow cytometric cell sorting.
All sorts were performed on stained cells fixed with 1% paraformaldehyde (Electron Microscopy Sciences, Ft. Washington, Pa.) with a modified FACS DIVA (BD Pharmingen). Instrument set-up was performed according to the manufacturer's instructions. All sorts were performed at 25 lb/in2. Instrument compensation was performed with antibody capture beads (BD Pharmingen) stained singly with individual antibodies used in the test samples.
Viral DNA.
HIV DNA was quantified by quantitative PCR with an ABI7700 (Perkin-Elmer, Norwalk, Conn.) as previously described (17). To quantify cell number in each reaction, quantitative PCR was performed simultaneously for albumin gene copy number as previously described (17). Standards were constructed for absolute quantification of Gag and albumin copy number and were validated with sequential dilutions of 8E5 and Ach2 cell lysates, which contain one copy of Gag per cell. Duplicate reactions were run and template copies were calculated with ABI7700 software. When no viral DNA was amplified from a given cell population, we report half the lower limit of detection. As the quantitative PCR for gag DNA is sensitive to a single copy of gag DNA, half the lower limit of detection is based on twice the number of cells put into each PCR (determined by albumin copy number).
Statistical analysis.
Correlations and statistical significance were determined by Spearman rank correlation analysis with Prism 3.0 software (Prism, San Diego, Calif.).
RESULTS
Polychromatic flow cytometry delineates highly purified T-cell subsets.
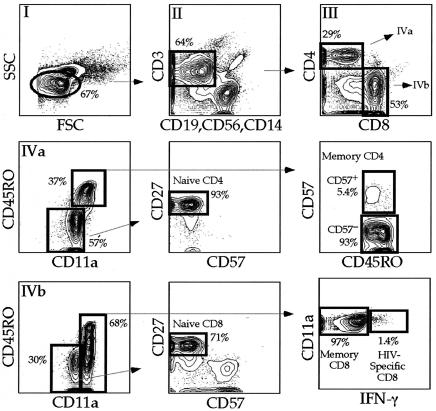
PBMC from 22 HIV-infected individuals (Table 1) were stained with a combination of 11 monoclonal antibodies, and seven T-cell populations were sorted by flow cytometry. We specifically chose this cohort of individuals because it represented a cross-section of HIV-infected individuals at different stages of HIV disease. We simultaneously measured expression of CD3, CD4, CD8, CD11a, CD27, CD45RO, CD57, and IFN-γ. In addition, to minimize background capture of antibodies, we designated one fluorochrome channel as the dump (27). This combination of surface and intracellular molecules was chosen based upon brightness of expression, stability after the freeze-thaw process, ease of individual antibodies to be conjugated to different fluorochromes, and ability to distinguish between naïve, memory, and terminally differentiated T-cell subsets. Using polychromatic flow cytometry technology (Fig. 1), we sorted naïve CD4+ T cells, naïve CD8+ T cells, memory CD8+ T cells, HIV-specific CD8+ T cells, and terminally differentiated and preterminally differentiated memory CD4+ T cells based on the phenotypic characteristics shown in Fig. 1 and described in Table 2.
FIG. 1.
Flow cytometric sorting strategy for T cells. PBMC from 16 subjects in the cohort were stimulated with overlapping HIV-peptides stained extracellularly with the antibody combination described in the text and intracellularly for IFN-γ. Lymphocytes were defined with forward and side scatter (I). CD3+ T cells were then defined based on expression of CD3 without expression of CD56, CD14, or CD19 (dump) (II). CD4+ T cells were then defined based on expression of CD4 without expression of CD8, CD8+ T cells were defined based on expression of CD8 without expression of CD4 (III). Naïve CD4+ T cells were defined based on dull expression of CD11a, no expression of CD45RO or CD57 with expression of CD27 (IV A). Memory CD4+ T cells were defined based on expression of CD45RO with high expression of CD11a. Memory CD4+ T cells were then separated based on expression of CD57. Naïve CD8+ T cells were defined under the same constraints as naïve CD4+ T cells (IV B). Memory CD8+ T cells were separated into HIV-specific (production of IFN-γ or tetramer binding) and other memory CD8+ T cells.
TABLE 2.
T-cell phenotypes
| T-cell subset | Phenotypea
|
||||||
|---|---|---|---|---|---|---|---|
| CD4 | CD8 | CD11a | CD45RO | CD27 | CD57 | IFN-γ | |
| Naïve CD4+ | + | − | Dull | − | + | − | |
| Naïve CD8+ | − | + | Dull | − | + | − | |
| Memory CD57− CD4+ | + | − | Bright | + | − | ||
| Memory CD57− CD4+ | + | − | Bright | + | + | ||
| HIV-specific CD8+ | − | + | Bright | − | |||
| Memory CD8+ | − | + | Bright | + | |||
All populations were also defined as lymphocytes (by forward and side scatter), CD3+, CD14−, CD16−, and CD19−. Blanks indicate that expression of that marker was not used to define the population.
Figure 1 illustrates the necessity of using multiple parameters; for example, naïve CD8+ T cells defined only as CD3+ CD8+ CD4− CD11adull CD45RO− can contain 30% nonnaïve CD8+ T cells. A representative postsort analysis for naïve CD4+ T cells (Fig. 2) revealed that less than 0.2% contaminating cells were present regardless of which phenotypic markers were examined. Postsort analyses of other cell populations were consistently as pure (≥99.8%, data not shown).
FIG. 2.
Postsort analysis of naïve CD4+ T cells. In order to ensure the purity of sorted populations, each sorted population (when possible) was reanalyzed on the same instrument with the same instrument settings. A representative example is shown. Sorted cells must be defined for lymphocytes because cellular debris results from high-speed sorting. The right four plots are only defined for lymphocytes based on characteristic forward and side scatter. All sorted populations were routinely ≥99.8% pure.
Memory CD4+ T cells are infected at highest frequency.
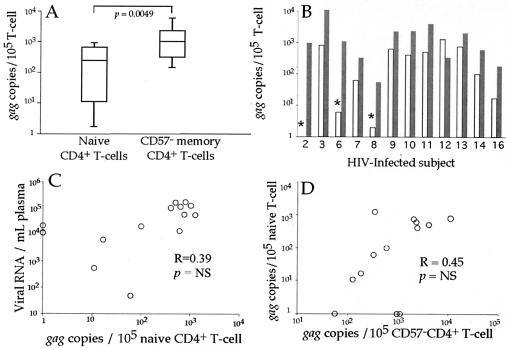
Following flow cytometric sorting, gag DNA frequencies within T-cell subsets were compared. The number of gag DNA copies amplified from each sample was normalized to the number of cells in each PCR and expressed as gag copies per 105 sorted T cells. In all individuals studied, we were able to amplify gag DNA from CD57− CD4+ memory T cells. Between 100 and 10,000 copies of gag DNA/105 sorted T cells were amplified from this population (Fig. 3a and b).
FIG. 3.
CD57+ CD4+ T cells have less viral DNA than memory CD57− CD4+ T cells. PBMC from HIV-infected individuals were stained with the antibody combination detailed in Fig. 2. Memory CD57− and CD57+ CD4+ T cells were sorted, and quantitative PCR for gag DNA and albumin was performed. Infection of CD57+ CD4+ T cells was compared to infection of CD57− memory CD4+ T cells in a subject-independent fashion (A) and a subject-dependent fashion, with white bars representing CD57+ memory CD4+ T cells and shaded bars representing CD57− memory CD4+ T cells (B). Asterisks mark individual subsets where no gag DNA was amplified, and the values listed are calculated based on half of the lower limit of detection. Corresponding subjects are listed along the x axis. The plasma viral load was compared to the number of infected CD57− memory CD4+ T cells (C). While there is a correlation between the number of infected CD57+CD4+ T cells and the number of infected CD57− memory CD4+ T cells (D), the CD57+ population contains significantly less HIV than the CD57− memory CD4+ T-cell subset (A and B).
Memory T cells are a heterogeneous population. In HIV infection this heterogeneity is particularly evident when studying CD57 expression by CD4+ T cells (CD57 marks terminally differentiated T cells) (8, 31, 53). The CD4+ CD57+ T-cell subset overlaps the effector memory T-cell subset (8, 45) and was dramatically expanded in many HIV-infected individuals (P = 0.002, data not shown). Hence, we sought to examine the infection history of memory CD57+ and CD57− memory CD4+ T cells. Collectively, memory CD57− CD4+ T cells contained more gag DNA than did CD57+ CD4+ T cells (Fig. 3a). Assuming virus copy number per cell is distributed similarly between infected cells, memory CD57− CD4+ T cells were always infected more frequently than CD57+ CD4+ T cells (Fig. 3b).
The number of infected CD57− CD4+ memory T cells correlated with plasma viral load (Fig. 3c). In addition, the number of infected CD57+ CD4+ T cells correlated with the number of infected CD57− memory CD4+ T cells (Fig. 3d). This indicates that the plasma virus pool and the pool of infected memory CD57+ and CD57− CD4+ T cells are intimately related.
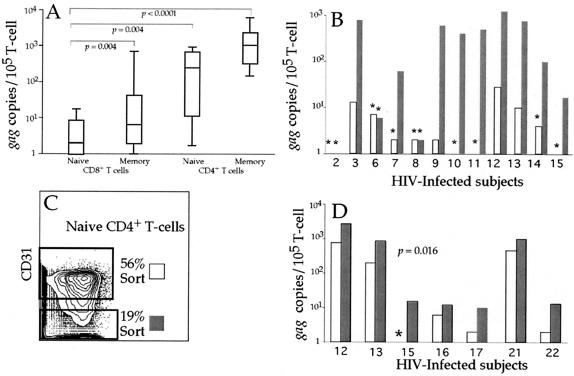
Memory CD8+ T cells are rarely infected by HIV.
Several reports show the presence of HIV in CD8+ T cells (25, 34, 44). Therefore, we initially compared the levels of gag DNA in bulk memory CD8+ T cells with that in memory CD57− CD4+ T cells (Fig. 4a and b). We found that although CD8+ T cells can harbor HIV DNA, the frequency was extremely low. Memory CD8+ T cells from many individuals had no detectable viral DNA (Fig. 4b). However, recently stimulated CD8+ T cells express low levels of CD4 and could be targets for HIV (25, 44). Therefore, we sorted CD8+ CD4dull T cells from five subjects in the cohort. There were more (5- to 100-fold) copies of viral DNA within the CD8+ CD4dull T cells than in memory CD8+ CD4− T cells (data not shown). These data suggest that memory CD8+ T cells are capable of becoming infected after activation-induced expression of CD4.
FIG. 4.
HIV infection of memory CD8+ T cells. The fraction of infected memory CD8+ T cells was compared to the number of infected memory CD57− CD4+ T cells for all subjects (A) and on an individual subject basis (B), with white bars representing memory CD8+ T cells and shaded bars representing CD57− CD4+ T cells. Asterisks mark individual subsets where no gag DNA was amplified, and the values listed are calculated based on half of the lower limit of detection. Corresponding subjects are listed along the x axis. Infection of HIV-specific CD8+ T cells (based on production of IFN-γ following HIV peptide stimulation) was then compared to infection of other memory CD8+ T cells, and no significant differences were observed (C). The infection frequency of HIV-specific CD8+ T cells was then compared to the infection frequency of other memory CD8+ T cells in a subject-dependent fashion (white bars represent HIV-specific CD8+ T cells, and shaded bars represent memory CD8+ T cells) (D). Corresponding subjects are listed along the x axis.
HIV-specific CD8+ T cells are not preferentially infected by HIV.
We hypothesized that HIV-specific CD8+ T cells might become preferentially infected as they respond to HIV antigens and become activated in vivo. Thus, we sorted HIV-specific CD8+ T cells identified by production of IFN-γ following HIV peptide stimulation from seven subjects in the cohort (17, 40). Only in four of these seven individuals were we able to amplify any gag DNA. Thus, while HIV-specific CD8+ T cells were capable of becoming HIV-infected, this population was clearly not preferentially infected (Fig. 4c and d). We also used tetramer binding instead of IFN-γ production to define HIV-specific CD8+ T cells. This confirmed that their infection was extremely rare (subjects 17 to 22, data not shown) and thus unlikely to account for functional defects within this population (1, 11, 36, 39, 51).
Naïve CD4+ T cells are infected at low frequency by HIV.
Naïve T cells may become infected during maturation as thymocytes or as mature naïve T cells in the periphery. To address each possibility, we studied infection of naïve CD4+ and CD8+ T cells. Sufficient naïve CD4+ T cells were sorted from 11 HIV-infected individuals. Naïve CD4+ T cells were found to have significantly less viral DNA (on average 10 times less) than CD57− memory CD4+ T cells (P = 0.005, Fig. 5a and b). In three of the individuals, no gag DNA was amplified from sorted naïve CD4+ T cells. However, in one subject there was more viral DNA in the naïve CD4+ T cells. We found no significant relationship between infection of naïve CD4+ T cells and plasma viral load (Fig. 5c, R = 0.39, P = not significant). In addition, the frequency of infection of naïve CD4+ T cells did not correlate with the frequency of infection of CD57−CD4+ memory T cells (Fig. 5d, R = 0.45, P = not significant). Hence, while naïve CD4+ T cells were capable of becoming infected by HIV, infected naïve CD4+ T cells did not significantly contribute to the pool of infected memory CD4+ T cells. Furthermore, the cellular and viral factors that influence the ability of memory T cells to become infected by HIV may not similarly influence the ability of naïve CD4+ T cells to become HIV infected (17, 19, 43, 47, 54).
FIG. 5.
Naïve CD4+ T cells have less viral DNA than memory CD57− CD4+ T cells. PBMC from HIV-infected individuals were stained with the antibody combination detailed in Fig. 2. Memory CD57− and naïve CD4+ T cells were sorted, and quantitative PCR for gag DNA and albumin was performed on sorted T cells. Infection of naïve CD4+ T cells was compared to infection of CD57− memory CD4+ T cells in a subject-independent fashion (A) and a subject-dependent fashion (B). White bars represent naïve CD4+ T cells, and shaded bars represent CD57− memory CD4+ T cells (B). Corresponding subjects are listed along the x axis. The plasma viral load was compared to the number of infected naïve CD4+ T cells (C). There was no correlation between the number of infected naïve CD4+ T cells and the number of infected CD57− memory CD4+ T cells (D).
Naïve CD8+ T cells are not infected by HIV.
Infection of CD8+CD4+ thymocytes could result in the export of infected mature naïve CD4+ and CD8+ T cells to the periphery. As naïve CD4+ T cells can contain HIV in vivo (Fig. 5), we wanted to determine whether infection of naïve CD8+ T cells could be observed. We sorted sufficient numbers of naïve CD8+ T cells from 12 individuals in the cohort, and were able to amplify gag DNA from only three of the naïve CD8+ T-cell subsets (Fig. 6a and b). Furthermore, more than 107 highly purified naïve CD8+ T cells were sorted in total (cumulative for all 12 subjects) but only six copies of gag DNA were detected. Even at our level of sorting precision (≥99.8%) at this extremely low level of gag DNA we cannot exclude contamination by other T-cell populations, and it is likely that these few copies of gag DNA actually reside in contaminating cells. This suggests that naïve CD8+ T cells carry no HIV and that the thymus exports no infected naïve T cells.
FIG. 6.
Infection of naïve CD8+ T cells and peripheral infection of naïve CD4+ T cells. Infection of highly purified naïve CD8+ T cells was compared to infection of naïve CD4+ T cells, memory CD8+ T cells, and CD57− memory CD4+ T cells (A). Naïve CD8+ T cells are significantly less likely to carry HIV than any other subset studied. Comparison of infection of naïve CD8+ T cells (white bars, B) and naïve CD4+ T cells (shaded bars, B) demonstrates that naïve CD8+ T cells rarely contain detectable viral DNA (asterisks mark individual subsets where no gag DNA was amplified and values are calculated as half the lower limit of detection). Corresponding subjects are listed along the x axis. Naïve CD4+ T cells were stained and defined as before with side scatter, forward scatter, CD3, dump, CD4, CD8, CD45RO, CD11a, CD27, and CD57 (Fig. 2) from four subjects in the cohort (13 to 16) and were then separated on the basis of surface CD31 expression (C). Sorted T cells were then assayed for gag DNA by quantitative PCR. The number of infected naïve CD31+ CD4+ T cells (white bars) was compared to the number of infected naïve CD31− CD4+ T cells (shaded bars) (D).
Naïve CD4+ T cells are infected by HIV in the periphery.
Our observation that naïve CD8+ T cells rarely, if ever, contain gag DNA suggested that naïve CD4+ T cells are likely to have become infected in the periphery. Recently it has been shown that naïve T cells which have proliferated without T-cell receptor-mediated stimulation lose surface expression of CD31 (29). We used CD31 expression to differentiate between naïve CD4+ T cells that had and had not proliferated. In 7 subjects, after gating for naïve CD4+ T cells as before (Fig. 1), we sorted naïve CD4+ T cells based on CD31 expression (Fig. 6c). We initially confirmed that the CD31+ naïve CD4+ T cells had undergone fewer rounds of proliferation. CD31+ naïve CD4+ T cells had, on average fivefold more copies of T-cell receptor excision circle than CD31− naïve CD4+ T cells (data not shown). In addition, in all 7 subjects the frequency of gag DNA was higher in the CD31− than the CD31+ subset (P = 0.016, Fig. 6d). Taken together, these data suggest that infection of naïve CD4+ T cells occurs primarily in the periphery within naïve CD4+ T cells that have or are proliferating and that infection of double positive thymocytes rarely, if ever, leads to infection within the naïve T-cell population.
DISCUSSION
It is generally accepted that activated memory CD4+ T cells are the predominant targets for HIV infection (5, 12). However, it remains unclear what other sources of infected cells exist, what factors lead to their infection, and to what extent these cells contribute to the total pool of infected cells. Understanding which T-cell subsets contain HIV in vivo could establish a mechanistic framework to explain the loss of CD4+ T cells and the inability of the HIV-specific immune response to control HIV replication. Here, we examined in vivo HIV infection of multiple highly purified and stringently defined T-cell subsets by quantifying viral DNA without further in vitro manipulations. The major findings to emerge from these studies are that central memory CD4+ T cells contain the highest frequency of viral DNA; terminally differentiated effector memory CD57+ CD4+ T cells contain, on average, 10 times fewer copies of viral DNA than central memory CD4+ T cells; memory CD8+ T cells rarely contain viral DNA unless activated to express CD4; HIV-specific CD8+ T cells are not preferentially infected by HIV; naïve CD4+ T cells that proliferate, or have proliferated, in the periphery contain more viral DNA than other naïve T cells; and naïve CD8+ T cells are probably never infected. Importantly, these trends are exactly the same regardless of disease state or treatment status.
Taken together, our data show that the T-cell subsets most likely to become infected are those CD4+ T cells with a history of proliferation: CD31− naïve T cells and, to a greater extent, resting memory T cells. However, our data also reveal that infection history itself influences proliferative and maturation capacity in vivo. First, it has been well documented that developing thymocytes can be infected by HIV (2, 4, 7, 37, 48), suggesting that they might give rise to infected naïve CD4+ and CD8+ T cells (19, 34). However, our results show that infection of developing thymocytes is unlikely to lead to infected naïve T cells in the periphery because we were able to find virtually no infected naïve CD8+ T cells in any HIV-infected individuals. This is supported by our observation that most infected naïve CD4+ T cells are of the CD31− phenotype, suggesting that they were probably infected while proliferating in the periphery. Our data do not suggest that developing thymocytes are not infected by HIV in vivo, rather that such infected thymocytes do not become infected naïve T cells. The importance of thymic infection would therefore be one of depleting the supply of new naïve T cells, and not that of supplying HIV-infected naïve T cells. The ability of HIV to infect naïve CD4+ T cells in the periphery suggests the potential ability of the virus to maintain long-lived latency due to the long life span of naïve T cells and that the probability of stimulating an infected naïve CD4+ T-cell by cognate major histocompatibility complex-peptide is extremely low (35).
Second, the lack of correlation between the infected naïve CD4+ T-cell pool and the infected memory CD4+ T-cell pool implies that infected naïve T cells do not significantly contribute to the pool of infected memory CD4+ T cells, but that they die following antigenic stimulation. This suggests a mechanism by which HIV infection can adversely affect maintenance of the memory CD4+ T-cell pool and shows that the predominant way of producing infected memory CD4+ T cells is by their direct infection.
Finally, although the memory CD4+ T-cell pool as a whole is the most frequently infected, we have shown that CD57+ memory CD4+ T cells, which have undergone the most rounds of proliferation to achieve terminal differentiation, are in fact 10-fold less likely to have been infected by HIV. These terminally differentiated memory CD4+ T cells are expanded in HIV infection (15, 31), in part, due to polyclonal T-cell activation (3, 21).
One interpretation of the marked disparity in frequency of infection is that if T cells become infected at an earlier stage in their proliferative history (when they are CD57−), they are less likely to survive and/or divide to become terminally differentiated CD57+ T cells. This would provide direct evidence that infection of memory CD4+ T cells in vivo prevents them from undergoing the normal homeostatic processes that contribute to the maintenance of the resting memory CD4+ T-cell pool. It is also possible that the CD57+ subset contains the same frequency of infected cells as the CD57− subset, but with on average 10-fold fewer copies of virus per cell. Studies with single-cell PCR to detect HIV DNA could help to clarify this possibility but are difficult because the frequency of infected cells is so low (17, 26). It is unlikely that the reason for the difference in infection frequency is simply that terminally differentiated CD57+ T cells are less infectible than other memory T cells. CD57+ T cells express the same levels of CD4 and CCR5/CXCR4 as CD57− T cells (data not shown). Furthermore, both subsets contain an equally small frequency of T cells which express activation markers such as CD69 and CD25, and CD57+ T cells die without proliferating after activation (8); thus, our analysis largely detects infection events that occurred before T cells became terminally differentiated CD57+. However, we also found that there are virtually no CD57+ memory CD4+ T cells that express Ki67 in the periphery. This finding might also contribute to the greater infection within the CD57− memory CD4+ T-cell subset.
Alternatively, the differences we observed in infectivity could arise due to infection of different T-cell subsets by viral subspecies with distinctive tropism or replicative capacity. Of particular interest is whether naïve CD4+ T cells are infected with CXCR4-or CCR5-tropic virus. As naïve CD4+ T cells do not express CCR5, we would speculate that naïve CD4+ T cells infected in the periphery would be infected with CXCR4-tropic virus.
We have previously shown that HIV-specific CD4+ T cells are preferentially infected by HIV (17). Since stimulated CD8+ T cells have been shown to express CD4 transiently following stimulation, leading to marginal infection of CD8+ T cells by HIV (25, 30, 44), we hypothesized that HIV-specific CD8+ T cells might also become preferentially infected by HIV. However, while we show that memory CD8+ T cells are occasionally infected by HIV, we did not find that the virus preferentially infected HIV-specific CD8+ T cells. In fact, we found relatively few copies of HIV gag DNA within HIV-specific CD8+ T cells, implying that infection of this subset neither contributes to the inability to control viral replication nor accounts for the observed defects within this subset (1, 11, 39, 50). Lack of preferential infection of HIV-specific CD8+ T cells might be explained by a number of possibilities. It is possible that upregulation of CD4 by stimulated HIV-specific CD8+ T cells is not sufficient to allow HIV infection. Alternatively, HIV-specific CD8+ T cells may produce enough β chemokines upon stimulation to prevent HIV infection (1, 41, 52). In addition, HIV-specific CD8+ and CD4+ T cells may be stimulated by different cell types or in different locations in vivo.
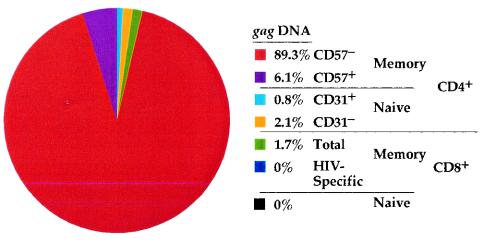
In summary, our data show which T-cell subsets are infected in vivo and to what extent each compartment contributes to the total pool of cellular associated virus (Fig. 7) and suggest what circumstances can lead to their infection and the consequences of that infection. Specifically, this approach allowed us to demonstrate the importance of cellular activation and proliferation in allowing HIV replication in vivo and further to show that infection of these cells in vivo leads to an altering of their life span, decreasing their likelihood of reaching terminal differentiation. Collectively, these findings support a mechanism by which HIV infection exacerbates depletion of CD4+ T cells in the context of homeostatic strain imposed by chronic T-cell activation.
FIG. 7.
T cells that harbor HIV. A pie chart averaged from four subjects in the cohort demonstrates the individual contributions of all T-cell subsets studied to the total pool of infected T cells. The magnitude of infection within each subset and the contribution of each subset to the pool of PBMC were used in the calculation.
Acknowledgments
We thank Steven De Rosa for guidance in polychromatic flow cytometry, Joanne Yu for antibody conjugation, and Steven Perfetto for assistance in instrument operation.
REFERENCES
- 1.Appay, V., D. F. Nixon, S. M. Donahoe, G. M. Gillespie, T. Dong, A. King, G. S. Ogg, H. M. Spiegel, C. Conlon, C. A. Spina, D. V. Havlir, D. D. Richman, A. Waters, P. Easterbrook, A. J. McMichael, and S. L. Rowland-Jones. 2000. HIV-specific CD8(+) T cells produce antiviral cytokines but are impaired in cytolytic function. J. Exp. Med. 192:63-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beltz, L. 1999. Thymic involution and HIV progression. Immunol. Today 20:429. [DOI] [PubMed] [Google Scholar]
- 3.Bentwich, Z., A. Kalinkovich, Z. Weisman, and Z. Grossman. 1998. Immune activation in the context of HIV infection. Clin. Exp. Immunol. 111:1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkowitz, R. D., K. P. Beckerman, T. J. Schall, and J. M. McCune. 1998. CXCR4 and CCR5 expression delineates targets for HIV-1 disruption of T-cell differentiation. J. Immunol. 161:3702-3710. [PubMed] [Google Scholar]
- 5.Blankson, J. N., D. Persaud, and R. F. Siliciano. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 53:557-593. [DOI] [PubMed] [Google Scholar]
- 6.Boaz, M. J., A. Waters, S. Murad, P. J. Easterbrook, and A. Vyakarnam. 2002. Presence of HIV-1 Gag-specific IFN-gamma+IL-2+ and CD28+IL-2+ CD4 T-cell responses is associated with nonprogression in HIV-1 infection. J. Immunol. 169:6376-6385. [DOI] [PubMed] [Google Scholar]
- 7.Bonyhadi, M., L. Rabin, S. Salimi, D. Brown, J. Kosek, J. McCune, and H. Kaneshima. 1993. HIV induces thymus depletion in vivo. Nature 363:728-732. [DOI] [PubMed] [Google Scholar]
- 8.Brenchley, J. M., N. J. Karandikar, M. R. Betts, D. R. Ambrozak, B. J. Hill, L. E. Crotty, J. P. Casazza, J. Kuruppu, S. A. Migueles, M. Connors, M. Roederer, D. C. Douek, and R. A. Koup. 2003. Expression of CD57 defines replicative senescence and antigen-induced apoptotic death of CD8+ T cells. Blood 101:2711-2720. [DOI] [PubMed] [Google Scholar]
- 9.Brooks, D. G., S. G. Kitchen, C. M. Kitchen, D. D. Scripture-Adams, and J. A. Zack. 2001. Generation of HIV latency during thymopoiesis. Nat. Med. 7:459-464. [DOI] [PubMed] [Google Scholar]
- 10.Cayota, A., F. Vuillier, D. Scott-Algara, V. Feuillie, and G. Dighiero. 1993. Differential requirements for HIV-1 replication in naive and memory CD4 T cells from asymptomatic HIV-1 seropositive carriers and AIDS patients. Clin. Exp. Immunol. 91:241-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Champagne, P., G. S. Ogg, A. S. King, C. Knabenhans, K. Ellefsen, M. Nobile, V. Appay, G. P. Rizzardi, S. Fleury, M. Lipp, R. Forster, S. Rowland-Jones, R. P. Sekaly, A. J. McMichael, and G. Pantaleo. 2001. Skewed maturation of memory HIV-specific CD8 T lymphocytes. Nature 410:106-111. [DOI] [PubMed] [Google Scholar]
- 12.Chun, T. W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y. H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183-188. [DOI] [PubMed] [Google Scholar]
- 13.Chun, T. W., D. Engel, S. B. Mizell, C. W. Hallahan, M. Fischette, S. Park, R. T. Davey, Jr., M. Dybul, J. A. Kovacs, J. A. Metcalf, J. M. Mican, M. M. Berrey, L. Corey, H. C. Lane, and A. S. Fauci. 1999. Effect of interleukin-2 on the pool of latently infected, resting CD4+ T cells in HIV-1-infected patients receiving highly active anti-retroviral therapy. Nat. Med. 5:651-655. [DOI] [PubMed] [Google Scholar]
- 14.Demoustier, A., B. Gubler, O. Lambotte, M. G. De Goer, C. Wallon, C. Goujard, J. F. Delfraissy, and Y. Taoufik. 2002. In patients on prolonged highly active antiretroviral therapy, a significant pool of HIV infected CD4 T cells are HIV-specific. AIDS 16:1749-1754. [DOI] [PubMed] [Google Scholar]
- 15.De Paoli, P., A. Carbone, S. Battistin, M. Crovatto, N. Arreghini, and G. Santini. 1987. Selective depletion of the OKT 4+ 4B4+ subset in lymph nodes from HIV+ patients. Immunol. Lett. 16:71-73. [DOI] [PubMed] [Google Scholar]
- 16.De Rosa, S. C., L. A. Herzenberg, and M. Roederer. 2001. 11-color, 13-parameter flow cytometry: identification of human naive T cells by phenotype, function, and T-cell receptor diversity. Nat. Med. 7:245-248. [DOI] [PubMed] [Google Scholar]
- 17.Douek, D., J. Brenchley, M. Betts, D. Ambrozak, B. Hill, Y. Okamoto, J. Casazza, J. Kuruppu, K. Kunstman, S. Wolinsky, Z. Grossman, M. Dybul, A. Oxenius, D. Price, M. Connors, and R. A. Koup. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417:95-98. [DOI] [PubMed] [Google Scholar]
- 18.Douek, D. C., L. J. Picker, and R. A. Koup. 2003. T cell dynamics in HIV-1 infection. Annu. Rev. Immunol. 21:265-304. [DOI] [PubMed]
- 19.Eckstein, D. A., M. L. Penn, Y. D. Korin, D. D. Scripture-Adams, J. A. Zack, J. F. Kreisberg, M. Roederer, M. P. Sherman, P. S. Chin, and M. A. Goldsmith. 2001. HIV-1 actively replicates in naive CD4(+) T cells residing within human lymphoid tissues. Immunity 15:671-682. [DOI] [PubMed] [Google Scholar]
- 20.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 21.Grossman, Z., M. B. Feinberg, and W. E. Paul. 1998. Multiple modes of cellular activation and virus transmission in HIV infection: a role for chronically and latently infected cells in sustaining viral replication. Proc. Natl. Acad. Sci. USA 95:6314-6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grossman, Z., M. Meier-Schellersheim, A. E. Sousa, R. M. Victorino, and W. E. Paul. 2002. CD4+ T-cell depletion in HIV infection: are we closer to understanding the cause? Nat. Med. 8:319-323. [DOI] [PubMed] [Google Scholar]
- 23.Grossman, Z., and W. E. Paul. 2000. The impact of HIV on naive T-cell homeostasis. Nat. Med. 6:976-977. [DOI] [PubMed] [Google Scholar]
- 24.Harari, A., G. P. Rizzardi, K. Ellefsen, D. Ciuffreda, P. Champagne, P. A. Bart, D. Kaufmann, A. Telenti, R. Sahli, G. Tambussi, L. Kaiser, A. Lazzarin, L. Perrin, and G. Pantaleo. 2002. Analysis of HIV-1- and CMV-specific memory CD4 T-cell responses during primary and chronic infection. Blood 100:1381-1387. [DOI] [PubMed] [Google Scholar]
- 25.Imlach, S., S. McBreen, T. Shirafuji, C. Leen, J. E. Bell, and P. Simmonds. 2001. Activated peripheral CD8 lymphocytes express CD4 in vivo and are targets for infection by human immunodeficiency virus type 1. J. Virol. 75:11555-11564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jung, A., R. Maier, J. P. Vartanian, G. Bocharov, V. Jung, U. Fischer, E. Meese, S. Wain-Hobson, and A. Meyerhans. 2002. Multiply infected spleen cells in HIV patients. Nature 418:144. [DOI] [PubMed] [Google Scholar]
- 27.Kantor, A., and M. Roederer. 1997. FACS analysis of lymphocytes, p. 49.1-49.13. In L. A. Herzenberg, D. M. Weir, L. A. Herzenberg, and C. Blackwell (ed.), Handbook of experimental immunology, 5th ed., vol. 2. Blackwell Science, Cambridge, England.
- 28.Keir, M. E., M. G. Rosenberg, J. K. Sandberg, K. A. Jordan, A. Wiznia, D. F. Nixon, C. A. Stoddart, and J. M. McCune. 2002. Generation of CD3+CD8low thymocytes in the HIV type 1-infected thymus. J. Immunol. 169:2788-2796. [DOI] [PubMed] [Google Scholar]
- 29.Kimmig, S., G. K. Przybylski, C. A. Schmidt, K. Laurisch, B. Mowes, A. Radbruch, and A. Thiel. 2002. Two subsets of naive T helper cells with distinct T-cell receptor excision circle content in human adult peripheral blood. J. Exp. Med. 195:789-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitchen, S. G., Y. D. Korin, M. D. Roth, A. Landay, and J. A. Zack. 1998. Costimulation of naive CD8+ lymphocytes induces CD4 expression and allows human immunodeficiency virus type 1 infection. J. Virol. 72:9054-9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Legac, E., B. Autran, H. Merle-Beral, C. Katlama, and P. Debre. 1992. CD4+CD7-CD57+ T cells: a new T-lymphocyte subset expanded during human immunodeficiency virus infection. Blood 79:1746-1753. [PubMed] [Google Scholar]
- 32.Lieberman, J., P. Shankar, N. Manjunath, and J. Andersson. 2001. Dressed to kill? A review of why antiviral CD8 T lymphocytes fail to prevent progressive immunodeficiency in HIV-1 infection. Blood 98:1667-1677. [DOI] [PubMed] [Google Scholar]
- 33.Livingstone, W. J., M. Moore, D. Innes, J. E. Bell, and P. Simmonds. 1996. Frequent infection of peripheral blood CD8-positive T-lymphocytes with HIV-1. Edinburgh Heterosexual Transmission Study Group. Lancet 348:649-654. [DOI] [PubMed] [Google Scholar]
- 34.McBreen, S., S. Imlach, T. Shirafuji, G. R. Scott, C. Leen, J. E. Bell, and P. Simmonds. 2001. Infection of the CD45RA+ (naive) subset of peripheral CD8+ lymphocytes by human immunodeficiency virus type 1 in vivo. J. Virol. 75:4091-4102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michie, C. A., A. McLean, C. Alcock, and P. C. Beverley. 1992. Lifespan of human lymphocyte subsets defined by CD45 isoforms. Nature 360:264-265. [DOI] [PubMed] [Google Scholar]
- 36.Mueller, Y. M., S. C. De Rosa, J. A. Hutton, J. Witek, M. Roederer, J. D. Altman, and P. D. Katsikis. 2001. Increased CD95/Fas-induced apoptosis of HIV-'pecific CD8(+) T cells. Immunity 15:871-882. [DOI] [PubMed] [Google Scholar]
- 37.Namikawa, R., H. Kaneshima, M. Lieberman, I. L. Weissman, and J. M. McCune. 1988. Infection of the SCID-hu mouse by HIV-1. Science 242:1684-1686. [DOI] [PubMed] [Google Scholar]
- 38.Napolitano, L. A., J. C. Lo, M. B. Gotway, K. Mulligan, J. D. Barbour, D. Schmidt, R. M. Grant, R. A. Halvorsen, M. Schambelan, and J. M. McCune. 2002. Increased thymic mass and circulating naive CD4 T cells in HIV-1-infected adults treated with growth hormone. AIDS 16:1103-1111. [DOI] [PubMed] [Google Scholar]
- 39.Oxenius, A., A. K. Sewell, S. J. Dawson, H. F. Gunthard, M. Fischer, G. M. Gillespie, S. L. Rowland-Jones, C. Fagard, B. Hirschel, R. E. Phillips, and D. A. Price. 2002. Functional discrepancies in HIV-specific CD8+ T-lymphocyte populations are related to plasma virus load. J. Clin. Immunol. 22:363-374. [DOI] [PubMed] [Google Scholar]
- 40.Pitcher, C. J., C. Quittner, D. M. Peterson, M. Connors, R. A. Koup, V. C. Maino, and L. J. Picker. 1999. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat. Med. 5:518-525. [DOI] [PubMed] [Google Scholar]
- 41.Price, D. A., A. K. Sewell, T. Dong, R. Tan, P. J. Goulder, S. L. Rowland-Jones, and R. E. Phillips. 1998. Antigen-specific release of beta-chemokines by anti-HIV-1 cytotoxic T lymphocytes. Curr. Biol. 8:355-358. [DOI] [PubMed] [Google Scholar]
- 42.Riley, J. L., B. L. Levine, N. Craighead, T. Francomano, D. Kim, R. G. Carroll, and C. H. June. 1998. Naive and memory CD4 T cells differ in their susceptibilities to human immunodeficiency virus type 1 infection following CD28 costimulation: implications for transmission and pathogenesis. J. Virol. 72:8273-8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roederer, M., P. A. Raju, D. K. Mitra, and L. A. Herzenberg. 1997. HIV does not replicate in naive CD4 T cells stimulated with CD3/CD28. J. Clin. Investig. 99:1555-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saha, K., J. Zhang, A. Gupta, R. Dave, M. Yimen, and B. Zerhouni. 2001. Isolation of primary HIV-1 that target CD8+ T lymphocytes with CD8 as a receptor. Nat. Med. 7:65-72. [DOI] [PubMed] [Google Scholar]
- 45.Sallusto, F., D. Lenig, R. Forster, M. Lipp, and A. Lanzavecchia. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401:708-712. [DOI] [PubMed] [Google Scholar]
- 46.Shankar, P., M. Russo, B. Harnisch, M. Patterson, P. Skolnik, and J. Lieberman. 2000. Impaired function of circulating HIV-specific CD8(+) T cells in chronic human immunodeficiency virus infection. Blood 96:3094-3101. [PubMed] [Google Scholar]
- 47.Spina, C. A., H. E. Prince, and D. D. Richman. 1997. Preferential replication of HIV-1 in the CD45RO memory cell subset of primary CD4 lymphocytes in vitro. J. Clin. Investig. 99:1774-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stoddart, C. A., T. J. Liegler, F. Mammano, V. D. Linquist-Stepps, M. S. Hayden, S. G. Deeks, R. M. Grant, F. Clavel, and J. M. McCune. 2001. Impaired replication of protease inhibitor-resistant HIV-1 in human thymus. Nat. Med. 7:712-718. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan, Y. B., A. L. Landay, J. A. Zack, S. G. Kitchen, and L. Al-Harthi. 2001. Upregulation of CD4 on CD8+ T cells: CD4dimCD8bright T cells constitute an activated phenotype of CD8+ T cells. Immunology 103:270-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trimble, L. A., L. W. Kam, R. S. Friedman, Z. Xu, and J. Lieberman. 2000. CD3zeta and CD28 down-modulation on CD8 T cells during viral infection. Blood 96:1021-1029. [PubMed] [Google Scholar]
- 51.Trimble, L. A., P. Shankar, M. Patterson, J. P. Daily, and J. Lieberman. 2000. Human immunodeficiency virus-specific circulating CD8 T lymphocytes have down-modulated CD3zeta and CD28, key signaling molecules for T-cell activation. J. Virol. 74:7320-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagner, L., O. O. Yang, E. A. Garcia-Zepeda, Y. Ge, S. A. Kalams, B. D. Walker, M. S. Pasternack, and A. D. Luster. 1998. Beta-chemokines are released from HIV-1-specific cytolytic T-cell granules complexed to proteoglycans. Nature 391:908-911. [DOI] [PubMed] [Google Scholar]
- 53.Weyand, C. M., J. C. Brandes, D. Schmidt, J. W. Fulbright, and J. J. Goronzy. 1998. Functional properties of CD4+ CD28- T cells in the aging immune system. Mech. Ageing Dev. 102:131-147. [DOI] [PubMed] [Google Scholar]
- 54.Woods, T. C., B. D. Roberts, S. T. Butera, and T. M. Folks. 1997. Loss of inducible virus in CD45RA naive cells after human immunodeficiency virus-1 entry accounts for preferential viral replication in CD45RO memory cells. Blood 89:1635-1641. [PubMed] [Google Scholar]
- 55.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213-222. [DOI] [PubMed] [Google Scholar]