Abstract
The nucleoid-associated proteins Hha and YdgT repress the expression of the toxin α-hemolysin. An Escherichia coli mutant lacking these proteins overexpresses the toxin α-hemolysin encoded in the multicopy recombinant plasmid pANN202-312R. Unexpectedly, we could observe that this mutant generated clones that no further produced hemolysin (Hly-). Generation of Hly- clones was dependent upon the presence in the culture medium of the antibiotic kanamycin (km), a marker of the hha allele (hha::Tn5). Detailed analysis of different Hly- clones evidenced that recombination between partial IS91 sequences that flank the hly operon had occurred. A fluctuation test evidenced that the presence of km in the culture medium was underlying the generation of these clones. A decrease of the km concentration from 25 mg/l to 12.5 mg/l abolished the appearance of Hly- derivatives. We considered as a working hypothesis that, when producing high levels of the toxin (combination of the hha ydgT mutations with the presence of the multicopy hemolytic plasmid pANN202-312R), the concentration of km of 25 mg/l resulted subinhibitory and stimulated the recombination between adjacent IS91 flanking sequences. To further test this hypothesis, we analyzed the effect of subinhibitory km concentrations in the wild type E. coli strain MG1655 harboring the parental low copy number plasmid pHly152. At a km concentration of 5 mg/l, subinhibitory for strain MG1655 (pHly152), generation of Hly- clones could be readily detected. Similar results were also obtained when, instead of km, ampicillin was used. IS91 is flanking several virulence determinants in different enteric bacterial pathogenic strains from E. coli and Shigella. The results presented here evidence that stress generated by exposure to subinhibitory antibiotic concentrations may result in rearrangements of the bacterial genome. Whereas some of these rearrangements may be deleterious, others may generate genotypes with increased virulence, which may resume infection.
Introduction
Pathogenic bacteria incorporate in their genomes DNA stretches that have been horizontally acquired (HGT DNA) and that contain genes required for the colonization of their hosts. The term “pathogenicity island” (PAI) refers to DNA regions that can be unstable, carry virulence determinants and are usually HGT [1], [2]. These DNA regions can also include, or be flanked by, insertion elements (IS elements) which, in turn, may facilitate integration in different regions of the chromosome. Genes other than those specifically required for virulence can be present in these islands (i.e., antibiotic resistance determinants, catabolic genes or even paralogues of genes encoded in the core genome). Several pathogenicity islands can be spontaneously excised from the chromosome at detectable rates. In most cases, the instability of PAIs is due to their precise excision from the chromosome via recombination between identical directed repeated sequences that flank the element. These short (9 to 20 bp) repeats are analogous to phage att sites. Upon PAI deletion, only one copy of the directed repeat remains on the chromosome [3]. In other instances, IS elements that flank some PAIs mediate deletion of the flanked DNA sequences [4]. As an example, recombination between two flanking IS100 elements has been shown to account for deletion of the high-pathogenicity island (HPI) from Yersinia pestis [5].
IS91 was first discovered in plasmids encoding the toxin α-hemolysin (Hly) in Escherichia coli [6]. IS91 is usually associated to various plasmid and chromosomal pathogenicity islands that harbor the α-hemolysin operon [7], [8], [9], and it was suggested that this element was involved in the dissemination of these pathogenicity determinants [10]. Although initially considered a rarity among IS elements, new examples have evidenced this element flanking several virulence determinants [11]. In addition to the hly genes, IS91 and closely related isoforms have also been located adjacent to several other virulence determinants in enteropathogenic, enterohemolytic and enterotoxigenic strains of E. coli [12], [13]. IS91 like elements differ from other IS elements in that they lack terminal inverted repeats and are considered to transpose by a mechanism termed rolling circle transposition [14].
Bacterial cells have developed several strategies to cope with sudden changes in environmental conditions that result in stress. Several stress-responsive mechanisms rely on regulatory circuits that modify the gene expression pattern [15]. In addition to altering DNA expression, it is also known that stress may account for changes in the DNA sequences. The SOS response, triggered by the accumulation of ssDNA, is a well-characterized mechanism by which bacterial cells, in response to environmental factors that cause DNA damage, increase the mutation or DNA rearrangement rates [16]. Stress-induced adaptative amplifications of DNA have been reported in living organisms [17] and, within the bacterial kingdom, are most studied in E.coli [18], [19], [20]. This microorganism can respond to starvation stress by amplifying specific DNA sequences that allow cells to adjust to these conditions. The general stress response regulator RpoS appears to be required for that response [19]. In enteric bacteria deletions of large DNA stretches, such as those flanked by directed repeats or IS elements, are known to occur. It has been reported that these DNA rearrangements occur spontaneously, at fixed rates [21]. However, in the plant pathogen Pseudomonas syringae pv. Phaseolicola, stress generated by host defenses leads to excision of certain genomic islands and other DNA rearrangements [22]. In this work, experimental data that correlate environmental stress with induction of DNA deletions in Enterobacteria are presented. Our results let us to conclude that exposure to subinhibitory concentrations of certain antibiotics accounts for deletions of genomic islands flanked by partial IS91 sequences.
Results
Kanamycin-dependent deletion of a DNA fragment including the operon encoding the toxin α-hemolysin in Escherichia coli
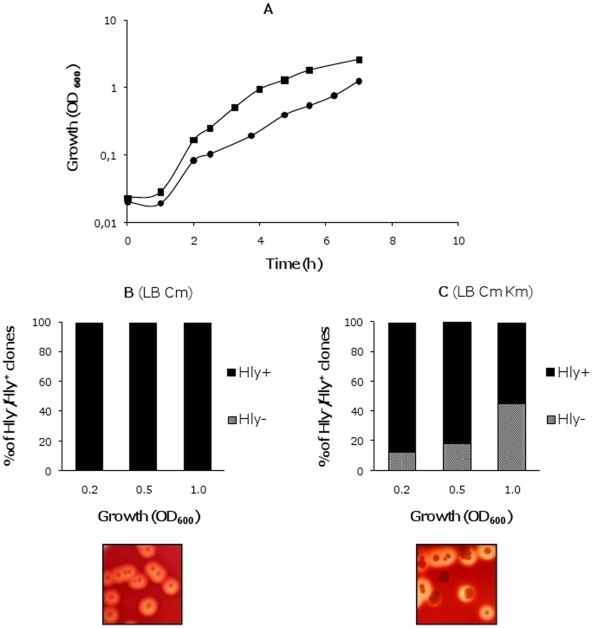
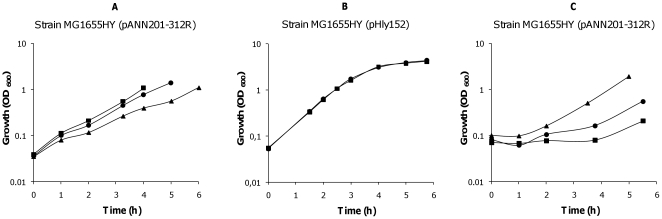
Production of E. coli α-hemolysin is tightly regulated. The nucleoid-associated proteins H-NS and Hha interact to silence expression of the toxin under several environmental conditions, such as low temperature and high osmolarity [23]. Proteins of the Hha family mimic the oligomerization domain of H-NS and form complexes with this latter protein to repress the expression of several virulence determinants (as reviewed in [24]). hns/hha mutants upregulate hemolysin expression, which can be readily visualized on blood agar plates because of their large hemolysis haloes (see Figure 1B). In the E. coli chromosome paralogues to both H-NS and Hha proteins are found: the StpA and YdgT proteins respectively [25]. These protein paralogues can compensate for the lack of either H-NS or Hha, partially attenuating the mutant phenotype. Hence, double hha ydgT mutants show a higher derepression of the hemolysin expression than single hha mutants [26]. The characterization studies of a double hha ydgT mutant from the E. coli strain MG1655 (strain MG1655HY) raised an unexpected phenotype. When strain MG1655HY was transformed with the plasmid pANN202312R (a medium-copy plasmid containing a 17010 bp SalI/SalI fragment with the complete hly operon from the wt plasmid pHly152 [27]), non-hemolytic (Hly-) colonies could be detected on blood agar plates. Interestingly, detection of non-hemolytic derivatives was dependent upon the presence of kanamycin (km) (a marker of the hha mutation) at a concentration of 25 mg/l in the culture medium. We decided to characterize the non-hemolytic derivatives and their relationship to the presence of km in the medium. Overnight cultures of the strain MG1655HY (pANN202-312R) in LB containing chloramphenicol (cm, the plasmid marker) were used to inoculate (1∶100) fresh LB cm and LB cm km media. Growth was monitored and the presence of non-hemolytic clones was quantified. When compared to the growth rate in km-free LB medium, the growth rate in medium containing km was significantly reduced (Figure 1A). Moreover, in LB cm km cultures but not in cultures in LB cm, Hly- clones could be detected. Their proportion increased at the later growth stages (Figures 1B and C).
Figure 1. Generation of non-hemolytic colonies from strain MG1655HY (pANN202-312R).
A. Growth curves of strain MG1655HY (pANN202-312R) in the absence ( ) and presence (
) and presence ( ) of km; B and C, proportion of hemolytic and non-hemolytic clones in both cultures at different stages of the growth curve. Inlets show blood agar plates inoculated with cells collected from both cultures.
) of km; B and C, proportion of hemolytic and non-hemolytic clones in both cultures at different stages of the growth curve. Inlets show blood agar plates inoculated with cells collected from both cultures.
Hly- clones from strain MG1655HY harbor pANN202-312R plasmid derivatives exhibiting two different deletion patterns
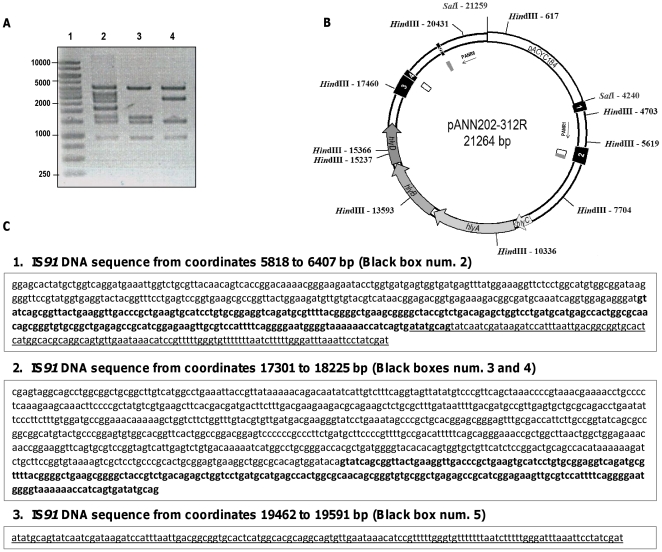
To gain insight into the origin of the Hly- clones, plasmid DNA from thirty Hly- colonies isolated in 6 independent experiments and was characterized by restriction analysis. Interestingly, two deletion patterns were found (Figure 2A). The complete sequence of the 17010 bp SalI/SalI fragment of plasmid pANN202-312R was obtained (GenBank accession number BankIt1460463 Seq1 JN130365), and the sites affected in both types of deletions were precisely determined (Figure 2B internal boxes in white and grey). The DNA sequence obtained complemented previous hybridization studies [6] and evidenced that five incomplete IS91 elements flank the hemolytic determinant of plasmid pHly152. Two DNA motifs, located in IS91 incomplete elements 2, 4 and 5 are repeated in direct orientation flanking both ends of the hly genes (Figure 2C, Figure S1). These sequences are the targets for the deletions that generate the two different restriction patterns observed.
Figure 2. Mapping of the IS91 directed repeats that generate the two deletion patterns observed.
A. HindIII restriction analysis of pANN202-312R plasmid DNA (lane 2) and plasmid DNA isolated from two Hly- clones exhibiting the two different deletion patterns identified (lanes 3 and 4, deletions 1 and 2 respectively). Lane 1 corresponds to the molecular mass marker. B. Physical map of plasmid pANN202-312R. HindIII and SalI restriction sites and their corresponding coordinates are shown. Black boxes 1 to 5 correspond to the five partial IS91 sequences that flank the hly genes. Internal grey boxes correspond to IS91 direct repeats generating deletion 1. Internal white boxes correspond to IS91 direct repeats generating deletion 2. C. DNA sequences of both IS91 direct repeats that flank the deletions. Box 1 shows the DNA sequence corresponding to IS91-2 from coordinates 5818 to 6407. The direct repeat generating deletion 1 is shown underlined. The direct repeat generating deletion 2 is shown in bold. Box 2 corresponds to the DNA sequences of IS91-3 and 4. The direct repeat generating deletion 2 is shown in bold. Box 3 corresponds to the DNA sequence of IS91-5. The direct repeat generating deletion 1 is shown underlined.
Kanamycin accounts for the generation of Hly- clones
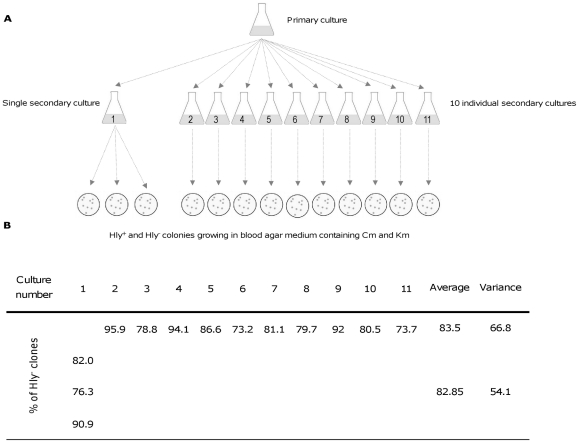
The fact that Hly- clones are only detected when MG1655HY (pANN202-312R) cells are grown in LB medium containing km could be interpreted as either the presence of km in the culture directly accounts for the generation of non-hemolytic derivatives, or it just selects preexisting Hly- clones because of their higher fitness in the presence of the antibiotic. To discern among these two possibilities, a fluctuation test was performed [28] (Figure 3). The mean and variance in the percentage of Hly- clones on the blood agar plates was similar for replicates taken from a single secondary culture and for individual samples taken from different secondary cultures (10). Hence, the fluctuations observed were due to random sampling only, and not to the selection of a preexisting population of Hly- clones. If this latter hypothesis should have been the case, significant fluctuations in the percentage of Hly- clones in the ten independent cultures should have been observed, with a consequently drastic increase in the variance when compared to that of replicates from the same secondary culture. The fluctuation test clearly indicates that the presence of the antibiotic in the culture medium is the underlying cause of deletion of the hly operon and does not account for the selection of preexisting spontaneous deletions.
Figure 3. Fluctuation test.
A. Shows the protocol used. The test was designed to determine if Hly- clones arose prior to exposure to km or specifically in response to exposure. A primary culture of strain MG1655HY (pANN202-312R) was grown in LB medium containing cm but lacking km. Eleven secondary cultures were performed by transferring small amounts of the primary culture (final cell concentration in the secondary culture, 103 cells/ml) into the same culture medium. These cultures underwent many rounds of cell division. From one of the cultures, three replicate subsamples were plated on blood agar plates containing cm and km. From the rest of the cultures, a single sample was plated onto identical plates. B. The percentage of Hly- clones in the different viable counts on blood agar medium containing cm and km.
The role of RecA, RpoS and the SOS response in generating Hly- clones in the strain MG1655HY (pANN202-312R)
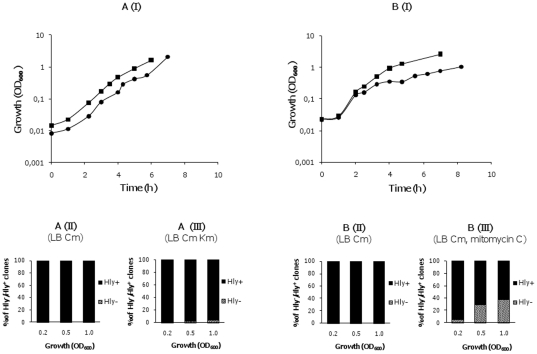
We tested the dependence of the deletions leading to the Hly- phenotype on the RecA protein. A recA mutant of strain MG1655HY was constructed by following the Datsenko&Wanner protocol (see Materials and Methods). recA knockout was confirmed both by PCR and by evidencing that P1-mediated transduction of different markers from strain MG1655 was not possible (data not shown). The strain MG1655HYrecA was transformed with the plasmid pANN202-312R and the transformants grown in LB medium with and without km. Growth of strain MG1655HYrecA (pANN202-312R) was monitored and the proportion of Hly- clones was determined (Figure 4A). km significantly affected the growth rate of strain MG1655HYrecA (pANN202-312R) and Hly- derivatives could be isolated, but at a lower frequency than in the MG1655HY strain. Restriction analysis of plasmid DNA isolated from Hly- clones derived from the recA strain indicated the presence of the two types of deletions previously identified (see Figure 2A). Hence, RecA function appears to facilitate recombination processes between the homologous IS91 sequences, but deletions can also occur in the absence of this protein.
Figure 4. RecA protein and the SOS system play a role in the generation of Hly- derivatives.
A. I. Growth curves of the strain MG1655HYrecA (pANN202-312R) in the absence ( ) and presence (
) and presence ( ) of km; II and III, the proportion of hemolytic and non-hemolytic clones in both cultures at different stages of the growth curve. B. I. Growth curves of the strain MG1655HY (pANN202-312R) in the absence (
) of km; II and III, the proportion of hemolytic and non-hemolytic clones in both cultures at different stages of the growth curve. B. I. Growth curves of the strain MG1655HY (pANN202-312R) in the absence ( ) and presence (
) and presence ( ) of mitomycin C. II and III, the proportion of hemolytic and non-hemolytic clones in both cultures at different stages of the growth curve.
) of mitomycin C. II and III, the proportion of hemolytic and non-hemolytic clones in both cultures at different stages of the growth curve.
RpoS has been shown as a requirement for stationary-phase mutation and DNA amplification [19]. We assessed if deletion of the hly operon required RpoS function. To test this, an rpoS deletion mutant from the strain MG1655HY was constructed and the effect of km on the growth rate and generation of Hly- derivatives was studied. The growth rate of the strain MG1655HYrpoS (pANN202-312R) was also reduced by the presence of km, and Hly- clones harboring the previously characterized deletions were also detected (data not shown).
A well-characterized effect of several bactericidal antibiotics, including km, is the generation of highly deleterious hydroxyl radicals, which leads, among other responses, to the induction of the SOS response [29]. We decided to test if the SOS inducer mitomycin C might also account for the generation of Hly- clones in cultures of the strain MG1655HY (pANN202-312R) in LB medium containing cm. Mitomycin C altered the growth rate and Hly- derivatives could also be isolated (Figure 4B).
A combination of a high kanamycin concentration (25 mg/l) and a high-level of hemolysin production are required to generate non-hemolytic derivatives from the strain MG1655HY
To gain insight into the mechanism underlying km-dependent deletion of the hemolytic determinant of plasmid pANN202-312R, we tested if the concentration of km in the culture medium influenced the generation of Hly- clones. Strain MG1655HY (pANN202-312R) was grown in LB medium with cm (50 mg/l) and different concentrations of km (0, 12.5 and 25 mg/l). Growth and generation of Hly- clones were monitored (Figure 5A). Remarkably, when the km concentration was reduced to 12.5 mg/l the growth rate was similar to that obtained in LB medium and the generation of Hly- clones was no longer observed.
Figure 5. Effect of km concentration, copy number of the hemolytic plasmid and growth temperature on growth rate and generation of Hly- clones.
A. Growth curves of the strain MG1655HY (pANN202-312R) in LB medium containing either no km ( ), 12.5 mg/l km (
), 12.5 mg/l km ( ) or 25 mg/l km (
) or 25 mg/l km ( ). B. Growth curves of the strain MG1655HY (pHly152) in LB medium containing either no km (
). B. Growth curves of the strain MG1655HY (pHly152) in LB medium containing either no km ( ) or 25 mg/l km (
) or 25 mg/l km ( ). C. Growth curves of strain MG1655HY (pANN202-312R) in LB medium at 25 (
). C. Growth curves of strain MG1655HY (pANN202-312R) in LB medium at 25 ( ), 30 (
), 30 ( ) or 37°C (
) or 37°C ( ). Hly- derivatives were only detected in the culture of strain MG1655HY (pANN202-312R) grown in LB medium containing 25 mg/l km.
). Hly- derivatives were only detected in the culture of strain MG1655HY (pANN202-312R) grown in LB medium containing 25 mg/l km.
Next we studied if the copy number of the hly operon would influence the sensitivity to 25 mg/l of km and the generation of Hly- derivatives. Strain MG1655HY harboring the parental low-copy number plasmid pHly152 was grown in LB medium containing no km and LB medium supplemented with 25 mg/l of km (Figure 5B). When growing in the presence of a km concentration of 25 mg/l, no Hly- derivatives were obtained from strain MG1655HY (pHly152). Hence, if hemolysin production is decreased by reducing the copy number of the hemolytic plasmid, no Hly- derivatives are obtained in LB medium containing 25 mg/l of km.
We also determined if plasmids pHly152 or pANN202-312R would modify the sensitivity to km of strain MG1655HY. The minimal inhibitory concentration (MIC) was determined. When comparing the km sensitivity of strains MG1655HY and MG1655HY (pHly152) no significant differences were observed. The presence of the plasmid did not alter the MIC value (data not shown). However, the km MIC was determined for the strains MG1655HY, MG1655HY (pANN202-312R) and MG1655HY (pANN202-312R' (Δhly)), and a very important reduction (10 fold) in the MIC was observed when the strain carries the plasmid pANN202-312R (1000 µg/ml versus 100 µg/ml). Nevertheless, cells harboring plasmid pANN202-312R' exhibited a km MIC similar to that of plasmid-free cells.
Taking into account that km (25 mg/l) influences the growth rate of strain MG1655HY (pANN202-312R), we decided to rule out that the generation of Hly- clones is a consequence of an alteration in the growth rate. Strain MG1655HY (pANN202-312R) was grown in LB medium containing cm (50 mg/l) and no km, at 37, 30 and at 25°C. Growth at 25°C resulted in a significant reduction in the growth rate (Figure 5C) but no Hly- derivatives could be observed in the absence of km.
The above reported results suggest that generation of Hly- derivatives takes place in the hemolysin overproducing strain MG1655HY (pANN202-312R) when it grows in medium containing a km concentration of at least 25 mg/l. Whereas that km concentration is not bactericidal for these cells (km MIC is 100 mg/l), it causes significant effects on the growth rate and also accounts for the generation of Hly- derivatives.
Subinhibitory concentrations of km and other antibiotics result in the deletion of the hemolysin operon in the wild-type MG1655 (pHly152) strain
The biochemical basis underlying the generation of Hly- derivatives in MG1655HY (pANN202-312R) cells growing in medium containing 25 mg/l of km remains to be determined. Nevertheless, the studies described above suggest that 25 mg/l of km is subinhibitory in the strain MG1655HY (pANN202-312R). Growth in the presence of 12.5 mg/l km results both in a significant increase in the growth rate and in the absence of Hly- clones. A feasible explanation for the high sensitivity to km showed by the strain MG1655HY (pANN202-312R) relies on the fact that the ATP-demanding process of producing such high levels of hemolysin would render cells inefficient in completely phosphorylating (inactivating) km at this concentration.
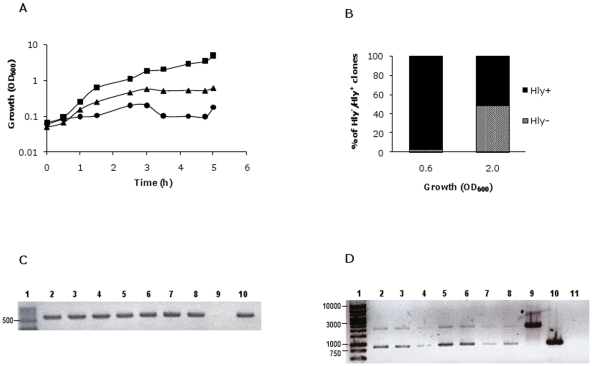
Assuming as correct the hypothesis that a km concentration of 25 mg/l is subinhibitory for strain MG1655HY (pANN202-312R) and that this results in the stress-induced deletion of the hly sequences, it should be expected that the deletion of the hly operon could be also detected by growing the wt strain MG1655 harboring the parental low-copy number hemolytic plasmid pHly152 in the presence of km concentrations that result inhibitory for an E. coli strain lacking a kmR determinant. To assess this, the strain MG1655 (pHly152) was grown in LB medium containing 0, 1, 2, 5 and 10 mg/l of km (Figure 6A). A km concentration of 5 mg/l was the highest antibiotic concentration that allowed cells to grow. Thus, cells growing in liquid LB medium containing 5 mg/l of km were plated on blood agar at different stages of the growth curve. Several non-hemolytic colonies could be detected (48% at an OD600 of 2.0, Figure 6B). The Hly- phenotype could be due either to plasmid curing or to the deletion of hly sequences. To distinguish between these cases, a PCR analysis of several of the Hly- clones was performed using the primers PANR8 and NBM18 (Table 1, Figure 6C). The presence of the hemolytic plasmid was observed in several independent experiments. To determine which deletion of the hly sequences had taken place, different Hly- clones were tested with primer pairs PANR1 and PANR6 (Table 1, Figure 2B). It was observed that both previously described types of deletion occurred, even within the same colony (Figure 6D). These results verify that subinhibitory km concentrations significantly increase the deletion rate of the hly operon of the plasmid pHly152.
Figure 6. Subinhibitory km concentrations result in deletion of Hly sequences in strain MG1655 (pHly152).
A. Growth curves of strain MG1655 (pHly152) in LB medium containing no km ( ), 5 (
), 5 ( ) and 10 (
) and 10 ( ) mg/l of km. B. The percentage of Hly- clones in the culture grown in LB km (5 mg/l). C. PCR analysis using primers PANR8 and NBM18 to confirm the presence of pHly152 in seven Hly- clones (lanes 2 to 8). Lane 1, molecular mass marker. Lane 9, negative control (strain MG1655). Lane 10, positive control (strain MG1655 (pHly152)). D. PCR analysis to confirm deletions 1 and 2 in the Hly- clones. Primers used were PANR1 and PANR6, and the fragments amplified were of 875 bp and 2126 bp for the deletions 1 and 2 respectively.
) mg/l of km. B. The percentage of Hly- clones in the culture grown in LB km (5 mg/l). C. PCR analysis using primers PANR8 and NBM18 to confirm the presence of pHly152 in seven Hly- clones (lanes 2 to 8). Lane 1, molecular mass marker. Lane 9, negative control (strain MG1655). Lane 10, positive control (strain MG1655 (pHly152)). D. PCR analysis to confirm deletions 1 and 2 in the Hly- clones. Primers used were PANR1 and PANR6, and the fragments amplified were of 875 bp and 2126 bp for the deletions 1 and 2 respectively.
Table 1. Oligonucleotides used in this study.
| Name | Sequence (5′ 3′) | Purpose |
| PANR8 | TCTGCGTGGAAGTATGAGC | To determine the presence/absence of pHly152 |
| NBM18 | CCATGCTGATGTGGCGCTTA | |
| PANR1 | GATGATGCCACAAAATGGAT | To determine which recombination took place |
| PANR6 | TTCCTGACACAGAACCGTAA | |
| NBM1 | ACTCAGGAAACCGTAGTACCT | To sequence pANN202-312R |
| NBM3 | GTAAAAGCCGCGAGGACAACG | |
| NBM4 | GAAGCCAAGTCAAACAACAG | |
| NBM5 | GCTGCTTCCTTCAACTGCCA | |
| NBM6 | GGATTGCGCACCGGAAACCC | |
| NBM8 | GATGCTTCCCCGTTTTGCCG | |
| NBM12 | TATGACGCCACCCTACCAGT | |
| NBM14 | CATTGCCATTGAAGCGGAGC | |
| RECAH1 | CAGAACATATTGACTATCCGGTATTACCCGGCATGACAGGAGTAAAAATGGTGTAGGCTGGAGCTGCTTC | To construct recA mutants |
| RECAH2 | ATGCGACCCTTGTGTTACAAACAAGACGATTAAAAATTTCGTTAGTTTCCATATGAATATCCTCCTTAGT | |
| RECAUP | CTTGTGGCAACAATTTCTACA | To check for recA mutation |
| RECADOWN | TCATGGCATATCCTTACAACT | |
| RPOSUP | TGCCGCAGCGATAAATCG | To check for rpoS mutation |
| RPOSDOWN | CGGACCTTTTATTGTGCACAG |
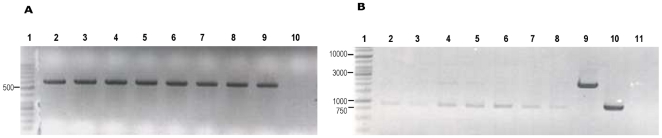
Finally, the effect of subinhibitory concentrations of other antibiotics such as ampicillin (ap) was also tested. In this case the maximal antibiotic concentration allowing MG1655 (pHly152) cells to grow was of 10 mg/l. Again, Hly- clones could readily be detected when cells were grown in LB ap (10 mg/l) medium, and those harboring the plasmid pHly152 exhibited the previously characterized deletions of the hly genes (Figure 7).
Figure 7. Deletion of the hly operon in strain MG1655 (pHly152) growing with a subinhibitory ampicillin concentration.
Analysis of seven Hly- clones isolated from strain MG1655 (pHly152) grown in LB medium containing ap (10 mg/l). A. PCR analysis using primers PANR8 and NBM18 to confirm the presence of pHly152. B. PCR analysis to confirm deletions 1 and 2 in the Hly- clones. Primers used were PANR1 and PANR6, and the fragments amplified were of 875 bp and 2126 bp for the deletions 1 and 2 respectively.
Discussion
Several studies about the effect of antibiotics on bacterial cells have been performed using antibiotic concentrations known to either kill or inhibit the growth of their target cells. The knowledge of the effect on bacteria of subinhibitory antibiotic concentrations has been significantly enhanced in the last years (as reviewed by Davies et al. [30] and Couce and Blazquez [31]). Among other effects, exposure to subinhibitory concentrations of antibiotics leads to increased mutagenesis rates [31], [32], alterations in the global transcriptional pattern [33], [34], lateral gene transfer [35] and intrachromosomal recombination [36], [37]. In this report we provide experimental data that further support the relationship between antibiotics and bacterial genetic variability. Our interest in studying an unexpected observation led us to demonstrate that exposure to subinhibitory concentrations of antibiotics may result in genomic rearrangements that significantly modify the bacterial phenotype. As stated by Shapiro [38], serendipity has led to a more complete view of the capacity that the bacterial cells exhibit for restructuring their genomes in response to several stimuli.
IS91 was first characterized in α-hemolytic plasmids of E. coli [6], [39]. Presently IS91 is the prototype of an expanding family of insertion elements (the ISCR elements, [40]) that have been found adjacent to both virulence determinants ([7], [12], [13], [41]) and antibiotic resistance genes [40]. Remarkably, the ISCR family is widespread among bacteria, but IS91 elements are mainly restricted to E. coli and Shigella strains. In both E. coli and Shigella IS91 has previously been associated to genetic rearrangements. The presence of either complete or partial IS91 sequences flanking the genes encoding heat-labile enterotoxins (eltAB) of enterotoxigenic E. coli strains resulted, among other effects, in spontaneous transpositions of the eltAB genes to the target plasmid pSU2600 [41]. In S. flexneri strain 2a, recombination between two flanking IS91 elements underlies one of the three mechanisms that result in deletion of the SLR pathogenicity island [21]. Our results are in accordance with these data. Remarkably, the 17,010 bp SalI/SalI DNA fragment that includes the Hly operon of plasmid pHly152 cloned in pACYC184 contains only incomplete IS91 copies and, in fact, some of these sequences, flanking the hly operon in a directed repeat orientation, are responsible for the recombinational events that lead to the deletion of two different DNA fragments. In some circumstances, RecA-dependent homologous recombination may be the mechanism underlying deletion of the hly genes (e.g. when the SOS system is induced), but RecA-independent recombination also takes place. RecA-independent IS91-mediated genetic rearrangements were also described for the eltAB genes of enterotoxigenic E. coli strains [41]. The mechanism that accounts for the deletion of the hly operon from plasmid pHly152 is necessarily independent of the IS91 TnpA transposase, since no complete copies of the tnpA gene are present either in the plasmid pANN202-312R or in the chromosome of the E. coli strain MG1655 (our unpublished results). We hypothesized that ORF121, the second and yet uncharacterized ORF encoded by IS91, which is present in one of the partial IS91 copies of the plasmid pANN202-312R, might play a role in these recombinational processes. However, deletion of that sequence did not influence loss of the hemolysin operon in the strain MG1655HY (pANN202-312R) (our unpublished results). Replication is a key process involved in RecA-independent instability of repetitive DNA sequences [42]. Hence, antibiotic-mediated alterations in DNA replication might account for deletion of the hly sequences in the absence of RecA.
Genomic analysis data reveal that IS91 is predominantly present in plasmids isolated from enterohemorragic E. coli strains (serotypes O111, O26 and O157), or in the chromosome and/or plasmids from Shigella isolates (our unpublished results). Interestingly, the six IS91 copies present in the chromosome of the S. flexneri strain 2457T cluster together and flank several virulence determinants [43]. Local hopping was proposed to explain the clustering pattern of these determinants. Nevertheless, if these IS91 elements can under certain circumstances be responsible for genomic rearrangements, then their random distribution in the Shigella chromosome would in many cases result in lethality. The fact that IS91 flanked virulence determinants are restricted to certain E. coli/Shigella strains may indicate that these strains display highly variable pathogenic phenotypes. Therefore, strains that do not incorporate IS91-like elements prevent genomic instability due to the presence of these elements in their genomes.
Several antibiotics such as trimethoprim, quinolones and β-lactams induce the SOS response [44]. It has recently been shown that induction of the SOS response can lead to integrons recombination and the expression of antibiotic resistance determinants in E. coli and Vibrio cholerae [44]. In Staphylococcus aureus, induction of the SOS response leads to dissemination of pathogenicity islands that are packaged in bacteriophages [45]. In this paper we present further evidence demonstrating that induction of the SOS response underlies the excision of a virulence determinant in E. coli. Hence, factors resulting in SOS induction (among them, several antibiotics) may also account for different types of genomic rearrangements in bacteria.
It is likely that bacteria in natural environments encounter subinhibitory concentrations of antibiotics more often than inhibitory concentrations. Moreover, clinical situations where bacteria may be exposed to low levels of antibiotics can occur due to several reasons (e.g. incomplete antibiotic treatment of an infection or the differential pharmacological response of certain tissues to antibiotics). In the case of the E. coli/Shigella pathogenic strains that harbor virulence determinants flanked by IS91 elements, exposure to low levels of antibiotics may result in genetic rearrangements and the deletion of IS91-flanked determinants. Whereas some changes might be deleterious, others may confer new phenotypes that potentiate their virulence in specific niches, hence increasing their invasiveness. Therefore, noncompliance with or incomplete antibiotic treatments should be avoided. Furthermore, a better understanding of the mechanisms underlying IS91-mediated deletion of virulence determinants in response to antibiotic exposure may help to better define additional targets for antimicrobial therapy.
Materials and Methods
Bacterial strains, plasmids, and media
The bacterial strains and plasmids used in this study are listed in Table 2. The plasmid pANN202-312R carries the hlyCABD gene cluster from the low-copy-number hemolytic plasmid pHly152, and was previously described by Godessart et al. [27]. E. coli strains were grown on Luria–Bertani (LB) medium (10 g NaCl, 10 g tryptone and 5 g yeast extract per liter) at 37°C under aerobic conditions. Blood agar was LB medium supplemented with a 5% of sheep blood defibrinated (Oxoid). In the growth medium the following antibiotics were used: chloramphenicol (cm), 50 µg/ml; kanamycin (km), 25 µg/ml (unless otherwise stated) and ampicillin (ap), 10 µg/ml. To study generation of Hly- derivatives from strain MG1655HY as well as from its recA and rpoS derivatives, the corresponding strain was transformed first with plasmid pANN202-312R. Transformant colonies were selected in blood agar containing cm. Single colonies were used to inoculate LB-cm cultures that were grown overnight and used to inoculate (1∶100) erlenmeyer flasks containing LB cm and LB cm km. Five independent colonies from five independent transformation experiments were studied.
Table 2. Strains and plasmids used in this study.
| Strain or plasmid | Description | Source or reference |
| E. coli K-12 laboratory strains: | ||
| MG1655 | F- lambda- ilvG- rfb-50 rph-1 | [50] |
| BSN26H | MC4100 trp::Tn10 hha::Tn5 | [47] |
| BSN26Y | MC4100 trp::Tn10 ΔydgT | [25] |
| MG1655HY | F- lambda- ilvG- rfb-50 rph-1 hha ydgT | This work |
| MG1655HYΔrecA | F- lambda- ilvG- rfb-50 rph-1 hha ydgT recA | This work |
| MG1655HYΔrpoS | F- lambda- ilvG- rfb-50 rph-1 hha ydgT rpoS | This work |
| RH90 | MC4100, rpoS359::Tn10, TcR | [48] |
| Plasmids: | ||
| pHly152 | hlyR hlyCABD | [51] |
| pANN202-321R | hlyR hlyCABD cloned in pACYC184, cmr | [27] |
| pANN202-321R' | pANN202-321R Δhly | This work |
Molecular techniques
Plasmid DNA preparations were carried out from 5 ml of overnight cultures according to the Qiagen kit protocol. Restriction analysis of the plasmid pANN202-312R isolated from hemolytic and non-hemolytic colonies was performed by standard techniques. PCR amplifications of the plasmid pHly152 were performed with a MJ Mini Gradient Thermal Cycler (BioRad®) using the oligonucleotides PANR1-PANR6 and PANR8-NBM18 (listed in table 1) at a Tm of 58 and 56°C respectively.
DNA sequencing
Sequencing was performed with the ABI PRISM Big Dye II Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) in an ABI PRISM 3700 DNA Analyzer (Applied Biosystems), according to the manufacturer's instructions. The sequencing primers used are listed in Table 1.
Genetic manipulations
P1vir mediated transduction was performed as previously described (Miller, [46]) to construct MG1655hha ydgT (HY) using lysates obtained from the strains BSN26hha [47] and BSN26ydgT [25]. An MG1655HYrpoS mutant was obtained by P1vir transduction using a lysate from the RH90 strain [48]. The P1vir mediated transduction was also used to check the recA deletion. A recA deletion mutant was obtained by one-step inactivation using PCR products, as previously described [49]. We used the sequence of the recA gene to define the corresponding amplification primers. The antibiotic resistance of the plasmid pKD3 (cm) was amplified using primers RECAH1 and RECAH2 (Table 1), corresponding to sequences P1 and P2 of plasmid pKD3, with homology extensions corresponding to nucleotides 2821789-2821833, and 2820701-2820752 of the complete genome of E. coli K-12 MG1655 (NC_000913). The PCR product was DpnI digested, purified and used to electroporate strain MG1655HY carrying the plasmid pKD46 grown at 30°C in the presence of arabinose 10 mM, conditions where the expression of Red recombinase was induced. Recombinants were selected at 37°C in LB medium containing chloramphenicol (cm) and then tested for the presence of pKD46. All the recombinants obtained were checked with RECAUP and RECADOWN primers (Table 1) and only one was found correct. We termed this mutant MG1655HYrecA.
Determination of hemolysin production
Strains carrying hemolytic plasmids (pANN202-312R or pHly152) were grown in presence and in absence of the antibiotic km. We checked hemolysin production by visualizing hemolysin haloes on blood agar plates.
Fluctuation test (Luria–Delbrück experiment)
An overnight culture of the strain MG1655HY (pANN202-312R) was used to inoculate eleven erlenmeyer flasks containing 20 ml of LB cm medium (final cell concentration after inoculation, 103 cells/ml). Cultures were incubated at 37°C to a final OD600 = 1. Serial dilutions were then made. From one of the subcultures, three independent serial dilutions were made. From the other ten cultures only one serial dilution was made. Appropriate dilutions were then plated on blood agar containing cm and km and the proportion of Hly- clones was determined.
Supporting Information
Location within the complete IS 91 sequence of the five partial IS 91 sequences present in plasmids pANN202-312R and pHly152.
(TIF)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by funds from the Spanish MICINN-FEDER (Bio2010-15683, BFU2010-21836-C02-01 and CSD2008-00013) and the Generalitat de Catalunya (2009SGR1352). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Blum G, Ott M, Lischewski A, Ritter A, Imrich H, et al. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 1994;62:606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 3.Dozois CM, Curtiss R., III Pathogenic diversity of Escherichia coli and the emergence of “exotic” islands in the gene stream. Vet Res. 1999;30:157–179. [PubMed] [Google Scholar]
- 4.Hacker J, Kaper JB. The concept of pathogenicity islands. In: Kaper JB, Hacker J, editors. Pathogenicity islands and other mobile virulence elements. Washington, DC: American Society for Microbiology; 1999. [Google Scholar]
- 5.Fetherston JD, Schuetze P, Perry RD. Loss of the pigmentation phenotype in Yersinia pestis is due to the spontaneous deletion of 102 kb of chromosomal DNA which is flanked by a repetitive element. Mol Microbiol. 1992;6:2693–2704. doi: 10.1111/j.1365-2958.1992.tb01446.x. [DOI] [PubMed] [Google Scholar]
- 6.Zabala JC, de la Cruz F, Ortiz JM. Several copies of the same insertion sequence are present in alpha-hemolytic plasmids belonging to four different incompatibility groups. J Bacteriol. 1982;151:472–476. doi: 10.1128/jb.151.1.472-476.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcillan-Barcia MP, de la Cruz F. Distribution of IS91 family insertion sequences in bacterial genomes: evolutionary implications. FEMS Microbiol Ecol. 2002;42:303–313. doi: 10.1111/j.1574-6941.2002.tb01020.x. [DOI] [PubMed] [Google Scholar]
- 8.Garcillan-Barcia MP, Bernales I, Mendiola MV, de la Cruz F. IS91 rolling-circle transposition. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Washington, DC: American Society for Microbiology; 2002. Mobile DNA II ASM Press. [Google Scholar]
- 9.Hacker J, Knapp S, Goebel W. Spontaneous deletions and flanking regions of the chromosomally inherited hemolysin determinant of an Escherichia coli O6 strain. J Bacteriol. 1983;154:1145–1152. doi: 10.1128/jb.154.3.1145-1152.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knapp S, Hacker J, Then I, Muller D, Goebel W. Multiple copies of hemolysin genes and associated sequences in the chromosomes of uropathogenic Escherichia coli strains. J Bacteriol. 1984;159:1027–1033. doi: 10.1128/jb.159.3.1027-1033.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz-Aroca E, de la Cruz F, Zabala JC, Ortiz JM. Characterization of the new insertion sequence IS91 from an alpha-hemolytic plasmid of Escherichia coli. Mol Gen Genet. 1984;193:493–499. doi: 10.1007/BF00382089. [DOI] [PubMed] [Google Scholar]
- 12.Burland V, Shao Y, Perna NT, Plunkett G, Sofia HJ, et al. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli 0157:H7. Nucleic Acids Res. 1998;26:4196–4204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolfe MK, de Haan LA, Cassels FJ, Willshaw GA, Warren R, et al. The CS6 colonization factor of human enterotoxigenic Escherichia coli contains two heterologous major subunits. FEMS Microbiol Lett. 1997;148:35–42. doi: 10.1111/j.1574-6968.1997.tb10263.x. [DOI] [PubMed] [Google Scholar]
- 14.Bernales I, Mendiola MV, de la Cruz F. Intramolecular transposition of insertion sequence IS91 results in second-site simple insertions. Mol Microbiol. 1999;33:223–234. doi: 10.1046/j.1365-2958.1999.01432.x. [DOI] [PubMed] [Google Scholar]
- 15.Sharma UJ, Chatterji D. Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity. FEMS Microbiol Rev. 2010;34:646–657. doi: 10.1111/j.1574-6976.2010.00223.x. [DOI] [PubMed] [Google Scholar]
- 16.Baharoglu Z, Bikard D, Mazel D. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet. 2010;6:e1001165. doi: 10.1371/journal.pgen.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hastings PJ. Adaptive amplification. Crit Rev Biochem Mol Biol. 2007;42:271–283. doi: 10.1080/10409230701507757. [DOI] [PubMed] [Google Scholar]
- 18.Hastings PJ, Bull HJ, Klump JR, Rosenberg SM. Adaptive amplification: an inducible chromosomal instability mechanism. Cell. 2000;103:723–731. doi: 10.1016/s0092-8674(00)00176-8. [DOI] [PubMed] [Google Scholar]
- 19.Lombardo MJ, Aponyi I, Rosenberg SM. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics. 2003;166:669–680. doi: 10.1534/genetics.166.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet. 2007;2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turner SA, Luck SN, Sakellaris H, Rajakumar K, Adler B. Nested deletions of the SRL Pathogenicity Island of Shigella flexneri 2a. J Bacteriol. 2001;183:5535–5543. doi: 10.1128/JB.183.19.5535-5543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnold DL, Jackson RW, Waterfield NR, Mansfield JW. Evolution of microbial virulence: the benefits of stress. Trends Genet. 2007;23:293–300. doi: 10.1016/j.tig.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 23.Madrid C, Nieto JM, Paytubi S, Falconi M, Gualerzi CO, et al. Temperature- and H-NS-dependent regulation of a plasmid-encoded virulence operon expressing Escherichia coli hemolysin. J Bacteriol. 2002;184:5058–5066. doi: 10.1128/JB.184.18.5058-5066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madrid C, Balsalobre C, García J, Juárez A. The novel Hha/YmoA family of nucleoid-associated proteins: use of structural mimicry to modulate the activity of the H-NS family of proteins. Mol Microbiol. 2007;63:7–14. doi: 10.1111/j.1365-2958.2006.05497.x. [DOI] [PubMed] [Google Scholar]
- 25.Paytubi S, Madrid C, Forns N, Nieto JM, Balsalobre C, et al. YdgT, the Hha paralogue in Escherichia coli, forms heteromeric complexes with H-NS and StpA. Mol Microbiol. 2004;54:251–263. doi: 10.1111/j.1365-2958.2004.04268.x. [DOI] [PubMed] [Google Scholar]
- 26.Vivero A, Baños RC, Mariscotti JF, Oliveros JC, García-del Portillo F, et al. Modulation of horizontally acquired genes by the Hha-YdgT proteins in Salmonella enterica serovar Typhimurium. J Bacteriol. 2008;190:1152–1156. doi: 10.1128/JB.01206-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Godessart N, Munoa FJ, Regue M, Juárez A. Chromosomal mutations that increase the production of a plasmid-encoded haemolysin in Escherichia coli. J Gen Microbiol. 1988;134:2779–2787. doi: 10.1099/00221287-134-10-2779. [DOI] [PubMed] [Google Scholar]
- 28.Luria SE, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohanski MA, Dwyer DA, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 30.Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9:445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Couce A, Blázquez J. Side effects of antibiotics on genetic variability. FEMS Microbiol Rev. 2009;33:531–538. doi: 10.1111/j.1574-6976.2009.00165.x. [DOI] [PubMed] [Google Scholar]
- 32.Kohanski MA, DePristo MA, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell. 2010;37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goh E, Yim G, Tsui W, McClure J, Surette MG, et al. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci USA. 2002;99:17025–17030. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yim G, de la Cruz F, Spiegelman GB, Davies J. Transcription modulation of Salmonella enterica serovar Typhimurium promoters by Sub-MIC levels of rifampin. J Bacteriol. 2006;188:7988–7991. doi: 10.1128/JB.00791-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427:72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 36.López E, Elez M, Matic I, Blazquez J. Antibiotic-mediated recombination: ciprofloxacin stimulates SOS-independent recombination of divergent sequences in Escherichia coli. Mol Microbiol. 2007;64:83–93. doi: 10.1111/j.1365-2958.2007.05642.x. [DOI] [PubMed] [Google Scholar]
- 37.López E, Blázquez J. Effect of subinhibitory concentrations of antibiotics on intrachromosomal homologous recombination in Escherichia coli. Antimicrob Agents Chemoter. 2009;53:3411–3415. doi: 10.1128/AAC.00358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shapiro JA. Letting Escherichia coli teach me about genome engineering. Genetics. 2009;183:1205–1214. doi: 10.1534/genetics.109.110007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zabala JC, García-Lobo JM, Díaz-Aroca E, de la Cruz F, Ortiz JM. Escherichia coli alpha-haemolysin synthesis and export genes are flanked by a direct repetition of IS91-like elements. Mol Gen Genet. 1984;197:90–97. doi: 10.1007/BF00327927. [DOI] [PubMed] [Google Scholar]
- 40.Toleman MA, Bennett PM, Walsh TR. ISCR elements: novel gene-capturing systems of the 21st Century? Microbiol Mol Biol Rev. 2006;70:296–316. doi: 10.1128/MMBR.00048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlor S, Reidl S, Blass J, Reidl J. Genetic arrangements of the regions adjacent to genes encoding heat-labile enterotoxins (eltAB) of enterotoxigenic Escherichia coli strains. Appl Environ Microbiol. 2000;66:352–358. doi: 10.1128/aem.66.1.352-358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bzymek M, Lovett ST. Instability of repetitive DNA sequences: The role of replication in multiple mechanisms. Proc Natl Acad Sci USA. 2001;98:8319–25. doi: 10.1073/pnas.111008398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zaghloul L, Tang C, Chin HY, Bek EJ, Lan R, et al. The distribution of insertion sequences in the genome of Shigella flexneri strain 2457T. FEMS Microbiol Lett. 2007;277:197–204. doi: 10.1111/j.1574-6968.2007.00957.x. [DOI] [PubMed] [Google Scholar]
- 44.Guerin E, Cambray G, Sanchez-Alberola N, Campoy S, Erill I, et al. The SOS Response controls integron recombination. Science. 2009;324:1034. doi: 10.1126/science.1172914. [DOI] [PubMed] [Google Scholar]
- 45.Úbeda C, Maiques E, Knecht E, Lasa I, Novick RP, et al. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol Microbiol. 2005;56:836–844. doi: 10.1111/j.1365-2958.2005.04584.x. [DOI] [PubMed] [Google Scholar]
- 46.Miller JH. New York: Cold Spring Harbor Laboratory Press; 1992. A Short Course in Bacterial Genetics A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria.456 [Google Scholar]
- 47.Nieto JM, Madrid C, Prenafeta A, Miquelay E, Balsalobre C, et al. Expression of the hemolysin operon in Escherichia coli is modulated by a nucleoid-protein complex that includes the proteins Hha and H-NS. Mol Gen Genet. 2000;263:349–358. doi: 10.1007/s004380051178. [DOI] [PubMed] [Google Scholar]
- 48.Hengge-Aronis R, Fischer D. Identification and molecular analysis of glgS, a novel growth-phase-regulated and rpoS-dependent gene involved in glycogen synthesis in Escherichia coli. Mol Microbiol. 1992;6:1877–1886. doi: 10.1111/j.1365-2958.1992.tb01360.x. [DOI] [PubMed] [Google Scholar]
- 49.Datsenko KA, Wanner LB. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bachmann BJ. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Ingraham JL, Low KB, Magasanik B, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella typhimurium:cellular and molecular biology. Washington, DC: American Society for Microbiology; 1987. [Google Scholar]
- 51.Noegel A, Rdest U, Goebel W. Determination of the functions of hemolytic plasmid pHlyl52 of Escherichia coli. J Bacteriol. 1981;145:233–247. doi: 10.1128/jb.145.1.233-247.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Location within the complete IS 91 sequence of the five partial IS 91 sequences present in plasmids pANN202-312R and pHly152.
(TIF)