Abstract
Four cDNAs, one encoding an α-subunit and three encoding β-subunits of the mitochondrial pyruvate dehydrogenase, were isolated from maize (Zea mays L.) libraries. The deduced amino acid sequences of both α- and β-subunits are approximately 80% identical with Arabidopsis and pea (Pisum sativum L.) homologs. The mature N terminus was determined for the β-subunit by microsequencing the protein purified from etiolated maize shoot mitochondria and was resolved by two-dimensional gel electrophoresis. This single isoelectric species comprised multiple isoforms. Both α- and β-subunits are encoded by multigene families in maize, as determined by Southern-blot analyses. RNA transcripts for both α- and β-subunits were more abundant in roots than in young leaves or etiolated shoots. Pyruvate dehydrogenase activity was also higher in roots (5-fold) compared with etiolated shoots and leaves. Both subunits were present at similar levels in all tissues examined, indicating coordinated gene regulation. The protein levels were highest in heterotrophic organs and in pollen, which contained about 2-fold more protein than any other organ examined. The relative abundance of these proteins in nonphotosynthetic tissues may reflect a high cellular content of mitochondria, a high level of respiratory activity, or an extra plastidial requirement for acetate.
The mitochondrial PDC is composed of multiple copies of three catalytic components, PDH [E1, EC 1.2.4.1], dihydrolipoamide acetyltransferase [E2, EC 2.3.1.12], and dihydrolipoamide dehydrogenase [E3, EC 1.8.1.4], and catalyzes the overall reaction: pyruvate + CoA + NAD+ → acetyl-CoA + NADH + CO2.
The PDH component is composed of nonidentical α- and β-subunits, forming an α2β2 heterotetramer. In mammals, 20 to 30 of these heterotetramers attach to the core of the complex, which is formed by 20 homotrimers of dihydrolipoamide acetyltransferase (for review, see Patel and Roche, 1990).
PDH decarboxylates pyruvate and forms a hydroxyethylidene-TPP intermediate. The C2 group is then transferred from TPP to the lipoyl moiety of the dihydrolipoamide acetyltransferase component by reductive acetylation. Dihydrolipoamide acetyltransferase catalyzes the acetyl transfer to CoA and dihydrolipoamide dehydrogenase reoxidizes the dihydrolipoamide moiety using NAD+ (Reed, 1973).
Most eukaryotic mitochondrial PDCs are regulated by reversible phosphorylation of the E1α-subunit by a specific PDH kinase and phosphatase (Patel and Roche, 1990, and refs. therein). Phosphorylation of E1α reduces its affinity for TPP and prevents pyruvate binding (Korotchkina et al., 1995), effectively inactivating the complex. Although the phosphorylation of one conserved Ser inactivates PDC, two other phosphorylation sites are present on mammalian E1α (Yeaman et al., 1978; Sugden et al., 1979). Plant E1α-subunits are also phosphorylated on Ser residues, but the number and location of the phosphorylation site(s) have not yet been established (Randall et al., 1996).
We have purified the maize (Zea mays L.) mitochondrial PDC, determined its kinetic properties, and established that the E1α- and E1β-subunits are 43 and 40 kD, respectively (Thelen et al., 1998a). The E1α-subunit has at least five isoelectric species, whereas the β-subunit has predominantly one. Although most of the Km and Ki values for maize PDC were typical of those for other plant PDCs, differences were noted that could be important for PDC regulation in C4 plants. For example, the Km for TPP was approximately 10-fold higher in maize than in pea (Pisum sativum L.) PDC. Conversely, the Km for Mg was almost 10-fold lower in maize compared with pea mitochondrial PDC. To better understand these kinetic differences and to begin to establish the molecular properties of PDC in C4 plants, we have undertaken an examination of the E1α- and E1β-subunits of the maize complex at the molecular level.
MATERIALS AND METHODS
Plant Materials
Maize (Zea mays L. cv B73; Illinois Seed Foundation, Urbana, IL) seedlings were grown in a growth chamber (10-h photoperiod) for about 14 d and etiolated maize seedlings were grown in a darkened growth chamber (30°C) for 5 d. Pea (Pisum sativum L. cv Little Marvel), Arabidopsis (cv Columbia), and Kalanchöe daigremontianum seedlings (provided by J. Ringbauer, University of Missouri, Columbia) were grown in a growth chamber (10-h photoperiod, 18°C) for about 14 d (pea), 40 d (Arabidopsis), and 25 d (K. daigremontianum). Mitochondria were isolated from etiolated maize shoots according to the method of Hayes et al. (1991) and from the other tissues using the procedure of Fang et al. (1987).
Radiochemicals
[1-14C]Pyruvate was purchased from New England Nuclear in the solid crystalline form and was dissolved in 6 mL of 20 mm sodium pyruvate containing 3 mm HCl; aliquots (50 μCi; 1 Ci = 37 GBq) were stored at −20°C.
Isolation of cDNAs and Sequencing Strategies
A maize cDNA clone obtained from the Maize EST Stock Center (Columbia, MO) encoded a polypeptide with homology to the C-terminal half of the Arabidopsis PDH E1β-subunit (accession no. U80186). The partial maize cDNA was used as a probe to screen a maize 2-week-old seedling λZAP (Stratagene) expression library (generously provided by Professor Alice Barkan, University of Oregon, Eugene). A screen of more than 3 million transformants yielded approximately 50 positive plaques. DNA was excised and prepared according to the manufacturer's instructions. Nested Exo III deletions were prepared for the longest cDNA according to the “Erase-A-Base” protocol (Promega). The deletion constructs were sequenced using the dye-deoxynucleotide chain termination method using AmpliTaq polymerase (Perkin-Elmer Cetus). Reaction products were analyzed on an automated sequencer (model 373, ABI, Foster City, CA) at the University of Missouri DNA Core Facility.
Two additional unique cDNAs encoding E1β polypeptides were obtained from the Pioneer Hi-Bred International (Johnston, IA) maize EST facility. These cDNAs were prepared and sequenced as described previously. Sequence alignment and phylogenetic analysis were performed with GeneWorks software (IntelliGenetics, Mountain View, CA). The Pioneer Hi-Bred International maize EST database was also examined for cDNAs encoding the E1α-subunit. One cDNA long enough to encode the entire E1α-subunit was found, along with several partial E1α clones. The full-length E1α cDNA was sequenced as described previously.
Protein Microsequencing
Approximately 200 mg of mitochondrial protein from etiolated maize shoots was used to obtain 0.4 mg of highly enriched PDC (as described by Thelen et al., 1998a). For N-terminal protein microsequencing, 100 μg of enriched PDC was resolved by two-dimensional gel electrophoresis according to standard procedures with the following modifications. Sodium thioglycolate (0.25 mm) and glutathione (0.1 mm) were added to the first- and second-dimension gels. The polyacrylamide gel for the second dimension was prerun for 30 min to remove any nonpolymerized acrylamide and persulfate. The protein was blotted to a PVDF membrane using transfer buffer (10 mm 3-[cyclohexylamino]-1-propanesulfonic acid [APS]-NaOH, pH 11.0, and 10% [v/v] methanol). After transfer the protein was stained with amido black (0.1% [w/v] amido black, 40% [v/v] methanol, and 1% [v/v] acetic acid), washed with 50% (v/v) methanol, and then dried. The immobilized proteins were excised and submitted to the University of Nebraska Protein Core Facility (Lincoln, NE) for sequencing by Edman degradation.
Antibodies
Monoclonal antibodies to the α-subunit of the mitochondrial PDH (E1α), the β-subunit to the ATPase, and the heat-shock chaperone (HSP70) were raised in mice immunized with total maize mitochondrial proteins (Luethy et al., 1993, 1995b; Lund et al., 1998). Polyclonal antibodies to the β-subunit of the mitochondrial PDH (E1β) were raised in rabbits immunized with purified recombinant Arabidopsis E1β-maltose-binding fusion protein (M.H. Luethy, unpublished data).
Nucleic Acid Analyses
Total genomic DNA was isolated from 10-d-old green maize leaves according to the method of Sambrook et al. (1989). Digested DNA (20 μg) was separated by electrophoresis on a 0.8% (w/v) agarose gel and then transferred to a Nytran membrane (Schleicher & Schuell). Following transfer, the membrane was UV cross-linked and rinsed in 2× SSC, 0.1% (w/v) SDS prior to prehybridization. Prehybridization was performed at 65°C for 6 h in 2.5× SSPE, 1% (w/v) SDS, 1% nonfat dry milk (Schnuck's, St. Louis, MO), and 0.025% (w/v) denatured salmon-sperm DNA. Hybridization was performed in the same solution and temperature with the entire E1α (EcoRI-XbaI) or E1β isoform 2 (XbaI) cDNA fragment labeled by random hexamer extension (specific activity = 2500 mCi/mg). Subsequent to hybridization, the membrane was washed twice with 2× SSC, 0.1% (w/v) SDS for 1 h, twice with 0.2× SSC, 0.1% (w/v) SDS for 2 h, and was then dried for autoradiographic exposure.
Total RNA was isolated from fresh maize organs using guanidinium extraction (Sambrook et al., 1989). The RNA (40 μg) samples were separated by electrophoresis on a 1.0% (w/v) agarose gel containing 2.2 m formaldehyde and subsequently transferred to a Nytran membrane. Following transfer the membrane was UV cross-linked and stained with 0.03% (w/v) methylene blue and 0.3 m sodium acetate, pH 6.0, to visualize RNA markers and rRNA. Hybridization and washing were carried out as described for the Southern analysis.
RT-PCR of Maize RNA
Approximately 60 μg of maize total RNA isolated from various organs was treated with 5 units of RNase-free DNase (Boehringer Mannheim) for 2 h at 37°C in 10 mm MgCl2, 1 mm DTT, and 50 units of RNase inhibitor (RNasin, Promega). The RNA was then extracted with phenol, precipitated with ethanol, and resuspended in nuclease-free water. The RNA was quantitated by A260 and diluted to 10 ng/μL for use in RT-PCR.
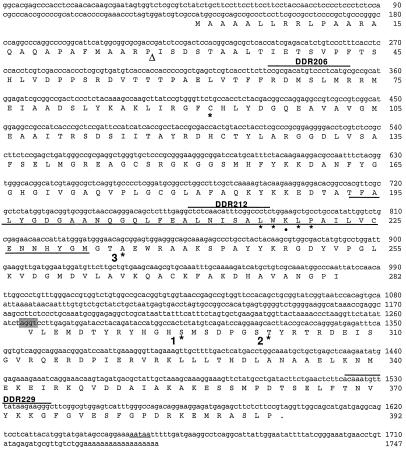
Each RT-PCR reaction contained the following RNase-free reagents: 1.5 mm magnesium sulfate, 0.2 mm deoxyribonucleotide triphosphates, 1.5 pmol/μL oligonucleotides, 0.1 unit/μL avian myeloblastosis virus RT, 0.1 unit/μL Tfl DNA polymerase (Promega), 1× avian myeloblastosis virus RT buffer, 2.5 ng/μL DNase-free RNA. Reverse transcription proceeded for 45 min at 48°C. The PCR cycling was as follows: 2 min at 94°C (one cycle); 30 s at 94°C, 1 min at 60°C, 2 min at 68°C (40 cycles); 7 min at 68°C (one cycle). The oligonucleotides used for RT-PCR span the putative intron of the E1α cDNA (Fig. 2) and are denoted: DDR206, 5′-ccgcgacatgtccctcatgc-3′ (sense oligonucleotide); DDR212, 5′-ctctcaacatttcggccctc-3′ (sense oligonucleotide); and DDR 229, 5′-gcccttcttataaacatttgt-3′ (antisense oligonucleotide).
Figure 2.
Nucleotide and deduced amino acid sequence for maize E1α cDNA. Amino acids are denoted by the single-letter abbreviations. The stop codon is indicated by a period. Underlined region indicates the putative TPP-binding domain. The targeting peptide-processing site, indicated by the triangle, is putative and based on characteristics of cleavage sites (von Heijne et al., 1989). The putative intron splice site is shaded and the polyadenylation signal is underlined. Oligonucleotides used as primers for RT-PCR in Figure 6B are overlined. Numbers to the right indicate base pairs or amino acid number. Asterisks and • denote residues referred to in the text.
Isolation of Proteins and Assay for PDC Activity
Total proteins from various maize organs were prepared by homogenization with a mortar and pestle in liquid nitrogen. Homogenized powder was immediately suspended in SDS-PAGE sample buffer (8 m urea, 4% [v/v] 2-mercaptoethanol and 4% [w/v] SDS) containing 0.01% (w/v) bromphenol blue, mixed thoroughly by vortexing, and incubated at 70°C for 30 min.
In vivo steady-state PDC activity was determined by the rapid-sampling technique of Budde and Randall (1990). Minus-enzyme and acid-precipitated protein controls were performed for each data point to determine background rates. Activity was linear and stable for up to 15 min. Each data point represents the mean of five determinations.
RESULTS
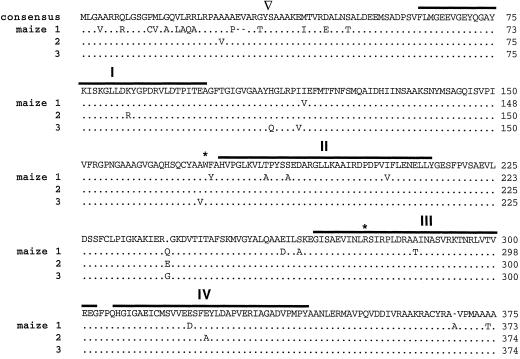
Three E1β cDNA clones were obtained from immature ear (AP9 inbred), green seedling (B73 inbred), and callus culture (B73 inbred) maize libraries and were designated isoforms 1, 2, and 3, respectively. These three cDNAs shared 65% (isoforms 1 and 2), 70% (1 and 3), and 89% (2 and 3) identity at the nucleotide level, whereas 90% identity was observed at the amino acid level (Fig. 1). The three E1β cDNAs were 1511, 1679, and 1655 bp in length, with ORFs starting at bases 82, 258, and 233 and in-frame stop codons at bases 1200, 1379, and 1354 for isoforms 1, 2, and 3, respectively. They encoded polypeptides 373, 374, and 374 amino acids in length with calculated Mrs of 39,767, 39,982, and 39,917. The calculated pI values were 5.3 and 4.8 for the precursor and mature proteins, respectively.
Figure 1.
Amino acid sequence comparison for maize E1β isoforms. Consensus sequence is noted at the top. Dots indicate identity. Gaps, indicated by dashes, were inserted to maximize homology. Overlines indicate the four conserved domains as first pointed out by Wexler et al. (1991). Asterisks denote residues referred to in text. The targeting peptide-processing site is indicated by the inverted triangle. GeneWorks (IntelliGenetics) software was used to perform the alignment algorithm.
The E1α cDNA was obtained from an immature ear (B73 inbred) maize library and was 1747 bp in length with a 930-bp ORF (Fig. 2). However, amino acid identity with the Arabidopsis E1α stopped at base 980 with Ile-282. A second ORF was found 285 bp downstream of Ile-282. This ORF started with Val-283 at base 1267 and had an in-frame stop codon at base 1597. This second ORF encoded a polypeptide with strong amino acid homology to the C-terminal 109 amino acids of Arabidopsis E1α. When joined, the two ORFs encoded a full-length E1α polypeptide 392 amino acids in length with a mass of 42,867 D. The calculated pI values were 8.26 and 6.89 for the precursor and mature protein, respectively. The 285-bp region connecting the two ORFs was likely an unspliced intron. One polyadenylation signal was observed in the E1α cDNA, 52 bases downstream of the stop codon (Fig. 2).
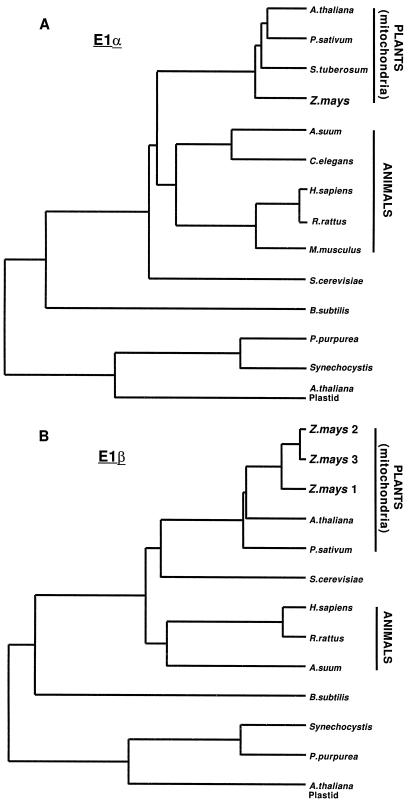
Among all eukaryotes, three distinct classes of E1α polypeptides were apparent in dendrogram analyses (Fig. 3A). The maize E1α polypeptides shared 77% amino acid identity with other plant mitochondrial copies. The E1α-subunits from plants were more closely related to the yeast (48% amino acid identity) than to the animal (43%–47%) polypeptides and were more distantly related to the plastidial (36%), cyanobacterial (30%), and Bacillus subtilis (23%) homologs. The maize mitochondrial E1β was also closely related to other plant mitochondrial E1β-subunits (80% amino acid identity). As is the case for E1α, the plant mitochondrial E1β was more similar to yeast (58% amino acid identity) than to the animal (55%), plastidial (36%), cyanobacterial (36%), or B. subtilis (32%) homologs (Fig. 3B).
Figure 3.
Dendrogram analysis of E1α (A) and β (B) subunits. Clustal alignments were performed using the GeneWorks software package (IntelliGenetics). The length of the horizontal lines indicates inverse degree of relatedness. Accession numbers to the sequences are: Porphyra purpurea (U38804); Solanum tuberosum (Z26949); Synechocystis sp. (D90915); Arabidopsis (U21214, U09137); Mus musculus (M76727); Arabidopsis plastid (U80185, U80186); Caenorhabditis elegans (Z47812); P. sativum (U51918, U56697); Homo sapiens (L13318, D90086); Rattus rattus (Z12158, P49432); Saccharomyces cerevisiae (P16387, M98476); Ascaris suum (M76554, M38017); B. subtilis (M31542).
By comparison with other TPP-binding enzymes (Hawkins et al., 1989; Robinson and Chun, 1993), a potential TPP-binding domain for E1α can be predicted (Fig. 2, underlined). This potential TPP-binding domain was highly conserved and contained one of only two Trp residues present in plant mitochondrial E1α-subunits (L-216–WKLP-220). The three Ser residues phosphorylated in mammalian E1α-subunits are indicated by asterisks in Figure 2. Phosphorylation site 1 (Ser-292), conserved in plastidial and mitochondrial E1α polypeptides, was the site responsible for inactivation (Sugden et al., 1979). Ser-300 at site 2 was conserved only in animal sequences, although the plant mitochondrial proteins contained a Ser one residue upstream of this site. Phosphorylation site 3 (corresponding to Ser-232 of human sequence) is an Ala in all plant mitochondrial E1α-subunits described to date, but is conserved in the mammalian and plastidial subunits.
A comparison of the primary sequence of E1β polypeptides from various organisms (Fig. 1, overlined) revealed the four regions of homology first noted by Wexler et al. (1991). Although conserved, the functional significance of these four regions is uncertain. Regions 2 and 3 are proposed to be involved in oxidative decarboxylation, and chemical covalent modification studies of mammalian PDH have shown that Trp and Arg residues are essential for catalytic activity (Ali et al., 1995; Eswaran et al., 1995). Surprisingly, Trp-173 (Fig. 1, asterisk) was conserved in the mitochondrial but not in the plastidial or bacterial E1β-subunits, whereas Arg-277 was conserved in all organisms except cyanobacteria.
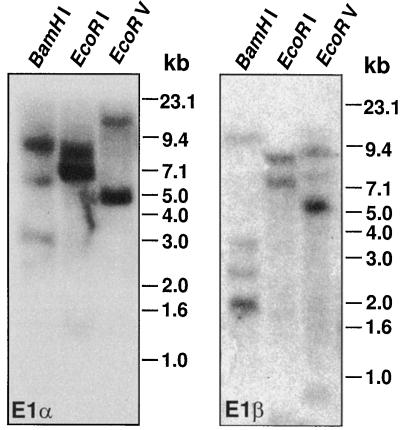
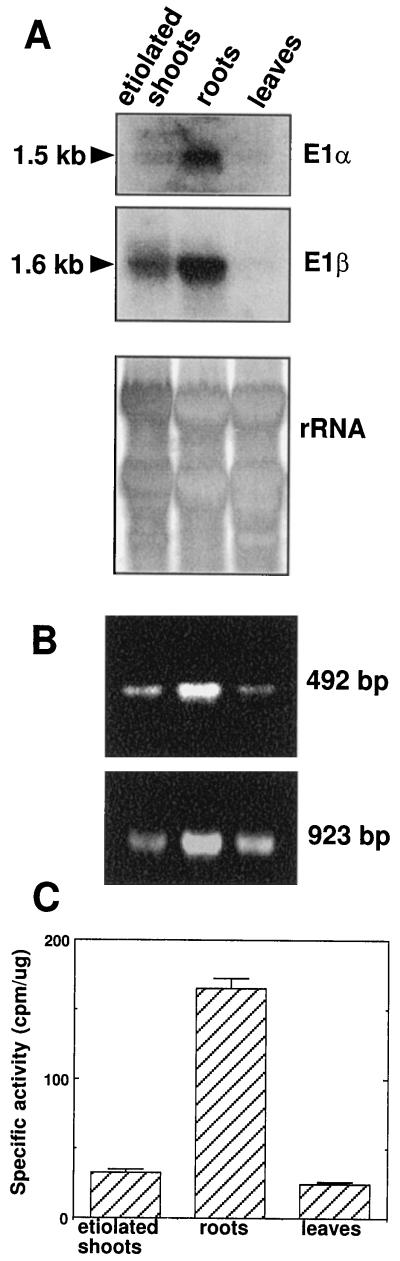
Genomic Southern analysis revealed that the maize genome contains multiple copies of both E1α and β genes (Fig. 4). The multigenic nature of maize E1β was confirmed by the cloning of three unique cDNAs. Northern analysis indicated that the E1α and β transcripts are approximately 1.5 and 1.6 kb in length, respectively (Fig. 5A).
Figure 4.
Genomic Southern analysis of maize E1α (left) and E1β (right). Genomic DNA isolated from maize leaves was digested with the indicated restriction enzymes. Approximately 30 μg of DNA was fractionated by electrophoresis on an agarose gel, transferred to a Nytran membrane, and probed with random-prime-labeled DNA. Marker sizes are indicated to the right in kilobases.
Figure 5.
RNA, RT-PCR, and activity analysis of PDH from maize tissues. A, Approximately 30 μg of total RNA isolated from dark-grown seedlings, roots, or light-adapted leaves was fractionated on an agarose formaldehyde gel, transferred to a Nytran membrane, and probed with the entire E1α or E1β cDNA. RNA marker sizes are indicated. As a gel-loading control, rRNA is included for comparison in the bottom panel. B, Oligonucleotides specific for the E1α cDNA were used for RT-PCR analysis with maize RNA isolated from dark-grown seedlings, roots, or light-adapted leaves as the template. The top panel displays the product obtained with primers DDR212 and DDR229. The product in the bottom panel was obtained with primers DDR206 and DDR229 (Fig. 2). C, In vivo PDH specific activity was determined from maize organs (dark-grown seedlings, roots, or light-adapted leaves) using a radioisotopic assay. Values are the means of five independent reactions. Error bars indicate sd.
Immunoblot Analysis and Protein Microsequencing
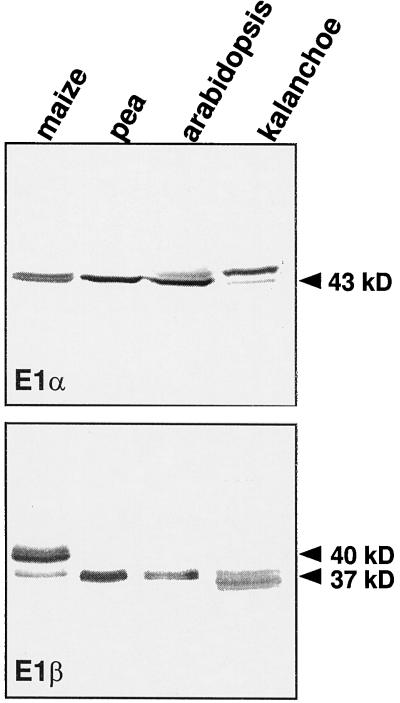
Monoclonal antibodies to maize mitochondrial E1α recognized a 43-kD protein in mitochondria isolated from maize, pea, Arabidopsis, and K. daigremontianum, representing C4 monocot (maize), C3 dicot (pea and Arabidopsis), and CAM plants (K. daigremontranum; Fig. 6). Antibodies raised against recombinant Arabidopsis E1β recognized a 37-kD protein from all plants; however, they primarily recognized a 40-kD polypeptide from maize. On two-dimensional electrophoresis the maize E1β from purified PDC preparations appeared predominantly as a single isoelectric species with a mass of 40 kD (Thelen et al., 1998a).
Figure 6.
Immunoblot analysis of mitochondria purified from various plants. Approximately 10 μg of purified mitochondrial protein was loaded in each lane. Maize mitochondria were obtained from etiolated shoots, whereas pea, Arabidopsis, and K. daigremontianum mitochondria were isolated from light-grown leaves. Molecular masses of polypeptides are indicated.
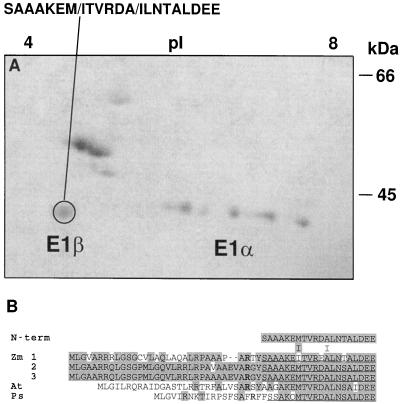
The 40-kD maize E1β-subunit was subjected to N-terminal polypeptide sequencing (Fig. 7A) and yielded 20 residues nearly identical to the deduced amino acid sequence of all three E1β cDNAs, corresponding to Ser-35 through Glu-54 (Fig. 7B). The only difference between the sequenced polypeptide and the deduced sequence of isoforms 2 and 3 (both derived from B73 inbreds) occurred at residue 49, where a Thr instead of a Ser was present. This discrepancy could be the result of a protein-sequencing error or multiple residues at this cycle, some of which went undetected. Comparing the deduced sequence of all three isoforms showed that isoform 1 has an Ile, whereas isoforms 2 and 3 have Met at position 41, indicating that the microsequenced polypeptide was likely a mixture of isoforms.
Figure 7.
N-terminal microsequencing of PDH subunits. A, Coomassie-blue-stained two-dimensional gel electrophoresis of highly purified maize mitochondrial PDC from etiolated shoots. The pI is indicated at the top and the size in kilodaltons is indicated to the right. The circled polypeptide was microsequenced from a replica-blot transferred to a PVDF membrane. At cycles 7 and 12 two residues were obtained (indicated by the slash). B, Comparison of the deduced amino acid sequence for the plant E1β subunits. Zm, Z. mays; At, Arabidopsis; Ps, P. sativum. Shading indicates amino acid identity. Gaps denoted by dashes were inserted to maximize homology. The N termini of the mature maize and pea polypeptides are underlined. Conserved Arg residues involved with peptide processing are indicated in bold type.
Organ-Specific Expression of PDH Subunits
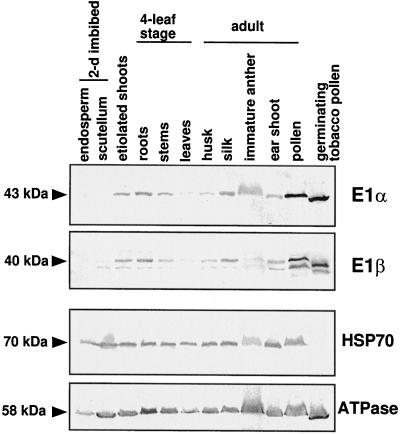
The transcripts for both subunits are more abundant in roots than in etiolated shoots or in light-adapted leaves (Fig. 5, A and B). This expression pattern is also supported by PDH-specific activity measurements showing a 5- and 7-fold higher specific activity in root compared with etiolated shoot and light-adapted leaves, respectively (Fig. 5C). The relative amount of each subunit, determined by immunoblot analysis, revealed that E1α and E1β proteins are abundant in roots, particularly compared with light-adapted leaves (Fig. 8). Both PDH subunits were expressed in similar amounts from all organs examined (Fig. 8). Overall, the PDH subunits are most abundant in nonphotosynthetic organs such as etiolated shoots, roots, flower silks, immature anthers, ear shoots, and, particularly, pollen. Neither subunit was detectable in the endosperm from 2-d-imbibed seeds.
Figure 8.
Immunoblot analyses showing the expression of PDH subunits from various organs. Maize kernels were allowed to soak for 2 d prior to isolating the endosperm and scutellum. Etiolated shoots were grown for 5 d in complete darkness. The remaining samples were obtained from light-grown plants, grown in either a growth chamber (four-leaf stage) or a greenhouse (adult). As a control, the blots were reprobed with antibodies specific to other mitochondrial proteins, HSP70 and the β-subunit to ATP synthase. Approximately 25 μg of a total protein preparation was loaded per lane. Molecular masses of polypeptides are indicated on the left (in kilodaltons).
DISCUSSION
Three unique cDNAs encoding three isoforms of the β-subunit of PDH and at least three hybridized bands on Southern analyses indicate that the maize β-subunit is encoded by a multigene family. Although the amino acid identity among the three isoforms is high, the percentage of identity at the nucleotide level is 65%, 70%, and 89%. Since the cDNAs are quite different, it is unlikely that the differences are due to sequencing errors or artifacts of library construction. The present data indicate that the maize genome is also multigenic for E1α. This multigenic nature suggests the possibility for complex regulation of PDH expression in maize.
The insertion within the ORF of the E1α cDNA can be parsimoniously explained as an unspliced intron. In favor of this is the higher AU content in this region (51%) compared with the coding region (42%) and an AU–UG splice sequence at the 3′ end of the intron, both characteristic of introns (Brown, 1986; Goodall et al., 1989). Furthermore, the transcript size for E1α suggests that this putative intron is normally excised. To confirm this, RT-PCR was performed with primers that overlap the region containing the putative intron to determine whether the intron sequence is typically removed. The PCR products shown in Figure 5B are the proper size amplicons provided that the E1α transcript lacks this 285-bp intron. Perhaps the excision efficiency of this intron is reduced because of the lack of a canonical splice site at the 5′ end.
The first 40 amino acids of the maize E1α and E1β isoforms are enriched with Ala, Arg, and Val residues (about 45%), typical of mitochondrial targeting peptides (von Heijne, 1989; Sjöling and Glaser, 1998). When the first 40 amino acids are modeled as an α-helix, the positively charged Arg residues cluster to one side, whereas the hydrophobic residues cluster on the other side, forming an amphipathic helix (data not shown), another characteristic of mitochondrial targeting peptides (von Heijne, 1989; Sjöling and Glaser, 1998). The mature N terminus of the maize E1β polypeptide was determined by sequencing the purified polypeptide (Fig. 7). Processing of the E1β polypeptides occurs between Tyr-32,34 and Ser-33,35 for maize, between Tyr-29 and Ala-30 (by analogy) for Arabidopsis (accession no. U09137), and between Phe-20 and Ser-21 for pea (accession no. U56697; N.R. David, unpublished results; Fig. 7). The plant E1β polypeptides all have a conserved Arg three residues upstream from the cleavage site, a characteristic of mitochondrial precursor processing (von Heijne, 1989).
We reported previously (Thelen et al., 1998a) that the kinetic properties of PDH from maize are similar to those of other plant species, except for the Km values for TPP and divalent cations. In light of the high amino acid conservation among plant PDH subunits, the discrepancy in Km values might be explained by differences in enzyme preparations (i.e. purity, buffer composition). However, the 10-fold higher Km for TPP is intriguing and might be due to nonconserved substitutions within the TPP-binding site. One particular substitution (Asp-215 [pea] to Lys-218 [maize]; Fig. 2, •) in the TPP-binding domain of E1α is adjacent to an essential hydrophobic residue (Trp-217). Perhaps a basic residue in place of an acidic residue within this conserved region weakens the association with the TPP moeity. Direct determination will require site-directed mutagenesis of the recombinantly expressed E1 subunit.
The E1α and E1β transcripts are most abundant in roots, followed by etiolated shoots and light-adapted leaves (Fig. 5, A and B). This order is the same as the abundance of E1α and E1β transcripts in Arabidopsis organs, i.e. roots contain the most (Luethy et al., 1994, 1995a). The order of the relative abundance for E1α and E1β protein (Fig. 8) follows that of their transcripts, roots > etiolated shoots >> leaves. The specific activities of PDH (Fig. 5C) from roots and leaves also correlate with transcript and protein levels. The specific activity in etiolated shoots was slightly lower than predicted by transcript and immunoblot analysis. However, caution must be used when interpreting PDH activity for three reasons. At present, the contribution of plastidial PDH to the total activity is difficult to estimate. Therefore, to keep plastid activity to a minimum, the assay was performed at pH 7.4 with 0.5 mm MgCl2, which is optimal for the mitochondrial but not for the plastidial PDH (Camp and Randall, 1985). Second, PDH is regulated by reversible phosphorylation. The maize PDH kinase is more abundant in leaves than in roots (Thelen et al., 1998b), which might confound the interpretation of PDH activity. Also, cell breakage necessary for enzyme release might alter the phosphorylation status of PDH due to mixing of subcompartmental ATP. To control for the latter, the time between organ disruption and assay initiation was kept to a minimum of about 15 s.
The steady-state level of both PDH subunits is highest in dry pollen (Fig. 8). To investigate whether PDH is highly expressed in pollen of other species, germinating tobacco pollen was also analyzed (provided by Dr. Stefano Caveniscini, University of Missouri) and the result was the same for both PDH subunits. From these data it is evident that PDH is highly expressed in both dry and germinating pollen. This finding is quite surprising in light of a recent observation that tobacco pollen was able to germinate in the presence of a mitochondrial PDC inhibitor (op den Camp and Kuhlemeier, 1997), which prompted this group to propose the absence of PDC and that an alternative pathway for acetyl-CoA production from pyruvate was operational. Our results suggest that both PDH subunits are abundant in pollen, which points to multiple routes to acetyl-CoA or to an intriguing regulatory difference in pollen PDC.
Immunoblot analysis was used to examine whether PDH polypeptides were differentially expressed in various organs relative to other mitochondrial proteins (Fig. 8). The results show that PDH expression does not correlate with the overall abundance of mitochondria in these organs, which indicates that PDH and perhaps PDC are spatially expressed in a manner different from total mitochondria. Why is PDH spatially regulated, and, in particular, why is it so low in green leaf tissue? Pyruvate is a potent inhibitor of PDH kinase, and in maize total pyruvate levels are relatively high (3–4 mm in leaves; Stitt and Heldt, 1985), which is favorable for PDH activity. Pyruvate generated from the decarboxylation of malate in bundle-sheath cells is recycled to PEP in mesophyll cells for another round of carboxylation (Hatch, 1987, and refs. therein). If the pyruvate intermediate were consumed in either cell type by PDH, pyruvate would need to be synthesized de novo. Since all indications are that pyruvate is recycled, PDH from maize must either have unique properties or be down-regulated in leaves. Except for the unusually high Km for TPP and the lower Km for Mg, the properties of the maize PDH are similar to PDHs from other C3 plants (Thelen et al., 1998a). Perhaps down-regulation of the mitochondrial PDH in light-adapted leaves allows the photosynthetic C3-C4-shuttling mechanism to proceed without consuming the pyruvate intermediate.
The apparent absence of PDH in scutellum and yet the relative abundance of mitochondrial ATPase suggests that carbon for oxidative phosphorylation may be supplied in a form other than through pyruvate. This is surprising in that mobilization of starch from the endosperm would seemingly proceed through glycolysis and the Kreb's cycle. The Kreb's cycle cannot function without acetyl-CoA input. Perhaps ATP needs are met by the oxidative pentose pathway supplying reducing equivalents to the electron transport chain, or acetyl-CoA for the Kreb's cycle is provided by β-oxidation of storage lipids.
In conclusion, we have identified cDNAs encoding both PDH subunits from maize. Both subunits are encoded by multigene families in maize, which may contribute to the surprisingly complex spatial regulation observed by immunoblot analysis. Transcript and protein levels indicate that the two subunits for PDH are coordinately regulated. Based on transcript, protein, and activity measurements, PDH is least abundant in light-adapted leaves, which might be advantageous for C4 metabolic function.
The accession numbers for the maize PDH isoforms reported in this article are AF069911 (E1α) and AF069908, AF069909, and AF069910 (for E1β isoforms 1, 2, and 3, respectively).
ACKNOWLEDGMENTS
The authors are grateful to Dr. Michael Muszynski for his assistance with the database searches and also to Pioneer Hi-Bred International for supplying the cDNA clones. The authors thank Nancy R. David for isolating mitochondria from the various plant species. We also thank Professor Thomas E. Elthon and Dr. Gautum Sarath for the protein microsequencing performed at the Protein Core Facility (University of Nebraska, Lincoln).
Abbreviations:
- EST
expressed sequence tag
- ORF
open reading frame
- PDC
pyruvate dehydrogenase complex
- PDH
pyruvate dehydrogenase
- RT
reverse transcriptase, TPP, thiamin PPi
Footnotes
This research was supported by National Science Foundation grant no. IBN-9419489 and by a Maize Training Grant Fellowship awarded to J.J.T. This is journal report 12,744 from the Missouri Agricultural Experiment Station.
LITERATURE CITED
- Ali MS, Shenoy BC, Eswaran D, Andersson LA, Roche TE, Patel MS. Identification of the tryptophan residue in the thiamin pyrophosphate binding site of mammalian pyruvate dehydrogenase. J Biol Chem. 1995;270:4570–4574. doi: 10.1074/jbc.270.9.4570. [DOI] [PubMed] [Google Scholar]
- Brown J. A catalogue of splice junction and putative branch point sequences from plant introns. Nucleic Acids Res. 1986;14:9549–9559. doi: 10.1093/nar/14.24.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budde RJA, Randall DD. Pea leaf mitochondrial pyruvate dehydrogenase complex is inactivated in vivo in a light-dependent manner. Proc Natl Acad Sci USA. 1990;87:673–676. doi: 10.1073/pnas.87.2.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp PJ, Randall DD. Purification and characterization of the pea chloroplast pyruvate dehydrogenase complex. Plant Physiol. 1985;77:571–577. doi: 10.1104/pp.77.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaran D, Ali MS, Shenoy BC, Korotchkina LG, Roche TE, Patel MS. Arginine-239 in the beta subunit is at or near the active site of bovine pyruvate dehydrogenase. Biochim Biophys Acta. 1995;1252:203–208. doi: 10.1016/0167-4838(95)00119-f. [DOI] [PubMed] [Google Scholar]
- Fang TK, David NR, Miernyk JA, Randall DD. Isolation and purification of functional pea leaf mitochondria free of chlorophyll contamination. Curr Top Plant Biochem Physiol. 1987;6:175. [Google Scholar]
- Goodall GJ, Filipowicz W. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell. 1989;58:473–483. doi: 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- Hatch MD. C4 photosynthesis: a unique blend of modified biochemistry anatomy and ultrastructure. Biochim Biophys Acta. 1987;895:81–106. [Google Scholar]
- Hawkins CF, Borges A, Perham RN. A common structural motif in thiamin pyrophosphate-binding enzymes. FEBS Lett. 1989;255:77–82. doi: 10.1016/0014-5793(89)81064-6. [DOI] [PubMed] [Google Scholar]
- Hayes MK, Luethy MH, Elthon TE. Mitochondrial malate dehydrogenase from corn. Plant Physiol. 1991;97:1381–1387. doi: 10.1104/pp.97.4.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotchkina LG, Khailova LS, Severin SE. The effect of phosphorylation on pyruvate dehydrogenase. FEBS Lett. 1995;364:185–188. doi: 10.1016/0014-5793(95)00382-j. [DOI] [PubMed] [Google Scholar]
- Luethy MH, David NR, Elthon TE, Miernyk JA, Randall DD. Characterization of a monoclonal antibody recognizing the E1α subunit of plant mitochondrial pyruvate dehydrogenase. J Plant Physiol. 1995a;145:443–449. [Google Scholar]
- Luethy MH, Horak A, Elthon TE. Monoclonal antibodies to the α- and β-subunits of the plant mitochondrial F1-ATPase. Plant Physiol. 1993;101:931–937. doi: 10.1104/pp.101.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luethy MH, Miernyk JA, Randall DD. The nucleotide and deduced amino acid sequences of a cDNA encoding the E1β-subunit of the Arabidopsis thaliana mitochondrial pyruvate dehydrogenase complex. Biochim Biophys Acta. 1994;1187:95–98. doi: 10.1016/0005-2728(94)90171-6. [DOI] [PubMed] [Google Scholar]
- Luethy MH, Miernyk JA, Randall DD. The mitochondrial pyruvate dehydrogenase complex: nucleotide and deduced amino-acid sequences of a cDNA encoding the Arabidopsis thaliana E1α subunit. Gene. 1995b;164:251–254. doi: 10.1016/0378-1119(95)00465-i. [DOI] [PubMed] [Google Scholar]
- Lund AA, Blum PH, Bhattramakki D, Elthon TE. Heat-stress response of maize mitochondria. Plant Physiol. 1998;116:1097–1110. doi: 10.1104/pp.116.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- op den Camp RGL, Kuhlemeier C (1997) Aldehyde dehydrogenase in tobacco pollen. Plant Mol Biol 35: 355–365 [DOI] [PubMed]
- Patel MS, Roche TE. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J. 1990;4:3224–3233. doi: 10.1096/fasebj.4.14.2227213. [DOI] [PubMed] [Google Scholar]
- Randall DD, Miernyk JA, David NR, Gemel J, Luethy MH. Regulation of leaf mitochondrial pyruvate dehydrogenase complex activity by reversible phosphorylation. Proc Phytochem Soc Eur. 1996;39:87–103. [Google Scholar]
- Reed LJ. Multienzyme complexes. Acc Chem Res. 1973;7:40–56. [Google Scholar]
- Robinson BH, Chun K. The relationships between transketolase, yeast pyruvate decarboxylase and pyruvate dehydrogenase of the pyruvate dehydrogenase complex. FEBS Lett. 1993;328:99–102. doi: 10.1016/0014-5793(93)80973-x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sjöling S, Glaser E. Mitochondrial targeting peptides in plants. Trends Plant Sci. 1998;3:136–140. [Google Scholar]
- Stitt M, Heldt HW. Generation and maintenance of concentrating gradients between the mesophyll and bundle sheath in maize leaves. Biochim Biophys Acta. 1985;808:400–414. [Google Scholar]
- Sugden PH, Kerbey AL, Randle PJ, Waller CA, Reid KBM. Amino acid sequences around the sites of phosphorylation in the pig heart pyruvate dehydrogenase complex. Biochem J. 1979;181:419–426. doi: 10.1042/bj1810419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen JJ, Miernyk JA, Randall DD. Partial purification and characterization of the maize mitochondrial pyruvate dehydrogenase complex. Plant Physiol. 1998a;116:1443–1450. doi: 10.1104/pp.116.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelen JJ, Miernyk JA, Randall DD. Molecular analysis of two pyruvate dehydrogenase kinases from maize. J Biol Chem. 1998b;273:26618–26623. doi: 10.1074/jbc.273.41.26618. [DOI] [PubMed] [Google Scholar]
- von Heijne G, Steppuhn J, Herrmann RG. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- Wexler ID, Hemalatha SG, Patel MS. Sequence conservation in the α and β subunits of pyruvate dehydrogenase and its similarity to branched-chain α-keto acid dehydrogenase. FEBS Lett. 1991;282:209–213. doi: 10.1016/0014-5793(91)80479-m. [DOI] [PubMed] [Google Scholar]
- Yeaman SJ, Hutcheson ET, Roche TE, Pettit FH, Brown JR, Reed LJ, Watson DC, Dixon GH. Sites of phosphorylation on pyruvate dehydrogenase from bovine kidney and heart. Biochemistry. 1978;17:2364–2370. doi: 10.1021/bi00605a017. [DOI] [PubMed] [Google Scholar]