Abstract
Poxviruses and gamma-2 herpesviruses share the K3 family of viral immune evasion proteins that inhibit the surface expression of glycoproteins such as major histocompatibility complex class I (MHC-I), B7.2, ICAM-1, and CD95(Fas). K3 family proteins contain an amino-terminal PHD/LAP or RING-CH domain followed by two transmembrane domains. To examine whether human homologues are functionally related to the viral immunoevasins, we studied seven membrane-associated RING-CH (MARCH) proteins. All MARCH proteins located to subcellular membranes, and several MARCH proteins reduced surface levels of known substrates of the viral K3 family. Two closely related proteins, MARCH-IV and MARCH-IX, reduced surface expression of MHC-I molecules. In the presence of MARCH-IV or MARCH-IX, MHC-I was ubiquitinated and rapidly internalized by endocytosis, whereas MHC-I molecules lacking lysines in their cytoplasmic tail were resistant to downregulation. The amino-terminal regions containing the RING-CH domain of several MARCH proteins examined catalyzed multiubiquitin formation in vitro, suggesting that MARCH proteins are ubiquitin ligases. The functional similarity of the MARCH family and the K3 family suggests that the viral immune evasion proteins were derived from MARCH proteins, a novel family of transmembrane ubiquitin ligases that seems to target glycoproteins for lysosomal destruction via ubiquitination of the cytoplasmic tail.
Ubiquitination plays a central role in diverse cellular functions, many of which are the consequence of ubiquitin-mediated proteasomal degradation (19). However, ubiquitination also regulates the sorting of proteins along the endocytic route to lysosomes (21). Protein targets are selected for ubiquitination by ubiquitin ligases, also called E3s. E3s simultaneously interact with a substrate and ubiquitin conjugating enzymes (E2s), receiving activated ubiquitin from the ubiquitin activating enzyme (E1). The two major families of E3s contain either HECT domains (for homologous to E6 AP C terminus) or RING domains (for really interesting new gene) (26). The RING domain belongs to a large class of zinc-finger motifs that is characterized by a conserved series of cysteines and histidines: the RING-finger (C3HC4), the RING-H2-finger (C3H2C3), the LIM-finger (C2HC5), and the TRIAD-finger (C6HC). A motif structurally related to the RING-finger is the plant homeodomain (PHD) or the leukemia-associated protein domain (LAP) (1, 32), which is characterized by the C4HC3 sequence. A subfamily of the PHD/LAP domain, termed the BKS domain, was discovered in gamma-2 herpesviruses and poxvirus, as well as several eukaryotic genomes (30). Yeast contains a single protein with this motif, SSM4 or DOA10. SSM4 is responsible for the endoplasmic reticulum (ER)-associated degradation of a subclass of hydrophobic proteins by the proteasome (35). The BKS domain of this protein was further shown to act as a ubiquitin ligase in vitro. Given both the sequence and the functional similarity to the RING and RING-H2 domains, it has been proposed to rename the BKS subtype of PHD/LAP motifs as RING-CH (35), a nomenclature adopted here.
The two RING-CH proteins, K3 and K5, in the genome of Kaposi's sarcoma associated herpesvirus (KSHV), as well as the single K3 gene of murine gamma-2 herpesvirus 68 (MHV68), were determined by several groups to encode proteins that inhibit the surface expression of MHC-I molecules (9, 24, 34). In addition to MHC-I, KSHV-K5 was found to downregulate surface expression of the costimulatory molecules ICAM-1 and B7.2 (10, 23). Sequences homologous to the gamma-2 herpesvirus RING-CH proteins are present in poxvirus genomes (30), and the myxomavirus homologue M153R targets MHC-I and CD4 by a mechanism similar to that used by the herpesviral proteins (17, 28). Thus, it seems that gamma-2 herpesviruses and poxvirus share a family of immune evasion proteins. Several names have been suggested for this family such as modulators of immune recognition (MIR) (11), Scrapins (17), or the K3 family (13).
Target molecules for the K3 family are rapidly internalized from the cell surface and destroyed in the lysosomes (9, 24, 28). A notable exception is MHV68-K3, which associates with MHC-I during peptide loading and causes MHC-I to be degraded by the proteasome (6, 27, 36). K3 family proteins require lysines in the cytoplasmic tail of their target molecules for downregulation, and MHC-I, CD4, or B7.2 molecules are ubiquitinated in cells transfected with K3 proteins (6, 11, 20, 28). Moreover, since the isolated RING-CH domain of several K3 family proteins was shown to display ubiquitin ligase activity in vitro, it was proposed that this family of RING-CH proteins act as E3 enzymes that mediate the ubiquitination of the cytosolic tails of target transmembrane proteins (6, 11, 28).
Since poxviruses and herpesviruses are unrelated viral families, it seemed likely that the viral RING-CH proteins originated from eukaryotic hosts. Several examples exist for host-related immune evasion genes shared between herpesviruses and poxviruses (29). Moreover, the existence of genes homologous to KSHV-K3 and -K5 in the genomes of lower eukaryotes was noted by Nicholas et al. (30) upon completion of the sequence for the corresponding region of the KSHV genome. Furthermore, several investigators described K3-related genes in mammalian genomes that are related to K3 family of viral proteins (22, 25). One of these homologous proteins, termed c-MIR, was recently shown to downregulate B7.2 in a manner very similar to that of KSHV-K5 (16). In contrast to the viral K3 family proteins, however, c-MIR was unable to reduce the surface expression of MHC-I (16). Besides c-MIR, we identified eight additional RING-CH proteins related to the viral proteins. The characterization of several members of this family, termed membrane-associated RING-CH proteins (MARCH), revealed that MARCH-IV and -IX effectively reduced the surface expression of MHC-I but not B7.2. We further report that MHC-I is ubiquitinated and internalized in a lysine-dependent manner in the presence of MARCH-IV and MARCH-IX. To our knowledge, these are the first cellular gene products identified that downregulate the surface expression of MHC-I. These data suggest that the viral K3 family was derived from MARCH-related proteins, a protein family that seems to regulate endocytosis of cell surface receptors via ubiquitination.
MATERIALS AND METHODS
Reagents.
Conanamycin A (Sigma) and lactacystin (Boston Biochem) were used at 50 nM and 20 μM, respectively. Protein A/G beads were from Santa Cruz Biotechnology. The antibodies included AP-1 (Sigma), K455 (from Per A. Peterson), anti-Fas (Pharmingen), anti-TfR (US Biologicals), anti-B7.1 (Pharmingen), anti-B7.2 (Pharmingen), anti-FLAG (Sigma), P4D1 (Santa Cruz), anti-EEA-1 (BD Transduction Laboratories), anti-Golgin-97 (Molecular Probes), anti-Calnexin (Stressgen), anti-Flag:FitC (Sigma), and anti-LAMP-1 (University of Iowa). Hybridomas for W6/32, BB7.2 and OKT4 were obtained from the American Type Culture Collection and were grown in house. HeLa cells used in the present study were stably transfected with tetracycline transactivator (Clontech).
Plasmids.
CDNA for B7.1 was isolated from B7.1/pRMHa3 (from Lars Karlsson) and inserted into pUHD10.1. B7.2 (16) was obtained from Satoshi Ishido, A2.1 and CD4 wild type and lysine mutants were as described previously (28), and vps4 wild type and green fluorescent protein (GFP)-Vps4-E228Q mutant were obtained from Phillip Woodman (5). MARCH-I cDNA was a gift from Sumio Sugano (cDNA FLJ20668, clone KIAA5851). cDNAs for MARCH-II, MARCH-V, MARCH-VII, and MARCH-VIII and both splice variants of MARCH-IX were obtained via the IMAGE consortium (clones 24525, 3905766, 3449089, 4830278, 4811410, and 3936171, respectively). MARCH-IV cDNA was obtained from the Kazusa DNA Research Institute (clone KIAA1399). Each MARCH gene was amplified via PCR with or without a C-terminal FLAG epitope tag by using suitable primers. The PCR products were digested with appropriate restriction enzymes and ligated into either the pUHD10-1 or tetracycline-regulatable pUHG10-3 vector (15). The amino-terminal portions, including the RING-CH domains of selected MARCH-I, -II, -IV, -VIII, and -IX proteins, were fused to glutathione S-transferase (GST) by using pGEX-4T-1 vector (AP Biotech).
Deletion constructs of MARCH-IV were created by amplifying the corresponding region of the MARCH-IV mRNA via PCR by using suitable primers with or without a C-terminal FLAG epitope tag. PCR products were digested with appropriate restriction enzymes and ligated into the pUHD10-1 vector.
Real-time PCR.
Short (50- to 100-bp) fragments from MARCH-I, MARCH-II, MARCH-IV, and MARCH-VIII and two splice variants of MARCH-IX cDNA were amplified via PCR with appropriate primers. A panel of first-strand cDNAs from human tissues (human MTC panel I; Clontech, Palo Alto, Calif.) was used as a template. The concentration of cDNA samples were normalized by using alpha-tubulin, beta-actin, G3PH, and phospholipase A2. The PCR was carried out in buffer composed of 1× SYBR Green PCR buffer (PE Biosystems), 3 mM MgCl2, 0.8 mM concentrations of deoxynucleoside triphosphates, 0.625 U of AmpliTaq Gold (PE Biosystems), 0.01 μl of Amperase (PE Biosystems), and 50 nM concentrations of primers. A total of 23 μl of buffer was added to 2 μl of template cDNA, and the mixture was run under the following conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Amplification was tracked via SYBR Green (PE Biosystems) incorporation by using an ABI Prism 7700 sequence detection system (Applied Biosystems, Foster City, Calif.). Absolute standards were generated by using known quantities of cloned MARCH cDNAs.
Uptake assay.
Uptake of MHC-I was measured as previously described (28). In short, at 24 h posttransfection, HeLa cells were washed, and the polyclonal anti-human MHC-I antiserum K455 (1:100) was allowed to bind for 30 min at 4°C. Cells were washed with phosphate-buffered saline (PBS) and either fixed immediately in 2% paraformaldehyde or incubated in fresh growth medium at 37°C for 120 min to allow uptake of antibody bound MHC-I. K455 was visualized with Alexafluor 594-conjugated goat anti-rabbit secondary antibody (Molecular Probes).
Immunofluorescence analysis and flow cytometry.
For flow cytometry, cells were treated with trypsin, washed with ice-cold PBS, and incubated with appropriate antibodies for 30 min at 4°C. After several washes in PBS, cells were incubated with phycoerythrin-conjugated goat anti-mouse secondary antibody (Dako) and washed again before analysis with a FACScalibur flow cytometer (BD Biosciences).
Immunofluorescence analysis was as described previously (28). In brief, cells were plated on glass coverslips (Fisher), transfected for 24 h, fixed with 2% paraformaldehyde, permeabilized with 0.2% Triton X-100, and blocked with 3% bovine serum albumin and 0.5% fish gelatin. After being stained with primary antibody and secondary antibodies for at least 30 min at 37°C, coverslips were mounted on slides and covered with Vectashield H-1200 plus DAPI (4′,6′-diamidino-2-phenylindole; Vector Laboratories). All pictures were taken in monochrome, contrast enhanced, and artificially colored by using Openlab software (Improvision).
Metabolic labeling, immunoprecipitation, and Western blotting.
After starving in methionine-free medium for 30 min, HeLa transfectants were metabolically labeled with [35S]cysteine-[35S]methionine (250 μCi; Amersham) for the indicated times. Labeled cells were washed twice with PBS and lysed immediately in PBS containing 1% Triton X-100 and protease inhibitors (Roche). Lysates were precleared with protein A/G-agarose beads and incubated with 3 μg of antibody for 1 h, followed by 2 h of incubation with protein A/G-agarose beads. Immunoprecipitated proteins were washed five times with 0.1% Triton X-100 in PBS. Reimmunoprecipitation experiments were as described previously (28).
For immunoblotting of unlabeled immunoprecipitates, we used the WesternBreeze chemiluminescence detection system (Invitrogen) following semidry transfer to polyvinylidene difluoride membranes (Millipore Corp.).
Ubiquitination assay.
UbcH2 and UbcH3 were obtained from Boston Biochem. His6-UbcH5a was obtained from Roger Everettt, expressed in BL21(DE3) cells (Stratagene), and purified as previously described (8). Purified E2 proteins were stored at −80°C. E3 proteins were purified as described previously (28) and stored at 4°C. In vitro reaction mixtures (20 μl total) included 50 nM rabbit E1 (Boston Biochem), 28 μM ubiquitin, 5 mM ATP, 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, and 2 mM dithiothreitol. E2 enzymes were used at concentrations varying from 0.1 to 2.3 μM, and E3 concentrations ranged from 3 to 30 μM. After 90 min of incubation at 30°C, reactions were stopped by boiling them in sodium dodecyl sulfate (SDS) sample buffer. Samples were electrophoresed through SDS-12% polyacrylamide gels and immunoblotted with anti-ubiquitin antibody P4D1.
RESULTS
Mammalian K3 family homologues.
The hallmarks of the viral K3 family are an amino-terminal RING-CH domain, followed by two transmembrane domains. Using the RING-CH domain of KSHV-K3 and KSHV-K5, we searched the GenBank database by using BLAST 2.0. Nine hypothetical human proteins of unknown function were identified that display RING-CH domains, followed by two or more predicted transmembrane domains (Fig. 1). We propose to name this mammalian gene family membrane-associated RING-CH (MARCH) proteins. MARCH-VI, also called TEB-4, contained 12 predicted transmembrane domains and is the predicted homologue of yeast SSM4/DOA10 (35). MARCH-V is predicted to contain four transmembrane domains. In the remaining seven predicted proteins, the RING-CH domain is followed by two predicted transmembrane domains. MARCH-VII has an amino-terminal extension of about 500 amino acids prior to the RING-CH domain and is only distantly related to the other MARCH proteins. Phylogenetic analysis reveals that the remaining six two-transmembrane proteins occur as three pairs that are closely related: MARCH-I and -VIII, MARCH-II and -III, and MARCH-IV and -IX. MARCH-VIII is identical to the previously characterized c-MIR (16). Although all six MARCH proteins form a branch of the phylogenetic tree that is clearly separate from the viral K3 family (not shown), they share the primary structure and topology of the viral K3 proteins, which all display short amino termini containing the RING-CH domain succeeded by two transmembrane domains.
FIG. 1.
Human homologues of viral K3 family proteins. The RING-CH domain from the nine identified human MARCH proteins (I to IX) and the known viral K3 family proteins were aligned by using Vector NTI 7.0 software. The RING-CH domain is shown boxed in dark gray, whereas other conserved amino acids are shown boxed in light gray. The hydropathy plots (middle panel) for each MARCH protein were generated by using TMPRED (www.ch.embnet.org). The location of the RING-CH domain is shown by a black box under the hydropathy plot, whereas the number of transmembrane domains predicted is shown to the right of the RING-CH alignment. Phylogenetic relationships for the full-length sequences (bottom) were generated by using Vector NTI suite 7 software.
Tissue-specific expression of MARCH proteins.
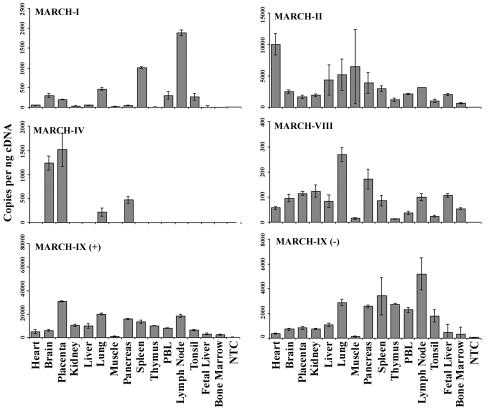
To investigate whether MARCH protein expression is ubiquitous or tissue specific, we performed real-time reverse transcription-PCR with primers specific for MARCH-I, -II, -IV, and -VIII and two splice variants of MARCH-IX on a panel of human tissue samples (Fig. 2). The most abundant expression was observed for MARCH-II and MARCH-IX, which were expressed in every tissue analyzed at high copy numbers (note the different scales in Fig. 2). The most restricted expression was observed for MARCH-IV, which was mainly expressed in brain and placenta. Transcripts for the other three MARCH proteins tested were detected in only some tissues and at generally lower levels. These data indicate both a tissue-specific and a widespread expression of MARCH proteins.
FIG. 2.
Tissue distribution of MARCH proteins. Expression of MARCH-I, -II, -IV, -VIII, and -IX was measured by real-time PCR in a panel of human tissue cDNAs (Clontech). MARCH-IX occurs in two splice variants, one of them missing the RING-CH domain. Both variants were included in this analysis. Expression of each gene was quantified by using absolute standards created from plasmid DNA. Concentrations of cDNAs were normalized by using the housekeeping genes GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and β-actin.
Subcellular localization of MARCH proteins.
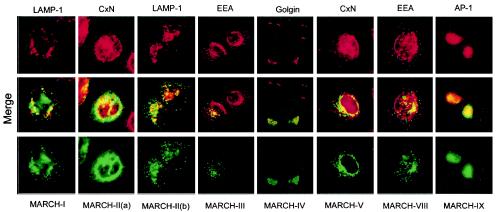
The predicted transmembrane domains implied that MARCH proteins localize to membranous compartments. To determine their subcellular localization, we transfected HeLa cells with FLAG-tagged cDNA constructs for MARCH-I, -II, -III, -IV, -V, -VIII, and -IX. Cells were analyzed by immunofluorescence with anti-FLAG, together with antibodies to various markers of subcellular compartments (Fig. 3). MARCH-I, MARCH-III, and MARCH-VIII showed a distinct punctate staining pattern that partially overlapped with endocytic or lysosomal vesicles. MARCH-V and MARCH-II showed an ER-like staining pattern that overlapped with calnexin staining (Fig. 3, staining pattern a). In about 30 to 40% of the cells however, MARCH-II showed a vesicular staining that did not overlap with calnexin but costained partially with Lamp (staining pattern b). The reason for these mutually exclusive patterns of MARCH-II is currently not known but does not seem to correlate with expression levels or cell cycle status (results not shown). MARCH-IV and MARCH-IX costained with the Golgi markers Golgin or the trans-Golgi marker AP-1, respectively. The staining pattern observed for most of the MARCH proteins is thus quite distinct from the ER-staining observed for the gamma-2 herpesvirus and poxvirus proteins (11, 28, 31) but is consistent with MARCH proteins being associated with subcellular membranes.
FIG. 3.
Localization of MARCH proteins to intracellular membranous organelles. HeLa cells were transfected with plasmids encoding C-terminally Flag-tagged versions of the indicated MARCH proteins. At 20 h posttransfection, cells were stained with antibodies against indicated cellular markers (top panel in red) or anti-FLAG (bottom panel in green). Colocalization is visualized as yellow in the merged panel (middle). All cellular markers were examined against all MARCH proteins. However, only results that indicated a significant overlap are shown. The markers of subcellular compartments were calnexin (Cxn) for ER, Golgin-97 for Golgi, Adaptor protein 1 (AP-1) for the trans-Golgi network, early endosomal antigen (EEA) for endosomes, and LAMP-1 for lysosomes.
Ubiquitin ligase activity of the RING-CH domains of MARCH proteins.
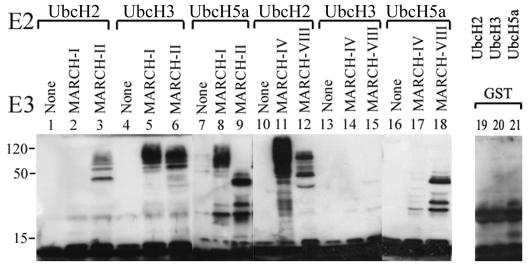
The predicted RING-CH domain implies ubiquitin ligase activity of MARCH proteins. RING-E3s are known to catalyze the formation of ubiquitin adducts in vitro when coincubated with ubiquitin, ATP, E1, and an appropriate E2 (26). Using purified GST fusion proteins, we examined whether the RING-CH domains of MARCH-I, -II, -IV, -VIII, and -IX act as ubiquitin ligases in vitro. These RING-CH domains were selected because the corresponding MARCH proteins downregulated cell surface glycoproteins as discussed in the next paragraph. The purified proteins were incubated with ubiquitin, ATP, and E1 together with E2 enzyme. Immunoblotting with antibodies against ubiquitin revealed that the RING-CH-GST fusion proteins of four MARCH proteins were able to catalyze the formation of high-molecular-weight molecular ubiquitin complexes in the presence of ubcH2, ubcH3, or ubcH5a (Fig. 4). Despite its high homology to MARCH-IV, MARCH-IX did not mediate ubiquitination with any of the E2s shown in Fig. 4 or with UbcH6, UbcH7, MmUbc6, or MmUbc7. However, since MARCH proteins showed a distinct specificity for individual E2 enzymes even when their RING-CH domains were closely related (e.g., MARCH-I and -VIII), the latter observation does not contradict a ubiquitin ligase function for MARCH-IX. More likely, MARCH-IX was unable to function with the panel of E2 tested. Ubiquitin complexes were not detected in the absence of MARCH proteins or in the presence of purified GST (Fig. 4 and unpublished data). The finding that four of five RING-CH domains examined in this assay demonstrated E3 ligase activity strongly suggests that all MARCH proteins act as ubiquitin ligases but differ in their E2 specificity.
FIG. 4.
MARCH proteins act as ubiquitin ligases in vitro. GST fusion proteins of the amino-terminal domains of individual MARCH proteins were incubated with ubiquitin, ATP, E1, and the indicated ubiquitin-conjugating enzymes (ubc, E2) and separated by SDS-polyacrylamide gel electrophoresis prior to immunoblotting with the ubiquitin-specific antibody P4D1. Ubiquitin ligase activity is indicated by the appearance of high-molecular-weight ubiquitinated species. Ubiquitinated proteins were not detected in the absence of E3s (None) or with GST alone (unpublished data).
Downregulation of surface receptors by MARCH proteins.
All viral K3 family members downregulate expression of MHC-I. Additional substrates are ICAM-1 and B7.2 (but not B7.1) for KSHV-K5, as well as Fas and CD4 for M153R. To examine whether MHC-I or any of these other glycoproteins are also affected by MARCH proteins, we transiently transfected HeLa cells with expression plasmids for all two-transmembrane MARCH proteins (except MARCH-VII) or the four-transmembrane MARCH-V and monitored the surface expression of selected glycoproteins. The following known substrates for viral K3 family members were tested: Fas (CD95) and MHC-I, both of which are endogenously expressed in HeLa cells, as well as transfected HLA-A2.1, CD4, and B7.2. We used endogenously expressed transferrin receptor (TfR) and transfected B7.1 as controls, neither of which is downregulated by the viral proteins. The results are plotted as the fluorescence of the respective surface markers versus the fluorescence of cotransfected GFP (Fig. 5A). Interestingly and unexpectedly, the expression of every surface glycoprotein was affected by at least one of the MARCH proteins, with the notable exception of B7.1. The four-transmembrane MARCH-V, as well as the two-transmembrane MARCH-III, did not downregulate any of the protein tested, although these proteins were clearly expressed as revealed by immunoprecipitation and immunoblotting of their FLAG-tagged versions (Fig. 5B and C). MARCH-II, which is ca. 60% identical to MARCH-III, downregulated TfR and B7.2. For the remaining four proteins, a distinct pattern emerged, whereby MARCH-I and -VIII downregulated TfR, B7.2, and Fas, whereas MARCH-IV and -IX downregulated MHC-I and CD4. The downregulation of B7.2 by MARCH-VIII is consistent with previous reports (16). Our results indicate an intrinsic substrate specificity correlating with sequence homology (Fig. 1). Some overlap in substrate specificity, however, seems to exist between these two pairs of proteins since surface levels of transfected A2.1 were also weakly reduced by MARCH-VIII and the levels of transfected B7.2 were weakly reduced by MARCH-IV. TfR downregulation was not previously reported for any of the viral proteins, and TfR is the only type II transmembrane protein included in our panel. Taken together, these data suggest that the human homologues of viral immune evasion proteins downregulate a distinct, partially overlapping, panel of cell surface glyocoproteins.
FIG. 5.
Downregulation of surface glycoproteins by the MARCH family. (A) HeLa cells were transfected with the respective MARCH expression plasmid and GFP. Surface expression of indicated surface proteins was measured via flow cytometry with specific primary antibodies and a phycoerythrin-conjugated secondary at 24 h posttransfection (unpublished data). Black arrows indicate significant downregulation. Expression of FLAG-tagged versions of MARCH proteins was confirmed with anti-FLAG antibody by immunoprecipitation (B), whereas steady-state levels were measured by immunoblot (C). Introduction of the FLAG tag at the carboxy terminus of MARCH proteins did not affect the ability of these proteins to downregulate their substrates (data not shown).
Functional domains of MARCH-IV and -IX.
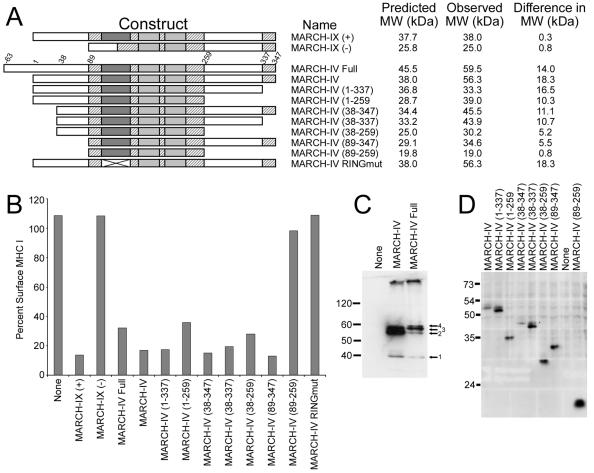
The distinct substrate specificity of MARCH-I/VIII and MARCH-IV/IX correlates with homologies in their primary structures. MARCH-I and -VIII are closely related (>90% identity) in their RING-CH domains and transmembrane domains, whereas their amino-terminal and carboxy-terminal regions are <20% identical. A similar relationship is also observed between MARCH-IV and MARCH-IX which are >90% identical in their RING-CH domain and transmembrane domain but generally <20% in the remaining sequence (Fig. 6). In contrast, the RING-CH and transmembrane domains of MARCH-II and -III, which differ in their ability to downregulate surface molecules, are only about 60% identical. Thus, it seems that the substrate specificities are largely determined by the RING-CH and transmembrane domains. To test this assumption, we performed deletion analysis of MARCH-IV. Expression of the predicted full-length MARCH-IV yields four different protein bands, of which the full-length protein (product 4) is only a minor fraction. In contrast, protein bands of the same size as those observed in a construct that lacks 63 amino acids in the amino-terminal region are predominant (Fig. 5). Since both the full-length and truncated version downregulate MHC-I (Fig. 6) and since MARCH-IX does not display an amino-terminal extension, it seems that the truncated version of MARCH-IV is the functional form with respect to MHC-I and CD4 downregulation. Therefore, the truncated form was used and denoted as MARCH-IV in all other experiments. Interestingly, the observed molecular mass of (truncated) MARCH-IV is 55 kDa (Fig. 5), whereas its predicted molecular mass is 38 kDa. In contrast, the observed mass of all other MARCH proteins, including MARCH-IX, was as predicted (Fig. 5 and 6). We deleted the amino-terminal and carboxy-terminal domains of MARCH-IV to map substrate-determining regions, as well as to determine the region of MARCH-IV responsible for this shift in apparent molecular mass. MHC-I downregulation was monitored by fluorescence-activated cell sorting (FACS) by using nontagged versions of the truncated proteins, whereas the apparent molecular mass was determined by immunoblot by using FLAG-tagged constructs. Truncation of the amino-terminal region preceding the RING-CH domain or deletion of the carboxy-terminal end following the transmembrane regions did not abolish the ability to downregulate MHC-I (Fig. 6). Therefore, it seems that the amino-terminal or carboxy-terminal regions, including small stretches conserved between MARCH-IV and MARCH-IX, are not essential for MHC-I downregulation consistent with the hypothesis that substrate specificity is determined by the RING-CH and transmembrane domains. However, a truncated version of MARCH-IV lacking both the amino terminus and the carboxy terminus was unable to downregulate MHC-I (Fig. 6). Thus, it seems that at least parts of the nonconserved regions are required for a functional molecule but not for substrate specificity. The apparent molecular weight of the shortest MARCH-IV construct containing only the RING-CH and transmembrane domains corresponded to the predicted molecular weight. In contrast, all other constructs that contained the MARCH-IV terminal regions showed an increase in apparent size. At present, we do not know the nature of this modification, but we ruled out N-linked glycosylation or ubiquitination (data not shown). Additional experiments to identify this modification are currently under way. Mutations in the RING-CH domains also established that this domain is essential for MHC downregulation of MARCH-IV and -IX. MARCH-IX occurs in two splice variants, one lacking the RING-CH domain. Forced expression of this RING-CH-deleted form does not downregulate MHC-I (Fig. 6). Likewise, point mutations in the RING-CH domain of MARCH-IV abolish MHC downregulation (Fig. 6), a finding consistent with an essential role of the E3 ligase activity for MHC downregulation.
FIG. 6.
Functional domains of MARCH-IV and MARCH-IX. (A) Diagram showing the homology between MARCH-IV and MARCH-IX, as well as the deletion mutants in MARCH-IV. The RING-CH domain (dark gray), the two transmembrane domains (light gray) and additional regions (hatched) are > 90% identical, whereas the remaining parts of the molecules (white) are <20% conserved. MARCH-IX (−) represents a splice variant lacking the RING-CH domain. Two conserved cysteines in the RING-CH domain were replaced with serines in the MARCH-IV RING mutant. N- and C-terminal deletion constructs of MARCH-IV are indicated; the predicted full-length construct contains a 63-amino-acid N-terminal extension. (B) Ratio (in percent) of the mean fluorescence of surface MHC-I detected by flow cytometry with W6/32 after 24 h of MARCH-transfected versus nontransfected HeLa cells gated for GFP. Both RING-CH mutants were unable to downregulate MHC surface expression. All MARCH-IV deletion constructs reduced MHC-I surface expression except MARCH-IV (89-259) containing the RING-CH and transmembrane regions only. (C) Immunoprecipitation, followed by immunoblotting with anti-FLAG, reveals the presence of two major protein bands of ca. 55 kDa (arrows 2 and 3) in cells transfected with both full-length MARCH-IV and MARCH-IV (1-347), whereas the predicted full-length protein (arrow 4) is only a minor species. (D) Immunoblot with anti-FLAG reveals the observed MW of the truncated MARCH-IV proteins. The difference between observed and predicted molecular weight is shown in panel A. Only MARCH-IV (89-259) migrated at the predicted position.
MHC-I downregulation by MARCH-IV and -IX occurs via endocytosis.
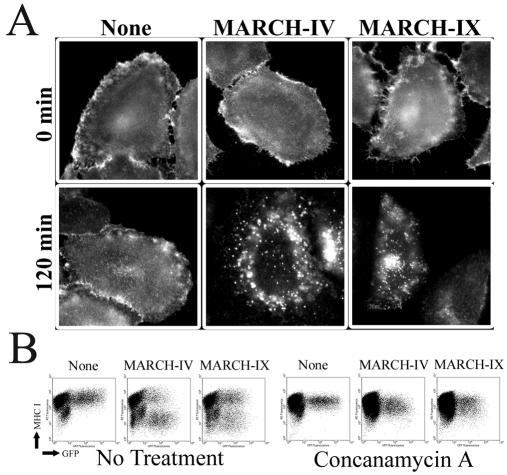
Our data indicate that the MARCH proteins are not only structurally but also functionally related to the viral K3 family of immune evasion proteins. Most members of the viral K3 family reduce expression of their target substrates by inducing a rapid internalization from the cell surface. The only known exception is MHV68-K3, which mediates proteasomal degradation of MHC-I prior to its exit from the ER (6). To study whether MHC-I was internalized by MARCH-IV and MARCH-IX, we compared the internalization of MHC-I in MARCH-IV- or MARCH-IX-transfected and nontransfected cells. MHC-I on the cell surface of transfectants was labeled by incubating intact cells with anti-MHC-I antibody at 4°C. The cells were then either fixed immediately for control or transferred to 37°C for 2 h to allow uptake of MHC-I. After fixation and visualization with a secondary antibody, a distinct punctuate staining was observed for MHC-I in MARCH-IV- or -MARCH-IX-transfected cells after 120 min, a finding consistent with rapid uptake of MHC-I (Fig. 7A). In contrast, uniform staining of the cell surface was observed in nontransfected or control cells. These results suggest that MHC-I travels to the cell surface prior to endocytosis.
FIG. 7.
Endocytosis and lysosomal degradation of MHC-I by MARCH-IV and MARCH-IX. (A) Uptake of MHC-I in HeLa cells transfected with vector control, MARCH-IV or MARCH-IX. At 24 h posttransfection, MHC-I was stained with antiserum K455 and either fixed after 30 min at 4°C (top panel) or transferred to 37°C for 2 h (bottom panel) prior to fixation and staining with Alexafluor 594 anti-rabbit secondary antibody. (B) Flow cytometry of MHC-I (W6/32 staining) on MARCH-IV or IX-transfected HeLa cells in the presence or absence of concanamycin A.
A role of the endocytic pathway in the removal of MHC-I from the cell surface by MARCH-IV and -IX was also suggested by experiments with inhibitors of endosome acidification such as concanamycin A. Treatment of MARCH-IV or -IX with concanamycin A restored MHC-I surface expression as measured by cytofluorometry (Fig. 7B). These results are consistent with HLA-A2.1 exiting the ER, being transported to the cell surface, and then rapidly internalized by endocytosis and ultimately destroyed, most likely in lysosomes.
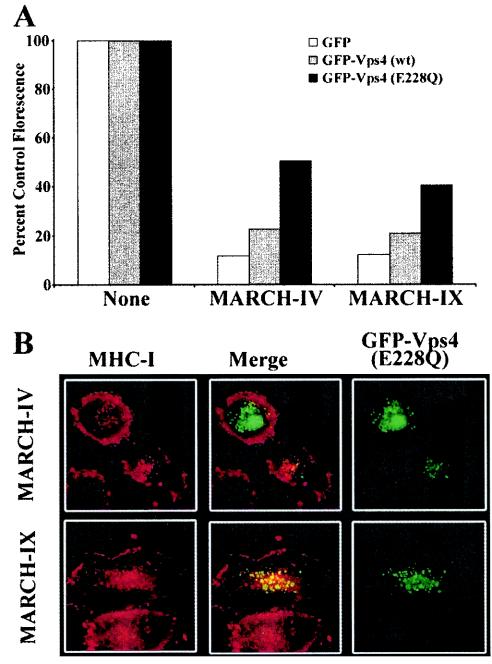
The proposed function of this protein family as ubiquitin ligases implies a role for ubiquitination in the internalization process. Ubiquitinated cell surface receptors are internalized and sorted to lysosomes via multivesicular bodies (MVBs), a process that is regulated by several sequential multiprotein ESCRT complexes (3). After completion of the MVB sorting, the AAA-type ATPase Vps4 (for vacuolar protein sorting protein 4) is required for the dissociation of the ESCRT complex from the membrane (3). Using an ATPase-deficient, GFP-tagged form of Vps4 (GFP-Vps4-E228Q), which is dominant negative (DN) (5), we examined whether the internalization of MHC-I in MARCH-IV- and MARCH-IX-transfected cells involves MVB formation. Surface expression of MHC-I was monitored on HeLa cells, cotransfected with MARCH-IV and either wild type or GFP-Vps4-E228Q. Neither DN nor wild-type Vps4 changed the surface levels of MHC-I in HeLa cells in the absence of MARCH-IV or MARCH-IX (data not shown). In contrast, cotransfection of GFP-Vps4-E228Q with MARCH-IV or -IX partially restored surface MHC-I expression, as shown by an increase in the mean fluorescence of transfected cells (Fig. 8A). In a separate line of experiments, we studied the subcellular localization of internalized MHC-I in cells transfected with GFP-Vps4-E228Q. It was shown previously that GFP-Vps4-E228Q generated and bound to aberrant vacuolated endosomes, whereas wild-type Vps4 stained the cytosol (5). Uptake of MHC-I molecules in MARCH-IV- and -MARCH-IX-transfected cells was assayed as described above, except that cells were also cotransfected with GFP-Vps4-E228Q. MHC-I remained at the cell surface in cells that stained brightly for GFP, indicating large amounts of GFP-Vps4-E228Q (Fig. 8B), a finding consistent with the FACS analysis. However, MHC-I and GFP-Vps4-E228Q colocalized in MARCH-IV- and MARCH-IX-transfected cells that expressed low levels of mutant vps4. We interpret this as evidence that MHC-I is taken up by endocytosis and sorted via the MVB pathway.
FIG. 8.
Vps4 restores MHC-I surface levels and colocalizes with MHC-I in HeLa cells transfected with MARCH-IV and MARCH-IX. (A) HeLa cells were transfected with GFP only or GFP-tagged wild-type Vps4 or the dominant-negative, ATP-hydolysis-deficient mutant GFP-Vps4(E228Q) (5), together with vector control MARCH-IV or MARCH-IX. At 24 h after transfection, cells were stained with W6/32, and the mean fluorescence of GFP-positive cells was determined. The ratio of the mean fluorescence of MARCH-transfected versus the corresponding vector-transfected cells is shown as a percentage. Cotransfection of GFP-Vps4(E228Q) partially restored MHC-I surface levels in MARCH-IV- and MARCH-IX-transfected cells compared to cells that were only transfected with GFP-Vps4(E228Q) and vector. (B) Uptake and colocalization of MHC-I molecules with GFP-Vps4(E228Q). Internalization of MHC-I by MARCH-IV and -IX was monitored as for Fig. 6, except that cells were cotransfected with GFP-Vps4(E228Q). Punctate staining of Vps4 (right panel, green) is consistent with its localization to endosomes (5). MHC-I (K455 staining, red, right panel) remained mostly at the cell surface in cells expressing high levels of GFP-Vps4(E228Q) (upper left cell in upper panel), whereas MHC-I and Vps4 colocalized in cells with lower GFP fluorescence, indicating lower Vps4 levels (cell in the center of upper and lower panel; merged fluorescence is shown in the middle column). No colocalization of MHC-I and Vps4 was observed in the absence of MARCH-IV and -IX, and wild-type Vps4 stained the cytoplasm (data not shown).
Ubiquitination is essential for MHC-I internalization.
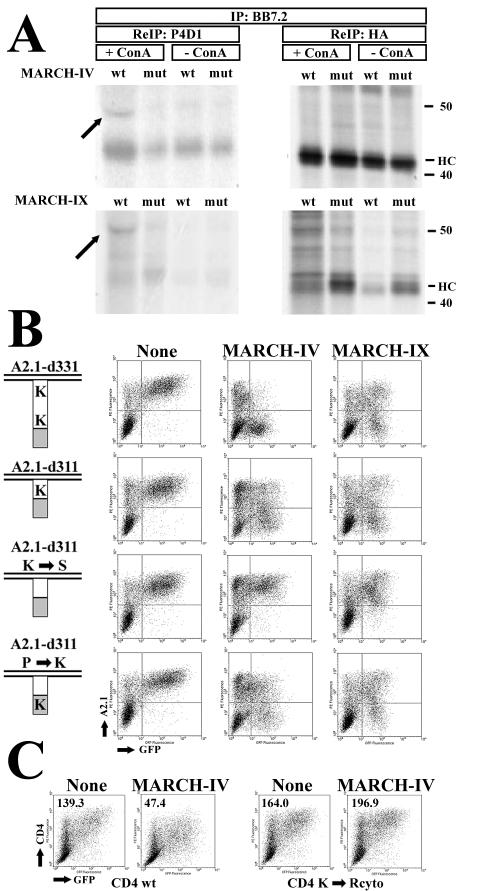
To examine whether MHC-I molecules are ubiquitinated in the presence of MARCH-IV and -IX, we cotransfected HeLa cells with a hemagglutinin (HA)-tagged truncated version of HLA-A2.1, d331 (31), together with MARCH-IV or -IX. For a control, we also transfected the RING-CH mutants of MARCH-IV and -IX described above. To slow down MHC-I turnover, cells were also treated with concanamycin A. Transfectants were labeled for 6 h, and HLA-A2.1 molecules were immunoprecipitated with antibody-BB7.2 (data not shown). The immunoprecipitates were denatured in SDS, and 10% of the sample was reprecipitated with anti-HA. MARCH-IV transfectants were kept in the presence of small amounts of tetracycline to reduce MARCH-IV expression levels so that MHC-I was not efficiently degraded during the 6-h labeling period. The remaining 90% of the sample was reprecipitated with ubiquitin-specific antibody P4D1 prior to SDS-polyacrylamide gel electrophoresis (Fig. 9A). Due to the high level of expression of HLA-A2.1, there was some carryover of heavy chain in the P4D1 immunoprecipitation. In addition, a single higher-molecular-mass species (∼49 kDa) was precipitated by P4D1 from the concanamycin A-treated cells transfected with wild-type MARCH-IV and -IX but not with their mutant forms. The size of this protein band is approximately equivalent to the 50 kDa expected for monoubiquitinated HLA-A2.1 d331 (note that the heavy chain of antibody P4D1 migrates to a similar position thus slightly displacing the labeled protein). The observed stabilization of the ubiquitinated intermediate by concanamycin A is also consistent with our previous observations in myxomavirus M153R-transfected cells (28). The most likely targets for ubiquitination on transmembrane glycoproteins are lysines in the cytoplasmic tail. To examine the role of lysines for substrate downregulation by MARCH-IV and MARCH-IX, we used a panel of constructs derived from HLA-A2.1 and a lysine-deleted version of CD4 (28). Flow cytometry revealed that removal of lysines from the tail of HLA-A2.1 renders these molecules resistant to MARCH-IV or MARCH-IX downregulation (Fig. 9B). This was also observed for CD4 and MARCH-IV (Fig. 9C). To rule out the possibility that these mutations changed the target molecules in such a way that they are no longer recognized by MARCH-IV or -IX, we introduced a lysine residue into an HA tag at the carboxy-terminal end of HLA-A2.1. This reintroduction of a lysine residue into a sequence not related to HLA-A2.1 restored downregulation both by MARCH proteins. Therefore, we conclude that lysines in the tail are an absolute requirement for MHC downmodulation by MARCH-IV and -IX, suggesting that ubiquitination is essential for internalization.
FIG. 9.
Ubiquitination of MHC-I and role of C-terminal lysines for MHC-I downregulation by MARCH-IV and MARCH-IX. (A) HeLa cells were transiently transfected with HA-tagged HLA-A2.1 d331 (31) and MARCH-IV, MARCH-IX, or their respective RING-CH mutants. MARCH-IX expression was fully induced by removal of tetracycline, whereas MARCH-IV expression was partially repressed with 5 ng of tetracycline since high levels of expression were not compatible with the long labeling protocol. Therefore, there is only a slight reduction of MHC expression levels in MARCH-IV transfectants compared to MARCH-IX transfectants. Where indicated, cells were treated with concanamycin A prior to and during labeling for 6 h. HLA-A2.1 was immunoprecipitated with BB7.2 and, after denaturation with SDS, reimmunoprecipitated with ubiquitin-specific antibody P4D1 (90% of the lysate, left panel) or anti-HA antibody (10% of the lysate, right panel). Due to the large amount of MHC heavy chain in the primary immunoprecipitation, some of it was carried over in the P4D1 reimmunoprecipitation. The arrow indicates ubiquitinated heavy chain. (B) Downregulation of HLA-A2.1 lysine mutants by MARCH-IV and MARCH-IX. Cytoplasmic tails of HLA-A2.1 are schematically depicted on the left (28). The number of lysines in the A2.1 tail (white) or the HA tag (gray) is shown. Surface expression of the individual constructs was monitored by flow cytometry with antibody BB7.2. Transfection with vector (left), MARCH-IV (middle) or MARCH-IV was tracked by cotransfection of GFP. (C) Surface expression of wild-type CD4 or CD4 lacking lysines in its cytoplasmic tail in the presence of MARCH-IV. Mean fluorescence values for GFP-positive cells are shown in the upper left.
DISCUSSION
We addressed the question whether MARCH family proteins were functionally related to viral immune evasion proteins of the K3 family. The overall sequence homology of these predicted proteins is rather low and limited mostly to the RING-CH domain (Fig. 1). The primary structure was thus suggestive but insufficient for us to conclude which of these proteins were functionally related to the viral immunomoldulators. Using a panel of potential substrates selected as known substrates of viral K3 family proteins, we observed an efficient and specific downregulation of various cell surface glycoproteins by the cellular homologues. The partially overlapping, partially distinct substrate specificity observed for the MARCH proteins is also typical of the viral proteins. The molecular reason for this specificity is not known, but substrates seem to interact transiently with the viral proteins (20) and substrate recognition seems to reside in the transmembrane regions of the targets (11). Consistently, we observed that sequence correlation in the RING-CH and transmembrane domains correlated with substrate specificity, whereas deletion of the amino-terminal or carboxy-terminal domains did not affect substrate downregulation.
Our results also confirm the previously reported downregulation of B7.2 by MARCH-VIII (c-MIR) (16). However, we observed that MARCH-I, which is homologous to MARCH-VIII, also efficiently downregulated B7.2 and that both MARCH-I and VIII also downregulated Fas and Tfr. Moreover, the unrelated MARCH-II also downregulated B7.2. In contrast to the findings of Goto et al., we did not observe a preferential expression of MARCH-VIII in lymph nodes. Rather, MARCH-VIII mRNA was found at low levels in most tissues examined (Fig. 2). Also different was our observation of a moderate, but significant downregulation of transfected HLA-A2.1 by MARCH-VIII. This could be caused by a higher level of expression achieved in our study with the tetracycline-inducible system which might facilitate a weak interaction. In general, however, a pattern emerged whereby MARCH-VIII and MARCH-I targeted a distinct set of surface glycoproteins compared to MARCH-IV and -IX. This pattern mimics the dichotomy of function for the K3 and K5 genes of KSHV. Although K3 downregulates a broader spectrum of MHC-I alleles compared to K5 (24), K5 but not K3 also downregulates B7.2 (10, 23).
The essential, sequence-independent role of lysines in the cytoplasmic tails of substrates together with ubiquitinated HLA-A2.1 strongly suggests that ubiquitination is required for downregulation by the MARCH family. In contrast to the study by Goto et al., who reported multiple ubiquitination of B7.2, we observed only a single species of ubiquitinated HLA-A2.1. This difference could be caused by the fact that B7.2 contains 11 lysines in its cytoplasmic tail (10, 11), whereas the HA-tagged A2.1 form used in our experiments contained only two lysines (31). Recently, it was shown that multiply ubiquitinated receptor tyrosine kinases did not contain polyubiquitin but were monoubiquitinated at multiple sites (18). Thus, it is possible that B7.2 contains monoubiquitins at multiple sites, whereas only one of the two lysines is used for ubiquitination of the HA-tagged version of HLA-A2.1. Notably, only two ubiquitinated forms of murine MHC-I molecule Db, which contains three lysines, were found in MHV68-K3 transfected cells (6). A central role for ubiquitin is further corroborated by the observed ubiquitin ligase activity of the RING-CH domain in vitro and by the partial restoration of MHC surface expression by GFP-Vps4-E228Q. Very similar observations were reported for both herpesviral and poxviral K3 family proteins, which were shown to require at least one lysine in the tail (6, 11, 28), and substrate internalization was inhibited by interference with the MVB pathway (20, 28). For both KSHV-K5 and MV-M153R, it was also shown that the isolated RING-CH domain acts as ubiquitin ligase (11, 28). The current model for the function of the viral proteins is that they transiently interact with their substrates and mediate the ubiquitination of the cytosolic tail. Our data, as well as those of Goto et al. (16), are consistent with the cellular homologues acting in a manner very similar to that of their viral relatives.
Although MARCH proteins induced the internalization of glycoproteins at the cell surface, subcellular localization studies revealed that these proteins all localize to intracellular membrane compartments. Similarly, all members of the viral K3 family localize to intracellular membranes, mostly the ER. It is therefore unclear at what stage of intracellular transport target glycoproteins become ubiquitinated. It is possible that ubiquitination occurs en route to the cell surface or target proteins could be ubiquitinated during recycling. The localization of MARCH-IV and -IX in the Golgi and trans-Golgi network, respectively, is consistent with either possibility.
The MARCH family represents a novel family of ubiquitin ligases with a noncanonical RING domain for which we adopted the name RING-CH-domain, as previously proposed for the SSM4/DOA10 protein (35). A close relationship between the RING-CH domain and other RING domains is also suggested by structural predictions, whereas other PHDs are predicted to fold into structures that are clearly distinct from the RING domain (2). The RING-CH domain thus expands the RING-E3 family, already the most abundant E3 family, with potentially several hundred members (26). In contrast to the MARCH family, however, most RING-E3s are either cytosolic, nuclear, or peripheral membrane proteins (26). Two examples for transmembrane RING-E3s that are not MARCH proteins are Der3/Hrd1 (7, 14) and p78 (12). These E3s have all been shown to be involved in the degradation of misfolded proteins in the ER or ER-associated protein degradation. MARCH-VI, the human homologue of DOA10/SSM4, is also likely to be involved in ER-associated protein degradation. In contrast, MARCH-IV and -IX (the present study) and MARCH-VIII (16) direct the internalization of their substrates. It seems likely that endocytosis, rather than proteasomal degradation, will also be the mechanism of the other two transmembrane MARCH proteins, since most of them seem to locate to the endosomal-lysosomal compartment.
The membrane association of MARCH proteins implies that their corresponding ubiquitin-conjugating enzymes are also membrane associated or need to be recruited to the membrane. Indeed, experimental evidence suggests that SSM4/DOA10, p78, and Der3/Hrd1 cooperate with ubc6 and ubc7 (7, 12, 14, 35). Ubc6 and ubc7 are bound to the ER membrane by a transmembrane domain and by interacting with another protein, respectively (4, 33). It is thus likely that MARCH-VI will interact with the human homologs of ubc6 or ubc7. MARCH proteins that leave the ER and regulate internalization, however, are more likely to interact with different E2s in vivo. Although the E2s interacting with MARCH proteins in vivo have yet to be identified, our in vitro ubiquitination experiments suggest that the amino-terminal regions of each MARCH protein dictate a preference for certain E2s.
At present we can only speculate about the physiological role of the MARCH proteins. The results obtained with forced expression of MARCH proteins in our experiments strongly suggest that several members of this family regulate the degradation of glycoproteins at the cell surface. MARCH-IV and MARCH-IX could be involved in the internalization of free heavy chains upon dissociation of peptide and β2-microglobulin. However, gene knockdown of MARCH-IX by small interfering RNA did not affect the turnover of MHC heavy chains in HeLa cells (data not shown). Alternatively, it might be that MARCH-IV and -IX have to be upregulated by external stimuli before they affect MHC-I turnover, whereas they are involved in the turnover of other cell surface protein at basal levels. Since we only screened a small number of cell surface proteins, it is highly likely that MARCH proteins will be involved in the internalization of other, yet-to-be-identified transmembrane proteins. Many cell surface receptors are internalized upon engagement with ligands, either soluble or bound to the surface of neighboring cells (26). The involvement of ubiquitin in these processes is just beginning to emerge and the regulation of ubiquitination is not known. We therefore speculate that MARCH proteins will be involved in regulating receptor internalization as well as protein turnover.
The functional relationship between MARCH proteins and the viral K3 protein family suggests that the viral proteins were derived from their cellular homologues. There is ample precedence for common host-related immunomodulators in both herpesviral and poxviral genomes (29). These host-derived proteins, however, often assume a new or modified function, possibly even acting as inhibitors of their host counterparts. Whereas in most of these cases the known function of the host-protein served as a road map to elucidate the function of the viral protein, the reverse strategy has been used here to discover the function of a novel eukaryotic protein family.
Acknowledgments
We are grateful to Sumio Sugano for providing the MARCH-I cDNA clone, to Philip Woodman for the vps4 constructs, to Lars Karlsson for B7.1, to Per Peterson for K455, and to Satoshi Ishido for B7.2.
This study was supported by grant NIH CA94011 and by the Medical Research Foundation of Oregon to K.F.
REFERENCES
- 1.Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20:56-59. [DOI] [PubMed] [Google Scholar]
- 2.Aravind, L., L. M. Iyer, and E. V. Koonin. 2003. Scores of RINGS but no PHDs in ubiquitin signaling. Cell Cycle 2:123-126. [DOI] [PubMed] [Google Scholar]
- 3.Babst, M., D. J. Katzmann, E. J. Estepa-Sabal, T. Meerloo, and S. D. Emr. 2002. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3:271-282. [DOI] [PubMed] [Google Scholar]
- 4.Biederer, T., C. Volkwein, and T. Sommer. 1997. Role of Cue1p in ubiquitination and degradation at the ER surface. Science 278:1806-1809. [DOI] [PubMed] [Google Scholar]
- 5.Bishop, N., and P. Woodman. 2000. ATPase-defective mammalian VPS4 localizes to aberrant endosomes and impairs cholesterol trafficking. Mol. Biol. Cell 11:227-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boname, J. M., and P. G. Stevenson. 2001. MHC class I ubiquitination by a viral PHD/LAP finger protein. Immunity 15:627-636. [DOI] [PubMed] [Google Scholar]
- 7.Bordallo, J., R. K. Plemper, A. Finger, and D. H. Wolf. 1998. Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol. Biol. Cell 9:209-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coscoy, L., and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc. Natl. Acad. Sci. USA 97:8051-8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coscoy, L., and D. Ganem. 2001. A viral protein that selectively downregulates ICAM-1 and B7-2 and modulates T-cell costimulation. J. Clin. Investig. 107:1599-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coscoy, L., D. J. Sanchez, and D. Ganem. 2001. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J. Cell Biol. 155:1265-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang, S., M. Ferrone, C. Yang, J. P. Jensen, S. Tiwari, and A. M. Weissman. 2001. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 98:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Früh, K., E. Bartee, K. Gouveia, and M. Mansouri. 2002. Immune evasion by a novel family of viral PHD/LAP-finger proteins of gamma-2 herpesviruses and poxviruses. Virus Res. 88:55-69. [DOI] [PubMed] [Google Scholar]
- 14.Gardner, R. G., G. M. Swarbrick, N. W. Bays, S. R. Cronin, S. Wilhovsky, L. Seelig, C. Kim, and R. Y. Hampton. 2000. Endoplasmic reticulum degradation requires lumen to cytosol signaling. Transmembrane control of Hrd1p by Hrd3p. J. Cell Biol. 151:69-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goto, E., S. Ishido, Y. Sato, S. Ohgimoto, K. Ohgimoto, M. Nagano-Fujii, and H. Hotta. 2003. c-MIR, a human E3 ubiquitin ligase, is a functional homolog of herpesvirus proteins MIR1 and MIR2 and has similar activity. J. Biol. Chem. 278:14657-14668. [DOI] [PubMed] [Google Scholar]
- 17.Guerin, J. L., J. Gelfi, S. Boullier, M. Delverdier, F. A. Bellanger, S. Bertagnoli, I. Drexler, G. Sutter, and F. Messud-Petit. 2002. Myxoma virus leukemia-associated protein is responsible for major histocompatibility complex class I and Fas-CD95 down-regulation and defines scrapins, a new group of surface cellular receptor abductor proteins. J. Virol. 76:2912-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haglund, K., S. Sigismund, S. Polo, I. Szymkiewicz, P. P. Di Fiore, and I. Dikic. 2003. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat. Cell Biol. 5:461-466. [DOI] [PubMed] [Google Scholar]
- 19.Hershko, A., and A. Ciechanover. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425-479. [DOI] [PubMed] [Google Scholar]
- 20.Hewitt, E. W., L. Duncan, D. Mufti, J. Baker, P. G. Stevenson, and P. J. Lehner. 2002. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J. 21:2418-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicke, L. 1997. Ubiquitin-dependent internalization and down-regulation of plasma membrane proteins. FASEB J. 11:1215-1226. [DOI] [PubMed] [Google Scholar]
- 22.Holzerlandt, R., C. Orengo, P. Kellam, and M. M. Alba. 2002. Identification of new herpesvirus gene homologs in the human genome. Genome Res. 12:1739-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishido, S., J. K. Choi, B. S. Lee, C. Wang, M. DeMaria, R. P. Johnson, G. B. Cohen, and J. U. Jung. 2000. Inhibition of natural killer cell-mediated cytotoxicity by Kaposi's sarcoma-associated herpesvirus K5 protein. Immunity 13:365-374. [DOI] [PubMed] [Google Scholar]
- 24.Ishido, S., C. Wang, B. S. Lee, G. B. Cohen, and J. U. Jung. 2000. Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J. Virol. 74:5300-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jenner, R. G., and C. Boshoff. 2002. The molecular pathology of Kaposi's sarcoma-associated herpesvirus. Biochim. Biophys. Acta 1602:1-22. [DOI] [PubMed] [Google Scholar]
- 26.Joazeiro, C. A., and A. M. Weissman. 2000. RING finger proteins: mediators of ubiquitin ligase activity. Cell 102:549-552. [DOI] [PubMed] [Google Scholar]
- 27.Lybarger, L., X. Wang, M. R. Harris, H. W. T. Virgin, and T. H. Hansen. 2003. Virus subversion of the MHC class I peptide-loading complex. Immunity 18:121-130. [DOI] [PubMed] [Google Scholar]
- 28.Mansouri, M., E. Bartee, K. Gouveia, B. T. Hovey Nerenberg, J. Barrett, L. Thomas, G. Thomas, G. McFadden, and K. Fruh. 2003. The PHD/LAP-domain protein M153R of myxomavirus is a ubiquitin ligase that induces the rapid internalization and lysosomal destruction of CD4. J. Virol. 77:1427-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McFadden, G., and P. M. Murphy. 2000. Host-related immunomodulators encoded by poxviruses and herpesviruses. Curr. Opin. Microbiol. 3:371-378. [DOI] [PubMed] [Google Scholar]
- 30.Nicholas, J., V. Ruvolo, J. Zong, D. Ciufo, H. G. Guo, M. S. Reitz, and G. S. Hayward. 1997. A single 13-kilobase divergent locus in the Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J. Virol. 71:1963-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paulson, E., C. Tran, C. Collins, and K. Früh. 2001. KSHV-K5 inhibits phosphorylation of the major histocompatibility complax class I tail. Virology 288:369-378. [DOI] [PubMed] [Google Scholar]
- 32.Saha, V., T. Chaplin, A. Gregorini, P. Ayton, and B. D. Young. 1995. The leukemia-associated-protein (LAP) domain, a cysteine-rich motif, is present in a wide range of proteins, including MLL, AF10, and MLLT6 proteins. Proc. Natl. Acad. Sci. USA 92:9737-9741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sommer, T., and S. Jentsch. 1993. A protein translocation defect linked to ubiquitin conjugation at the endoplasmic reticulum. Nature 365:176-179. [DOI] [PubMed] [Google Scholar]
- 34.Stevenson, P. G., S. Efstathiou, P. C. Doherty, and P. J. Lehner. 2000. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc. Natl. Acad. Sci. USA 97:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swanson, R., M. Locher, and M. Hochstrasser. 2001. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 15:2660-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu, Y. Y., M. R. Harris, L. Lybarger, L. A. Kimpler, N. B. Myers, H. W. T. Virgin, and T. H. Hansen. 2002. Physical association of the K3 protein of gamma-2 herpesvirus 68 with major histocompatibility complex class I molecules with impaired peptide and β2-microglobulin assembly. J. Virol. 76:2796-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]