Abstract
Considerable controversy surrounds the impact of hepatitis C virus (HCV) protein expression on viability of host cells and regulation of the cell cycle. Both promotion of cellular proliferation and apoptosis have been observed in different experimental systems. To determine whether expression of the entire complement of HCV proteins in the context of ongoing viral RNA replication significantly alters the host cell transcriptome and cell cycle regulatory processes, we carried out high-density oligonucleotide microarray studies and analyzed cell cycle distributions and S-phase entry in Huh7 cell clones harboring selectable, full-length, replicating HCV RNAs that express the entire genotype 1b, HCV-N polyprotein, and clonally related cells in which all viral RNA was eliminated by prior treatment with alpha interferon. Oligonucleotide microarray analyses revealed only subtle, coordinated differences in the mRNA profiles of cells containing replicating viral RNA and their interferon-cured progeny, with variation between different cell clones having a greater influence on the cellular transcriptome than the presence or absence of replicating HCV RNA. Flow cytometric analysis demonstrated no significant differences in cell cycle distribution among populations of asynchronously growing cells of both types. Cell lines containing replicating viral RNA and their interferon-cured progeny were able to reenter the cell cycle similarly after transient G1 arrest. In contrast, although viral protein expression and genome replication did not alter cell cycle control in these cells, HCV genome replication was highly dependent on cellular proliferation, with viral RNA synthesis strongly decreased in poorly proliferating, confluent, or serum-starved cells and substantially enhanced in the S phase of the cell cycle.
Hepatitis C is the most common cause of clinically significant liver disease in the United States. Although acute infection with hepatitis C virus (HCV) is usually subclinical, persistent infection develops in up to 80% of initially infected patients (2). The majority of persistently infected individuals develop evidence of chronic liver injury and are at risk for progressive hepatic fibrosis leading to cirrhosis and ultimately death due to liver failure or hepatocellular carcinoma. Approximately 10,000 individuals die each year with chronic liver disease associated with HCV infection in the United States alone (1), rendering this virus a very significant public health problem.
HCV is classified within a distinct genus, Hepacivirus, in the family Flaviviridae (15). Its genome consists of positive sense, single-stranded RNA of approximately 9.5 kb in length (9). The genomic RNA contains a single open reading frame, from which one large polyprotein is expressed, and flanking 5′ and 3′ untranslated regions. Translation of the viral RNA occurs through a cap-independent mechanism via an internal ribosomal entry site (IRES) located in the 5′ untranslated region. A combination of cellular and virally encoded protease activities leads to processing of the polyprotein into individual structural and nonstructural proteins. Three structural proteins have been identified with the core protein forming a poorly defined viral capsid, and E1 and E2, both glycosylated envelope proteins, forming heterodimers. These structural proteins are processed by host signal peptidases. C terminal to E2, further proteolytic processing by host peptidases yields a short hydrophobic peptide, p7, which may function as a membrane channel (44), followed by the nonstructural (NS) proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B. NS2 and the amino-terminal protease domain of NS3 are responsible for proteolytic cleavage at the NS2-NS3 junction (20, 52), whereas other proteolytic cleavage events involved in the maturation of the NS proteins are carried out by the major viral serine protease, NS3, in association with its cofactor, NS4A (5, 7, 35). In addition to its function as a protease, NS3 also contains helicase and nucleoside triphosphatase activities within its C-terminal domain (28). Specific replicative functions have not been identified for NS4B and NS5A, but both are likely to contribute to the viral replicase complex. NS5B constitutes the HCV RNA-dependent RNA polymerase and is thus the catalytic core of the replicase.
In addition to functions related to RNA replication, both NS3/4A and NS5A appear to play important roles in modulating antiviral responses within infected cells and in promoting long-term persistence of the virus. The NS3/4A protease blocks viral activation of interferon (IFN) regulatory factor 3 (IRF-3), by directing the proteolytic cleavage of an unidentified, cellular virus-activated kinase, or other signaling molecule (14). NS5A appears to interfere with the function of the double-stranded RNA (dsRNA)-induced protein kinase R (16-18). NS5A has also been suggested to influence host signaling processes, such as phosphatidylinositol 3-kinase activation (42) and apoptotic pathways (30, 38), and to possess transcriptional transactivation properties (10). In addition to these reported effects of NS proteins on cellular processes, a large and often contradictory body of literature exists concerning the impact of expression of the viral structural proteins on cellular processes. Most prominent and best documented of these is the effect of core protein expression on fatty acid metabolism and storage, resulting in the accumulation of excess numbers of lipid-laden droplets in vitro, and steatosis in transgenic mice (4, 22, 24, 25, 40, 45). All of these studies suggest the possibility that one or more manifestations of hepatitis C may reflect direct disruption of normal cellular control processes by the viral proteins.
Several previous studies suggest that one or more HCV proteins have the potential to modulate the normal regulation of the cell cycle. These findings could have important implications for hepatitis C pathogenesis and the development of hepatocellular carcinoma, in particular. HCV proteins that have been described to have such effects include core, NS3, NS4B, and NS5A (3, 13, 19, 30, 64). However, these reports contain many inconsistencies. Core, for example, has been described to both enhance and repress p21 function (11, 21, 23, 27, 29, 37, 43, 50), actions that would be expected to exert opposing effects on cell cycle progression. The same is true for studies that have examined the impact of core protein on the regulation of apoptosis (12, 23, 49, 56, 65). The discrepancies in these observations are likely to arise from the use of different experimental systems, including different cell lines and viral sequences, and the overexpression of only partial segments of the viral polyprotein.
Until recently, investigation of HCV replication mechanisms and the cellular consequences of HCV infection have been hampered significantly by the absence of a robust in vitro infection-replication system, as well as the lack of a suitable small animal model. However, recent progress has led to the establishment of autonomously replicating subgenomic HCV RNAs, expressing only NS proteins (6, 8, 36, 47), as well as genome-length, selectable HCV RNAs that express all of the viral proteins (26, 46). These RNAs replicate efficiently and stably under selective pressure in Huh7 cells, a human hepatoma-derived cell line. However, their replication is strongly influenced by the proliferation status of the host cells, with the abundance of viral RNA rapidly declining when cells reach high density (47). The mechanism that underlies this requirement for host cell proliferation has not been identified yet, but it is possible that HCV RNA replication is more permissive in certain phases of the cell cycle than in others.
Here we describe a detailed investigation of the impact of the replication of HCV RNA, with concomitant expression of the complete viral polyprotein, on the homeostasis of the host cell, including the cellular mRNA transcription profile and regulation of the cell cycle. We show that these effects are minimal, despite many published reports in the literature that might suggest the contrary. In contrast, the cell cycle has an important effect on viral RNA synthesis, which is enhanced in S-phase cells.
MATERIALS AND METHODS
Cell lines.
Huh7 cell lines containing autonomously replicating, genome-length, dicistronic, selectable HCV RNAs derived from the genotype 1b HCV-N strain of HCV have been described previously (26). The G418-selected clonal cell lines used in the present study were NNeo/C5B clones 2-3 and 3, respectively. Cells were routinely maintained in Dulbecco modified Eagle medium-H supplemented with antibiotics, 10% heat-inactivated fetal bovine serum, and 250 μg of G418/ml. To cure the cells of the replicating HCV RNA, cells were treated with 100 U of IFN-α2b/ml in the absence of G418 for 2 weeks. The absence of HCV RNA after treatment was confirmed by Northern blot, reverse transcription-PCR (RT-PCR), and the failure of IFN-α-cured cells to survive G418 selection. The HCV RNA-free cured cell lines were designated 2-3c and 3c, respectively.
Cell cycle distribution and flow cytometry.
To determine the proportion of S-phase cells actively synthesizing DNA, the nucleoside analog bromodeoxyuridine (BrdU) was added to the culture medium at a concentration of 10 μg/ml for 30 min. Cells were washed, treated with trypsin, and processed for BrdU and 7-amino-actinomycin D (7-AAD) staining with the BrdU flow kit (Pharmingen) according to the manufacturer's specifications. For G1 arrest experiments, 10 μg of aphidicolin (Sigma)/ml were added to the culture medium for 24 h. The drug was removed by three washes with phosphate-buffered saline (PBS), and the cells were refed with complete medium and allowed to progress through the cell cycle as indicated prior to harvesting.
Flow cytometric analysis was carried out within the University of Texas Medical Branch Flow Cytometry Core Facility with a FACScan flow cytometer. Fluorescein isothiocyanate (FITC) fluorescence was monitored at 530 nm to detect the BrdU label, and 7-AAD fluorescence was monitored at 695 nm to monitor the DNA content. The percentages of cells in various stages of the cell cycle were determined by using the WinMDI 2.8 flow cytometry software package. For flow cytometric analysis of HCV antigen expressing cells, clone 2-3 cells were arrested in the G1 phase of the cell cycle with aphidicolin and released as described above. Cells were fixed by using reagents supplied with the BrdU flow kit, and NS5A detected by staining with a specific MAb (7022P; Maine Biotech) at a dilution of 1:50, followed by incubation with FITC-conjugated secondary anti-mouse immunoglobulin (Sigma).
High-density oligonucleotide microarrays.
Affymetrix oligonucleotide microarray analyses compared the gene expression profiles of Huh7 cell clones containing replicating NNeo/C5B RNA with isoclonal progeny that had been cured of HCV RNA by IFN treatment. As a positive control, a similar analysis compared the cured 2-3c cells with 2-3c cells infected 20 h prior to RNA extraction with Sendai virus (SenV; 100 hemagglutinin units/ml of culture medium) as described previously (14). For these assays, total cellular RNA was extracted from cells by using the RNAqueous kit (Ambion) and quantified by spectrophotometry. Then, 25 μg of total RNA was used as a template for first-strand cDNA synthesis, which was converted to double-stranded DNA and used subsequently to synthesize biotinylated cRNA probes. The latter were fragmented and hybridized to Affymetrix human GeneChip oligonucleotide microarrays (HG-U95A or HG-U133A) containing 12,626 or 22,283 oligonucleotide probe sets, respectively, representing known human genes, according to the manufacturer's protocol (Affymetrix, Santa Clara, Calif.). The GeneChip arrays were scanned by using an Affymetrix confocal scanner (Agilent), and the data were analyzed according to the absolute and comparative expression algorithms contained within the Affymetrix microarray suite (version 5.0) software package. In comparative analyses of arrays, a call of “decreased” was considered significant only if the probe set was scored as “present” in the absolute expression analysis of the baseline array, whereas a call of “increased” was considered significant only if the probe set was scored as “present” in the experimental array. A two-way analysis of variance (ANOVA) identified probe sets with signal intensities varying according to the specific Huh7 clone (2-3 versus 3) and/or the presence or absence of replicating HCV RNA, and not random variation, at a 95% confidence level [i.e., Pr(F) ≤ 0.05]. For this ANOVA, the input was prefiltered to exclude probe sets with an absolute call of “absent” in all arrays, or an absolute fold change of <2.0 across all pairwise comparisons (2-3, 2-3c, 3, and 3c cells). Heat maps of the selected probe sets were produced by using the Spotfire DecisionSite 7.1.1 software package.
Real-time RT-PCR.
Total RNA was extracted from Huh7 cells with TRIzol S reagent (Invitrogen). The purified RNA pellet was dissolved in 50 μl of diethyl pyrocarbonate-treated water (Ambion) containing 0.1 U of RNase inhibitor (Ambion)/μl and stored at −80°C. Quantification of HCV RNA was carried out by TaqMan RT-PCR amplification, normalized to human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA (Perkin-Elmer) or 18S RNA. The RNA standard was in vitro-transcribed HCV RNA, with RNA concentrations determined by spectrophotometry at 260 nm. Copy numbers for the HCV target sequence and GAPDH or 18S RNA controls were estimated by comparing threshold crossing (Ct) values of samples with those of the appropriate standard curve. The HCV probe and primers were identical to those reported by Takeuchi et al. (57). TaqMan RT-PCRs were carried out by using reagents provided with the EZ RT-PCR core reagent kit (Perkin-Elmer) on an ABI Prism 7700 instrument and used the following thermal cycling conditions: one cycle of 50°C for 2 min, 60°C for 30 min, and 95°C for 5 min, followed by 40 cycles of 95°C for 20 s and 60°C for 1 min. The sensitivity for HCV RNA was 10 copies.
Northern blots.
Total RNA was extracted by using TRIzol S reagent (Invitrogen) and RNA concentration was determined by agarose gel electrophoresis and spectrometry at 260 nm. Equal amounts of RNA were electrophoretically separated on denaturing agarose gels and transferred to positively charged Hybond N+ nylon membranes (Amersham-Pharmacia) by using reagents provided with the Northern Max kit (Ambion) according to the manufacturer's instructions. RNAs were immobilized by UV cross-linking (Stratagene). HCV RNA was hybridized to a digoxigenin-labeled, negative-sense NS5B specific riboprobe. Detection of bound riboprobes was with anti-digoxigenin-alkaline phosphatase-conjugated Fab2 fragments, followed by reaction with CSPD (Roche) and exposure to X-ray film.
Immunoblots.
Cell lysates were prepared in NP-40 lysis buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 10% glycerol, 1 mM EDTA, 1% Nonidet P-40) supplied with 2 μg of aprotinin/ml. Cell lysates were incubated for 15 min on ice prior to debris removal by centrifugation at 14,000 rpm at 4°C for 30 min. Protein concentrations were determined by the bicinchoninic acid method. Lysate samples containing 50 μg of protein were separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis, transferred to Hybond enhanced chemiluminescence (ECL) nitrocellulose membranes (Amersham-Pharmacia), blocked with 3% nonfat dry milk in PBS, and incubated with mouse MAbs against core (Maine Biotech), E2 (a kind gift of T. Miyamura), NS3 (Novocastra), and actin (Sigma), respectively, diluted in 3% nonfat dry milk-PBS. After being washed in PBS, membranes were probed with an anti-mouse horseradish peroxidase-conjugated secondary antibody, and bound antibodies were visualized by using reagents provided with the ECL Western blot detection kit (Amersham).
In vivo HCV RNA labeling.
Cells were arrested in the G1 phase of the cell cycle with aphidicolin as described above. At 4 h prior to harvesting, the medium was changed to phosphate-free DMEM plus 5% FBS to deplete intracellular phosphate and, for inhibition of cellular RNA synthesis, actinomycin D was added at a concentration of 5 μg/ml. At 3 h prior to harvesting, [32P]orthophosphate was added at a concentration of 500 μCi/ml. Cells were harvested in TRIzol S, and total RNA was extracted and quantitated by agarose gel electrophoresis. Equal amounts of RNA were loaded on nondenaturing agarose gels and separated by electrophoresis, and radioactively labeled HCV RNA was detected by autoradiography. Bands were quantitated by densitometry of autoradiograms. For in vivo labeling during serum starvation, the cells were phosphate starved in the presence of actinomycin D as described above and then labeled for 15 h prior to harvesting. RNA extraction and analysis were carried out as described above.
RESULTS
Huh7 cells supporting HCV RNA replication and isoclonal, “cured” progeny cell lines.
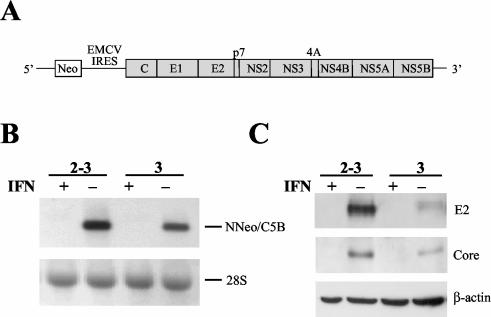
To determine the impact of viral RNA replication and expression of the complete HCV polyprotein on the homeostasis of host Huh7 cells, we studied two independently selected, clonal Huh7 cell lines containing autonomously replicating dicistronic RNAs encoding the entire viral polyprotein (Fig. 1A), NNeo/C5B clone 2-3 and NNeo/C5B clone 3. These G418-resistant cell clones, and the full-length replicating RNAs they harbor, have been described previously (26). We compared these cell lines with clonally related progeny cells in which the HCV RNA had been eliminated by prior treatment with IFN-α in the absence of G418 (designated 2-3c and 3c, respectively; see Materials and Methods). The absence of replicating RNA in these cured cell lines was confirmed by Northern analysis with a probe specific for the HCV NS5B sequence (Fig. 1B), TaqMan PCR; by Western blot analysis for HCV protein expression (Fig. 1C); and by the absence of selectable neomycin resistance when G418 was added back to the cell culture media (data not shown). Such IFN-α-cured cell lines are well suited to serve as negative controls for the experiments described below because they are of the same clonal origin as the Huh7 cells harboring the replicating viral RNAs. In addition, since the concentration of IFN-α used is minimally toxic to the cells, there was little pressure for further clonal selection during the 2-week treatment period.
FIG. 1.
Elimination of replicating N-Neo/C5B RNAs by IFN-α treatment. (A) Diagram of full-length dicistronic selectable HCV RNA. The HCV IRES drives expression of the neomycin resistance gene (Neo) as a selectable marker. Expression of the full-length HCV polyprotein is under control of the encephalomyocarditis virus (EMCV) IRES. (B) Huh7 cells harboring dicistronic full-length replicating HCV RNAs (NNeo/C5B, clones 2-3 and 3) were cultured in the presence or absence of 100 U of IFN-α2b (IFN)/ml for 2 weeks. The RNA status of the cells was determined by Northern blot with a probe specific for NS5B (top panel). 28S rRNA is shown as a loading control (bottom panel). Cells treated with IFN-α had no detectable HCV RNA. (C) Absence of HCV protein expression after IFN-α treatment was verified by immunoblot with antibodies specific for E2 and the core protein. An immunoblot of β-actin is shown as a loading control.
Host cell transcription is not markedly altered in the presence of replicating HCV RNAs.
As indicated above, a number of previous studies suggest that the expression of several HCV proteins may significantly influence host cell homeostasis (3, 10, 11, 13, 19, 21-23, 30, 32, 37, 39, 42, 50, 51, 63-65). To directly assess the impact of HCV polyprotein expression and viral RNA replication on cellular gene expression, high-density Affymetrix oligonucleotide microarrays were hybridized with probes prepared from cells containing replicating HCV RNA and their cured, clonally related progeny. The influence of HCV RNA replication on cellular mRNA profiles was assessed by a pairwise comparison of the results obtained with the 2-3 and 2-3c cell lines and with the 3 and 3c cell lines, respectively. Studies were carried out with both the Affymetrix HG-U95A and HG-U133A GeneChip microarrays, which contain oligonucleotide probe sets representing 12,626 and 22,283 known human genes, respectively. RNAs prepared from 2-3 and 2-3c cells were analyzed in two independent experiments by using the HG-U95A GeneChip and once with the HG-U133A Gene-Chip; clone 3 and 3c cells were compared once by using each GeneChip type. Independently prepared RNA preparations were tested in each of these analyses.
These results demonstrated that HCV RNA replication with accompanying high-level expression of the entire HCV polyprotein has remarkably little impact on the abundance of cellular mRNA transcripts (Table 1). Huh7 cells contain transcripts that generate probes hybridizing with approximately 50% of the probe sets represented on either the HG-U95A or HG-U133A GeneChips. However, a comparison of the transcripts present in 2-3 cells with those in 2-3c cells (or 3 cells with 3c cells) indicated that no more than 20 of the >5,300 transcripts recognized by the HG-U95A GeneChip were either decreased or increased in abundance by >2-fold. Remarkably, only three transcripts differed in abundance by >3-fold (Table 1). Similar results were obtained with the higher density HG-U133A GeneChip when 2-3 cells were compared to 2-3c cells (only 25 transcripts of 10,698 identified by the GeneChip differed in absolute abundance by >2-fold, and 7 transcripts differed in absolute abundance by >3-fold) or when type 3 cells were compared to 3c cells (43 of 10,359 transcripts differed by >2-fold, and only 14 transcripts differed by >3-fold) (Table 1).
TABLE 1.
Comparative analysis of the transcriptional profiles of Huh7 cell clones containing replicating NNeo/C5B RNA and their isoclonal, IFN-α-cured progeny
| Probe seta | No. of probe sets
|
||||
|---|---|---|---|---|---|
| 2-3c vs. clone 2-3 with GeneChip:
|
3c vs. clone 3 with GeneChip:
|
2-3c vs. 2-3c + SenV with GeneChip U133A | |||
| U95A | U133A | U95A | U133A | ||
| Total | 12,625 | 22,283 | 12,625 | 22,283 | 22,283 |
| “Present” | 5,350b | 10,698 | 5,102 | 10,359 | 10,698 |
| I >2-fold | 10 ± 0.5b | 5 | 8 | 38 | 307 |
| I >3-fold | 1.5 ± 0.5 | 2 | 2 | 12 | 152 |
| D >2-fold | 9.0 ± 1.0 | 20 | 1 | 5 | 70 |
| D >3-fold | 0.5 ± 0.5 | 5 | 0 | 2 | 12 |
“Present,” probe sets with a call of “present” in the absolute expression analysis of the baseline array (IFN-treated 2-3c or 3c cells that are free of HCV RNA), I, increased in the experimental array (2-3 or 3 cells containing HCV RNA) relative to the baseline array; D, decreased in the experimental array (2-3 or 3 cells containing HCV RNA) relative to the baseline array.
Mean of results obtained in two independent experiments ± the standard deviation.
Since each of the microarray experiments summarized in Table 1 was carried out with independent cellular RNA preparations, we looked for identity among the transcripts determined to be increased or decreased in abundance in each experiment. For the most part, these transcripts were unique to each experiment, suggesting that the difference in abundance was a random event and not linked to the presence or absence of HCV gene expression and RNA replication. However, transcripts encoding bone morphogenetic protein 2 (GenBank NM_001200), a member of the transforming growth factor β family (61), were among those identified as having increased >2-fold in abundance in two of the three microarray experiments comparing 2-3c with 2-3 cells. These transcripts were also scored as “increased” in comparisons of the 3c and 3 cells with both the HG-U95A and HG-U133A GeneChips, but in each case <2-fold (1.6- and 1.4-fold, respectively). Similarly, transcripts for natriuretic peptide precursor B (GenBank NM_002521) (55) were among the few identified as increased >2-fold in two of three experiments comparing 2-3c with 2-3 cells (HG-U95A and HG-U133A GeneChips) and one of two microarray experiments (HG-U133A GeneChip) comparing 3c and 3 cells. The increased abundance of these transcripts in both the 2-3 and the 3 cells compared to their clonally related, IFN-cured counterparts suggests that they may be upregulated in response to HCV RNA replication. In contrast, transcripts encoding Drosophila delta-like homolog (GenBank U15979), which encodes a transmembrane protein belonging to the epidermal growth factor-like superfamily (31), were identified as decreased in abundance by >2-fold in both comparisons of the 3c cells with clone 3 cells but were unchanged in comparisons of the 2-3c and 2-3 cells, making the significance of this observation less certain.
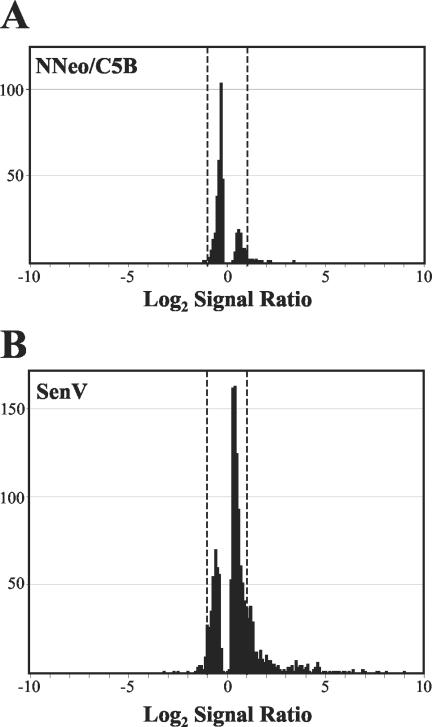
Because the differences in the transcript profiles of cells containing replicating HCV RNAs and their clonally related, IFN-α-cured progeny were so minimal and lacking in the reproducible induction of dsRNA- or IFN-stimulated genes (ISGs), we carried out a similar comparison of the transcript profiles of cured 2-3c cells before and after infection with SenV, which is known to induce transcription of a large number of ISGs (34, 41). In contrast to the minimal changes in transcripts associated with HCV RNA replication, SenV infection resulted in >2-fold increases or decreases in the abundance of 377 of the 10,698 transcripts recognized by the HG-U133A GeneChip (and >3-fold changes in 164 genes) (Table 1). A total of 1,460 transcripts were scored increased or decreased in abundance (to any extent) in this comparison, compared to only 404 in the comparison of the 2-3c with 2-3 cells (Fig. 2). Since many of the transcripts that were increased in abundance in the SenV-infected 2-3 cells were ISGs (data not shown), we conclude that the cured 2-3c cells retain the capacity for IFN-mediated responses but that these responses do not occur or are substantially muted in cells containing replicating HCV RNA and expressing a high abundance of the HCV polyprotein.
FIG. 2.
HG-U133A Affymetrix high-density oligonucleotide microarray analyses comparing global mRNA transcript profiles in IFN-α-treated 2-3c cells that do not contain HCV RNA with clonally related 2-3 cells supporting replication of NNeo/C5B RNA (A) or 2-3c cells infected 20 h previously with SenV (B). Shown are the number of probe sets distributed across the range of differences in signal intensities between baseline 2-3c cells and the “experimental” data set: HCV RNA-containing clone 2-3 cells or SenV-infected 2-3c cells. Differences in signal intensities are expressed as the log2 of the ratio of the experimental to baseline value. A positive value indicates an increase in transcript abundance in the experimental data set; a negative value indicates a decrease in abundance. Probe sets falling outside the dotted lines (log2 signal ratio of >1 or <−1) represent transcripts with >2-fold differences in abundance from the baseline.
A two-way ANOVA was carried out on data derived from the 2-3, 2-3c, 3, and 3c cells by using each type of GeneChip, after we eliminated transcripts with <2-fold absolute variation and those that were scored “absent” on all of the GeneChips, in an effort to identify those transcripts that varied in abundance according to the cell clone and/or presence or absence of HCV RNA replication. Combining data from the replicate experiments, 462 genes were found to be differentially regulated with the HG-U95A GeneChip (data not shown), and 557 genes were found to be differentially regulated with the HG-U133A GeneChip (Fig. 3). Surprisingly, clustering analyses carried out with these transcripts (using either the HG-U95A or HG-U133A data sets) indicated that the differences in transcript abundance were significantly greater between the 2-3c (or 2-3) and 3c (or 3) cell clones than between cells containing replicating viral RNA and their clonally related IFN-α-cured counterparts (Fig. 3). Although it is possible that the minimal changes observed in the mRNA expression profiles could play a role in altering the homeostasis of HCV-infected cells and contribute to pathogenesis, these observations are remarkable given numerous prior reports describing profound effects of HCV protein expression on cellular gene expression (11, 13, 29, 37, 39, 48, 50, 51, 53).
FIG. 3.
Affymetrix microarray profiling of mRNA transcripts in cells containing replicating NNeo/C5B RNA. Shown is an intensity matrix plot (“heat map”) produced by a hierarchical clustering analysis of those oligonucleotide probe sets representing mRNA transcripts identified by two-way ANOVA as being differentially regulated either as a result of cell clone (i.e., clone 2-3 versus clone 3) or the presence or absence of HCV RNA. Microarray analyses were carried out with the Affymetrix HG-U133A GeneChip with total cellular RNA isolated from two different HCV RNA-bearing Huh7 clones—2-3 and 3—and their IFN-treated, clonally related cell progeny (2-3c and 3c). After we eliminated probe sets marked “absent” in HCV-RNA-positive cells and transcripts with a difference in the hybridization intensity signal of <2-fold, the data from 557 probe sets were subjected to clustering analysis. The results demonstrate that the variation between the clone 2-3 and clone 3 Huh7 cell lines influences cellular transcription profiles much more than the presence or absence of replicating HCV RNA.
Cell cycle control in cells containing autonomously replicating HCV RNA.
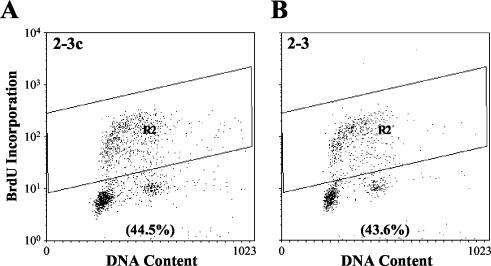
The results described above suggest that the replication of the HCV genome has little impact on the homeostasis of Huh7 cells. However, it is possible that significant effects on host cell metabolism and/or regulation of the cell cycle could result from posttranscriptional events induced by HCV proteins. Thus, to determine whether HCV RNA replication and polyprotein expression functionally influences the proportion of cells that are in the S phase of the cell cycle, asynchronously growing cells were labeled with the nucleoside analog BrdU and then analyzed by flow cytometry after being stained with a BrdU-specific antibody (Fig. 4). The percentages of cells in the G1, S, and G2/M phases did not differ significantly among the four cell lines examined (2-3, 2-3c, 3, and 3c [data not shown]) in several independent experiments. We conclude that the presence or absence of replicating HCV RNA in these cells, coupled as it is with the expression of the complete viral polyprotein, does not grossly influence the distribution of the cells across the various steps in the cell cycle.
FIG. 4.
Flow cytometry determination of the S-phase distribution of asynchronously growing Huh7 2-3c cells (A) and clonally related 2-3 cells (B) containing replicating, full-length HCV RNA. Cells were labeled with BrdU for 30 min and then analyzed by flow cytometry after being stained for BrdU incorporation and DNA content. The percentage of BrdU-positive cells in the two cell lines is indicated.
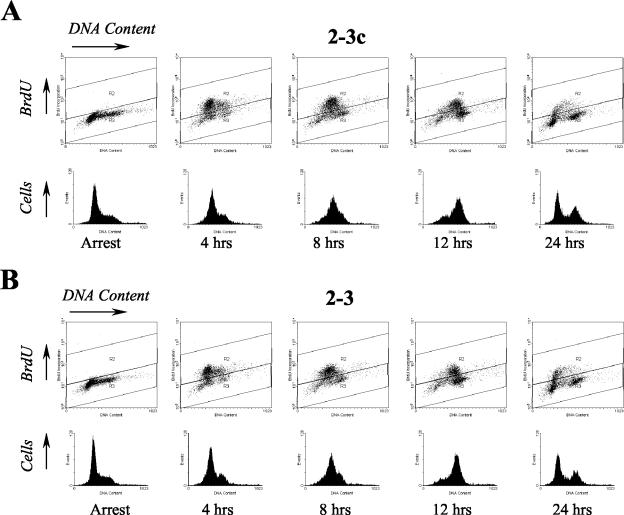
To further investigate the possible effects of HCV RNA and protein expression on host cell cycle regulation, we carried out a series of cell cycle arrest and release experiments. Huh7 cells harboring full-length HCV RNAs and their IFN-α-cured counterparts were arrested in the G1 phase of the cell cycle by treatment with aphidicolin, a DNA polymerase inhibitor. Cells were released from this arrest and allowed to progress through the cell cycle for various amounts of time and then labeled with BrdU to detect the S-phase population. Cells were stained with the BrdU-specific antibody, as well as 7-AAD, to measure total cellular DNA content prior to flow cytometry. The results from a typical experiment carried out with 2-3 cells and their IFN-α-cured progeny, 2-3c cells, are shown in Fig. 5. Neither the 2-3 nor the 2-3c cells were completely synchronized in G1 by aphidicolin treatment (as indicated by a small proportion of cells with >2n DNA content). However, aphidicolin treatment resulted in almost complete arrest of DNA synthesis as evidenced by the virtual absence of BrdU incorporation (<2%) in arrested cells (Table 2 and Fig. 5).
FIG. 5.
Flow cytometry analysis of cell cycle entry in populations of Huh7 2-3c cells (A) and clonally related 2-3 cells (B) containing full-length, replicating NNeo/C5B RNA. Cells were arrested in the G1 phase of the cell cycle with aphidicolin and released from the cell cycle block after 24 h. Prior to harvesting at the indicated time points, cells were labeled with BrdU for 30 min. BrdU incorporated into S-phase cells was stained with a monoclonal FITC-conjugated antibody, and the total DNA content was determined by staining with 7-AAD.
TABLE 2.
Percent BrdU-positive cells during and after aphidicolin arrest
| Cell type | % Brd-positive cells at:
|
||||
|---|---|---|---|---|---|
| Arrest | 4 h | 8 h | 12 h | 24 h | |
| 2-3c cells | 19 | 65.5 | 58.7 | 44.5 | 19.3 |
| Clone 2-3 | 1.8 | 66.3 | 58.5 | 38.8 | 23.5 |
After release from cell cycle arrest, both 2-3 and 2-3c cells rapidly progressed into S phase, with the BrdU labeling evident in similar proportions of cells within 4 to 8 h (compare Fig. 5). By 12 h some cells had already completed the cell cycle as indicated by an increase in the G1 population, and by 24 h the cell cycle distribution had returned to almost that of an asynchronous population. When the percentage of BrdU-positive S-phase cells was determined at multiple time points, there were no significant differences between the HCV RNA-positive 2-3 cells and the HCV-negative 2-3c cells (Table 2). Similar results were obtained in studies carried out with the clone 3 cells and their IFN-α-cured progeny, 3c cells (data not shown). We conclude that the presence of HCV RNA in Huh7 cells does not interfere with reentry of cells into S phase after transient G1 arrest. These results are consistent with the absence of changes in the expression levels of cell cycle regulatory genes in the microarray analyses described above.
HCV RNA replication depends on actively growing host cells.
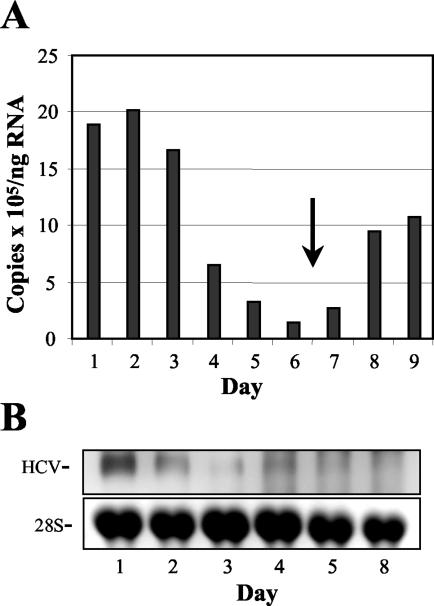
Earlier studies with replicons derived from the sequence of the Con1 isolate of HCV indicated that the replication of HCV RNA requires actively growing host cells (47). To determine whether the replication of NNeo/C5B RNA was similarly dependent upon cell growth, Huh7 cells containing the replicating RNA and their IFN-α-cured counterparts were cultured until near confluence, with samples taken just before cells reached confluence and every day thereafter. Total cellular RNA was extracted and analyzed by real-time RT-PCR and Northern blot with primers and probes specific for the HCV sequence (Fig. 6). Within 48 h of reaching confluence, the abundance of intracellular HCV RNA (copies per nanogram total cellular RNA) dropped approximately 20-fold, recovering when the confluent cells were split and resumed exponential growth (Fig. 6A). These results were confirmed by Northern analysis in a separate experiment (Fig. 6B).
FIG. 6.
HCV RNA abundance declines in Huh7 2-3 cells as cells reach confluence. (A) Real-time RT-PCR quantitation of HCV RNA normalized to cellular GAPDH mRNA abundance. Cells were approximately 80% confluent on day 1 and 100% confluent on day 2. The cells were split on day 6 (arrow). The HCV RNA levels declined rapidly as cells grew more and more confluent and rebounded once logarithmic growth was resumed. (B) Northern blot detection of HCV RNA in a similar experiment with an NS5B-specific probe. Cells were near 100% confluence on day 1. HCV RNA levels decreased with increasing cell confluence, in agreement with the RT-PCR data.
To corroborate these results and relate these findings to growth arrest induced by different means, we monitored HCV RNA levels under conditions of serum starvation. Clone 2-3 cells were incubated with increasing concentrations of fetal bovine serum (0 to 10%) for 3 days and analyzed by Northern blot (Fig. 7A). HCV RNA abundance was not affected at serum concentrations as low as 5% but decreased significantly with lower concentrations in serum. To determine whether this decrease in HCV RNA abundance was due to inhibition of RNA synthesis, in vivo labeling studies were performed. Cells that had been serum starved for 72 h were refed with 10% serum, and HCV RNA synthesis monitored at intervals by labeling with [32P]orthophosphate (in the presence of actinomycin D to inhibit cellular RNA synthesis) (Fig. 7B). Incorporation of label was analyzed by gel electrophoresis of extracted RNAs and autoradiography, followed by densitometric quantitation (Fig. 7C). This experiment demonstrated that HCV RNA synthesis was significantly decreased in serum-starved cells compared to untreated cells. HCV RNA synthesis returned to normal within 48 h of adding back serum to the starved cells. These data demonstrate that prevention of cell growth by different means leads to substantial inhibition of HCV RNA synthesis.
FIG. 7.
HCV RNA abundance in 2-3 cells during serum starvation. (A) Northern blot detection of HCV RNA in clone 2-3 cells grown in medium containing the indicated percentage of serum for 3 days. Total RNA was extracted and hybridized to a probe specific for NS5B. HCV RNA abundance declined with decreasing concentrations in serum. (B) Analysis of HCV RNA synthesis during serum starvation by in vivo labeling. Normal Huh7 cells (lane 1) or clone 2-3 cells (lanes 2 to 5) were incubated in the presence (lanes 1 and 2) or absence of serum for 72 h (lanes 3 to 5). Serum was added back for 48 and 72 h, respectively, after the 72-h period of serum starvation for the samples shown in lanes 4 and 5. HCV RNA synthesis was monitored by incorporation of [32P]orthophosphate for 15 h, followed by gel electrophoresis and autoradiography of the isolated RNA (top panel). Also shown is the 26S RNA band from the same gel visualized with ethidium bromide (bottom panel). (C) Results of densitometric quantitation of the 2-3 cell data shown in panel B, with values for HCV RNA normalized to the amount of 26S RNA and the values for cells growing in 10% serum set arbitrarily to 100%.
Steady-state abundance of HCV RNA does not change during transient G1 arrest.
The previous results suggest that HCV RNA levels decline in cells that are not growing exponentially. It was therefore of interest to determine whether the abundance of HCV RNA is similarly modulated during transient G1 arrest and subsequent cell cycle release. Clone 2-3 cells were arrested in G1 by aphidicolin treatment as described above and subsequently released from the cell cycle block. Total RNA was extracted, and the abundance of HCV RNA at progressive times after release was monitored by real-time RT-PCR and Northern analysis (Fig. 8A). Although the intracellular abundance of HCV RNA varied somewhat as the cells progressed through the cell cycle, with the abundance slightly elevated at time points corresponding to the majority of cells being in S phase, the RNA abundance remained relatively unchanged during a 24-h period of cell cycle arrest, followed by cell cycle progression. Monitoring of HCV protein expression under these conditions by immunoblot analysis demonstrated that the intracellular abundance of NS3 is not influenced by the cell cycle (Fig. 8B). After 2 days of arrest with aphidicolin, only a slight decline in the abundance of NS3 was observed.
FIG. 8.
HCV RNA and protein abundances during cell cycle arrest and release. (A) Huh7 2-3 cells harboring replicating NNeo/C5B RNA were G1 arrested with aphidicolin for 24 h and subsequently released from the cell cycle block. Cells were harvested at the indicated times and total RNA extracted and analyzed by real-time RT-PCR (top panel) and Northern blot (bottom panel). The levels of HCV RNA were not altered significantly between G1 arrested and released cells and varied only slightly as the cells progressed though the cell cycle. (B) Immunoblot analysis of NS3 expression in clone 2-3 cells during and after aphidicolin release. Total cell lysates containing 50 μg of protein were analyzed by immunoblot with an MAb specific for NS3 (top panel) or β-actin (lower panel).
To determine whether individual cells express a greater abundance of HCV proteins depending on the specific stage of the cell cycle, we used flow cytometry to analyze aphidicolin-arrested and released cells stained with an antibody specific for NS5A. Virtually no fluorescence was detected with parental Huh7 cells, whereas approximately 70% of the clone 2-3 cells contained an abundance of NS5A sufficient for detection in this system. The percentage of HCV antigen-positive cells did not vary significantly with arrest or with subsequent release and progression through the cell cycle (data not shown). We obtained similar results with G418-resistant Huh7 cell clones supporting the replication of subgenomic replicons (NNeo/3-5B) or replicons expressing all of the HCV polyprotein with the exception of core (NNeo/E1-5B) (data not shown).
Synthesis of HCV RNAs is stimulated during S phase.
Although major differences in HCV RNA and protein abundance were not observed during cell cycle arrest and release, these experiments do not rule out the possibility that viral RNA synthesis might be stimulated during a specific phase of the cell cycle. The apparent constant abundance of steady-state HCV RNA might be due to a balance between RNA degradation and newly synthesized RNA or a relatively slow rate of turnover of viral RNA, making the contributions of an increased rate of synthesis difficult to detect over a brief period. To address this question, we assessed HCV RNA synthesis by monitoring [32P]orthophosphate incorporation during and after aphidicolin arrest. As in the [32P]orthophosphate incorporation experiments described previously, these measurements required the inhibition of cellular RNA synthesis by actinomycin D. Although HCV RNA synthesis remained detectable in G1-arrested cells, the rate of incorporation of [32P]orthophosphate was increased ∼4-fold by 8 h after release from cell cycle arrest (Fig. 9). At this point in time, the majority of cells are in S phase (Fig. 5). The subsequent decline in HCV RNA synthesis observed at later time points with these synchronized cells, after most cells have completed S phase (Fig. 9, 24 h), provides further support for the notion that HCV RNA synthesis is increased during this specific phase of the cell cycle.
FIG. 9.
In vivo labeling of replicating full-length HCV RNA during cell cycle arrest and release. Huh7 2-3 cells were arrested in G1 by treatment with aphidicolin for 24 h and subsequently released from the cell cycle block. At 4 h before being harvested, the cells were starved in phosphate-free medium for 1 h, followed by the addition of [32P]orthophosphate at a concentration of 500 μCi/ml. Total RNA was extracted and analyzed by electrophoresis, and radioactively labeled, newly synthesized HCV RNA was quantitated by densitometry. There was a reproducible increase in HCV RNA synthesis as the cells progressed through S phase, with HCV RNA synthesis peaking approximately 8 h after release from the G1 block.
DISCUSSION
In contrast to the findings reported here, many previous reports suggest that both the structural and the NS proteins of HCV may have strong effects on host cell homeostasis and cell cycle control (3, 11, 19, 21, 23, 27, 30, 37, 39, 43, 56, 62-64). Most describe studies involving single gene expression experiments, often with highly active promoters resulting in significant protein overexpression to nonphysiologic levels and usually involving only a single viral protein, sometimes with a potentially critical domain deleted. In addition, these previous studies used a variety of cellular backgrounds, including cells of nonhepatic origin, or hepatic cells that are incapable of supporting HCV RNA replication. All of these factors may have contributed to the different and occasionally conflicting phenotypes that have been described in the past with cells expressing individual HCV proteins. In particular, protein-protein interactions among different HCV NS proteins have been reported and are likely important for replicase activity (33, 58). It is plausible that effects observed with overexpression of individual proteins might not occur in the context of expression of the complete complement of viral proteins, when replication complexes are formed and the appropriate protein-protein interactions take place. The studies we describe here were carried out in Huh7 cells, which are permissive for the replication of HCV RNA. Importantly, our observations have been drawn from cells containing autonomously replicating HCV RNAs encoding the entire viral polyprotein, within the context of actively replicating viral RNA.
Significantly, we observed no significant effects of HCV protein expression on cell cycle regulation, and only minimal effects on the cellular transcriptome. This absence of a disruption or perturbation of normal homeostatic cellular processes cannot be attributed readily to a lower level of expression, since clone 2-3 and 3 cells contain viral proteins that are readily detected in immunoblots and viral RNA that is easily visualized by Northern analysis (Fig. 1). Since this level of protein expression is likely to exceed that which occurs normally within the liver of most infected persons, these observations have important implications for the pathogenesis of hepatitis C.
The minimal differences we observed in the transcriptional profiles of cells containing these replicating HCV RNAs and their IFN-α-cured, clonally related progeny was surprising given the published literature referred to above. However, this finding was confirmed with two, independently selected G418-resistant Huh7 cell lines, each supporting the replication of HCV-N RNA expressing the entire complement of viral proteins. In interpreting these data, it is important to note that the clone 2-3 and 3 cell lines were selected from Huh7 cells because of their ability to support replication of the viral RNA in the absence of gross disturbances in cellular metabolism or cell cycle regulation that would impair cellular proliferation. Thus, the possibility cannot be excluded that a greater impact on host transcription might occur in nonselected hepatocytes, for example, those resident within the liver in vivo. The factor(s) that render Huh7 cells uniquely permissive for HCV replication remains uncertain, but it is possible that this permissiveness and the failure of HCV to alter gene expression and cell cycle regulation in Huh7 cells are functionally related.
Nonetheless, the clustering analysis carried out with Affymetrix data (Fig. 3) derived from both the clone 2-3 and clone 3 cells makes it clear that, although differences between the cell clones exceeded the impact of HCV polyprotein expression on the cellular transcriptome, there is a subtle, coordinated transcriptional response to HCV RNA replication and protein expression that involves minor changes in the abundance of a large number of cellular mRNAs (162 probe sets in the HG-U95 GeneChip experiments, and 176 probe sets with the HG-U133A GeneChips, as determined by ANOVA at a 95% confidence level [data not shown]). The nature and pathological significance of the transcriptional pathways that are influenced by HCV polyprotein expression and RNA replication remain to be more fully elucidated. However, it is interesting that transcripts encoding bone morphogenetic protein 2 were found to be increased in abundance in both clone 2-3 and 3 cells (see Results), since this member of the transforming growth factor β family has been shown to stimulate the synthesis of type 2 collagen (59).
The contrast we observed between the large number of ISG transcripts induced by SenV infection of 2-3c cells and the apparent absence of a significant ISG response in clone 2-3 cells (Fig. 2) was striking. This is unlikely to be due to an inadequate stimulus for an IFN response in the 2-3 cells, since they contain abundant replicating viral RNA, including almost certainly dsRNA species (Fig. 1). The failure to observe an ISG transcriptional response is thus most likely due to disruption of IRF-3 activation pathways by the NS3/4A protease expressed by the replicating HCV RNA (14). The robust response of ISGs to HCV infection that has been observed in the livers of chimpanzees studied by similar Affymetrix assays (54) also provides a sharp contrast to what we observed in these HCV RNA-containing Huh7 cell clones. These differences are likely to stem from the much more complex cellular milieu that is present in the chimpanzee liver, the presence of an active adaptive immune response, and many immune-stimulated cells that are not infected with the virus.
In analyzing the cell cycle regulation in the clone 2-3 and clone 3 cells, we focused on the G1-S transition since previous reports suggest effects of HCV proteins on p53 and p21 function (2, 11, 13, 19, 27, 29, 30, 39, 43, 50, 51, 60, 62, 63), which would be expected to predominantly affect the G1 checkpoint. We did not specifically address whether an HCV-dependent alteration occurs at the G2/M border, since the absence of any gross changes in cell cycle distribution among asynchronously growing cells were not suggestive of this possibility (Fig. 4). Interestingly, in other studies involving Huh7 cells with conditional expression of either the HCV structural proteins or the full-length polyprotein, we also failed to observe any effects of HCV protein expression on cellular regulation, including the previously reported core-dependent upregulation of p21, for example (29, 37, 43, 62; data not shown). This is in keeping with the findings reported here regarding the absence of cell cycle disruption by HCV, observations that are corroborated by the gene expression profile analyses.
As a complement to studying the impact of viral RNA replication on the cell cycle and cellular homeostasis, we investigated the impact of the cell cycle on viral RNA replication. In agreement with previously published results (47), we found that HCV RNA levels declined drastically when cells were no longer able to grow exponentially. The decrease in HCV RNA levels was most pronounced in experiments in which Huh7 cells were allowed to grow to confluence, after which HCV RNA levels decreased drastically within 48 h (Fig. 6). Serum starvation produced similar effects within 72 h (Fig. 7A). Although the abundance of HCV RNA was minimally affected during 24 h of cell cycle arrest with aphidicolin, HCV RNA synthesis was decreased both in aphidicolin-arrested cells and in serum-starved cells (Fig. 7B and C and 9). However, it is possible that decreased de novo synthesis is not the sole mechanism responsible for the decrease in viral RNA abundance in nonproliferating cells. Other mechanisms, such as destabilization or active degradation of viral RNA might contribute to the rapid decline observed in confluent cells.
Our results demonstrate that the steady-state abundance of HCV RNA does not vary dramatically in arrested cells, compared to cells in various phases of the cell cycle (Fig. 8). In contrast, HCV RNA synthesis is significantly stimulated in S-phase cells (Fig. 9). It is likely that in well-established cell clones supporting HCV RNA replication, a balance is reached between HCV RNA degradation and S-phase-stimulated new synthesis, resulting in stable, homeostatic HCV RNA expression throughout the cell cycle. The existence of a functional link between the cell cycle of the host cell and regulation of replication of a positive-strand viral RNA is interesting. The mechanisms underlying this connection are not understood at present and require further investigation.
Acknowledgments
This study was supported by grant U19-AI40035 from the National Institute of Allergy and Infectious Diseases. F.S. and K.L. were supported by J. W. McLaughlin Postdoctoral Fellowships. K.L. is the John Mitchell Hemophilia of Georgia Liver Scholar of the American Liver Foundation.
REFERENCES
- 1.Alter, M. 1997. Epidemiology of hepatitis C. Hepatology 26:62S-65S. [DOI] [PubMed] [Google Scholar]
- 2.Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. [DOI] [PubMed] [Google Scholar]
- 3.Arima, N., C. Y. Kao, T. Licht, R. Padmanabhan, and Y. Sasaguri. 2001. Modulation of cell growth by the hepatitis C virus nonstructural protein NS5A. J. Biol. Chem. 276:12675-12684. [DOI] [PubMed] [Google Scholar]
- 4.Barba, G., H. F. Harada, T. Kohara, M. Goulinet, S. Matsuura, Y. Eder, G. Z. Schaff, M. T. Chapman, and C. Brechot. 1997. Hepatitis C virus core protein shows a cytoplasmic localization and associates to cellular lipid storage droplets. Proc. Natl. Acad. Sci. USA 94:1200-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartenschlager, R., L. Ahlborn-Laake, J. Mous, and H. Jacobsen. 1994. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J. Virol. 68:5045-5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartenschlager, R., and V. Lohmann. 2001. Novel cell culture systems for the hepatitis C virus. Antivir. Res. 52:1-17. [DOI] [PubMed] [Google Scholar]
- 7.Bartenschlager, R., V. Lohmann, T. Wilkinson, and J. O. Koch. 1995. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J. Virol. 69:7519-7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 9.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina-Selby, P. J. Barr, et al. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung, K. M., O. K. Song, and S. K. Jang. 1997. Hepatitis C virus nonstructural protein 5A contains potential transcriptional activator domains. Mol. Cell 7:661-667. [PubMed] [Google Scholar]
- 11.Dubourdeau, M. T., Y. Matsuura, L. Alric, B. Pipy, and D. Rousseau. 2002. Infection of HepG2 cells with recombinant adenovirus encoding the HCV core protein induces p21WAF1 down-regulation: effect of transforming growth factor beta. J. Hepatol. 37:486-492. [DOI] [PubMed] [Google Scholar]
- 12.Dumoulin, F. L., A. van dem Bussche, J. Sohne, T. Sauerbruch, and U. Spengler. 1999. Hepatitis C virus core protein does not inhibit apoptosis in human hepatoma cells. Eur. J. Clin. Investig. 29:940-946. [DOI] [PubMed] [Google Scholar]
- 13.Feng, D. Y., R. X. Chen, Y. Peng, H. Zheng, and Y. H. Yan. 1999. Effect of HCV NS(3) protein on p53 protein expression in hepatocarcinogenesis. World J. Gastroenterol. 5:45-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 15.Francki, R. I. B., D. L. Knudson, and F. Brown. 1991. Classification and nomenclature of viruses: fifth report of the International Committee on Taxonomy of Viruses. Arch. Virol. 2:223. [Google Scholar]
- 16.Gale, M., Jr., C. M. Blakely, B. Kwieciszewski, S. L. Tan, M. Dossett, N. M. Tang, M. J. Korth, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1998. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol. Cell. Biol. 18:5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gale, M. J., Jr., M. J. Korth, and M. G. Katze. 1998. Repression of the PKR protein kinase by the hepatitis C virus NS5A protein: a potential mechanism of interferon resistance. Clin. Diagn. Virol. 10:157-162. [DOI] [PubMed] [Google Scholar]
- 18.Gale, M. J., Jr., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217-227. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh, A. K., R. Steele, K. Meyer, R. Ray, and R. B. Ray. 1999. Hepatitis C virus NS5A protein modulates cell cycle regulatory genes and promotes cell growth. J. Gen. Virol. 80:1179-1183. [DOI] [PubMed] [Google Scholar]
- 20.Grakoui, A., D. W. McCourt, C. Wychowski, S. M. Feinstone, and C. M. Rice. 1993. A second hepatitis C virus-encoded proteinase. Proc. Natl. Acad. Sci. USA 90:10583-10587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han, H. J., W. J. Lee, and K. L. Jang. 2002. Cooperative repression of cyclin-dependent kinase inhibitor p21 gene expression by hepatitis B virus X protein and hepatitis C virus core protein. FEBS Lett. 518:169-172. [DOI] [PubMed] [Google Scholar]
- 22.Honda, A., Y. Arai, N. Hirota, T. Sato, J. Ikegaki, T. Koizumi, M. Hatano, M. Kohara, T. Moriyama, M. Imawari, K. Shimotohno, and T. Tokuhisa. 1999. Hepatitis C virus structural proteins induce liver cell injury in transgenic mice. J. Med. Virol. 59:281-289. [DOI] [PubMed] [Google Scholar]
- 23.Honda, M., S. Kaneko, T. Shimazaki, E. Matsushita, K. Kobayashi, L. H. Ping, H. C. Zhang, and S. M. Lemon. 2000. Hepatitis C virus core protein induces apoptosis and impairs cell cycle regulation in stably transformed Chinese hamster ovary cells. Hepatology 31:1351-1359. [DOI] [PubMed] [Google Scholar]
- 24.Hope, R. G., D. J. Murphy, and J. McLauchlan. 2002. The domains required to direct core proteins of hepatitis C virus and GB virus-B to lipid droplets share common features with plant oleosin proteins. J. Biol. Chem. 277:4261-4270. [DOI] [PubMed] [Google Scholar]
- 25.Hope, R. G., and D. J. Murphy. 2000. Sequence motifs required for lipid droplet association and protein stability are unique to the hepatitis C virus core protein. J. Gen. Virol. 81:1913-1925. [DOI] [PubMed] [Google Scholar]
- 26.Ikeda, M., M. Yi, K. Li, and S. M. Lemon. 2002. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J. Virol. 76:2997-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jung, E. Y., M. N. Lee, H. Y. Yang, D. Yu, and K. L. Jang. 2001. The repressive activity of hepatitis C virus core protein on the transcription of p21waf1 is regulated by protein kinase A-mediated phosphorylation. Virus Res. 79:109-115. [DOI] [PubMed] [Google Scholar]
- 28.Kim, D. W., Y. Gwack, J. H. Han, and J. Choe. 1995. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem. Biophys. Res. Commun. 215:160-166. [DOI] [PubMed] [Google Scholar]
- 29.Kwun, H. J., and K. L. Jang. 2003. Dual effects of hepatitis C virus core protein on the transcription of cyclin-dependent kinase inhibitor p21 gene. J. Viral Hepat. 10:249-255. [DOI] [PubMed] [Google Scholar]
- 30.Lan, K. H., M. L. Sheu, S. J. Hwang, S. H. Yen, S. Y. Chen, J. C. Wu, Y. J. Wang, N. Kato, M. Omata, F. Y. Chang, and S. D. Lee. 2002. HCV NS5A interacts with p53 and inhibits p53-mediated apoptosis. Oncogene 21:4801-4811. [DOI] [PubMed] [Google Scholar]
- 31.Lee, Y. L., L. Helman, T. Hoffman, and J. Laborda. 1995. dlk, pG2 and Pref-1 mRNAs encode similar proteins belonging to the EGF-like superfamily: identification of polymorphic variants of this RNA. Biochim. Biophys. Acta 1261:223-232. [DOI] [PubMed] [Google Scholar]
- 32.Lerat, H., M. Honda, M. R. Beard, K. Loesch, J. Sun, Y. Yang, M. Okuda, R. Gosert, S. Y. Xiao, S. A. Weinman, and S. M. Lemon. 2002. Steatosis and liver cancer in transgenic mice expressing the structural and nonstructural proteins of hepatitis C virus. Gastroenterology 122:352-365. [DOI] [PubMed] [Google Scholar]
- 33.Lin, C., J. W. Wu, K. Hsiao, and M. S. Su. 1997. The hepatitis C virus NS4A protein: interactions with the NS4B and NS5A proteins. J. Virol. 71:6465-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lohmann, V., J. O. Koch, and R. Bartenschlager. 1996. Processing pathways of the hepatitis C virus proteins: in vitro studies on the activation of the hepatitis C virus NS3 proteinase by the NS4A cofactor. J. Hepatol. 24:11-19. [PubMed] [Google Scholar]
- 36.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 37.Lu, W., S. Y. Lo, M. Chen, K. Wu, Y. K. Fung, and J. H. Ou. 1999. Activation of p53 tumor suppressor by hepatitis C virus core protein. Virology 264:134-141. [DOI] [PubMed] [Google Scholar]
- 38.Majumder, M., A. K. Ghosh, R. Steele, X. Y. Zhou, N. J. Phillips, R. Ray, and R. B. Ray. 2002. Hepatitis C virus NS5A protein impairs TNF-mediated hepatic apoptosis, but not by an anti-FAS antibody, in transgenic mice. Virology 294:94-105. [DOI] [PubMed] [Google Scholar]
- 39.Majumder, M., R. Steele, R. Ray, and R. B. Ray. 2001. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. J. Virol. 75:1401-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLauchlan, J., M. K. Lemberg, G. Hope, and B. Martoglio. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 21:3980-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Megyeri, K., W. C. Au, I. Rosztoczy, N. B. Raj, R. L. Miller, M. A. Tomai, and P. M. Pitha. 1995. Stimulation of interferon and cytokine gene expression by imiquimod and stimulation by Sendai virus utilize similar signal transduction pathways. Mol. Cell. Biol. 15:2207-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakao, H., Y. He, S. Tareen, S. Vijaysri, B. Jacobs, M. Katze, and M. Itoh. 2000. HCV NS5A protein forms a complex with the multisite docking protein Gab1 and potentiates activation of the PI3K/AKT signaling pathway by epidermal growth factor. 7th International Meeting on Hepatitis C and Related Viruses, Queensland, Australia.
- 43.Otsuka, M., N. Kato, K. Lan, H. Yoshida, J. Kato, T. Goto, Y. Shiratori, and M. Omata. 2000. Hepatitis C virus core protein enhances p53 function through augmentation of DNA binding affinity and transcriptional ability. J. Biol. Chem. 275:34122-34130. [DOI] [PubMed] [Google Scholar]
- 44.Pavlovic, D., D. C. Neville, O. Argaud, B. Blumberg, R. A. Dwek, W. B. Fischer, and N. Zitzmann. 2003. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. USA 100:6104-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perlemuter, G., P. Letteron, G. Vona, A. Topilco, Y. Chretien, K. Koike, C. J. Pessayre, G. Barba, and C. Brechot. 2002. Hepatitis C virus core protein inhibits microsomal triglyceride transfer protein activity and very low density lipoprotein secretion: a model of viral-related steatosis. FASEB J. 16:185-194. [DOI] [PubMed] [Google Scholar]
- 46.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Polyak, S. J., D. M. Paschal, S. McArdle, M. J. Gale, Jr., D. Moradpour, and D. R. Gretch. 1999. Characterization of the effects of hepatitis C virus nonstructural 5A protein expression in human cell lines and on interferon-sensitive virus replication. Hepatology 29:1262-1271. [DOI] [PubMed] [Google Scholar]
- 49.Ray, R. B., K. Meyer, and R. Ray. 1996. Suppression of apoptotic cell death by hepatitis C virus core protein. Virology 226:176-182. [DOI] [PubMed] [Google Scholar]
- 50.Ray, R. B., R. Steele, K. Meyer, and R. Ray. 1998. Hepatitis C virus core protein represses p21WAF1/Cip1/Sid1 promoter activity. Gene 208:331-336. [DOI] [PubMed] [Google Scholar]
- 51.Ray, R. B., R. Steele, K. Meyer, and R. Ray. 1997. Transcriptional repression of p53 promoter by hepatitis C virus core protein. J. Biol. Chem. 272:10983-10986. [DOI] [PubMed] [Google Scholar]
- 52.Reed, K. E., A. Grakoui, and C. M. Rice. 1995. Hepatitis C virus-encoded NS2-3 protease: cleavage-site mutagenesis and requirements for bimolecular cleavage. J. Virol. 69:4127-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shrivastava, A., S. K. Manna, R. Ray, and B. B. Aggarwal. 1998. Ectopic expression of hepatitis C virus core protein differentially regulates nuclear transcription factors. J. Virol. 72:9722-9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Su, A. I., J. P. Pezacki, L. Wodicka, A. D. Brideau, L. Supekova, R. Thimme, S. Wieland, J. Bukh, R. H. Purcell, P. G. Schultz, and F. V. Chisari. 2002. Genomic analysis of the host response to hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 99:15669-15674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sudoh, T., K. Maekawa, M. Kojima, N. Minamino, K. Kangawa, and H. Matsuo. 1989. Cloning and sequence analysis of cDNA encoding a precursor for human brain natriuretic peptide. Biochem. Biophys. Res. Commun. 159:1427-1434. [DOI] [PubMed] [Google Scholar]
- 56.Takamatsu, M., T. Fujita, and H. Hotta. 2001. Suppression of serum starvation-induced apoptosis by hepatitis C virus core protein. Kobe J. Med. Sci. 47:97-112. [PubMed] [Google Scholar]
- 57.Takeuchi, T., A. Katsume, T. Tanaka, A. Abe, K. Inoue, K. Tsukiyama-Kohara, R. Kawaguchi, S. Tanaka, and M. Kohara. 1999. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology 116:636-642. [DOI] [PubMed] [Google Scholar]
- 58.Tu, H., L. Gao, S. T. Shi, D. R. Taylor, T. Yang, A. K. Mircheff, Y. Wen, A. E. Gorbalenya, S. B. Hwang, and M. M. Lai. 1999. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263:30-41. [DOI] [PubMed] [Google Scholar]
- 59.Valcourt, U., J. Gouttenoire, A. Moustakas, D. Herbage, and F. Mallein-Gerin. 2002. Functions of transforming growth factor-beta family type I receptors and Smad proteins in the hypertrophic maturation and osteoblastic differentiation of chondrocytes. J. Biol. Chem. 277:33545-33558. [DOI] [PubMed] [Google Scholar]
- 60.Wang, F., I. Yoshida, M. Takamatsu, S. Ishido, T. Fujita, K. Oka, and H. Hotta. 2000. Complex formation between hepatitis C virus core protein and p21Waf1/Cip1/Sdi1. Biochem. Biophys. Res. Commun. 273:479-484. [DOI] [PubMed] [Google Scholar]
- 61.Wozney, J. M., V. Rosen, A. J. Celeste, L. M. Mitsock, M. J. Whitters, R. W. Kriz, R. M. Hewick, and E. A. Wang. 1988. Novel regulators of bone formation: molecular clones and activities. Science 242:1528-1534. [DOI] [PubMed] [Google Scholar]
- 62.Yamanaka, T., T. Kodama, and T. Doi. 2002. Subcellular localization of HCV core protein regulates its ability for p53 activation and p21 suppression. Biochem. Biophys. Res. Commun. 294:528-534. [DOI] [PubMed] [Google Scholar]
- 63.Yoshida, I., K. Oka, R. Hidajat, M. Nagano-Fujii, S. Ishido, and H. Hotta. 2001. Inhibition of p21/Waf1/Cip1/Sdi1 expression by hepatitis C virus core protein. Microbiol. Immunol. 45:689-697. [DOI] [PubMed] [Google Scholar]
- 64.Zemel, R., S. Gerechet, H. Greif, L. Bachmatove, Y. Birk, A. Golan-Goldhirsh, M. Kunin, Y. Berdichevsky, I. Benhar, and R. Tur-Kaspa. 2001. Cell transformation induced by hepatitis C virus NS3 serine protease. J. Viral Hepat. 8:96-102. [DOI] [PubMed] [Google Scholar]
- 65.Zhu, N., A. Khoshnan, R. Schneider, M. Matsumoto, G. Dennert, C. Ware, and M. M. Lai. 1998. Hepatitis C virus core protein binds to the cytoplasmic domain of tumor necrosis factor (TNF) receptor 1 and enhances TNF-induced apoptosis. J. Virol. 72:3691-3697. [DOI] [PMC free article] [PubMed] [Google Scholar]