Abstract
Recent reports suggest that nuclear domain(s) 10 (ND10) is the site of papillomavirus morphogenesis. The viral genome replicates in or close to ND10. In addition, the minor capsid protein, L2, accumulates in these subnuclear structures and recruits the major capsid protein, L1. We have now used cell lines deficient for promyelocytic leukemia (PML) protein, the main structural component of ND10, to study the role of this nuclear protein for L2 incorporation into virus-like particles (VLPs). L2 expressed in PML protein knockout (PML−/−) cells accumulated in nuclear dots, which resemble L2 aggregates forming at ND10 in PML protein-containing cells. These L2 assemblies also attracted L1 and the transcriptional repressor Daxx, suggesting that they are functional in the absence of PML protein. In addition, L2-containing VLPs assembled in PML−/− cells. In order to analyze whether incorporation of L2 into VLPs requires any specific subcellular localization, an L1 mutant defective for nuclear transport and L2 mutants deficient in nuclear translocation and/or ND10 localization were constructed. Using this approach, we identified two independent L2 domains interacting with L1. Mutant L2 proteins not accumulating in ND10 were incorporated into VLPs. Mutant L1 protein, which assembled into VLPs in the cytoplasm, did not incorporate L2 defective for nuclear translocation. The same mutant L2 protein, which passively diffuses into the nucleus, is incorporated into wild-type L1-VLPs in the nucleus. Our data demonstrate that the incorporation of L2 into VLPs requires nuclear but not ND10 localization.
Human papillomaviruses (HPVs) comprise a large group of nonenveloped epitheliotropic viruses causing tumors of the skin and mucosa. They are composed of 360 copies of the major capsid protein, L1, organized in 72 capsomeres, probably 12 copies of the minor capsid protein, L2, and a closed circular, double-stranded DNA genome of approximately 8,000 bp (3, 35). The viral life cycle depends on terminally differentiating keratinocytes and includes infection of cells of the basal layer (stratum germinativum), vegetative DNA replication in nondividing cells of the stratum spinosum, and capsid protein synthesis and virion morphogenesis in fully differentiated cells of the stratum granulosum (reviewed in reference 21). Due to difficulties in generating HPVs in vitro and in mimicking the papillomavirus life cycle in cell culture, surrogate assays have been developed for the study of certain aspects of their life cycle, including the HPV infection process, viral DNA replication, and virion morphogenesis.
Heterologous expression of capsid protein L1 in eukaryotic cells results in spontaneous assembly into virus-like particles (VLPs) that incorporate L2 protein when coexpressed (13, 18, 28, 39). VLPs have been used to study cell binding and internalization of the papillomavirus capsid (24, 40). If present during capsid protein synthesis, extrachromosomal plasmids are also encapsidated to yield pseudovirions, a useful tool for the study of the HPV infection process (27, 32, 36).
Transfection of cell lines in vitro, using reporter plasmids harboring the papillomavirus origin of replication and expression cassettes for supplying virus-specific replication factors, revealed that papillomavirus replication occurs near nuclear domain(s) (ND10). These distinct subnuclear structures (i) have been proposed as transient deposition sites for nuclear proteins, (ii) are implicated in control of transcription (7), cell growth (12), and apoptosis (41), and (iii) are targeted and destroyed by early proteins of large DNA viruses (1, 2, 15, 33). HPV E1 and E2, both directly involved in DNA replication, were shown to contribute to ND10 association of the papillomavirus genome (34). Furthermore, L2 protein was shown to accumulate in these subnuclear structures, recruiting L1 protein into ND10 (5, 10). L2-induced reorganization of ND10, including attraction of the transcriptional repressor Daxx and dispersal of the transcriptional activator Sp100, was also reported. In addition, L2 protein transiently colocalizes with ND10 in HPV-induced lesions (9). Therefore, ND10 were proposed as sites of papillomavirus morphogenesis.
The promyelocytic leukemia (PML) protein is the main structural component of ND10 (14, 42). It was identified as fusion protein with the retinoic acid receptor in cell lines established from patients suffering from acute PML (6, 17). ND10 are disrupted in cells deficient for PML protein (42). Using PML protein knockout (PML−/−) cells, we have now investigated the role of PML protein in L1/L2 capsid formation. We found that PML protein is dispensable for L2 incorporation into VLPs. The L2 protein forms nuclear aggregates in PML−/− cells reminiscent of ND10, attracting L1 and Daxx and yielding L2-containing VLPs. Using different combinations of L1 and L2 mutants, we observed that nuclear localization but not nuclear L2 aggregate formation is required for L1/L2 capsid assembly.
MATERIALS AND METHODS
Mutagenesis and generation of recombinant vaccinia viruses.
C-terminal deletion mutants of HPV33 L2 were obtained by PCR with pCMV33L2 as a template and the oligonucleotide 5′-GGTGAATTCCATGAGACACAAACGATCTAC-3′ (ON-33L2-1-5′) as a forward primer. The following oligonucleotides were used as the reverse primer: 5′-GCGGGATCCCTAACGTTTACGCCTGCGACG-3′ (ON-33L2-456-3′), 5′-AAAGGATCCCTATGGGCTAGATGTGGGAA-3′ (ON-33L2-420-3′), 5′-AGAGGATCCCTAATCATTAATACTATAAGA-3′ (ON-33L2-360-3′), 5′-CTAGGATCCCTAAGGACTTAAATCCTGATA-3′ (ON-33L2-330-3′), and 5′-ACGGGATCCCTAACGCACAGTATGTCTACG-3′ (ON-33L2-300-3′). N-terminal deletion mutants were generated by PCR with the oligonucleotide 5′-GCGGGATCCCTAGGCCGCCACACGGACATC-3′ (ON-33L2-467-3′) as reverse primer and the oligonucleotides 5′-GGTGAATTCCATGGCATCTGCAACACAACT-3′ (ON-33L2-M12-5′), 5′-ACTGAATTCCATGCCGGTTACTGTAGACAC-3′ (ON-33L2-M91-5′), 5′-ATTGAATTCCATGTCATCTATTCAAACTAT-3′ (ON-33L2-M150-5′), and 5′-ATTGAATTCATGGATGGTTTGTATGATGTTTATGC-3′ (ON-33L2-M360-5′) as the forward primers. Upstream of the ATG and downstream of the stop codon recognition sites for EcoRI and BamHI, respectively, had been added (indicated in boldface). The resulting fragments were cloned into pCR2.1topo (Invitrogen). Fragments were cut out by using EcoRI and BamHI and cloned into pTM1 cut correspondingly to obtain pTM33L2-1/456, -1/420, -1/360, -1/330, -1/300, -12/467, -91/467, -150/467, -360/467, and -12/390. The numbers indicate the 33L2 amino acids still present in the construct. Corresponding recombinant vaccinia viruses were obtained after cotransfection with wild-type (wt) vaccinia virus DNA according to published procedures (36).
Cell lines and antiodies.
The mouse knockout cell line (PML−/−) and the parental cell line PEF (14, 16) were immortalized by using simian virus 40 (SV40) T antigen to yield PML−/−-T and PEF-T. The osteosarcoma cell line HuTK−143 B was obtained from Bernhard Moss (23). All cells were grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum and antibiotics at 37°C. L1- and L2-specific mouse monoclonal antibodies (MAbs) and rabbit polyclonal antibodies were as described elsewhere (29, 37, 38). Daxx-specific antibody was purchased from Santa Cruz.
Infection of cells.
Confluent cells were split 1:4 and grown for 24 h at 37°C in DMEM supplemented with 10% fetal calf serum and antibiotics (Life Technologies). Cells were washed once with phosphate-buffered saline (PBS) and subsequently infected with recombinant vaccinia viruses diluted in serum-free DMEM at a multiplicity of infection of 2 for each virus. After incubation for 1 h at room temperature, virus-containing medium was replaced by supplemented DMEM. Cells were processed for Western blotting, sucrose gradient analysis, and immunofluorescence after the indicated periods of time at 37°C, respectively.
Immunofluorescence.
Cells, grown on coverslips and infected with vaccinia viruses as described above, were fixed with methanol-0.02 M EGTA (−20°C) for at least 20 min, washed twice with PBS, and blocked in 5% goat serum dissolved in PBS. Coverslips were then incubated for 1 h at 37°C with the indicated antibodies. After an extensive wash with PBS, coverslips were again blocked for 30 min with 5% goat serum and subsequently incubated at 37°C with Cy3-conjugated Affinipure goat anti-rabbit immunoglobulin G (IgG) and Cy2-conjugated Affinipure goat anti-mouse IgG (Jackson Immunoresearch Products) for 1 h. Coverslips were subsequently washed with PBS and mounted onto slides by using Fluoprep mounting medium (bioMérieux). Pictures were taken by using a Zeiss Axiovert 200 M microscope and a Zeiss Axiocam digital camera. The Axiovision Software 3.0 was used for merging pictures.
Sucrose gradient analysis.
Cells infected with the indicated recombinant vaccinia viruses for 48 h were harvested by centrifugation and washed once with PBS. Cells were lysed in 10 mM HEPES-10 mM KCl-1.5 mM MgCl2 (pH 7.6) by freezing and thawing. Nuclei were collected at 1,000 × g and resuspended in 5 ml of PBS per 2 × 108 cells. VLPs were extracted by sonication (100 W, three times 45 s each time, 50% time interval). Insoluble material was removed by centrifugation (8,000 rpm [Sorvall SS34] for 10 min at 4°C). The supernatant was adjusted with cesium chloride to a density of 1.29 g/cm3 and subjected to buoyant density gradient centrifugation by using a Beckman VTi 65 rotor (16 h, 45,000 rpm, 10°C). VLP-containing fractions were identified by Western blotting, with MAbs 33L1-7 and 33L2-1 for detection of HPV33 capsid proteins, and by density measurements. VLPs were pooled, dialyzed against PBS for 2 h, and loaded onto a 10 to 60% sucrose gradient in PBS supplemented with 5 μg of bovine serum albumin/ml (Beckman SW40 rotor). After 150 min at 36,000 rpm and 4°C, the gradients were fractionated from the top (750 μl/fraction). The fractions were subjected to precipitation with trichloroacetic acid, and proteins were resuspended in sample buffer at 100°C for 5 min. Proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by immunoblotting with MAbs 33L1-7 and 33L2-1 and horseradish peroxidase-conjugated goat anti-mouse IgG by using enhanced chemiluminescence (Amersham). For quantification, films were scanned and analyzed by using the Kodak 1D image analysis software (version 3.5).
Electron microscopy.
Partially purified VLPs were directly spotted onto carbon-coated copper grids and negatively stained with 2% phosphotungstic acid. Grids were observed, and photomicrographs were obtained with a Zeiss EM900 transmission electron microscope at an instrumental magnification of ×50,000.
RESULTS
L2 accumulates in nuclear dots in PML−/− cells and attracts Daxx into these structures.
To analyze the subcellular distribution of L2, we expressed the wt HPV33 L2 protein, comprising 467 amino acids, by using recombinant vaccinia viruses in mouse PML−/− cells and in the parental PEF-T cells. L2 protein was detected by immunostaining (Fig. 1). At 6 h postinfection a diffuse nuclear staining was observed. In addition, L2 started to accumulate in nuclear aggregates in a manner reminiscent of ND10. It was almost exclusively found in these aggregates 8 h after addition of recombinant vaccinia viruses to the cells. No significant difference in the subcellular distribution of L2 was obvious in the two cell lines (Fig. 1A). Similar results were obtained when HPV16 L2 was expressed after transfection of codon-optimized (humanized) L2 (19) (data not shown), suggesting that accumulation of L2 (i) is not due to the vaccinia virus expression system and (ii) is not restricted to HPV33, as already implicated by previous studies (11, 19). These data indicate that L2 aggregate formation in the nucleus occurs independently of PML protein.
FIG. 1.
L2 forms nuclear aggregates in PML protein knockout cells and attracts Daxx into these structures. PEF-T and PML−/− cells were grown on coverslips and subsequently infected for the indicated periods of time with VTF7-3 and vac33L2 (A and C) or with VTF7-3 and vac33L1 (D). Cells were stained for L2 alone (A), for L2 and Daxx (C), and for L1 and Daxx (D). (B) Uninfected PEF-T and PML−/− cells stained for Daxx served as controls.
We then analyzed the L2-induced changes of Daxx. Daxx shows a diffuse and punctate nuclear staining pattern in PEF-T cells, a finding indicative of heterochromatin and ND10 association (20). A diffuse nuclear distribution was observed in PML−/− cells not expressing L2 (Fig. 1B). Upon expression of L2, Daxx accumulated in nuclear dots in both cell lines (Fig. 1C). At 12 h after infection, Daxx-positive nuclear dots became clearly visible. These dots expanded and the vast majority of Daxx localized to them 20 h after infection. At this time, Daxx dots completely overlapped with L2 dots in both cell lines (Fig. 1C). Daxx also accumulated in HPV16 L2 nuclear aggregates forming after transfection of PML−/− cells with codon-otimized HPV16 L2 (data not shown). In contrast, Daxx was not relocated in PML−/− cells expressing L1 via recombinant vaccinia virus (Fig. 1D). Under these conditions, L1 protein displayed a diffuse nuclear staining. These data suggest that vaccinia virus infection alone does not induce relocation of Daxx and that the observed effect is due to L2 and is not restricted to HPV33. This view is in line with previously published observations (10). We have recently shown by immunofluorescence and immunoprecipitation analysis that Daxx and L2 interact in PML protein-containing cells either directly or indirectly via an adapter protein (4, 10). The attraction of Daxx into L2-containing dots in PML−/− cells proves that the L2-Daxx interaction does not depend on the presence of PML protein.
L2 is incorporated into VLPs in PML−/− cells.
Whereas expression of L1 alone in PML−/− cells resulted in a diffuse nuclear staining of the major capsid protein, it accumulated into L2 aggregates when coexpressed with L2 (Fig. 2A). This suggested an interaction between L1 and L2 in PML−/− cells. To analyze whether L2 protein was incorporated into VLPs, capsid proteins were extracted from nuclei of L1- and L2-expressing PEF-T and PML−/− cells, and VLPs were partially purified by buoyant density gradient centrifugation. Extracts generated from cells expressing L2 alone served as controls. L2 incorporation was subsequently determined by velocity gradient centrifugation by using linear sucrose gradients. As measured by L1-specific Western blot, VLPs extracted from PEF-T cells sedimented as a broad peak in the sucrose gradient. Electron microscopy of the peak fractions confirmed the presence of VLPs (Fig. 3A). L2 protein comigrated with L1, a finding indicative of incorporation into VLPs (Fig. 2B). In addition, a significant portion of L2 was also found in the pellet of such gradients. L2 protein was exclusively found in the pellet when extracted from cells not expressing L1. This is due to L2 aggregate formation and has been observed with all cell lines analyzed thus far. Identical results were obtained with VLPs extracted from PML−/− cells (Fig. 2B and 3A), suggesting that PML protein is not required for L2 incorporation into VLPs. To determine the efficiency of VLP assembly and L2 incorporation in PML−/− cells, VLPs were further purified by cesium chloride step gradients to remove unassembled L1 molecules. The relative amount of L1 protein in VLP-containing fractions was then determined by Western blotting, and results were corrected for cell number used for expression (data not shown). The VLP yields in PEF-T and PML−/− cells did not differ significantly. Also, the percentage of unassembled L1 protein was similar in preparation from both cell lines, suggesting that VLP assembly is not impaired in PML−/− cells. This finding is in line with the report of efficient VLP assembly of L1 mutant proteins deficient for nuclear translocation (26). The possibility of late VLP formation during the extraction procedure was excluded by mixing experiments (data not shown). In addition, the L1/L2 ratio was essentially the same in VLPs generated in PEF-T and PML−/− cells (31.4 ± 18.55 and 25.77 ± 12.15, respectively). Since it is believed that 12 L2 molecules are incorporated into one VLP composed of 360 L1 molecules, a ratio of 30:1 is expected. We conclude that PML protein is neither required for efficient assembly of L1 into VLPs nor required for L2 incorporation into VLPs.
FIG. 2.
L1 protein is attracted into L2 nuclear aggregates in PML−/− cells. (A) PML−/− cells were grown on coverslips and subsequently coinfected for the indicated periods of time with VTF7-3, vac33L1, and vac33L2. Cells were fixed and stained for L1 and L2. (B) Nuclear extracts were prepared from PEF-T and PML−/− cells expressing L2 alone or both L1 and L2 and then subjected to buoyant cesium chloride density gradient centrifugation. Aliquots of the VLP-containing fractions (L1 and L2) and the fraction with corresponding density (L2 alone) were analyzed by sedimentation through linear sucrose gradients. The gradients were fractionated and analyzed by L1- and L2-specific Western blot. Twelvefold more protein was used for the detection of L2 compared to L1. Cosedimentation of L1 and L2 is indicative of L2 incorporation into VLPs.
FIG. 3.
Electron micrographs of VLPs. (A) wt VLPs generated in PML−/− or PEF-T cells. (B) Mutant VLPs composed of L1ΔNLS generated in HuTK− cells. Bar, 100 nm.
Two L2 domains independently interact with L1 in the nucleus.
To analyze whether L2 incorporation into VLPs requires any specific subcellular localization, we tested various L2 mutant proteins for incorporation into VLPs by cosedimentation analysis upon coexpression with wt L1 in the human osteosarcoma cell line HuTK−. Mutant proteins L2-1/456 and -1/420 (comprising L2 amino acids 1 to 456 and 1 to 420, respectively), accumulated in ND10, L2-1/360, and L2-1/330, displayed diffuse nucleoplasmic staining. In contrast, L2-1/300 mainly localized to the cytoplasm early in infection but was also detected in the nucleus after prolonged expression. This mutant lacks a functional nuclear localization signal (NLS) and passively translocates into the nucleus by diffusion (see inserts in Fig. 4A and reference 4). Western blot analysis of sucrose gradients revealed cosedimentation of wt L1 with the C-terminally truncated L2 proteins, since the capsid proteins were detected in the same fractions. This is indicative of L2 incorporation into wt VLPs (Fig. 4A and Table 1). Quantitative analyses suggest that, with the exception of L2-1/300, all C-terminal L2 deletion mutants were incorporated into VLPs at a ratio comparable to wt L2 (L1/L2-1/456, 33.97 ± 9.4; L1/L2-1/330, 25.3 ± 12.2). L2-1/300 was less efficiently incorporated into VLPs, yielding a much higher L1/L2 ratio. The reduced incorporation of L2-1/300 may be due to its lower nuclear concentration. Using immunofluorescence, we also tested L2 mutant proteins, which accumulate in ND10, for their ability to attract L1 and found that L2-1/456 and -1/420 still attracted L1 protein into these subnuclear structures (Fig. 4B and Table 1). Our data suggest that neither ND10 localization (L2-1/330) nor NLS-dependent nuclear transport of L2 (L2-1/300) is required for assembly of L2-containing VLPs.
FIG. 4.
Two L2 domains independently interact with L1 yielding L1/L2-VLPs. (A and C) Partially purified VLPs extracted from PML−/− cells expressing L2-1/456, -1/330, -1/300 (A), or -150/467 (C), together with wt L1, were subjected to sedimentation through linear sucrose gradients. Fractions were analyzed for presence of L1 and L2 by Western blot. Fourfold more protein was used for the detection of L2 compared to L1. Inserts show the subcellular localization of the indicated L2 mutant proteins 6 h (L2-1/456, -1/330, -1/300, and -150/467) and 12 h (L2-1/300) after infection. The result for L2-1/300 at 12 h postinfection is shown to demonstrate diffusion into the nucleus. (B and D) The indicated L2 mutants were expressed together with wt L1 in PML−/− cells by using recombinant vaccinia viruses. Cells were then immunostained for L1 and L2.
TABLE 1.
Identification of L2 domains interacting with L1a
| Domain | Interaction with wt L1
|
Interaction with L1ΔNLS
|
||
|---|---|---|---|---|
| VLP incorporation | ND10 attraction | VLP incorporation | ND10 attraction | |
| wt L2 (1/467) | Yes | Yes | Yes | Yes |
| L2-1/456 | Yes | Yes | No | No |
| L2-1/420 | Yes | Yes | NT | No |
| L2-1/360 | Yes | No* | NT | No* |
| L2-1/330 | Yes | No* | No | No* |
| L2-1/300 | Yes | No* | No | No* |
| L2-12/467 | NT | Yes | NT | No |
| L2-91/467 | NT | Yes | NT | No |
| L2-150/467 | Yes | Yes | No | No |
| L2-360/467 | NT | Yes | NT | NT |
*, mutants that do not accumulate in ND10; NT, not tested.
L2-150/467, lacking the N-terminal 149 amino acids, was also incorporated into wt VLPs (Fig. 4C). The interaction of larger N-terminal deletion mutant proteins of L2 with L1 was analyzed by L2-induced attraction of L1 into ND10, since L2-specific antibodies for the detection of C-terminal L2 fragments in Western blots were not available. As shown in Fig. 4D, L2-360/467 still localized to ND10 as indicated by the recruitment of L1. Taken together, these data demonstrate that the HPV33 L2 protein contains two domains interacting with L1 independently of each other: amino acids 1 to 300 as measured by its incorporation into VLPs and amino acids 360 to 467, which are sufficient to recruit L1 into ND10. Although we could not measure direct interaction between L2-360/467 and L1, the data by Finnen et al. (8), which have recently reported a small peptide within the C-terminal L2 sequence associating with capsomers, make this interaction highly likely.
L2 is not incorporated into VLPs assembling in the cytoplasm.
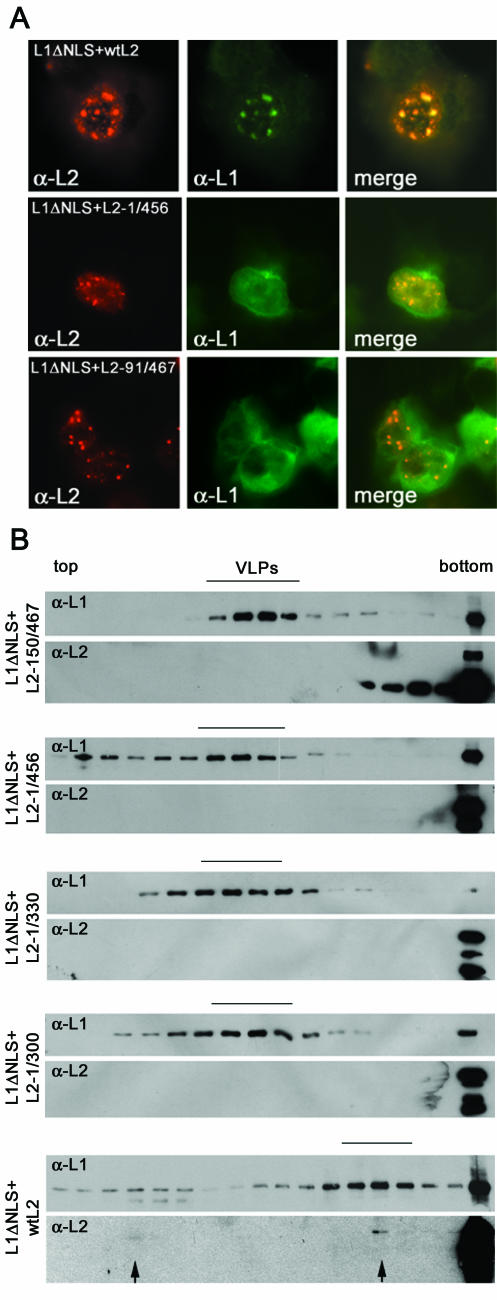
The data presented above suggest that L2 incorporation into VLPs does not require any specific subnuclear localization. To analyze possible interactions of the capsid proteins in the cytoplasm of HuTK− cells, we first tested rescue of nuclear transport-defective L1ΔNLS by mutant L2 proteins, by using immunofluorescence microscopy. This L1 mutant lacks 29 amino acids from the C terminus and contains no cryptic NLS as previously demonstrated (9). We also showed that wt L2 forms complexes with L1ΔNLS in the cytoplasm resulting in nuclear transport, ND10 accumulation of L1ΔNLS, and formation of L2-containing mutant VLPs (9, 30). Surprisingly, none of the L2 mutants tested was able to facilitate nuclear translocation of L1ΔNLS (Fig. 5A), although they attracted nucleoplasmic L1 into ND10 (Fig. 4B and D). Even mutant L2 proteins with short deletions (L2-1/456 and -12/467) failed to rescue L1ΔNLS.
FIG. 5.
wt L2 but not L2 mutant protein rescues L1ΔNLS for nuclear translocation and is incorporated into mutant VLPs. (A) The indicated L2 variants were coexpressed with L1ΔNLS in PML−/− cells for 12 h by using recombinant vaccinia viruses. L1 and L2 were visualized by immunofluorescence. (B) Incorporation of wt and mutant L2 proteins into VLPs composed of L1ΔNLS was analyzed by sedimentation through linear sucrose gradients (20 to 40% sucrose for wt L2 and 10 to 60% sucrose for mutant L2 protein) and Western blot. Fourfold more protein was used for the detection of L2 than for L1. With the exception of wt L2 no cosedimentation with L1ΔNLS was observed.
We next tested the incorporation of L2 proteins into VLPs composed of L1ΔNLS by cosedimentation analysis. The formation of VLPs by mutant L1 was confirmed by electron microscopy. Compared to VLPs composed of wt L1, mutant VLPs were more variable in size and morphology but were abundantly present (Fig. 3B). Similar observations have been made with C-terminally truncated L1 before (26). wt L2 is incorporated into mutant VLPs. Cosedimentation with capsomeres at the top of the gradient is also observed. The L2/L1 ratio is reduced compared to wt L1-VLPs probably due to an incomplete rescue of L1ΔNLS nuclear transport. As expected, L2-150/467, -1/456, and -1/330, which do not colocalize with L1ΔNLS, were not incorporated into mutant VLPs (Fig. 5B). Surprisingly, L2-1/300, which colocalizes with L1ΔNLS in the cytoplasm, was also not present in mutant VLPs. As shown above, all of these L2 mutant proteins were incorporated into VLPs, which assemble in the nucleus. Taken together, our data demonstrate that assembly of L2-containing VLPs requires nuclear localization.
DISCUSSION
Using PML−/− cells, we demonstrate that the main structural component of ND10, PML protein, is not required for L2 incorporation into the L1 capsid. In contrast to a variety of large DNA viruses, which partially destroy these subnuclear structures, papillomaviruses seem to use these sites for their replication without obvious destruction (34). Since capsid proteins, driven by L2, are also deposited in ND10 (5, 9), the resulting close vicinity and increased local concentration of all virus components may facilitate the papillomavirus morphogenesis. Even though L2 influences the composition of ND10 (10), PML protein is not obviously affected, suggesting that L2 does not directly interact with PML protein. This is in line with our observations (i) that L2 precipitates Daxx but not PML protein, (ii) that Daxx and L2 structures expand beyond PML protein-positive dots after prolonged L2-expression (10), and (iii) that L2 forms functional aggregates in PML−/− cells, attracting L1 and Daxx. It is still not known which factor induces the L2 accumulation in ND10. It seems possible that Daxx, which partially localizes to ND10, initially triggers deposition of L2 at these sites. However, it is also conceivable that other known (11) or yet-to-be-identified proteins are responsible for L2 attraction into ND10.
Even though wt L2 protein forms nuclear aggregates attracting L1 and Daxx, aggregate formation is not essential for all L2 functions. L2 deletion mutant proteins, displaying diffuse nuclear distribution, are still incorporated into L1 capsids, suggesting that L1-L2 interaction does not require accumulation of both capsid proteins in nuclear aggregates. Since these mutants no longer interact with Daxx (4), interaction with this factor, which has been implicated in transcriptional repression, chromatin modulation, and apoptosis (22), is also not essential for coordinate capsid assembly. Our quantitative analyses suggest that the efficacy of capsid formation in the absence of L2/Daxx aggregates is not reduced. Using expression of HPV11 wt L1 and L2 peptides in Escherichia coli, other researchers have recently also shown that L1-L2 interaction does not depend on nuclear factors (8). L2 aggregate formation and consequently Daxx may, however, be important for genome encapsidation. The interpretation of such an analysis would be difficult due to the introduction of large deletions into L2, which may influence additional functions of this protein. Due to the use of mouse cell lines, we were also not able to test DNA encapsidation in the absence of PML protein. In our hands, formation of pseudovirions requires high copy numbers of a target plasmid. This is obtained by SV40 T-antigen-driven amplification in the COS cell system (36). Even though the mouse cell lines used for the present study express SV40 T antigen, mouse cells are not permissive for SV40 DNA replication (31). We therefore failed to generate pseudovirions in both PEF-T and PML−/− cells.
The characterization of the L2 mutants also led to the identification of two L2 domains independently interacting with L1. This is in concordance with a previous report for BPV-1 L2 (25). wt L2 is able to interact functionally with L1 in the cytoplasm (L1ΔNLS) and in the nucleus (wt L1), resulting in the formation of L1/L2-VLPs. In both cases, the VLP assembly occurs in the nucleus, since the L2/L1ΔNLS complex is translocated into the nucleus and subsequently into ND10 (9, 30). In contrast, none of the L2 mutant proteins formed functional complexes with L1 in the cytoplasm, even if only few amino acids were deleted. These observations indicate that either L1 or L2 adopts different conformations in the cytoplasm and nucleus. Since L1ΔNLS readily assembles to VLPs in the cytoplasm, it is highly likely that L2 undergoes the structural change upon nuclear translocation. Members of the chaperone family are possible factors modulating the conformation of L2. They seem to interact with L2 in the cytoplasm resulting in their cotranslocation into the nucleus (L. Florin et al., unpublished observations).
Taken together, the incorporation of the minor capsid protein L2 into VLPs does not depend on the main structural ND10 component, PML protein, but is restricted to the nucleus. In the absence of PML protein a substructure of ND10 may be maintained to which L2 binds, possibly via Daxx.
Acknowledgments
We thank M. Müller for generously providing an expression plasmid for humanized HPV16 L2. We are grateful to R. E. Streeck for critical reading of the manuscript and for helpful suggestions.
This study was supported by grants to M.S. from Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Adamson, A. L., and S. Kenney. 2001. Epstein-barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J. Virol. 75:2388-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn, J. H., and G. S. Hayward. 1997. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J. Virol. 71:4599-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, T. S., W. W. Newcomb, N. H. Olson, L. M. Cowsert, C. Olson, and J. C. Brown. 1991. Structures of bovine and human papillomaviruses: analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys. J. 60:1445-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker, K. A., L. Florin, C. Sapp, and M. Sapp. 2003. Dissection of human papillomavirus type 33 L2 domains involved in nuclear domain (ND) 10 homing and reorganization. Virology 314:161-167. [DOI] [PubMed] [Google Scholar]
- 5.Day, P. M., R. B. Roden, D. R. Lowy, and J. T. Schiller. 1998. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J. Virol. 72:142-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de The, H., C. Chomienne, M. Lanotte, L. Degos, and A. Dejean. 1990. The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor alpha gene to a novel transcribed locus. Nature 347:558-561. [DOI] [PubMed] [Google Scholar]
- 7.Doucas, V. 2000. The promyelocytic (PML) nuclear compartment and transcription control. Biochem. Pharmacol. 60:1197-1201. [DOI] [PubMed] [Google Scholar]
- 8.Finnen, R. L., K. D. Erickson, X. S. Chen, and R. L. Garcea. 2003. Interactions between papillomavirus L1 and L2 capsid proteins. J. Virol. 77:4818-4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Florin, L., C. Sapp, R. E. Streeck, and M. Sapp. 2002. Assembly and translocation of papillomavirus capsid proteins. J. Virol. 76:10009-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Florin, L., F. Schäfer, K. Sotlar, R. E. Streeck, and M. Sapp. 2002. Reorganization of nuclear domain 10 induced by papillomavirus capsid protein L2. Virology 295:97-107. [DOI] [PubMed] [Google Scholar]
- 11.Görnemann, J., T. G. Hofmann, H. Will, and M. Müller. 2002. Interaction of human papillomavirus type 16 L2 with cellular proteins: identification of novel nuclear body-associated proteins. Virology 303:69-78. [DOI] [PubMed] [Google Scholar]
- 12.Gottifredi, V., and C. Prives. 2001. P53 and PML: new partners in tumor suppression. Trends Cell. Biol. 11:184-187. [DOI] [PubMed] [Google Scholar]
- 13.Hagensee, M. E., N. Yaegashi, and D. A. Galloway. 1993. Self-assembly of human papillomavirus type 1 capsids by expression of the L1 protein alone or by coexpression of the L1 and L2 capsid proteins. J. Virol. 67:315-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishov, A. M., R. M. Stenberg, and G. G. Maul. 1997. Human cytomegalovirus immediate-early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishov, A. M., O. V. Vladimirova, and G. G. Maul. 2002. Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71 at ND10 facilitates initiation of viral infection at these nuclear domains. J. Virol. 76:7705-7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kakizuka, A., W. H. Miller, Jr., K. Umesono, R. P. Warrell, Jr., S. R. Frankel, V. V. Murty, E. Dmitrovsky, and R. M. Evans. 1991. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell 66:663-674. [DOI] [PubMed] [Google Scholar]
- 18.Kirnbauer, R., F. Booy, N. Cheng, D. R. Lowy, and J. T. Schiller. 1992. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc. Natl. Acad. Sci. USA 89:12180-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leder, C., J. A. Kleinschmidt, C. Wiethe, and M. Müller. 2001. Enhancement of capsid gene expression: preparing the human papillomavirus type 16 major structural gene L1 for DNA vaccination purposes. J. Virol. 75:9201-9209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, H., C. Leo, J. Zhu, X. Wu, J. O'Neil, E. J. Park, and J. D. Chen. 2000. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol. Cell. Biol. 20:1784-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMurray, H. R., D. Nguyen, T. F. Westbrook, and D. J. McAnce. 2001. Biology of human papillomaviruses. Int. J. Exp. Pathol. 82:15-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michaelson, J. S. 2000. The Daxx enigma. Apoptosis 5:217-220. [DOI] [PubMed] [Google Scholar]
- 23.Moss, B., O. Elroy-Stein, T. Mizukami, W. A. Alexander, and T. R. Fuerst. 1990. New mammalian expression vectors. Nature 348:91-92. [DOI] [PubMed] [Google Scholar]
- 24.Müller, M., L. Gissmann, R. J. Cristiano, X. Y. Sun, I. H. Frazer, A. B. Jenson, A. Alonso, H. Zentgraf, and J. Zhou. 1995. Papillomavirus capsid binding and uptake by cells from different tissues and species. J. Virol. 69:948-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okun, M. M., P. M. Day, H. L. Greenstone, F. P. Booy, D. R. Lowy, J. T. Schiller, and R. B. Roden. 2001. L1 interaction domains of papillomavirus L2 necessary for viral genome encapsidation. J. Virol. 75:4332-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paintsil, J., M. Müller, M. Picken, L. Gissmann, and J. Zhou. 1996. Carboxyl terminus of bovine papillomavirus type-1 L1 protein is not required for capsid formation. Virology 223:238-244. [DOI] [PubMed] [Google Scholar]
- 27.Roden, R. B., H. L. Greenstone, R. Kirnbauer, F. P. Booy, J. Jessie, D. R. Lowy, and J. T. Schiller. 1996. In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype. J. Virol. 70:5875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rose, R. C., W. Bonnez, R. C. Reichman, and R. L. Garcea. 1993. Expression of human papillomavirus type 11 L1 protein in insect cells: in vivo and in vitro assembly of viruslike particles. J. Virol. 67:1936-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sapp, M., U. Kraus, C. Volpers, P. J. Snijders, J. M. Walboomers, and R. E. Streeck. 1994. Analysis of type-restricted and cross-reactive epitopes on virus-like particles of human papillomavirus type 33 and in infected tissues using monoclonal antibodies to the major capsid protein. J. Gen. Virol. 75:3375-3383. [DOI] [PubMed] [Google Scholar]
- 30.Schäfer, F., L. Florin, and M. Sapp. 2002. DNA binding of L1 is required for human papillomavirus morphogenesis in vivo. Virology 295:172-181. [DOI] [PubMed] [Google Scholar]
- 31.Stadlbauer, F., C. Voitenleitner, A. Bruckner, E. Fanning, and H. P. Nasheuer. 1996. Species-specific replication of simian virus 40 DNA in vitro requires the p180 subunit of human DNA polymerase α-primase. Mol. Cell. Biol. 16:94-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stauffer, Y., K. Raj, K. Masternak, and P. Beard. 1998. Infectious human papillomavirus type 18 pseudovirions. J. Mol. Biol. 283:529-536. [DOI] [PubMed] [Google Scholar]
- 33.Sternsdorf, T., T. Grotzinger, K. Jensen, and H. Will. 1997. Nuclear dots: actors on many stages. Immunobiology 198:307-331. [DOI] [PubMed] [Google Scholar]
- 34.Swindle, C. S., N. Zou, B. A. Van Tine, G. M. Shaw, J. A. Engler, and L. T. Chow. 1999. Human papillomavirus DNA replication compartments in a transient DNA replication system. J. Virol. 73:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trus, B. L., R. B. Roden, H. L. Greenstone, M. Vrhel, J. T. Schiller, and F. P. Booy. 1997. Novel structural features of bovine papillomavirus capsid revealed by a three-dimensional reconstruction to 9 Å resolution. Nat. Struct. Biol. 4:413-420. [DOI] [PubMed] [Google Scholar]
- 36.Unckell, F., R. E. Streeck, and M. Sapp. 1997. Generation and neutralization of pseudovirions of human papillomavirus type 33. J. Virol. 71:2934-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volpers, C., M. Sapp, C. A. Komly, P. Richalet-Secordel, and R. E. Streeck. 1993. Development of type-specific and cross-reactive serological probes for the minor capsid protein of human papillomavirus type 33. J. Virol. 67:1927-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Volpers, C., M. Sapp, P. J. Snijders, J. M. Walboomers, and R. E. Streeck. 1995. Conformational and linear epitopes on virus-like particles of human papillomavirus type 33 identified by monoclonal antibodies to the minor capsid protein L2. J. Gen. Virol. 76:2661-2667. [DOI] [PubMed] [Google Scholar]
- 39.Volpers, C., P. Schirmacher, R. E. Streeck, and M. Sapp. 1994. Assembly of the major and the minor capsid protein of human papillomavirus type 33 into virus-like particles and tubular structures in insect cells. Virology 200:504-512. [DOI] [PubMed] [Google Scholar]
- 40.Volpers, C., F. Unckell, P. Schirmacher, R. E. Streeck, and M. Sapp. 1995. Binding and internalization of human papillomavirus type 33 virus-like par-ticles by eukaryotic cells. J. Virol. 69:3258-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, Z. G., L. Delva, M. Gaboli, R. Rivi, M. Giorgio, C. Cordon-Cardo, F. Grosveld, and P. P. Pandolfi. 1998. Role of PML in cell growth and the retinoic acid pathway. Science 279:1547-1551. [DOI] [PubMed] [Google Scholar]
- 42.Zhong, S., S. Müller, S. Ronchetti, P. S. Freemont, A. Dejean, and P. P. Pandolfi. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748-2752. [PubMed] [Google Scholar]