Abstract
Herpesviruses acquire a primary envelope by budding of capsids at the inner leaflet of the nuclear membrane. They then traverse into the cytoplasm after fusion of the primary envelope with the outer leaflet of the nuclear membrane. In the alphaherpesvirus pseudorabies virus (PrV), the latter process is impaired when the US3 protein is absent. Acquisition of final tegument and envelope occurs in the cytoplasm. Besides the capsid components, only the UL31 and UL34 gene products of PrV have unequivocally been shown to be part of primary enveloped virions, whereas they lack several tegument proteins present in mature virions (reviewed by T. C. Mettenleiter, J. Virol. 76:1537-1547, 2002). Using immunoelectron microscopy, we show that the US3 protein is present in primary enveloped as well as in mature virions. It is also detectable in intracytoplasmic inclusions produced in the absence of other viral tegument components or envelope-associated glycoproteins. In particular, inclusions formed in the absence of the inner tegument protein UL37 contained the US3 protein. Thus, the US3 protein is a tegument component of both forms of enveloped alphaherpes virions. We hypothesize that US3 protein in primary virions modulates deenvelopment at the outer leaflet of the nuclear membrane and is either lost from primary virions during nuclear egress and subsequently reacquired early during tegumentation or is retained during transit of the nucleocapsid through the nuclear membrane.
Herpesvirus morphogenesis is a complex process. The viral DNA genome is replicated and packaged into preformed icosahedral capsids in the nucleus of the host cell. Nucleocapsids are then translocated through the nuclear membrane into the cytoplasm by an envelopment-deenvelopment process. To this end, intranuclear capsids obtain a first (primary) envelope by budding at the inner leaflet of the nuclear membrane, in this process also acquiring a primary tegument, resulting in the presence of primary enveloped herpes virions between the two leaflets of the nuclear membrane. The primary envelope then fuses with the outer leaflet of the nuclear membrane, and nucleocapsids are released into the cytoplasm. They then obtain their final set of tegument proteins, as well as their final envelope with mature viral glycoproteins, by budding into trans-Golgi vesicles. Release of free infectious virions is subsequently achieved by fusion of vesicle and plasma membrane (reviewed in reference 34).
For the formation of primary virions, the UL31 and UL34 proteins of pseudorabies virus (PrV), herpes simplex virus type 1 (HSV-1), and murine cytomegalovirus have been shown to play important roles (6, 13, 23, 35, 50). In their absence, primary envelopment at the inner leaflet of the nuclear membrane is blocked, and nucleocapsids accumulate in the nucleus. Since UL34 is predicted to be a type II transmembrane protein, it was proposed that it may represent a primary envelope protein and that the UL31 gene product may be part of the primary tegument. Both proteins physically interact which is in accordance with these assumptions (13, 35, 48).
Analysis of lipid composition first demonstrated that the envelope of mature HSV-1 particles is distinct from the nuclear membrane, indicating that the mature viral envelope may not be derived from the nuclear membrane (55). Subsequent electron microscopic data substantiated that primary enveloped and mature virions are also morphologically distinct. Whereas mature herpes virions are characterized by a fuzzy electron-dense tegument and an envelope with clearly visible projections (spikes), primary enveloped virions lack the typical spikes and their tegument is tightly juxtaposed to the envelope in a sharp bordered rim (17, 18). Since perinuclear virions have thus far not been purified to any extent to allow unequivocal analysis of their protein content, immunoelectron microscopy has been used to study their composition. These data clearly showed that the UL34 and UL31 proteins are constituents of primary virions (13, 23, 49). Since they also contain nucleocapsids, it is reasonable to assume that capsid proteins (the products of the UL18, UL19, UL35, and UL38 genes [52]) and capsid-associated proteins such as the UL6 portal protein (36) may also be part of these virus particles. Whether other envelope or tegument proteins are also present in primary virions is unclear.
In the subsequent fusion of primary envelope and outer leaflet of the nuclear membrane the US3 protein plays an important role. In its absence, primary enveloped virions accumulate in the perinuclear space, which is due to an apparent defect in deenvelopment (24, 49). Consequently, it had to be assumed that the US3 protein may be associated with primary virions. The HSV-1 and PrV US3 proteins are protein kinases (43, 47, 59), homologs of which are present in the alphaherpesviruses but not in the other herpesvirus subfamilies (1, 7, 32). In contrast, the UL13 gene products, which also represent protein kinases (10, 30, 37, 38, 51, 53), are conserved in all herpesvirus subfamilies. Neither the UL13 nor the US3 homologs are required for viral replication (10, 20, 24, 46, 47). Interestingly, although HSV-1 US3 has been proposed to phosphorylate the UL34 protein (44, 45), it has been shown that phosphorylation of the PrV UL34 protein was not altered in the absence of the US3 gene (24). Thus, lack of phosphorylation of the UL34 protein due to absence of US3 is not the reason for the observed de-envelopment defect. We hypothesized that US3 may play an important structural role in primary virions which is relevant for the fusion process.
MATERIALS AND METHODS
Viruses and cells.
PrV mutants were derived from the laboratory strain Kaplan (PrV-Ka [19]). Viruses were grown in rabbit kidney (RK13) or porcine kidney (PSEK) cells in minimal essential medium supplemented with 10 or 5% fetal calf serum, respectively. Isolation and characterization of PrV-ΔUS3 (24), PrV-ΔUL37 (25), PrV-ΔUL47 (27), PrV-ΔUL48 (14), PrV-ΔUL3.5 (16), PrV-ΔUL11 (28), and PrV-gEIM− (4) have been described. PrV strain Becker (PrV-Be [3]) was kindly provided by L. W. Enquist, Princeton University. An aliquot of the original isolate of strain Bartha K61 (PrV-Ba [2]) was generously provided by the late A. Bartha, Budapest, Hungary.
Western blot analysis.
Virus purification and preparation of infected cell lysates were done as described recently (28). Samples of infected cell lysates and purified virions were separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (29) and electrotransferred onto nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany [54]). Blots were blocked with 6% skim milk in phosphate-buffered saline and incubated with monospecific sera against the UL11 (1:10,000 [28]), UL31 (1:100,000 [13]), UL34 (1:100,000 [23]), UL37 (1:100,000 [25]), UL46 (1:100,000 [27]), UL47 (1:100,000 [27]), UL48 (1:100,000 [14]), UL49 (1:100,000 [5]), glycoprotein H (gH; 1:50,000 [22]), gD (1:10,000 [21]), gI (1:10,000 [4]), and gM (1:50,000 [28]) proteins or with monoclonal antibodies against gB (b43-b5 [40]), gC (B16-c8 [24]), gE (A9-b15 [21]), and gK (11). Bound antibody was detected with peroxidase-conjugated secondary antibodies (Dianova, Hamburg, Germany) and visualized by chemiluminescence (Super Signal; Pierce) recorded on X-ray films.
Immunoelectron microscopy.
Immunoelectron microscopy using specific antibodies and 10-nm gold-tagged secondary anti-rabbit antibodies (GAR10; British Biocell International) was performed as described recently (28). Ultrathin sections were counterstained and analyzed with an electron microscope (Tecnai 12; Philips, Eindhoven, The Netherlands). In addition to the antisera mentioned above, a UL36 specific polyclonal serum was used (26).
RESULTS
US3 protein is part of primary virions.
To analyze the association of US3 protein with primary virions, electron microscopic examinations were performed. RK13 cells infected with PrV-Ka were fixed 14 h after infection and analyzed by conventional and immunoelectron microscopy. Monospecific antisera against the UL31, UL34, US3, UL36, UL37, UL46, UL47, UL48, UL49, and US3 proteins were used. All stages of virus maturation could be identified, including the presence of perinuclear enveloped virions (Fig. 1A). As expected, these virions labeled with the anti-UL31 and anti-UL34 antisera (Fig. 1B and C) but failed to show reactivity with antisera directed against the UL36, UL37, UL46, UL47, UL48, and UL49 tegument proteins (Fig. 1D to I). In contrast, extracellular virions (Fig. 2A) labeled with the anti-tegument protein antisera (Fig. 2D to I) but failed to react with the anti-UL31 and anti-UL34 sera (Fig. 2B and C). Surprisingly, both perinuclear primary enveloped (Fig. 1J) and extracellular mature virions (Fig. 2J) were labeled by the anti-US3 antiserum. Thus, US3 apparently is part of both primary and mature virions. In contrast, the UL31 and UL34 proteins are only present in primary virions, and the major tegument components of mature virus particles UL36, UL37, UL46, UL47, UL48, and UL49 were not detectable in primary virions.
FIG. 1.
Immunoelectron microscopy of primary enveloped PrV virions. Rabbit kidney cells were infected with PrV-Ka and analyzed 14 h after infection by either conventional (A) or immunoelectron microscopy with antisera against the UL31 (B), UL34 (C), UL36 (D), UL37 (E), UL46 (F), UL47 (G), UL48 (H), UL49 (I), or US3 (J) proteins. Bar, 200 nm.
FIG. 2.
Immunoelectron microscopy of mature extracellular PrV virions. Rabbit kidney cells were infected with PrV-Ka and analyzed 14 h after infection by either conventional (A) or immunoelectron microscopy using antisera against the UL31 (B), UL34 (C), UL36 (D), UL37 (E), UL46 (F), UL47 (G), UL48 (H), UL49 (I), or US3 (J) proteins. Bar, 200 nm.
Since the anti-US3 antiserum was the only serum that reacted with primary enveloped and mature virus particles, we verified the specificity of the antiserum by using cells infected with a US3 deletion mutant of PrV (24). As previously observed, in the absence of the US3 protein primary envelopment occurred but deenvelopment was impaired, resulting in the accumulation of primary virions in the perinuclear space in large invaginations of the inner leaflet of the nuclear membrane (24, 49, 57) (Fig. 3A). These primary virions did not label with the US3 antiserum (Fig. 3B), whereas the anti-UL31 (Fig. 3C) and anti-UL34 (Fig. 3D) antisera detected their respective antigens in PrV-ΔUS3 primary enveloped virions.
FIG. 3.
Specificity of the US3 antiserum. Rabbit kidney cells were infected with PrV-ΔUS3 and analyzed 14 h after infection by either conventional (A) or immunoelectron microscopy using antisera against the US3 (B), UL31 (C), or UL34 (D) proteins. Bar, 200 nm.
Correlating with its incorporation into primary enveloped virions, US3 protein was also detectable at the nuclear membrane (Fig. 4A) in PrV-Ka-infected cells, although it appeared less prominent than the UL31 (Fig. 4B) or UL34 (Fig. 4C) proteins. To summarize, the PrV US3 protein is present in infected cells at the nuclear membrane and is incorporated into perinuclear virions during primary envelopment.
FIG. 4.
US3 is present at the nuclear membrane. Rabbit kidney cells infected with PrV-Ka were analyzed by immunoelectron microscopy using antisera against US3 (A), UL31 (B), or UL34 (C). Bar, 200 nm.
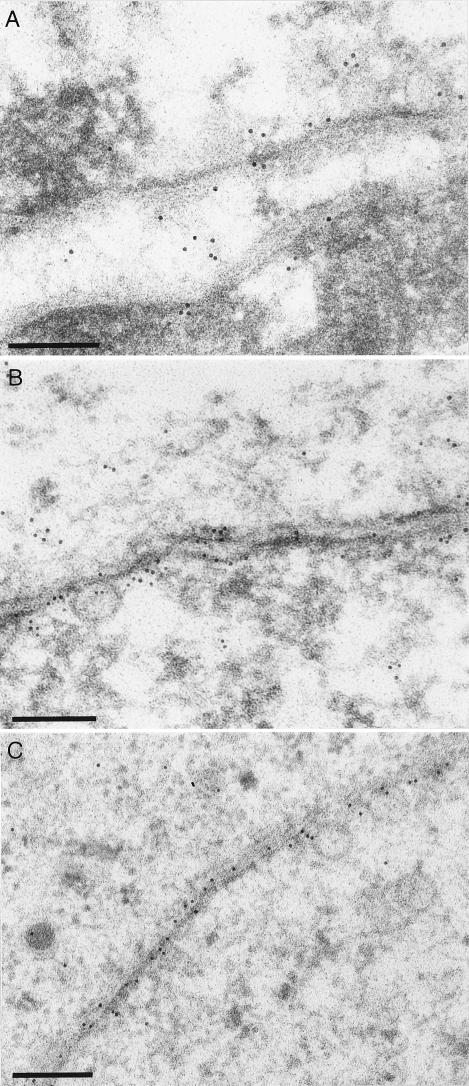
US3 protein is present at intracytoplasmic nucleocapsids that accumulate in the absence of tegument or envelope glycoproteins.
To analyze whether US3 is retained at the nucleocapsid during nuclear egress or whether it is reacquired in the cytosol, mutant viruses with defects at different stages of the intracytoplasmic virion maturation pathway were analyzed (Fig. 5). US3 protein was present in inclusions formed in the absence of the UL47 tegument protein (Fig. 5C and D) (27) or the UL11 tegument protein (Fig. 5I and J) (28) or in the simultaneous absence of envelope glycoproteins E, I, and M (Fig. 5K and L) (4). The anti-US3 serum also labeled intracytosolic capsids which accumulate in the absence of the UL48 (Fig. 5E and F) (14) or UL3.5 (Fig. 5G and H) (16) proteins. It has previously been shown that absence of these respective viral component(s) impairs virion formation prior to secondary envelopment (4, 5, 14, 16, 27, 28). In the absence of the UL37 protein, which presumably forms one of the inner layers of tegument around the nucleocapsid by physically interacting with capsid-bound UL36 protein (26, 60), nucleocapsids aggregate in the cytoplasm in an ordered arrangement (Fig. 5A) (25). Since these inclusions, besides the UL36 protein, do not contain major components of the mature tegument such as the UL49 protein (25), they most likely contain nucleocapsids blocked early in tegumentation. However, these nucleocapsids also labeled with the anti-US3 antiserum (Fig. 5B), indicating that the US3 protein is either retained during nuclear egress or reacquired very early during tegumentation.
FIG. 5.
Detection of US3 in cells infected with tegument protein deletion mutants of PrV. Rabbit kidney cells infected with PrV-ΔUL37 (A and B), PrV-ΔUL47 (C and D), PrV-ΔUL48 (E and F), PrV-ΔUL3.5 (G and H), PrV-ΔUL11 (I and J), and PrV-gEIM− (K and L) were analyzed by conventional electron microscopy (A, C, E, G, I, and K) or immunoelectron microscopy with a monospecific anti-US3 serum (B, D, F, H, J, and L). Bars: 500 nm (A, C, E, G, I, and K); 250 nm (B, D, F, H, J, and L).
Absence of US3 protein does not alter composition of mature virions.
During formation of mature virions intricate protein-protein interactions among tegument components and between tegument and envelope constituents occur (reviewed in reference 34). To analyze whether the absence of the US3 tegument component alters incorporation of other tegument or envelope proteins, purified PrV-ΔUS3 virions were analyzed by Western blotting with monospecific antisera or monoclonal antibodies. As shown in Fig. 6, in comparison to purified wild-type PrV-Ka virions (Fig. 6, lanes 1) the absence of the US3 protein in PrV-ΔUS3 (Fig. 6, lanes 2) did not result in an alteration in the incorporation of other tegument proteins such as UL37, UL46, UL47, UL48, UL49 (Fig. 6A), or UL11 (Fig. 6B) nor of the envelope glycoproteins gB, gC, gD, gE, gI, gH, gK, or gM (Fig. 6C). Thus, at least these tegument and envelope proteins are incorporated into mature virions independent of the US3 protein.
FIG. 6.
Protein composition of PrV-ΔUS3 virions. PrV-Ka (lanes 1) and PrV-ΔUS3 (lanes 2) virions were purified by sucrose gradient centrifugation and analyzed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis with 10% (A and C) or 15% (B) polyacrylamide. After electrotransfer onto nitrocellulose membranes, blots were analyzed with antibodies against the US3, UL37, UL46, UL47, UL48, UL49, UL11, gB, gC, gD, gE, gI, gH, gK, and gM proteins. The locations of molecular mass markers are indicated on the left.
US3 protein is packaged into mature PrV-Bartha virions.
It has previously been described that the attenuated live vaccine strain PrV-Bartha does not package US3 protein into mature virions (31). Among other mutations (reviewed in reference 33), PrV-Bartha contains a deletion in the Us genome region encompassing the gE, gI, and US9 genes (42). Since this could indicate a requirement for these proteins for virion incorporation of US3, we analyzed virions of PrV-Bartha (PrV-Ba) by using stock virus from the original Hungarian isolate. Virus was grown in RK13 cells and purified by centrifugation through a discontinuous 30 to 50% sucrose gradient as described previously (27). For comparison, wild-type strains PrV-Ka and PrV-Be were included. As shown in Fig. 7, PrV-Ka, PrV-Be, and PrV-Ba virions all contained similar amounts of tegument protein UL37, as well as gH, whereas gE, as expected, was absent from PrV-Ba. Surprisingly, PrV-Ba also contained unaltered amounts of US3. Therefore, the packaging of US3 is apparently not dependent on the presence of either gE or gI. In contrast to earlier descriptions (31), we also found that PrV-Ba virions contained the major tegument protein UL49.
FIG. 7.
US3 protein is a component of PrV-Ba virions. PrV-Ka, PrV-Be, and PrV-Ba virions were purified by sucrose gradient centrifugation and analyzed by immunoblotting with antisera against the US3, UL49, UL37, gE, and gH proteins. The locations of molecular mass markers are indicated on the left.
DISCUSSION
Our data provide the first exhaustive analysis of the composition of primary enveloped perinuclear virions by immunolabeling. Unfortunately, this is currently the only method available for analysis of these intermediates in herpesvirus maturation, since no reliable protocol for purification of perinuclear virions has yet been found. In our studies (see Fig. 1), perinuclear virions labeled with the UL31- and UL34-specific antisera but not with antisera directed against the UL36, UL37, UL46, UL47, UL48, or UL49 tegument proteins of mature virions. In contrast, mature virions (Fig. 2) were not recognized by the UL31 and UL34 specific antisera but readily labeled with antisera against the other tegument proteins. Thus, our data fully support the envelopment-deenvelopment-reenvelopment model of herpesvirus morphogenesis since they show that the two forms of nucleocapsid-containing enveloped herpesvirus particles, i.e., primary and mature virions, have a different protein composition. However, we cannot completely rule out that amounts of tegument proteins that are below the detection limit of our antisera may be present on primary virions, although this appears to be unlikely. Also, even posttranslationally altered forms of the proteins should have been detectable by our potent polyclonal monospecific antisera.
The only protein that we detected as part of the tegument of primary and mature PrV virions was US3. This has recently also been proposed for the homologous HSV-1 protein (49). The PrV US3 gene is transcribed into two mRNAs which are translated into two C-terminally identical US3 proteins (56, 58). Whereas both translation products of 41 and 53 kDa are present in infected cells, only the smaller protein of 41 kDa was detected in mature virus particles (31). Since our antiserum is directed against a part of the US3 protein common to both forms, it detects both US3 translation products. Therefore, we cannot differentiate at the moment whether different forms of the US3 protein may associate with virions during primary and secondary envelopment.
In the absence of the UL37 tegument protein, which interacts with the capsid-associated UL36 tegument protein (26), tegumentation is inhibited at an early step, resulting in the accumulation of intracytoplasmic clusters of capsids in an ordered arrangement (25). Our immunoelectron microscopic analyses proved that these clusters contained the US3 protein. Thus, the data indicate that the US3 protein is either retained during nuclear egress or reacquired early during tegumentation in the cytosol.
Recent data indicated that expression of gE and/or gI may be important for virion localization of US3 since the attenuated PrV vaccine strain Bartha (2), was found not to package the US3 or UL49 proteins (31). PrV-Bartha, among other defects, lacks the genes encoding gE and gI (reviewed in reference 33), and carries point mutations within the gM gene resulting in the elimination of an N-glycosylation site (12). Moreover, the UL49 tegument protein had previously been shown to interact with the cytoplasmic tail of gE and gM (15). Since the gE and gI genes are absent from the PrV-Ba genome and gM is modified, these defects could account for the observed lack of incorporation of UL49. However, our preparations of gradient-purified PrV-Ba virions obtained from an original seeding stock of this virus clearly contained both UL49 and US3 proteins. Whether this difference is due to a difference in the purity of virus preparations, time of sampling of the virus supernatant for purification, or differences in handling of the virion preparations is unclear at present.
The US3 protein, in addition to its role as a virion structural component, also exhibits protein kinase activity (47, 59). Although the UL34 protein has been described as one of the substrates for modification by US3 (44, 45) and the presence or absence of US3 has been shown to influence intracellular localization of the UL34 protein (24), no difference in the phosphorylation state of the PrV UL34 protein has been detected (24) irrespective of the presence or absence of US3. Thus, the protein kinase activity of US3 may not be relevant for the processes analyzed here. It is conceivable that during tegumentation in primary and secondary envelopment, the US3 protein primarily plays a structural role.
The two noncapsid proteins that have thus far been shown to play an important role in primary envelopment, the UL31 and UL34 gene products, are conserved throughout the herpesviruses (1, 7, 32), indicating that they execute a function which is fundamental to herpesvirus replication. We hypothesize that these two proteins may be part of an ancient herpesvirus fusion machinery which is simple (perhaps consisting of only these two proteins) and efficient (it is difficult to detect virions in the process of deenvelopment by electron microscopy). The nonconserved US3 protein may have evolved to play a modulatory role in this process since in its absence deenvelopment is inhibited but not completely blocked, and the resulting reduction of extracellular virus titers is only ∼10-fold. In contrast, absence of the UL31 or UL34 proteins reduces virus titers by 100- to 1,000-fold.
It is currently unclear whether there is redundancy in the functions US3 executes. The large subunit of the ribonucleotide reductase, expressed from homologs of the HSV UL39 gene, can function as a protein kinase (8, 39). Morever, herpesviruses from all subfamilies encode a protein kinase, homologous to the UL13 protein of HSV-1, whose exact function is not yet clear, although it has been demonstrated to phosphorylate several substrates, including gE/gI (38) and the UL49 tegument protein (9), and it modulates UL41 function (41). However, neither gE nor UL41 nor UL49 are conserved in the herpesvirus family, which results in a seeming paradox: a conserved viral protein has as major substrates nonconserved viral proteins. Thus, it may be conceivable that UL13 compensates in part for loss of US3, perhaps even in nuclear egress. It has indeed been shown that a PrV double mutant lacking US3 and UL13 exhibits a severe growth defect (10). It will be interesting to analyze whether the UL13 protein is also present in primary enveloped and mature virions and whether the concomitant absence of both kinases results in a more drastic effect on nuclear egress.
The observed presence of US3 protein in primary enveloped and mature virions correlates with its intracellular distribution during virus infection (24, 31). During infection it is detectable in both, the nucleus and the cytoplasm. However, so are other PrV tegument proteins such as UL47 (27) or UL48 (14), but these are apparently not incorporated into primary virions. Therefore, either a specific sorting mechanism exists which limits access to nascent primary virions for the “correct” constituents, or the intranuclear distribution of those proteins which are part of the mature virion but do not become part of the primary virus particle is different from that of US3 to prevent access to the nascent virus. It is also conceivable that differences in the temporal regulation of gene expression, e.g., early versus late kinetics, may influence localization of proteins in the virus particle.
In summary, our studies identify the US3 protein, a nonconserved herpesvirus protein, as the first tegument component described thus far that is part of the primary enveloped and of the mature virion. Whether the impairment in deenvelopment that is observed in the absence of the US3 protein reflects an important structural role for this protein in primary enveloped virions or whether it is due to effects of its kinase function remains to be analyzed.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (Me 854/5-2).
We thank Petra Meyer, Elke Zorn, Mandy Jörn, Uta Hartwig, and Nadine Müller for excellent technical assistance.
REFERENCES
- 1.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. F. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. Tuffnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. [DOI] [PubMed] [Google Scholar]
- 2.Bartha, A. 1961. Experimental reduction of virulence of Aujeszky's disease virus. Mag. Allat. Lap. 16:42-45. [Google Scholar]
- 3.Becker, C. H. 1967. Zur primären Schädigung vegetativer Ganglien nach Infektion mit dem Herpes suis Virus bei verschiedenen Tierarten. Experientia 23:109-217. [DOI] [PubMed] [Google Scholar]
- 4.Brack, A., J. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brack, A., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, Y. E., C. Van Sant, P. W. Krug, A. E. Sears, and B. Roizman. 1997. The null mutant of the UL31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J. Virol. 71:8307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchinson, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 8.Chung, T. D., J. P. Wymer, C. C. Smith, M. Kulka, and L. Aurelian. 1989. Protein kinase activity associated with the large subunit of herpes simplex virus type 2 ribonucleotide reductase (ICP10). J. Virol. 63:3389-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coulter, L., H. Moss, J. Lang, and D. J. McGeoch. 1993. A mutant of herpes simplex virus type 1 in which the UL13 protein kinase gene is disrupted. J. Gen. Virol. 74:387-395. [DOI] [PubMed] [Google Scholar]
- 10.De Wind, N., J. Domen, and A. Berns. 1992. Herpesviruses encode an unusual protein-serine/threonine kinase which is nonessential for growth in cultured cells. J. Virol. 66:5200-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dietz, P., B. G. Klupp, W. Fuchs, B. Köllner, E. Weiland, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein K requires the UL20 gene product for processing. J. Virol. 74:5083-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dijkstra, H., T. C. Mettenleiter, and B. G. Klupp. 1997. Intracellular processing of pseudorabies virus glycoprotein M (gM): gM of strain Bartha lacks N glycosylation. Virology 237:113-122. [DOI] [PubMed] [Google Scholar]
- 13.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The multifunctional UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs, W., B. G. Klupp, H. Granzow, H.-J. Rziha, and T. C. Mettenleiter. 1996. Identification and characterization of the pseudorabies virus UL3.5 protein, which is involved in virus egress. J. Virol. 70:3517-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Granzow, H., F. Weiland, A. Jöns, B. G. Klupp, A. Karger, and T. C. Mettenleiter. 1997. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J. Virol. 71:2072-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 13:78-92. [DOI] [PubMed] [Google Scholar]
- 20.Kimman, T. G., N. de Wind, T. de Bruin, Y. de Visser, and J. Voermans. 1994. Inactivation of glycoprotein gE and thymidine kinase or the US3-encoded protein kinase synergistically decreases in vivo replication of pseudorabies virus and the induction of protective immunity. Virology 205:511-518. [DOI] [PubMed] [Google Scholar]
- 21.Klupp, B. G., W. Fuchs, E. Weiland, and T. C. Mettenleiter. 1997. Pseudorabies virus glycoprotein L is necessary for virus infectivity but is dispensable for virion localization of glycoprotein H. J. Virol. 71:7687-7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klupp, B. G., and T. C. Mettenleiter. 1999. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J. Virol. 73:3014-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363-2371. [DOI] [PubMed] [Google Scholar]
- 25.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klupp, B. G., W. Fuchs, H. Granzow, R. Nixdorf, and T. C. Mettenleiter. 2002. The pseudorabies virus UL36 tegument protein physically interacts with the UL37 gene product. J. Virol. 76:3065-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopp, M., B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 76:8820-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion structural component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laemmli, U. K. 1970. Cleavage of the structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 30.Littler, E., A. D. Stuart, and M. S. Chee. 1992. Human cytomegalovirus UL97 open reading frame encodes a protein that phosphorylates the antiviral nucleoside analogue ganciclovir. Nature 358:160-162. [DOI] [PubMed] [Google Scholar]
- 31.Lyman, M. G., G. Demmin, and B. W. Banfield. 2003. The attenuated pseudorabies virus strain Bartha fails to package the tegument proteins US3 and VP22. J. Virol. 77:1403-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGeoch, D. J., M. B. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, and D. McNab. 1988. The complete DNA sequence of the unique long region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 33.Mettenleiter, T. C. 1995. Molecular properties of alphaherpesviruses used in transneuronal pathway tracing, p. 367-393. In M. G. Kaplitt and A. D. Loewy (ed.), Viral vectors. Academic Press, Inc., San Diego, Calif.
- 34.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muranyi, W., J. Haas, M. Wagner, G. Krohne, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297:854-857. [DOI] [PubMed] [Google Scholar]
- 36.Newcomb, W., R. M. Juhas, D. R. Thomsen, F. L. Homa, A. Burch, S. K. Weller, and J. C. Brown. 2001. The UL6 gene product forms the portal of entry of DNA into the herpes simplex virus capsid. J. Virol. 75:10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ng, T., and C. Grose. 1992. Serine protein kinase associated with varicella-zoster virus ORF 47. Virology 191:9-18. [DOI] [PubMed] [Google Scholar]
- 38.Ng, T., W. Ogle, and B. Roizman. 1998. UL13 protein kinase of herpes simplex virus 1 complexes with glycoprotein E and mediates the phosphorylation of the viral Fc receptor: glycoproteins E and I. Virology 241:37-48. [DOI] [PubMed] [Google Scholar]
- 39.Nikas, I., J. McLauchlan, A. J. Davison, W. R. Taylor, and J. B. Clements. 1986. Structural features of ribonucleotide reductase. Proteins Struct. Funct. Genet. 1:376-384. [DOI] [PubMed] [Google Scholar]
- 40.Nixdorf, R., B. G. Klupp, and T. C. Mettenleiter. 2001. Restoration of function of carboxy-terminally truncated pseudorabies virus glycoprotein B by point mutations in the ectodomain. J. Virol. 75:11526-11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Overton, H., D. McMillan, L. Hope, and P. Wong-Kai-In. 1994. Production of host shutoff-defective mutants of herpes simplex virus type 1 by inactivation of the UL13 gene. Virology 209:97-106. [DOI] [PubMed] [Google Scholar]
- 42.Petrovskis, E., J. G. Timmins, T. M. Giermann, and L. E. Post. 1986. Deletions in vaccine strains of pseudorabies virus and their effect on synthesis of glycoprotein gp63. J. Virol. 60:1166-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purves, F. C., A. Donella-Deana, F. Marchiori, D. Leader, and L. A. Pinna. 1986. The substrate specificity of the protein kinase induced in cells infected with herpesviruses: studies with synthetic substrates indicate structural requirements distinct from other protein kinases. Biochim. Biophys. Acta 889:208-215. [DOI] [PubMed] [Google Scholar]
- 44.Purves, F. C., D. Spector, and B. Roizman. 1991. The herpes simplex virus protein kinase encoded by the US3 gene mediates posttranslational modification of the phosphoprotein encoded by the UL34 gene. J. Virol. 65:5757-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purves, F. C., D. Spector, and B. Roizman. 1992. UL34, the target of the herpes simplex US3 protein kinase, is a membrane protein which in its unphosphorylated state associates with novel phosphoproteins. J. Virol. 66:4295-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purves, F. C., W. Ogle, and B. Roizman. 1993. Processing of the herpes simplex virus regulatory protein α22 mediated by the UL13 protein kinase determines the accumulation of a subset of α and γ mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. USA 90:6701-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purves, F. C., R. M. Longnecker, D. P. Leader, and B. Roizman. 1987. Herpes simplex virus 1 protein kinase is encoded by open reading frame US3 which is not essential for virus growth in cell culture. J. Virol. 61:2896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reynolds, A., B. Ryckman, J. Baines, Y. Zhou, L. Liang, and R. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reynolds, A., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roller, R., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, R. F., and T. F. Smith. 1989. Identification of new protein kinase-related genes in three herpesviruses, herpes simplex virus, varicella-zoster virus, and Epstein-Barr virus. J. Virol. 63:450-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steven, A. C., and P. G. Spear. 1996. Herpesvirus capsid assembly and envelopment, p. 312-351. In W. Chiu, R. M. Burnett, and R. Garcea (ed.), Structural biology of viruses. Oxford University Press, New York, N.Y.
- 53.Sullivan, V., C. L. Talarico, S. C. Stanat, M. Davis, D. M. Coen, and K. K. Biron. 1992. A protein kinase homologue controls phosphorylation of ganciclovir in the human cytomegalovirus-infected cells. Nature 358:162-164. [DOI] [PubMed] [Google Scholar]
- 54.Towbin, H., T. Staehelin, and J. Gordon. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76:4350-4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Genderen, I. L., R. Brandimarti, M. Torrisi, G. Campadelli, and G. van Meer. 1994. The phospholipid composition of extracellular herpes simplex virions differs from that of the host cell nuclei. Virology 200:831-836. [DOI] [PubMed] [Google Scholar]
- 56.van Zijl, M., H. van der Gulden, N. de Wind, A. Gielkens, and A. Berns. 1990. Identification of two genes in the unique short region of pseudorabies virus; comparison with herpes simplex virus and varicella-zoster virus. J. Gen. Virol. 71:1747-1755. [DOI] [PubMed] [Google Scholar]
- 57.Wagenaar, F., J. M. A. Pol, B. Peeters, A. L. J. Gielkens, N. de Wind, and T. G. Kimman. 1995. The US3-encoded protein kinase of pseudorabies virus affects egress from virions from the nucleus. J. Gen. Virol. 76:1851-1859. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, G., and D. P. Leader. 1990. The structure of the pseudorabies virus genome at the end of the inverted repeat sequences proximal to the junction with the short unique region. J. Gen. Virol. 71:2433-2441. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, G., R. Stevens, and D. P. Leader. 1990. The protein kinase encoded in the short unique region of pseudorabies virus: description of the gene and identification of its product in virions and infected cells. J. Gen. Virol. 71:1757-1765. [DOI] [PubMed] [Google Scholar]
- 60.Zhou, H., D. Chen, J. Jakana, F. J. Rixon, and W. Chiu. 1999. Visualization of tegument-capsid interactions and DNA in intact herpes simplex virus type 1 virions. J. Virol. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]