Abstract
Repair of DNA double-strand breaks (DSBs) by homologous recombination (HR) requires resection of 5′-termini to generate 3′-single-strand DNA tails1. Key components of this reaction are Exonuclease 1 and the bifunctional endo/exonuclease, Mre112-4. Mre11 endonuclease activity is critical when DSB termini are blocked by bound protein—such as by the DNA end-joining complex5, topoisomerases6, or the meiotic nuclease, Spo117-13—but a specific function for the Mre11 3′-5′ exonuclease activity has remained elusive. Here, we reveal a role for the Mre11 exonuclease during the resection of Spo11-linked 5′-DNA termini in vivo. We show that the residual resection observed in Exo1-mutant cells is dependent on Mre11, and that both exonuclease activities are required for efficient DSB repair. Previous work has indicated resection to traverse unidirectionally1. Using a combination of physical assays for 5′-end-processing, our results suggest an alternative mechanism involving bidirectional resection. First, Mre11 nicks the strand to be resected up to 300 nucleotides from the 5′-terminus of the DSB—much further away than previously assumed. Second, this nick enables resection in a bidirectional manner, using Exo1 in the 5′-3′ direction away from the DSB, and Mre11 in the 3′-5′ direction towards the DSB end. Finally, Mre11 exonuclease activity confers resistance to DNA damage in cycling cells, suggesting that Mre11-catalysed resection may be a general feature of various DNA repair pathways.
We sought to clarify Mre11-dependent processing reactions at DSBs with blocked termini. Blocked DSB ends are created in meiosis by the topoisomerase-like transesterase, Spo11, to initiate crossover recombination14 (Fig. S1a). Aside from the importance of meiotic recombination to sexual reproduction and genetic diversity, we reasoned that the molecular reactions pertaining to repair of covalently blocked Spo11-DSBs would be informative to the repair of DNA lesions with other types of end blockage, such as failed topoisomerase reactions and DSB ends competitively bound by the nonhomologous end-joining (NHEJ) machinery.
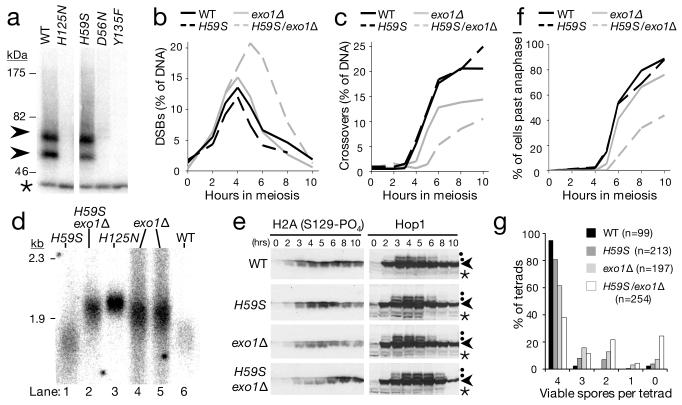
Mre11-dependent endonucleolytic processing (nicking) of Spo11-DSBs generates two classes of Spo11-oligonucleotide fragments originating from the DSB end15 (Fig. S1). Spo11-oligo complexes do not form in mre11 mutants completely abrogated for nuclease activity (mre11-D56N, and mre11-H125N; Fig. 1a) solidifying earlier conclusions that Mre11 directly processes Spo11-DSBs7-13. Recently, an allele of Mre11 that is exonuclease deficient, but mostly endonuclease proficient was described for the orthologous proteins in S. pombe and P. furiosus16. We generated the equivalent mutation in budding yeast Mre11 (Histidine 59 to Serine; Fig. S2a) and tested its function. During meiosis, Spo11-oligo complexes were readily detected in mre11-H59S cells (Fig. 1a), indicating that the allele is endonuclease proficient in vivo. Biochemical assays with recombinant ScMre11-H59S showed reduced, but not abolished, 3′-5′ exonuclease activity on linear duplex DNA, and retention of much of the ssDNA endonuclease activity (Fig. S2)—observations consistent with mre11-H59S partially separating the two nuclease functions.
Figure 1. Mre11 and Exo1-dependent resection and repair of meiotic DSBs.
a, Autoradiograph of Spo11-oligo formation (arrowheads) in Mre11 nuclease-defective cells: mre11-D56N, -H125N, -H59S, and in control spo11-Y135F cells where Spo11-DSBs do not form. Immunoprecipitated Spo11-oligo complexes are 3′ end-labelled using terminal transferase and separated on SDS-PAGE15 (see Fig. S1). Asterisk marks an unrelated labelling artifact; b-f, Timecourse of events during meiosis for the indicated genotypes: b-c, Quantitative analysis of DSB (b) and crossover (c) signals at HIS4::LEU2. DNA at each timepoint was digested with PstI (b) or XhoI (c), separated by agarose gel electrophoresis and blots hybridised with probes to LSB5 (b) or STE50 (c). DSB signals were quantified as a percentage of specific lane signal (see also Fig. S3); d, Relative extent of DSB resection at HIS4::LEU2 (at 4 hours in meiosis). DNA was digested with BglII and blots hybridised with a STE50 probe (see also Fig. S4); e, Western blot analysis of phosphorylated H2Ax and Hop1 (arrowhead) detected from TCA-precipitated whole cell lysates. Phosphorylated Hop1 indicated by dot. Asterisk marks cross-reacting band; f, Progression through anaphase I and II assessed by microscopic examination of DAPI-stained cells; g, Distribution of spore viabilities per tetrad (n; number of 4-spore tetrads dissected). The difference in distribution between mre11-H59S/exo1Δ and exo1Δ is highly significant (Chi-square test for goodness-of-fit; Chi value = 63.084, d.f. = 4, P < 0.0001). Absolute spore viabilities are: WT, 97%; mre11-H59S, 90%; exo1Δ, 76%, mre11-H59S/exo1Δ, 59%.
We assessed DSB repair kinetics at two meiotic recombination hotspots using southern blotting and probes for the relevant genomic loci (HIS4::LEU2 and ARE1; Fig. 1b; Fig. S3). In the mre11-H59S strain, DSBs formed at normal levels and repaired as crossovers with normal timing (Fig. 1b,c). Single-stranded DNA resection, which can be qualitatively assessed by the relative migration of the DSB band on native agarose gels, also appeared unaffected by the mre11-H59S mutation (Fig. 1d lanes 1 and 6; Fig. S4).
The lack of an obvious defect in ssDNA resection proficiency suggested that any potential contribution to ssDNA generation by Mre11 might be masked by the activity of another nuclease. During meiosis, the major resection pathway requires Exo117-21. We tested this idea by combining the mre11-H59S allele with an EXO1 deletion (exo1Δ), which we found itself to have slightly delayed DSB repair kinetics, with fewer DSBs repairing as crossovers (Fig. 1b,c). Migration of the DSB band on agarose gels revealed that ssDNA resection was measurably reduced in exo1Δ—but not entirely abolished relative to a mre11-H125N control (where the failure to remove Spo11 prevents all resection; Fig. 1d, lane 3 and 4-5; Fig. S4). Our data agree with those of others investigating a meiotic role for Exo117-21.
The combination of mre11-H59S with exo1Δ caused DSBs to transiently accumulate for a longer period, with DSBs detectable for two hours longer than in matched controls (Fig. 1b), and with formation of crossover recombinants reduced and delayed (Fig. 1c). These defects in DSB repair are correlated with a reduction in the mobility of DSB DNA on agarose gels (Fig. 1d, S4). Specifically, in comparison to the mre11-H59S or exo1Δ single mutants, we observed DSB signals to migrate similarly to DSBs from the mre11-H125N control (Fig. 1d, lanes 2 and 3). This reduction in mobility is indicative of less single-stranded resection in mre11-H59S/exo1Δ than of either single mutant. We conclude that Exo1 and Mre11 collaborate to enable efficient ssDNA generation at meiotic DSBs.
Unrepaired DSBs cause Tel1/Mec1(ATM/ATR)-dependent phosphorylation of histone H2Ax on serine 129 and hyper-phosphorylation of Hop1 (a meiosis-specific adaptor of the DNA damage response22). Phosphorylated H2Ax and Hop1 were detected in all strains indicating that significant resection is not essential for activation of Tel1/Mec1 (Fig. 1e). However, phospho-H2Ax accumulated and persisted until late timepoints only in mre11-H59S/exo1Δ, and Hop1 phosphorylation persisted for at least two hours longer than in matched controls. These observations are consistent with a genome-wide defect in DSB repair in mre11-H59S/exo1Δ. The Tel1 branch of the signaling pathway is primarily activated by unresected DSBs23. We observed that Hop1 phosphorylation in the mre11-H59S/exo1Δ background is highly dependent on Tel1 (Fig. S5), consistent with less resection occurring genome-wide.
To investigate if DSB processing defects were affecting meiotic chromosome segregation, we determined the efficiency of progression through anaphase I and II (Fig. 1f). Meiotic progression of wildtype and mre11-H59S was essentially identical, while exo1Δ was delayed by about 1 hour, with slightly reduced overall efficiency. In contrast, nuclear division in the mre11-H59S/exo1Δ double mutant was poor, with more than half of the cells having failed to complete even the first meiotic nuclear division after 10 hours in meiosis. Analysis of sporulation efficiency after 24 hours revealed increased incidence of aberrant tetrad maturation and/or nuclear packaging where orphaned chromosome fragments were observed outside the maturing spore wall (Fig. S6). These defects in meiotic chromosome segregation manifested as reduced spore viability in the double-mutant compared to controls (Fig. 1g).
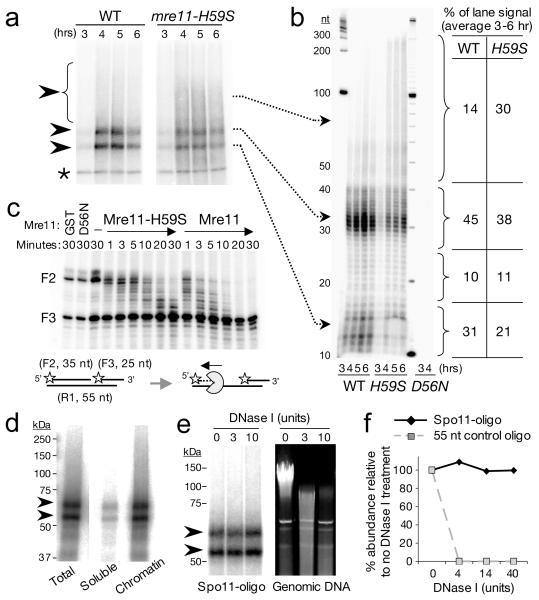
To characterise in greater detail the molecular defect caused by mre11-H59S, we looked carefully at the distribution of Spo11-oligonucleotide products generated in vivo by the 5′-DSB processing reaction (Fig. 2a,b). In wildtype cells, two major classes of Spo11-oligo are observed, which differ by the length of attached DNA15. By contrast, in mre11-H59S we observed a shift in this distribution towards higher molecular weight Spo11-oligo species (Fig. 2a and Fig. S7). Importantly, total Spo11-oligo formation was not itself delayed (Fig. S7), indicating that the Spo11-removal reaction initiates with normal timing in mre11-H59S cells, but is defective in forming shorter molecules. To clarify the precise size-distribution of the Spo11-oligo molecules, we fractionated deproteinised oligos using denaturing PAGE (Fig. 2b). In wild type cells, two peak areas of signal 10-17 nucleotides (nt) and 28-40 nt were apparent. These correspond to the shorter and longer Spo11-oligo classes detected on SDS-PAGE (Fig. 2a). We additionally detected signal (24 % of total) in molecules 18-27 and 41-300 nt long (Fig. 2b), indicating that the length-distribution of processed molecules is significantly more heterogeneous than previously assumed, and that nicks are made at up to 300 nt from the DSB end.
Figure 2. Mre11-exonucleolytic processing of DSB ends.
a-b, Spo11-oligo detection in wildtype (WT) and mre11-H59S during meiosis. 3′ end-labelled Spo11 complexes are fractionated by SDS-PAGE (a) or by nucleotide resolution urea/PAGE after proteolytic removal of Spo11 peptide (b); c, Mre11-H59S displays reduced 3′-5′ exonuclease activity on a nicked duplex. Reactions were performed as for Fig. S2c. Stars indicate 5′ label, F3 3′-end has 5 nt extension and is refractory to Mre11-mediated resection25; d, Chromatin association of Spo11-oligo complexes. Wildtype cell extracts were fractionated and the abundance of Spo11-oligo complexes assessed in soluble versus chromatin-enriched material; e-f, Nuclease resistance of Spo11-oligo complexes. Chromatin-enriched material from (d) was treated with DNase I, and abundance of Spo11-oligo complexes compared to the simultaneous degradation of genomic DNA (e), or of a control 55 nt oligonucleotide (f).
In the mre11-H59S mutant, the distribution was shifted such that the long oligo molecules of 41-300 nt, made up a third of the material detected (Fig. 2b). Although it is possible that the altered distribution of oligo molecules is caused by a reduction in the endonuclease activity of Mre11-H59S, our physical and genetic observations in both WT and mre11-H59S can be readily explained if resection begins at relatively distant nicks (up to 300 nt from the DSB) and traverses bidirectionally both away from (using Exo1) and towards (using Mre11) the DSB end21. Such a model is consistent with the opposing polarities of the Mre11 and Exo1 exonuclease activities24,25, and with the synergistic loss in resection we observe in mre11-H59S/exo1Δ. Moreover, the length distribution of Spo11-oligos is compatible with the extent of Exo1-independent resection reported recently by others21, and as predicted, Spo11-oligo length is unchanged by loss of EXO1 (Fig. S8). Finally, only a low background of Spo11-oligo complexes are detected in endonuclease-defective mre11-D56N, ruling out the possibility that the long oligo molecules arise via an alternative nuclease nicking the 5′ strand (Fig. 2b). To test this mechanism in vitro, we incubated Mre11 protein with a nicked duplex substrate designed to mimic this proposed in vivo reaction, and found Mre11-H59S to resect from the nick with lower efficiency than wildtype Mre11 (Fig. 2c).
Together, these observations led us to consider that the steady-state length of Spo11-oligo complexes might arise via the relative processivity of the 3′-5′ Mre11-exonuclease and the relative sensitivity to nucleolytic degradation of DNA close to the DSB end. Spo11-DSB formation requires at least ten factors14, suggesting that a large protein complex may reside at—and protect—the DSB end. If this model were correct, we expected Spo11-oligo complexes to be associated with chromatin and resistant to nucleolytic degradation. We tested this idea by incubating a chromatin-enriched nuclear pellet from meiotic yeast cells with DNase I, then assessed the amount of Spo11-oligo complexes remaining relative to both bulk DNA and to an exogenous protein-free oligonucleotide included in parallel reactions (Fig. 2d-f). Greater than 90 % of Spo11-oligos are found in the chromatin fraction and remarkably, no loss in signal was observed despite extensive nucleolytic degradation of both the chromosomal DNA and the control oligonucleotide. We conclude that Spo11-oligo complexes are occluded from degradation even by exogenous nucleases—a prediction of our model.
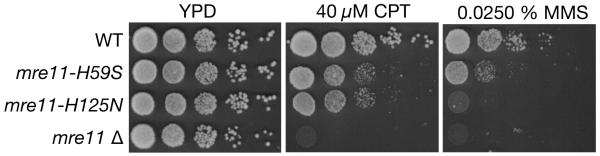
In cycling cells, Mre11 nuclease activity promotes the onset of resection—a requisite for repair of DSBs by homologous recombination1. To investigate a specific role for the Mre11 3′-5′ exonuclease during DNA repair in cycling cells, we challenged yeast cells with exposure to DNA damaging agents. Similar to complete abrogation of the endo/exonuclease activities (mre11-H125N), reduced Mre11 exonuclease activity (mre11-H59S) sensitised cells to the DNA alkylating agent methyl methanesulphonate (MMS), and to the topoisomerase poison camptothecin (CPT; Fig. 3). Compared to an MRE11 deletion however, mre11-H59S and mre11-H125N are themselves far less sensitive, consistent with physical interactions between the Mre11-complex being retained (Fig. S9). In agreement with Mre11 endonuclease activity being unaffected in mre11-H59S, and allowing redundant processing pathways, combining mre11-H59S with a deletion of EXO1 did not further sensitise cells to MMS (Fig. S10). Together these observations suggest that the exonuclease activity of Mre11 is involved in the repair of various classes of DNA lesion.
Figure 3. DNA damage sensitivity of exonuclease defective Mre11 cells.
Ten-fold serial dilutions of the indicated strains were spotted onto solid media containing the indicated compounds and incubated at 30°C for 2 days (CPT) or 3 days (MMS).
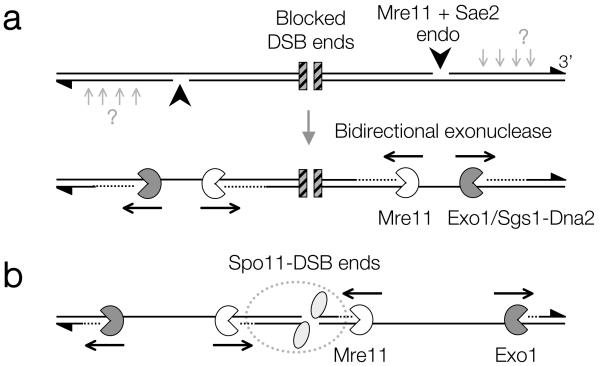
Understanding the regulation of DSB repair is a complex issue involving multiple factors with overlapping roles. Here, we propose a biological function for the 3′-5′ exonuclease activity of the evolutionarily conserved Mre11 protein. Previous work has indicated DSB resection to traverse unidirectionally1. Here, we propose a refined model that involves the coordination of two resection activities of opposing polarity: Exo1 away from the DSB and Mre11 towards the DSB end (Fig. 4). We favour the view that this exonuclease reaction begins at nicks created by the Mre11 endonuclease (in conjunction with Sae2) and which are positioned at variable distance from the DSB end, perhaps due to locus-specific chromatin architecture26. Although our assay detects only the site of incision closest to the DSB end, Mre11 and Sae2 may create multiple nicks on the resecting strand17,21,27 (A. S. H. Goldman, personal communication), which in combination with exonucleolytic processing might further enhance resection efficiency. Finally, the recent observation that the length and abundance of SPO11-oligonucleotide complexes is increased in Atm−/− mice (S. Keeney and M. Jasin personal communication), suggests that Mre11 exonuclease activity may be an evolutionarily conserved feature directly regulated by ATM.
Figure 4. Model for bidirectional processing of DSBs by Mre11 and Exo1.
a, Following DSB formation with blocked ends (hatched squares), Mre11/Sae2-dependent nicks flanking the DSB ends create initiation sites for bidirectional resection by Exo1/Sgs1-Dna2 away from the DSB, and by Mre11 towards the DSB end. Such terminal blocks could arise after base damage, trapping of a topoisomerase, or by avid binding of the NHEJ complex. 3′ ends marked with triangles. Mre11/Sae2 may make multiple nicks on the 5′ strand (light grey arrows) facilitating resection; b, In meiosis, the DSB ends are terminally blocked by covalently bound Spo11 protein (grey ellipses), and may be protected from Mre11-dependent exonuclease degradation by a large metastable multisubunit complex (dashed outline), thereby generating the observed size-distribution of Spo11-oligonucleotide complexes.
During meiosis, bidirectional processing may help to reinforce subsequent steps of repair, which at least in some cases appear to occur differentially on either side of the DSB26,28,29. Our observation that liberated Spo11-oligo complexes remain chromatin-bound and relatively protected provides a clue to potential mechanisms of end differentiation. For example, retention of proteins on one or both of the DSB ends could influence subsequent steps of repair15 (Fig. 4b). Finally, our observation that Mre11-dependent incision occurs at some distance from the DSB end suggests that DSB formation and processing reactions are coordinated over a considerable distance (300 bp of B-form DNA is ~100 nm). How these incision points are regulated—and restricted to only the 5′-ending strand—are fascinating questions for the future.
Although much of this work concerns specifics of meiotic DSB processing, we suggest that a similar pathway may occur whenever the DSB end is blocked by a lesion or protein complex that prevents direct loading of the 5′-3′ exonuclease machinery. Such DNA blockages may be crosslinked protein, damaged DNA ends, or simply the stable binding of high affinity proteins to the DSB. As such, the nucleolytic incision pathway may provide the key point of regulation that controls the balance between NHEJ and HR.
Methods summary
Yeast strains and culture methods
Meiotic cultures were prepared as described15. Strains were derived from SK1 using standard techniques. Spo11 protein is tagged by the HA3His6 epitope15. Mre11 mutations were introduced by pop-in/out using plasmids derived from pRS414-MRE1110. exo1Δ, tel1Δ, and sae2Δ are full replacements of the ORF with kanMX4, hphNT2, and kanMX6 respectively. A full strain list is available on request. For chronic DNA damage sensitivity, exponentially growing cultures were spotted in 10-fold dilutions onto freshly prepared media.
Molecular techniques
Spo11-oligonucleotide complexes were detected by two rounds of immunoprecipitation and end-labeling as described15. DSBs and crossover recombinants were detected by Southern blotting genomic DNA after fractionation on agarose gels using standard techniques28. Radioactive signals were collected on phosphor screens, scanned with a Fuji FLA5100, and quantified using ImageGauge software. Phosphorylated H2Ax and Hop1 protein were detected using Ab17353 (Abcam) and anti-Hop1 antisera (F. Klein) respectively. Subcellular fractionation of cell contents was performed by purifying hypotonically lysed spheroplasts through a sucrose cushion. Chromatin digests were performed at 25°C with DNase I (NEB). Reactions were split and processed for DNA extraction or Spo11-oligo purification. As a positive control, parallel reactions included a 5′-labeled 55 nt oligonucleotide. Recombinant GST-Mre11 alleles were purified and reacted with DNA substrates as described10.
Supplementary Material
Acknowledgements
We thank R. Cha, E. Hoffmann, N. Hunter, S. Keeney, and J. Nitiss for yeast strains; L. Symington for plasmids; F. Klein and J. Petrini for antisera. V.G. is supported by an MRC New Investigator Grant to M.J.N. M.J.N. is supported by a University Research Fellowship from the Royal Society, and a Career Development Award from the Human Frontiers Science Program Organisation.
Appendix
Methods
Meiotic cultures
YPD cultures (1% Yeast extract / 2 % Peptone / 2 % Glucose) were diluted 100-fold into YPA (1 % Yeast extract / 2 % peptone / 1 % K-acetate) and grown vigourously for 14 hours at 30 °C. Cells were collected by centrifugation, washed once in water, resuspended in an equal volume of prewarmed 2 % K-acetate containing diluted amino acid supplements, and shaken vigourously at 30 °C.
Construction of Mre11 nuclease mutant strains
The mre11-H59S mutant was constructed by site-directed mutagenesis PCR on the integrating plasmid pSM444 (pRS406::mre11-D56N), reversing the D56N mutation to wild type and introducing H59S (forward primer: GTACAGTCCGGTGATCTTTTTAGCGTGAATAAGCC; reverse primer: GGCTTATTCACGCTAAAAAGATCACCGGACTGTAC). Integration plasmids containing mre11-D56N and mre11-H125N nuclease dead mutant alleles were kindly provided by L. Symington (pSM444 and pSM438 respectively). To replace chromosomal MRE11 with mutant alleles, the plasmid containing mutations in the nuclease domains of MRE11 were linearized with SphI and transformed in the SK1 strain. Uracil-positive (Ura+) transformants were inoculated in rich medium overnight and 20 μl of culture were spread onto medium containing 5-fluoroorotic acid (5-FOA) in order to select pop-out events. The presence of the mre11-D56N and mre11-H125N alleles in the resulting 5-FOA resistant cells was assessed by sensitivity to DNA damaging agent: cells from single 5-FOA resistant colonies were patched onto plates containing high concentration of CPT (30-50 μM) supplemented with phloxin B. Because the introduction of the mre11-H59S mutation also alter a PmlI restriction site, the presence of the mre11-H59S allele was tested by sensitivity of PCR reactions covering the mutated region to PmlI restriction. Replacement of wildtype MRE11 by targeted mutations was confirmed by PCR amplification and sequencing.
Spot tests for DNA damage sensitivity
Cells were grown overnight in liquid YPD, diluted into fresh media and grown to log phase, adjusted to OD600 = 0.2, serially diluted 10-fold, and 5 μl spotted onto control and drug-containing YPD plates.
Spo11-oligonucleotide assays
Spo11-oligonucleotide complexes were detected by immunoprecipitation and end-labeling following established methods15. To reduce the coprecipitation of nonspecific genomic DNA, two rounds of immunoprecipitation were used. Specifically, 10-50 ml of sporulating culture was lysed in 10 % ice cold TCA using zirconia beads and a BioSpec 24. Precipitated material was dissolved in STE (2 % SDS / 0.5 M Tris pH 8.1 / 10 mM EDTA / 0.05 % bromophenol blue), and boiled for 5 minutes. Extracts were diluted two-fold in 2 x IP buffer (2% Triton X100, 300 mM NaCl, 30 mM Tris.HCl pH 8.1, 2 mM EDTA), centrifuged 10 minutes at 16,000 g at 4°C, and supernatant was diluted a further two-fold in 1 x IP buffer. Anti-HA antibody (F-7; Santa Cruz Biotechnology) was added at 1 in 500, protein-G-agarose matrix (Roche) at 1 in 50, and then incubated with rotation for 4 hours at 4°C. Immune complexes were collected by low speed centrifugation, and washed 3 times with 1 x IP buffer. Beads were boiled for 5 minutes in 250 μl STE, chilled on ice, and diluted two-fold as above with 2 x IP buffer, recentrifuged, and supernatant was dissolved further two-fold in 1 x IP buffer. Fresh antibody and beads were added at the above dilutions. The second IP was performed overnight at 4°C, and then washed as above. Two additional washes in 1 x TKAc (20 mM Tris.acetate pH 7.9 / 50 mM K.acetate) were performed before incubation with 10-20 units TdT (Fermentas) and 5-10 uCi CoTP (cordycepin triphosphate; Perkin Elmer) in 1x TKAc / 0.25 mM CoCl2 buffer at 37 °C for 1 hour. Labeled complexes were washed twice with 1 x IP buffer, and eluted in Laemmli loading buffer for direct analysis on 7.5 % SDS-PAGE. For nucleotide resolution analysis of Spo11-oligo lengths, eluted complexes were mixed with 1 μg glycogen (Roche) and 10 volumes of 100 % ethanol, and precipitated overnight at −20 °C. Precipitates were collected by centrifugation, dissolved in 15 μl of 10 x TE containing 0.5 μg/ml proteinase K, and incubated at 50 °C for 30 minutes. Eluted oligonucleotides were mixed with 3 volumes of loading dye (95 % formamide / 10 mM EDTA / 0.01 % bromophenol blue / 0.01 % xylene cyanol), and fractionated through a 28 cm tall, 0.5 mm thick 12 % polyacrylamide (19:1) / 6 M urea gel in 1 x TBE running buffer at approx 1200 V for 50-60 minutes. Gels were fixed in 10 % methanol / 7 % acetic acid / 5 % glycerol, vacuum dried and exposed to phosphor screens for imaging.
DSB and crossover analysis
Genomic DNA was isolated from aliquots of synchronously sporulating cultures using standard methods. Briefly, spheroplasts were prepared in 1 M sorbitol / 0.1 M EDTA / 0.1 M NaHPO4 pH 7.5 / 1 % BME and 200 μg/ml zymolyase 100T for 20-30 minutes at 37 °C, and lysed by adding SDS to 0.5 % and proteinase K to 200 μg/ml with incubation for 60 minutes at 60 °C. Protein was removed by mixing with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1), and nucleic acids precipitated by adding one-tenth volume of 3 M NaAc pH5.2 and an equal volume of 100 % ethanol. Precipitates were washed in 70 % ethanol and dissolved in 1 x TE containing 100 μg/ml RNAse, incubated for 60 minutes at 37 °C, reprecipitated with ethanol / NaAc and DNA pellets left to dissolve in 1 x TE overnight at 4 °C. Genomic DNA was digested with restriction enzymes following the manufacturer’s recommendations, fractionated on 0.6 %, 0.7 % or 0.9 % agarose gels in 1 x TAE or 1 x TBE buffer and ethidium bromide (as required), and blotted under vacuum to zetaprobe membrane (BioRad) in 0.5 M NaOH / 1.5 M NaCl. Blots were equilibrated in hybridization solution (0.5 M NaHPO4 pH 7.2 / 7 % SDS / 1 mM EDTA/ 1% BSA) at 65 °C, and random primed radioactive probes (High Prime; Roche) prepared form gel-purified PCR products were then added. Blots were washed 4 times in 40 mM NaHPO4 / 1 % SDS / 1 mM EDTA at 65 °C, then dried and exposed to phosphor screens for imaging.
Western blotting
TCA-denatured cell material dissolved in STE / 5% β-mercaptoethanol was fractionated in 7.5 % or 10 % SDS-PAGE, transferred to PVDF membrane (Millipore) in 1 x CAPS buffer (10 mM CAPS.NaOH pH 11 / 10 % methanol), and incubated in TBST + 3 % BSA with the requisite antibodies: Phosphorylated H2Ax was detected using Ab17353 diluted 1/5000 in TBST (Abcam) and Hop1 detected using anti-Hop1 antisera at a 1/4000 dilution (F. Klein).
DAPI staining
Cells were fixed in 100 % methanol, and aliquots mixed with 1 μg/ml DAPI in 75% glycerol. The number of cells with 0, 2 and 4 nuclei was scored under a fluorescence microscope. At least 200 cells were counted per timepoint. Sporulation efficiency was determined by observation of fixed tetrads (24 hours into meiosis, on 3 independent timecourses) under a Zeiss fluorescence microscope with both fluorescence and bright-field illumination. Samples were randomized and 150 to 300 cells were counted for each strain per timecourse. Sporulations were scored as abnormal when significant extranuclear material was detected by DAPI staining and spores walls morphology were abnormal under bright-field illumination. DAPI and DIC images were captured using a personalDV (DeltaVision) system (Applied Precision) using the softWoRx software.
Chromatin enrichment
Cell pellets corresponding to 100ml meiotic cultures were resuspended in 30 ml Sphero/CoHex buffer (1 M sorbitol / 50 mM HEPES pH 7.5 / 10 mM EDTA / 5 mM hexamine cobalt chloride) supplemented with 1% β-mercaptoethanol and zymolyase 100T to 50 μg/ml, and incubated for ~25 minutes at 37 °C. Spheroplasts were transferred onto 10 ml sucrose cushion, spun 5 minutes at 16,000 g at 4 °C. Spheroplast pellets were resuspended in 1 ml sucrose buffer (1 M sucrose / 50 mM MES.NaOH / 50 mM NaCl /1 mM EDTA / 0.5 mM MgCl2 / Roche protease inhibitors cocktail / AEBSF to 10 ug/ml), lysed in 30 ml lysis buffer (50 mM MES.NaOH / 1 mM MgCl2 / 5 mM EDTA, Roche protease inhibitors cocktail / AEBSF to 10 μg/ml), homogenized in a dounce homogenizer, and spun 5 minutes at 16,000 g at 4 °C. Pellets were suspended in 30 ml lysis buffer and subcellular fractionation of cell contents was performed by centrifugation through a 10 ml sucrose cushion. Pellets were washed once in the same buffer, suspended in sucrose buffer, and 300 ul aliquots were stored at −20 °C. Aliquots were incubated at 25 °C with DNase I (NEB) in 50 mM HEPES / 50 mM NaCl / 5 mM MgCl2 buffer supplemented with complete protease inhibitors tablet (Roche) and AEBSF. Reactions were split and processed for genomic DNA extraction or Spo11-oligo purification. As a positive control, parallel reactions included a 5′-labeled 55 mer oligonucleotide (R1) that was detected on 1 x TBE PAGE.
Recombinant protein purification
GST-Mre11 alleles were purified and reacted with DNA substrates as described10. Briefly, MRE11 wild-type, mre11-H59S, and mre11-D56N open reading frames were amplified by PCR, cloned with BamHI/SalI ends into pEG-KT plasmid and were transformed into the protease defective JEL1 mre11Δ strain (LSY1706; a gift from L. Symington). Galactose induction was performed as described in http://www.bio.brandeis.edu/haberlab/jehsite/protocol.html. Cells from a 100 ml culture were collected by centrifugation for 5 minutes at 5,000 rpm at 4°C, washed in cold lysis buffer (20 mM Tris.HCl pH 8 / 1 mM EDTA / 500 mM NaCl, 0.1 % Triton X100 / 10 % glycerol), and lysed in 500 μl lysis buffer supplemented with 50 mM NaF, 10 μg/ml AEBSF and complete protease inhibitor tablet (Roche) using zirconia beads and a BioSpec 24 apparatus. Lysates were cleared by centrifugation 10 minutes at 13,000 rpm at 4°C, and supernatants were incubated with 200 μl of Glutathione Sepharose beads slurry (pre-washed in lysis buffer) for 1 hour at 4°C. Beads were washed 3 times in lysis buffer and proteins were eluted over 30 minues at 4°C in elution buffer (50 mM Tris pH 8 / 1 mM EDTA / 20 mM glutathione reduced / 40 % glycerol, supplemented with protease inhibitors), and aliquots were frozen in liquid nitrogen and stored at −80°C.
Oligonucleotide nicked substrate and exonuclease assay
The nicked, double stranded substrate described in Figure 2c was made by 5′-[32P] end-labeling of oligonucleotides F2: GACCTGGCACGTAGGACAGCATGGGATCTGGCCTG and F3 TGTTACACAGTGCTACAGACatggt (lower case designate a 3′ overhang) with PNK, and annealing to the unlabeled reverse complement, R1: GTCTGTAGCACTGTGTAACACAGGCCAGATCCCATGCTGTCCTACGTGCCAGGTC. Annealing was verified by migration on 1 x TBE / 12 % polyacrylamide gel. ~0.66 pmoles of double-stranded nicked substrate was incubated with ~0.13 pmoles of recombinant protein in nuclease buffer (25 mM MOPS pH 7 / 60 mM KCl / 0.2 % Triton X100 / 2 mM DTT / 50 mM MnCl2) for indicated length of time at 37 °C. Reactions were stopped by adding SDS to 0.3 % and proteinase K to 0.5 mg/ml and incubated for 15 minutes at 37 °C. Reactions were denaturated in 2 volumes of formamide loading buffer (95 % formamide / 10 mM EDTA / 0.005 % bromophenol blue / 0.01 % xylene cyanol) at 95 °C and separated on a 6 M urea / 12 % polyacryamide gel in 1 x TBE. Gels were exposed to a phosphor screen for imaging.
Footnotes
Author Information. The authors declare no competing financial interests.
References
- 1.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiology and Molecular Biology Reviews. 1999;63:349. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–774. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicolette ML, Lee K, Guo Z, Rani M, et al. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat Struct Mol Biol. 2010;17:1478–1485. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–994. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29:3358–3369. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hartsuiker E, Neale MJ, Carr AM. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell. 2009;33:117–123. doi: 10.1016/j.molcel.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, et al. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartsuiker E, Mizuno K, Molnar M, Kohli J, et al. Ctp1CtIP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol Cell Biol. 2009;29:1671–1681. doi: 10.1128/MCB.01182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milman N, Higuchi E, Smith GR. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol Cell Biol. 2009;29:5998–6005. doi: 10.1128/MCB.01127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nairz K, Klein F. mre11S--a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 1997;11:2272–2290. doi: 10.1101/gad.11.17.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothenberg M, Kohli J, Ludin K. Ctp1 and the MRN-complex are required for endonucleolytic Rec12 removal with release of a single class of oligonucleotides in fission yeast. PLoS Genet. 2009;5:e1000722. doi: 10.1371/journal.pgen.1000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsubouchi H, Ogawa H. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol Cell Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 15.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams RS, Moncalian G, Williams JS, Yamada Y, et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell. 2008;135:97–109. doi: 10.1016/j.cell.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodgson A, Terentyev Y, Johnson RA, Bishop-Bailey A, et al. Mre11 and Exo1 contribute to the initiation and processivity of resection at meiotic double-strand breaks made independently of Spo11. DNA Repair (Amst) 2010 doi: 10.1016/j.dnarep.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Keelagher RE, Cotton VE, Goldman AS, Borts RH. Separable roles for Exonuclease I in meiotic DNA double-strand break repair. DNA Repair (Amst) 2010 doi: 10.1016/j.dnarep.2010.09.024. [DOI] [PubMed] [Google Scholar]
- 19.Manfrini N, Guerini I, Citterio A, Lucchini G, Longhese MP. Processing of meiotic DNA double-strand breaks requires cyclin-dependent kinase and multiple nucleases. J Biol Chem. 2010;285:11628–11637. doi: 10.1074/jbc.M110.104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsubouchi H, Ogawa H. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:2221–2233. doi: 10.1091/mbc.11.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zakharyevich K, Ma Y, Tang S, Hwang PY, et al. Temporally and Biochemically Distinct Activities of Exo1 during Meiosis: Double-Strand Break Resection and Resolution of Double Holliday Junctions. Mol Cell. 2010;40:1001–1015. doi: 10.1016/j.molcel.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carballo JA, Johnson AL, Sedgwick SG, Cha RS. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell. 2008;132:758–770. doi: 10.1016/j.cell.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 23.Usui T, Ogawa H, Petrini JH. A DNA damage response pathway controlled by Tel1 and the Mre11 complex. Mol Cell. 2001;7:1255–1266. doi: 10.1016/s1097-2765(01)00270-2. [DOI] [PubMed] [Google Scholar]
- 24.Szankasi P, Smith GR. A DNA exonuclease induced during meiosis of Schizosaccharomyces pombe. J Biol Chem. 1992;267:3014–3023. [PubMed] [Google Scholar]
- 25.Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 26.Pan J, Sasaki M, Kniewel R, Murakami H, et al. A Hierarchical Combination of Factors Shapes the Genome-wide Topography of Yeast Meiotic Recombination Initiation. Cell. 2011;144:719–31. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jazayeri A, Balestrini A, Garner E, Haber JE, Costanzo V. Mre11-Rad50-Nbs1-dependent processing of DNA breaks generates oligonucleotides that stimulate ATM activity. EMBO J. 2008;27:1953–1962. doi: 10.1038/emboj.2008.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 29.Kim KP, Weiner BM, Zhang L, Jordan A, et al. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell. 2010;143:924–937. doi: 10.1016/j.cell.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.