Abstract
Background
Giardia duodenalis and Entamoeba spp. are among the most common intestinal human protozoan parasites worldwide and they are frequently reported in captive non-human primates (NHP). From a public health point of view, infected animals in zoos constitute a risk for animal caretakers and visitors. In this study we carried out the molecular identification of G. duodenalis and Entamoeba spp. from nine species of primates housed in the zoological garden of Rome, to better ascertain their occurrence and zoonotic potential.
Results
G. duodenalis was found only in Lemur catta (47.0%). Entamoeba spp. were detected in all species studied, with the exception of Eulemur macaco and Varecia rubra. The number of positive pools ranged from 5.9% in L. catta to 81.2% in Mandrillus sphinx; in Pan troglodytes the observed prevalence was 53.6%. A mixed Entamoeba-Giardia infection was recorded only in one sample of L. catta. All G. duodenalis isolates belonged to the zoonotic assemblage B, sub assemblage BIV. Three Entamoeba species were identified: E. hartmanni, E. coli and E. dispar.
Conclusions
Our results highlight the importance of regularly testing animals kept in zoos for the diagnosis of zoonotic parasites, in order to evaluate their pathogenic role in the housed animals and the zoonotic risk linked to their presence. A quick detection of the arrival of pathogens into the enclosures could also be a prerequisite to limit their spread into the structure via the introduction of specific control strategies. The need for molecular identification of some parasite species/genotype in order to better define the zoonotic risk is also highlighted.
Background
Protozoa are the most common parasites in captive non-human primates (NHP). Amongst others, Giardia duodenalis and Entamoeba spp. are frequently reported [1-3]; the simplicity of their monoxenous life cycle, the low infective dose and the short prepatent period facilitate their dispersal among captive animals once they have entered the enclosures (cages, pens, etc.) [4,5].
The relevance of Giardia and Entamoeba infections in zoo animals goes beyond their clinical effects. From a public health point of view, these protozoa have high zoonotic potential, being among the most common intestinal human parasites worldwide [6-8]. Infected NHP in zoos constitute a risk for animal caretakers [9,10] and possibly people visiting the zoological gardens. On the other hand, infected people could be the source of infection for the captive NHP via water and/or food contamination. Hence, the epidemiological relevance of gaining a better understanding of the transmission patterns of these pathogens in and from zoo facilities.
From a management point of view, new threats have arisen in the last few years due to the tendency to reproduce habitats as similar as possible to the natural ones. In particular, the removal of the cement as flooring and the use of distributing food mixed in the litter to elicit natural seeking behaviours, has facilitated faecal contamination and made disinfection of cages and pens very difficult, resulting in the creation of habitats suitable for parasite survival and transmission.
G. duodenalis is known to provoke gastrointestinal disorders in NHP [10,11]. This species includes at least seven genotypes or assemblages (A→G), assemblages A and B being detected both in humans and NHP [12,13]. Regarding the genus Entamoeba, in a recent study [14] six species (E. histolytica, E. dispar, E. moshkovskii, E. hartmanni, E. coli and E. polecki-like organisms) were recorded in captive NHP in Belgium and The Netherlands. The clinical relevance of Entamoeba spp. is more difficult to ascertain in NHP. Several species are considered non-pathogenic, while E. histolytica and its virulent variant E. nuttalli are known to provoke severe and sometimes lethal intestinal and extra-intestinal disorders in monkeys and apes [2,15-17]. Problems arise in E. histolytica diagnosis, because of the impossibility via conventional microscopy to distinguish between this species and the non-pathogenic E. dispar and E. moshkovskii. Hence, in the presence of a copro-parasitological diagnosis of amoebiasis, there is justification for using PCR to rule out infection with the pathogenic and potentially lethal species [18-21].
In the literature, there is no data regarding intestinal protozoa occurrence and molecular identification in NHP from Italian zoos. Following detection of G. duodenalis and Entamoeba spp. during routine copro-parasitological examinations, a study was carried out aimed at the molecular identification of G. duodenalis assemblages and Entamoeba species in NHP housed in the zoological gardens of Rome, to better define their presence and to understand their zoonotic potential.
Methods
Study site and Sampling
The Bioparco is one of the oldest zoological gardens in Europe, founded in 1911. It is located in the city centre of Rome (Italy), covering an area of 18 ha and housing about 1000 specimens belonging to almost 200 species comprising mammals, birds and reptiles. It was designed in accordance with the new concept of "Zoo without bars" to improve animal welfare. Animals live in large spaces with reconstruction of the natural habitats suitable for each species. To avoid contact with people, glass screens or ditches border animal cages and pens.
Nine species and 3 families of NHP were involved in this study, the families being: Lemuridae (prosimians), Cercopithecidae (Old Word monkeys) and Hominidae (apes) (Table 1). At the Bioparco, monkeys and apes are kept in monospecific groups of 2-98 individuals sharing the same pens. All NHP are housed in cages and pens littered with natural materials such as ground bark and they are fed mixing the food with the litter.
Table 1.
Number of NHP specimens per cage; number of pools of faeces tested microscopically and number of pooled positive samples for Entamoeba spp. and Giardia duodenalis.
| Scientific name | Common name | N° of specimens/cage | N° pool of faeces | Entamoeba sp. N°positive (%) | G. duodenalis N°positive (%) |
|---|---|---|---|---|---|
| Lemuridae | |||||
| Lemur catta | Ring-tailed lemur | 10 | 17 | 1 (5.9) | 8 (47.0) |
| Eulemur macaco | Black lemur | 3 | 4 | 0 | 0 |
| Varecia rubra | Red ruffed lemur | 3 | 5 | 0 | 0 |
| Cercopithecidae | |||||
| Cercocebus torquatus | Collared mangabey | 5 | 10 | 6 (60.0) | 0 |
| Chlorocebus aethiops | Vervet monkeys | 2 | 5 | 4 (80.0) | 0 |
| Macaca fuscata | Japanese macaque | 98 | 22 | 12 (54.5) | 0 |
| Mandrillus sphinx | Mandrill | 18 | 16 | 13 (81.2) | 0 |
| Hominidae | |||||
| Pan troglodytes | Common chimpanzee | 5 | 41* | 22 (53.6**) | 0 |
| Pongo pygmaeus | Bornean orangutan | 3 | 13 | 6 (46.1) | 0 |
| Total | 133 | 64 (48.1) | 8 (6.0) | ||
* Individual faecal samples
** Prevalence
Due to the group housing, copro-parasitological analysis was performed on pools of faeces collected from the litter, with the exception of P. troglodytes, where individual sampling was possible.
Coprological examination
All the samples were sent to the laboratory of Parasitology of the IZSLT for routine copro-parasitological diagnosis. Faecal samples were examined for parasitic protozoa cysts and/or trophozoites using the wet mount Lugol's iodine staining method and the formol ethyl-acetate concentration technique [22,23]. In case of ambiguous results obtained using microscopy, G. duodenalis infection was confirmed via a commercial immunofluorescence kit, (MERIFLUOR® Cryptosporidium/Giardia, Meridian Bioscience, Inc.).
Samples positive for G. duodenalis or Entamoeba spp. cysts were transferred to the University of Rome Tor Vergata for molecular characterization.
Molecular analysis and sequencing
Genomic DNA was extracted from faecal samples using the QIAamp DNA Stool Mini Kit (Qiagen, Italy) following manufacturer's instructions. Molecular analysis was carried out by amplifying an 18S rRNA genus fragment both for G. duodenalis and Entamoeba spp.
For Giardia, a nested PCR procedure was performed to amplify a 130 bp region using the primers RH4 and RH11 [24] for the first step and the primers GiarR and GiarF in the second amplification round [25].
The amplification of the specific fragment of Entamoeba spp. was obtained using the primers JVC and DSPR2 [26], which are also able to detect the virulent variant of E. histolytica, E. nuttalli, observed in NHP [17]. The amplicons were from 622 to 667 bp long depending on the species.
To confirm Giardia 18S rRNA genetic identity of the samples, an additional analysis was performed by sequencing the triose phosphate isomerase (tpi) and glutamate dehydrogenase (gdh) loci. To amplify the tpi fragment, a nested PCR procedure was used to obtain a 530 bp region using the primers AL3543 and AL3546 for the first step and AL3544 and AL3545 for the second one [27]. For gdh, a semi-nested PCR was carried out to amplify a 432 bp fragment with the primers GDHeF and GDHiR in the primary reaction, and GDHiF and GDHiR in the secondary [28]. In all PCR reactions, positive and negative controls were added.
Amplicons were visualized by electrophoresis on SYBR Safe DNA-stained 1% agarose gel (Invitrogen). Bands of the predicted sizes were excised and DNA was purified with the NucleoSpin Extract II purification kit (Macherey-Nagel, GmbH & Co. KG, Germany) and sequenced in both directions by Bio-Fab Research (Italy).
Sequences were edited with FinchTV 1.4 Software (Geospiza, Inc, Seattle, WA) by the analysis of chromatograms. Consensus sequences were obtained using ClustalW2 Multiple Sequence Alignments and queried against known sequences of GenBank database using BLAST. Molecular characterization of Entamoeba spp. and G. duodenalis isolates was based on both sequence comparison and phenetic analysis. Phenetic analyses were conducted with the MEGA (version 4.0) software. A distance-based analysis was carried out using the maximum composite likelihood method, and trees were constructed by the Neighbour Joining (NJ) algorithm. Bootstrap values were calculated by analysing 500 replicates.
Results
Between January 2010 and March 2011, 133 faecal samples were analysed microscopically for parasites (Table 1). G. duodenalis was found only in L. catta, in 47% of tested pooled samples. Entamoeba spp. were detected in all the considered species, except for E. macaco and V. rubra; number of positive pooled samples ranged from 5.9% in L. catta to 81.2% in Mandrillus sphinx. In P. troglodytes the observed prevalence (individual sampling) was 53.6%.
A mixed Entamoeba-Giardia infection was recorded only in one sample of L. catta. All animals showed no symptoms.
Molecular analysis
Amplification of the 18S-rRNA, gdh and tpi G. duodenalis genes
Of the eight Giardia positive isolates from L. catta tested at the 18S-rRNA, gdh and tpi loci, all three genes were successfully amplified and sequenced from 4 isolates, while one or two genes were amplified from the other four. The combined results of the multilocus analyses are summarized in Table 2.
Table 2.
Summary of multilocus genotyping results of Giardia duodenalis samples from Lemur catta at the level of assemblage and sub-assemblage.
| Isolate code | 18S-rRNA | gdh | tpi |
|---|---|---|---|
| L1 | B | B(IV) | B(IV) |
| L2 | B | B(IV) | B(IV) |
| L8A | B | B(IV) | B(IV) |
| L8B | B | B(IV) | B(IV) |
| L8C | B | - | B(IV) |
| L8D | B | - | - |
| L10A | B | - | - |
| L10B | B | - | - |
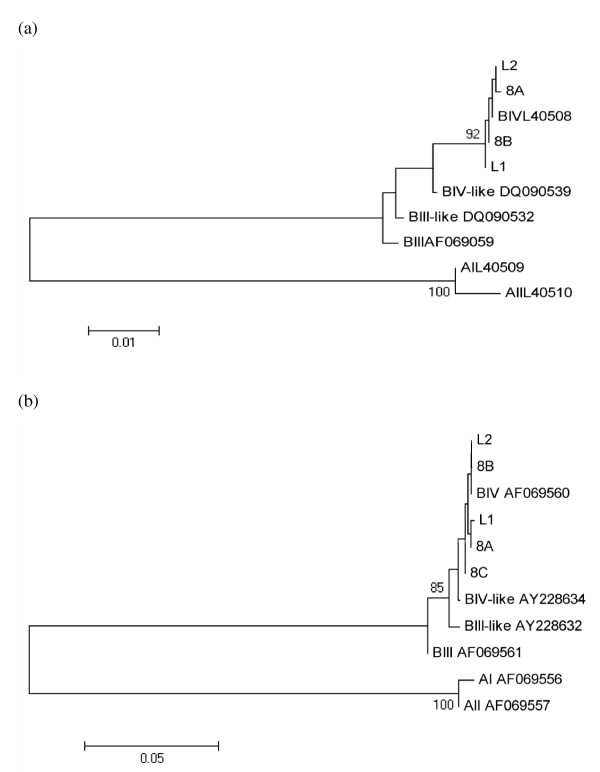
Sequences obtained were identified at the 18S-rRNA locus as assemblage B, by comparing to GenBank sequences of Giardia genotypes (Accession Numbers: AF199446 (assemblage A), AF199447 (assemblage B), AY775200 (assemblage C), AY775199 (assemblage D), AY297957 (assemblage E), AF199444 (assemblage F) and AF199450 (assemblage G). The bootstrap consensus trees for both gdh and tpi genes obtained by NJ method yielded one monophyletic group corresponding to the assemblage B, sub assemblage BIV which included Giardia sequences from L. catta (Figure 1). Mixed assemblages, showing double peaks at the diagnostic positions at the three loci in the chromatograms, were not observed. Consensus trees obtained by the phenetic analysis confirmed this result.
Figure 1.
Phenetic relationships of G. duodenalis inferred by NJ analysis of the gdh (a) and tpi (b) loci. Only bootstrap values > 70 are indicated. The Accession Numbers utilized for gdh were L40509 (AI), L40510 (AII), AF069059 (BIII), DQ090539 (BIII-like), L40508 (BIV), DQ090532 (BIV-like) and those ones for tpi were AF069556 (AI), AF069557 (AII), AF069561 (BIII), AY228632 (BIII-like), AF069560 (BIV), AY228634 (BIV-like).
Amplification of the 18S-rRNA Entamoeba genes
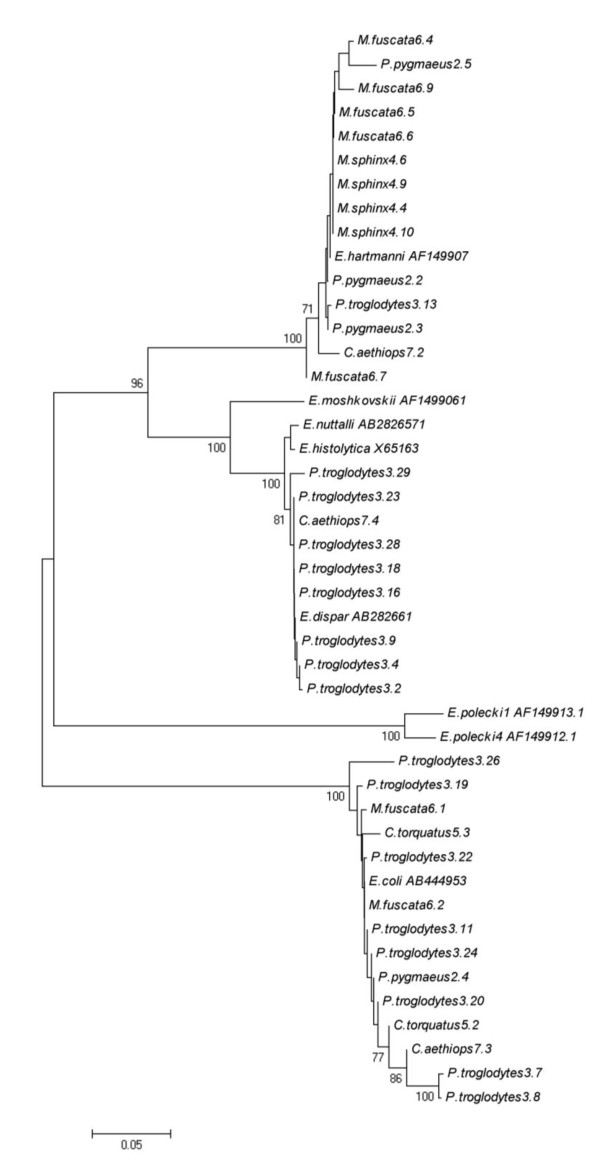
The results revealed the presence of Entamoeba DNA in 45 out of 64 samples. In total three Entamoeba species were identified: E. hartmanni (31.1%), E. coli (31.1%) and E. dispar (20.0%) (Table 3). The assignment of the Entamoeba species within the hosts is reported in the constructed phenetic tree (Figure 2). No mixed infections were detected. Non-interpretable sequences were obtained from 8 of the amplified samples, but showed homology to Entamoeba spp. For 4 samples, DNA amplification was unsuccessful; for 15 sequences homology was found to DNA sequences from plants, ascomycetes and zygomycetes (Malus domestica, Pyrus communis, Sordaria fimicola, Neurospora crassa, Helicostylum pulchrum, Sporodiniella umbellata).
Table 3.
Summary of molecular identification of Entamoeba species.
| Host species | E. hartmanni N°positive (%*) | E. dispar N°positive (%*) | E. coli N°positive (%*) | Entamoeba sp. N°positive (%*) |
|---|---|---|---|---|
| Cercocebus torquatus | 0 | 0 | 2 (66.7) | 1 (33.3) |
| Chlorocebus aethiops | 1 (33.3) | 1 (33.3) | 1 (33.3) | 0 |
| Macaca fuscata | 5 (62.5) | 0 | 2 (25.0) | 1 (12.5) |
| Mandrillus sphinx | 4 (80.0) | 0 | 0 | 1 (20.0) |
| Pan troglodytes | 1 (4.5) | 8 (36.3) | 8 (36.3) | 5 (22.7) |
| Pongo pygmaeus | 3 (75.0) | 0 | 1 (25.0) | 0 |
| Total | 14 (31.1) | 9 (20.0) | 14 (31.1) | 8 (17.8) |
* Calculated as follows: number of positive samples for the species/number of Entamoeba positive samples at molecular analysis
Figure 2.
Phenetic relationships of Entamoeba spp. inferred by NJ analysis of the 18S rRNA genus. Only bootstrap values > 70 are indicated. The Accession Numbers utilized for the identification of Entamoeba spp. were AB282657 (E. nuttalli), X65163 (E. histolytica), AB282661 (E. dispar), AB444953 (E. coli), AF149907 (E. hartmanni), AF149906 (E. moshkoskii), AF149913 (E. polecki-like variant 1) and AF149912 (E. polecki-like variant 4).
Discussion
To the best of our knowledge, these are the first data regarding G. duodenalis and Entamoeba spp. in NHP housed in a zoological garden in Italy. Of particular concern are the levels of infection recorded (47% for G. duodenalis in L. catta and up to 81.2% for Entamoeba spp. in M. sphinx) and the report of zoonotic assemblages and species for both taxa. These findings appear comparable with previous data available in the literature. Regarding Entamoeba spp., Levecke et al. [2] reported values of prevalence among Old World monkeys ranging from 30% and 100% and among apes ranging from 0% and 100%, with higher values observed in M. sphinx and P. pygmaeus. Lower levels of infection, with overall values ranging from 29.3% for monoinfections to 51.9% for mixed infections, were recorded by Levecke et al. [14] in 36 species of NHP housed in nine zoos in Belgium and The Netherlands.
With respect to G. duodenalis, its report from captive L. catta confirms previous studies, where values of prevalence as high as 94% were recorded [2,13]. Interestingly, Villers et al. [29], in a comparative study on intestinal parasites of wild and captive L. catta, found this parasite only in the captive ones. Also, lack of symptoms in L. catta infected with Giardia is a recorded occurrence [29,30]. From a molecular point of view, the identification of assemblage B confirms the report of Beck et al. [30] in L. catta from Zagreb zoological garden and of previous data about this parasite from NHP [31-33]. Zoonotic assemblages A and B have also been described in NHP by Graczyk et al. [34] and Vitazkova and Wade [12].
Captive animals in zoos are exposed to various stresses and pathogens, resulting in an increased probability of becoming infected with parasites and of developing high parasite burdens [35]. Exposure to strict non-natural contact with specimens of the same as well as of other species, humans included, enhances their probability of becoming infected with zoonotic agents [30]. Moreover, questions arise about the route of entry of these pathogens in a theoretically isolated enclosure. Identification of transmission routes is always difficult [30], and only a hypothesis can be postulated: monoxenous protozoa following the faecal-oral transmission route like G. duodenalis and Entamoeba spp. could arrive in a zoo via contaminated vegetables fed to the animals, via water supplies, carried (even mechanically) by free ranging mammals and birds entering cages and pens and with the intake of new animals arriving from other zoos. In the present study, the presence of Giardia only in L. catta could be related to the recent (May 2007) admission of the colony of Ring-tailed lemur in the Bioparco, probably including infected but asymptomatic specimens. The hypothesis of a single and confined intake event is supported by the molecular data obtained by the multi-locus sequencing analysis, never recording mixed Giardia assemblage infections. On the basis of the different nature of the genes (multicopy/single copy), a different amplification rate at the three loci is frequently reported in the literature. However, despite regarding only 5 isolates of the 8 obtained, all of them were identified as assemblage B, sub-assemblage BIV, in contrast to other studies where isolates from NHP were identified as BIII, BIII-like and/or BIV-like at either the gdh or the tpi gene [32,36]. The lack of symptoms in the animals made the intake of the parasite easier, despite the quarantine regulations adopted in the zoo. More difficult to hypothesize is the route of entry of Entamoeba spp., as these parasites were widespread among many different host species of NHP and their intake could have occurred a long time ago, giving rise to their establishment in the zoological garden and subsequent spread to many susceptible host species.
Regarding molecular analysis of Entamoeba isolates, discrepancy between microscopy and genetic analysis results could be due to both inhibitory problems of PCR and to low sensitivity/specificity of microscopy [14]. As expected, since all animals were asymptomatic, E. histolytica and the virulent variant observed in NHP, E. nuttalli, were not detected. The other species, E. hartmanni, E. dispar and E. coli, are usually recovered and considered commensals.
Mixed infections are frequently reported in the literature. For G. duodenalis, Levecke et al. [13] showed mixed assemblages in 32.7% of NHP samples, including Ring-tailed lemurs. Also for Entamoeba spp., in a later study by Levecke et al., [14], most of NHP samples (51.9%) carried more than one species. The absence of mixed infections in our results could be related to the molecular approach used in the study, since more sensitive methods have been recently suggested [37,14]. However, concerning G. duodenalis, our data could likely be related to the low number of positive samples and/or to the arrival of this parasite with L. catta in a single intake event.
Regardless of their pathogenic potential for humans, the presence of Entamoeba and Giardia raises questions about the risk linked to zoo keeping operations in primate enclosures. In particular, all parasites found in the present study (E. dispar, E. coli, E. hartmanni and, especially, G. duodenalis assemblage B sub-assemblage BIV) warrant the maximum attention regarding the possibility of transmission among animals and their caretakers and can be considered potentially zoonotic pathogens. Zoo animals infected with zoonotic parasites also pose a problem concerning habitat contamination with cysts and eggs potentially able to infect people or other animals, as demonstrated by the detection of Giardia cysts in water bodies in and close to a zoological garden in Malaysia [35].
It appears very difficult to control such a parasite once it gets into the structure. New systems in the pens' flooring and feeding modalities make the washing and disinfection of pens and cages more difficult and facilitate the parasite life cycle, with continuous re-infections of the housed animals. In the present study, repeated treatments of L. catta specimens were ineffective in erradicating the infection from the colony, with animals testing positive even immediately after treatments. In general, the pharmacological control of protozoa appears arduous, and, as a matter of fact, their presence in captive NHP, in the absence of clinical symptoms, is often accepted and they are managed rather than controlled.
The presence of parasites with direct life cycles in zoos raises many management problems, linked to the difficulty of preventing cyst/eggs transport from one enclosure to the other. The absence of Giardia positive samples in the examined species other than L. catta, allows us to suppose that the parasite was actually confined to the Ring-tailed lemur colony and that the prophylaxis measures implemented until now by the zoo veterinarians (disinfection of animal-keepers shoes, use of disposable gloves and instruments, etc.) were effective in avoiding the spreading of the infection to other cages and pens.
Finally, this study suggests the need to improve copro-parasitological diagnosis with molecular analysis, aimed at distinguishing among pathogenic/non pathogenic and zoonotic/non zoonotic species and assemblages and to improve sensitivity of tests carried out on animals kept in quarantine before their intake into zoo facilities, thus preventing the arrival of pathogens in these particular kinds of confined habitats.
Conclusions
Our results highlight the need for regularly testing of animals kept in zoo facilities for the diagnosis of zoonotic parasites, in order to point out their eventual arrival in the enclosures and evaluate their pathogenic role in the housed animals and the zoonotic risk linked to their arrival. A quick detection of the arrival of pathogens into the enclosures could also be a prerequisite to limit their spread in the structure via the implementation of specific control strategies and could permit the identification of intake routes, thus allowing the introduction of specific prophylactic measures. The need for molecular identification of some parasite species/genotype in order to better define the risk linked to their presence in the zoo animals is also highlighted.
List of abbreviations used
NHP: Non-human primates; IZSLT: Istituto Zooprofilattico Sperimentale delle Regioni Lazio e Toscana.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
FB designed and carried out molecular analysis and wrote the paper. CP carried out molecular analysis and drafted the manuscript. KGF and PDC carried out sample collection, contributed and commented on the paper. DDC drafted and commented on the manuscript. CDL designed sampling collection, carried out parasitological examinations and wrote the paper. All authors approved the final version of the manuscript.
Contributor Information
Federica Berrilli, Email: berrilli@uniroma2.it.
Cristina Prisco, Email: cristinaprisco@gmail.com.
Klaus G Friedrich, Email: klaus.friedrich@bioparco.it.
Pilar Di Cerbo, Email: pilar.dicerbo@libero.it.
David Di Cave, Email: dicave@uniroma2.it.
Claudio De Liberato, Email: claudio.deliberato@izslt.it.
References
- Munene E, Otsyula M, Mbaabu DAN, Mutahi WT, Muriuki SMK, Muchemi GM. Helminth and protozoan gastrointestinal tract parasites in captive and wild-trapped African non-human primates. Vet Parasitol. 1998;78:195–201. doi: 10.1016/s0304-4017(98)00143-5. [DOI] [PubMed] [Google Scholar]
- Levecke B, Dorny P, Geurden T, Vercammen F, Vercruysse J. Gastrointestinal protozoa in non-human primates of four zoological gardens in Belgium. Vet Parasitol. 2007;148:236–246. doi: 10.1016/j.vetpar.2007.06.020. [DOI] [PubMed] [Google Scholar]
- Pérez Cordòn G, Hitos Prados A, Romero D, Sànchez Moreno M, Pontes A, Osuna A, Rosales MJ. Intestinal parasitism in the animals of the zoological garden "Peña Escrita" (Almuñecar, Spain) Vet Parasitol. 2008;156:302–309. doi: 10.1016/j.vetpar.2008.05.023. [DOI] [PubMed] [Google Scholar]
- Tanyuksel M, Petri WA. Laboratory diagnosis of amebiasis. Clin Microbiol Rev. 2003;16:713–729. doi: 10.1128/CMR.16.4.713-729.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RCA, Monis PT. Variation in Giardia: implications for taxonomy and epidemiology. Adv Parasitol. 2004;58:69–137. doi: 10.1016/S0065-308X(04)58002-8. [DOI] [PubMed] [Google Scholar]
- O'Donoghue PJ. Cryptosporidium and cryptosporidiosis in man and animal. Int J Parasitol. 1995;25:139–195. doi: 10.1016/0020-7519(94)e0059-v. [DOI] [PubMed] [Google Scholar]
- Thompson RCA. Giardiasis as a re-emerging infectious disease and its zoonotic potential. Int J Parasitol. 2000;30:1259–1267. doi: 10.1016/s0020-7519(00)00127-2. [DOI] [PubMed] [Google Scholar]
- Stauffer W, Ravdin IJ. Entamoeba histolytica: an update. Cur Opin Infect Dis. 2003;16:479–485. doi: 10.1097/00001432-200310000-00016. [DOI] [PubMed] [Google Scholar]
- Miller RA, Brondson MA, Kuller L, Morton WR. Clinical and parasitologic aspects of cryptosporidiosis in nonhuman primates. Lab Anim Sci. 1990;40:42–46. [PubMed] [Google Scholar]
- Hamlen HJ, Lawrence JM. Giardiasis in laboratory-housed squirrel monkeys: a retrospective study. Lab Anim Sci. 1994;44:235–239. [PubMed] [Google Scholar]
- Kalishman J, Paul-Murphy J, Scheffer J, Thomson JA. Survey of Cryptosporidium and Giardia spp. in a captive population of common marmosets. Lab Anim Sci. 1996;46:116–119. [PubMed] [Google Scholar]
- Vitazkova SK, Wade SE. Parasites of free-ranging black howler monkeys (Alouatta pigra) from Belize and Mexico. Am J Primatol. 2006;68:1089–1097. doi: 10.1002/ajp.20309. [DOI] [PubMed] [Google Scholar]
- Levecke B, Geldhof P, Claerebout E, Dorny P, Vercammen F, Cacciò SM, Vercruysse J, Geurden T. Molecular characterization of Giardia duodenalis in captive non-human primates reveals mixed assemblages A and B infections and novel polymorphisms. Int J Parasitol. 2009;39:1595–1601. doi: 10.1016/j.ijpara.2009.05.013. [DOI] [PubMed] [Google Scholar]
- Levecke B, Dressen L, Dorny P, Verweij JJ, Vercammen F, Casaert S, Vecruysse J, Geldhof P. Molecular identification of Entamoeba spp. in captive nonhuman primates. J Clin Microbiol. 2010;48:2988–2990. doi: 10.1128/JCM.00013-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matz-Rensing K, Floto A, Kaup FJ, Schaller K. Gastric amebiasis due to Entamoeba histolytica infection in a mandrill (Mandrillus sphinx) Book of Abstracts of the 5th Scientific Meeting of the European Association of Zoo- and Wildlife Veterinarians, Ebeltoft, Denmark, 19-23 May 2004.
- Rivera WL, Yason JADL, Adao DEV. Entamoeba histolytica and E. dispar infections in captive macaques (Macaca fascicularis) in the Philippines. Primates. 2010;51:69–74. doi: 10.1007/s10329-009-0174-x. [DOI] [PubMed] [Google Scholar]
- Tachibana H, Yanagi T, Pandey K, Cheng XJ, Kobayashi S, Sherchand JB, Kanbara H. An Entamoeba sp. strain isolated from rhesus monkey is virulent but genetically different from Entamoeba histolytica. Mol Biochem Parasitol. 2007;153:107–114. doi: 10.1016/j.molbiopara.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Rivera WL, Tachibana H, Silva-Tahat MRA, Uemura H, Kanbara H. Differentiation of Entamoeba histolytica and E. dispar DNA from cysts present in stool specimens by polymerase chain reaction: its field application in the Philippines. Parasitol Res. 1996;82:585–589. doi: 10.1007/s004360050169. [DOI] [PubMed] [Google Scholar]
- Rivera WL, Tachibana H, Kanbara H. Field study on the distribution of Entamoeba histolytica and Entamoeba dispar in the northern Philippines as detected by the polymerase chain reaction. Am J Trop Med Hyg. 1998;59:916–921. doi: 10.4269/ajtmh.1998.59.916. [DOI] [PubMed] [Google Scholar]
- Tachibana H, Cheng XJ, Kobayashi S, Fujita Y, Udono T. Entamoeba dispar, but not E. histolytica, detected in a colony of chimpanzees in Japan. Parasitol Res. 2000;86:537–541. doi: 10.1007/s004360000205. [DOI] [PubMed] [Google Scholar]
- Takano J, Narita T, Tachibana H, Shimizu T, Komatsubara H, Terao H, Kujimoto K. Entamoeba histolytica and Entamoeba dispar infections in cynomolgus monkeys imported into Japan for research. Parasitol Res. 2005;97:255–257. doi: 10.1007/s00436-005-1415-6. [DOI] [PubMed] [Google Scholar]
- Ritchie LS. An ether sedimentation technique for routine stool examination. Bull US Army Med Dep. 1948;8:326. [PubMed] [Google Scholar]
- Young KH, Bullock SL, Melvin DM, Spruill CL. Ethyl acetate as a substitute for diethyl ether in the formalin-ether sedimentation technique. J Clin Microbiol. 1979;10:852–853. doi: 10.1128/jcm.10.6.852-853.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins RM, Meloni BP, Groth DM, Wetherall JD, Reynoldson JA, Thompson RC. Ribosomal RNA sequencing reveals differences between the genotypes of Giardia isolates recovered from humans and dogs living in the same locality. J Parasitol. 1997;83:44–51. [PubMed] [Google Scholar]
- Read C, Walters J, Robertson ID, Thompson RC. Correlation between genotype of Giardia duodenalis and diarrhoea. Int J Parasitol. 2002;32:229–231. doi: 10.1016/s0020-7519(01)00340-x. [DOI] [PubMed] [Google Scholar]
- Santos HL, Bandea R, Martins LA, de Macedo HW, Peralta RH, Peralta JM, Ndubuisi MI, da Silva AJ. Differential identification of Entamoeba spp. based on the analysis of 18S rRNA. Parasitol Res. 2010;106:883–888. doi: 10.1007/s00436-010-1728-y. [DOI] [PubMed] [Google Scholar]
- Sulaiman IM, Fayer R, Bern C, Gilman RH, Trout JM, Schantz PM, Das P, Lal AA, Xiao L. Triose phosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg Infect Dis. 2003;9:1444–1452. doi: 10.3201/eid0911.030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read CM, Monis PT, Thompson RC. Discrimination of all genotypes of Giardia duodenalis at the glutamate dehydrogenase locus using PCR-RFLP. Infect Genet Evol. 2004;4:125–130. doi: 10.1016/j.meegid.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Villers LM, Jang SS, Lent CL, Lewin-Koh S, Norosoarinaivo J. Survey and comparison of major intestinal flora in captive and wild ring-tailed lemur (Lemur catta) populations. Am J Primatol. 2008;70:175–184. doi: 10.1002/ajp.20482. [DOI] [PubMed] [Google Scholar]
- Beck R, Sprong H, Bata I, Lucinger S, Pozio E, Cacciò SM. Prevalence and molecular typing of Giardia spp. in captive mammals at the zoo of Zagreb, Croatia. Vet Parasitol. 2011;185:40–46. doi: 10.1016/j.vetpar.2010.09.026. [DOI] [PubMed] [Google Scholar]
- Smith HV, Cacciò SM, Cook N, Nichols RAB, Tait A. Cryptosporidium and Giardia as foodborne zoonoses. Vet Parasitol. 2007;149:29–40. doi: 10.1016/j.vetpar.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Cacciò SM, Ryan U. Molecular epidemiology of giardiosis. Mol Biochem Parasitol. 2008;160:75–80. doi: 10.1016/j.molbiopara.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Cacciò SM, Beck R, Lalle M, Marinculic A, Pozio E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int J Parasitol. 2008;38:1523–1531. doi: 10.1016/j.ijpara.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Graczyk TK, Bosco-Nizeyi J, Ssebide B, Thompson RC, Read C, Cranfield MR. Anthropozoonotic Giardia duodenalis genotype (assemblage) A infections in habitats of free-ranging human-habituated gorillas, Uganda. J Parasitol. 2002;88:905–909. doi: 10.1645/0022-3395(2002)088[0905:AGDGAA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Lim YA, Lai MM, Mahdy MAK, Mat Naim HR, Smith HV. Molecular detection of Giardia contamination in water bodies in a zoo. Environ Res. 2009;109:857–859. doi: 10.1016/j.envres.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Itagaki T, Kinoshita S, Aoki M, Itoh N, Saeki H, Sato N, Uetsuki J, Izumiyama S, Yagita K, Endo T. Genotyping of Giardia intestinalis from domestic and wild animals in Japan using glutamete dehydrogenase gene sequencing. Vet Parasitol. 2005;133:283–287. doi: 10.1016/j.vetpar.2005.05.061. [DOI] [PubMed] [Google Scholar]
- Ortega-Pierres G, Smith HV, Cacciò SM, Thompson RC. New tools provide further insights into Giardia and Cryptosporidium biology. Trends Parasitol. 2009;25:410–416. doi: 10.1016/j.pt.2009.06.002. [DOI] [PubMed] [Google Scholar]