Abstract
The AML1 (RUNX1) gene, one of the most frequent targets of translocations associated with human leukemias, encodes a DNA-binding protein that plays pivotal roles in myeloid differentiation through transcriptional regulation of various genes. Previously, we reported that AML1 is phosphorylated on two serine residues with dependence on activation of extracellular signal-regulated kinase, which positively regulates the transcriptional activity of AML1. Here, we demonstrate that the interaction between AML1 and the corepressor mSin3A is regulated by phosphorylation of AML1 and that release of AML1 from mSin3A induced by phosphorylation activates its transcriptional activity. Furthermore, phosphorylation of AML1 regulates its intranuclear location and disrupts colocalization of AML1 with mSin3A in the nuclear matrix. PEBP2β/CBFβ, a heterodimeric partner of AML1, was shown to play a role in protecting AML1 from proteasome-mediated degradation. We show that mSin3A also protects AML1 from proteasome-mediated degradation and that phosphorylation-induced release of AML1 from mSin3A results in degradation of AML1 in a time-dependent manner. This study provides a novel regulatory mechanism for the function of transcription factors mediated by protein modification and interaction with cofactors.
AML1 (RUNX1) was first identified on chromosome 21 as the gene that is disrupted in the (8;21)(q22;q22) translocation, which is one of the most frequent chromosome abnormalities associated with acute myelogenous leukemia (AML) (20, 24, 29). AML1 is also disrupted in t(3;21)(q26;q22), which is found in the blastic crisis phase of chronic myelogenous leukemia (23). It was also reported that the AML1 gene is rearranged in acute lymphoblastic leukemia carrying t(12;21)(p12;q22) and that this translocation generates the TEL-AML1 fusion protein (5, 34). Furthermore, point mutations in the Runt domain of the AML1 gene were found in patients with AML (31, 33), familial platelet disorder with predisposition to AML (35), or myelodysplastic syndrome (9). These findings suggest that the structural alterations of AML1 caused by translocations or point mutations trigger leukemic transformation of hematopoietic cells.
AML1 regulates the transcription of various genes that are important in hematopoiesis (27, 36, 37, 48). Furthermore, it has been revealed that AML1 can cause neoplastic transformation when overexpressed in fibroblasts, suggesting a potential role for AML1 in promoting cellular proliferation (17). From analyses of mice lacking AML1, it was shown that AML1 plays an important role in liver-derived hematopoiesis and angiogenesis (30, 38).
Previously, the regulatory mode of AML1 functions through signal transduction pathways was investigated, and it was revealed that AML1 is phosphorylated with dependence on the activation of extracellular signal-regulated kinase (ERK) (40). ERK-dependent phosphorylation enhances the transcriptional activity of AML1, and mutations of the phosphorylation sites reduce the transforming capacity of AML1 in fibroblasts (40). These results indicate that the functions of AML1 are potentially regulated by ERK-induced phosphorylation, which is activated by cytokine and growth factor stimuli. However, the mechanism by which the transcriptional ability of AML1 is activated by phosphorylation has not been clear.
A growing number of proteins have been shown to interact with AML1 to modify its function (2, 10, 12, 19, 21, 28, 44). Here, we have investigated whether these protein-protein interactions are affected by phosphorylation of AML1, and we found that the corepressor mSin3A is released from phosphorylated AML1, whereby the transcriptional activity of phosphorylated AML1 is activated. Furthermore, we demonstrated that mSin3A is a key regulator for the intranuclear localization and stability of AML1. This study reveals a previously unappreciated role of the corepressor mSin3A as a regulator of AML1.
MATERIALS AND METHODS
Plasmid construction.
The pME-AML1, pME-PEBP2β, pCMV-MK, and Tww-tk-Luc reporter plasmids were constructed as described previously (41). The AML1 mutants S249/266A and S249/266E were obtained by replacing the serine residues with alanines or glutamic acids by the site-directed mutagenesis method for AML1 cDNA (15). FLAG-tagged AML1 constructs were generated as described previously (10). The deletion mutant of S249/266A, Δ(181-210)SA, was created by PCR from S249/266A with the insertion of a BglII site to join the fragments. The carboxy-terminal deletion mutant, AML1 1-288, was generated as described previously (17).
Cell culture and DNA transfection.
COS-7 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS). HEL cells were cultured in RPMI 1640 supplemented with 10% FCS. P19 mouse embryonal carcinoma cells were cultured as described previously (40). COS-7 and P19 cells were transfected with expression plasmids using SuperFect Transfection Reagent (Qiagen Inc.). Mouse embryonic fibroblasts from normal and PEBP2β-deficient mice (26) were transfected with expression plasmids by using Effectene Transfection Reagent (Qiagen Inc.).
Immunoprecipitation and Western blotting.
COS-7 cells were cultured for 30 to 35 h after transfection in DMEM containing 10% FCS and then transferred to DMEM containing 0.1% FCS and incubated for 12 to 15 h. They were then either not treated or treated for 5 min with 10% FCS plus 100 ng of recombinant human epidermal growth factor (EGF) (Sigma) per ml and harvested in phosphate-buffered saline supplemented with 1 mM EDTA, 1.5 mg of iodoacetamide/ml, 0.2 mM phenylmethylsulfonyl fluoride, 0.1 trypsin inhibitory unit/ml of aprotinin, 25 mM β-glycerophosphate, and 0.5% Triton X-100. These cell lysates were precleared with protein G-Sepharose (Pharmacia), mixed with the anti-mSin3A antibody K-20 (Santa Cruz), and rotated for 2 h; this was followed by recovery of mSin3A on protein G-Sepharose beads. The beads were washed four times with the lysis buffer. Immunoprecipitates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting with the anti-FLAG M2 monoclonal antibody (Sigma).
HEL cells (107) were harvested in 1% Triton lysis buffer (10 mM Tris-HCl, pH 7.4, 5 mM EDTA, 150 mM NaCl, 1% Triton-X, 10% glycerol, 10 U of aprotinin/ml, 2 mM phenylmethylsulfonyl fluoride, 25 mM β-glycerophosphate), and the cell lysates were precleared with protein G-Sepharose. The same amount of cell lysates was mixed with rabbit polyclonal immunoglobulin G (IgG) (Santa Cruz) as a control or with anti-AML1 antibody (Ab-1; Oncogene), and rotated for 12 h; this was followed by recovery of AML1 on protein G-Sepharose beads. The beads were washed four times with the lysis buffer. Immunoprecipitates were subjected to SDS-PAGE and Western blotting with anti-mSin3A antibody.
We lysed the PEBP2β-deficient fibroblasts with 1% Triton lysis buffer, and the cell lysates were subjected to SDS-PAGE and Western blotting with anti-FLAG M2 monoclonal antibody, anti-PEBP2β antibody, or anti-actin monoclonal antibody (Chemicon International). The anti-PEBP2β antibody was prepared as described elsewhere (39). To block proteolysis, lactacystin (10 mM) dissolved in dimethyl sulfoxide was added to the cell culture medium overnight.
Chromatin and nuclear matrix fractions were prepared by sequential extraction with cytoskeleton buffer and digestion buffer followed by 0.25 M ammonium sulfate extraction in a biochemical-fractionation assay as described previously (47). To detect lamin B1 localized in the nuclear matrix, we used anti-lamin B1 antibody (M-20) (Santa Cruz).
Transcriptional-response assays.
Luciferase assays were performed as described previously (8). Briefly, reporter and expression plasmids were transfected into P19 or COS-7 cells by a SuperFect protocol. P19 cells were harvested and subjected to the luciferase assay after 30 to 36 h of transfection. COS-7 cells were starved in DMEM containing 0.1% FCS with or without lactacystin, harvested, and subjected to the luciferase assay after 12 h of transfection. The data were normalized using the internal control of transfection efficiency, as described previously (8).
Immunofluorescence and in situ nuclear matrix isolation.
COS-7 cells transfected with expression plasmids were fixed in 3.7% formaldehyde in phosphate-buffered saline. They were treated with the anti-FLAG and anti-mSin3A antibodies and then incubated with Texas-red-conjugated donkey anti-mouse IgG and fluorescein isothiocyanate-conjugated donkey anti-rabbit IgG (Jackson Immuno Research Laboratories, Inc.), respectively, as secondary antibodies, as described previously (8, 16). In situ nuclear matrices were prepared as described elsewhere (46, 47).
RESULTS
ERK-mediated phosphorylation disrupts the interaction of AML1 with mSin3A.
It has been demonstrated that AML1 is phosphorylated on serines 249 and 266 by ERK and that phosphorylation of AML1 activates the transcriptional activity (40). To elucidate the mechanism for phosphorylation-induced activation of AML1, we analyzed the interaction of AML1 with other proteins when it is phosphorylated.
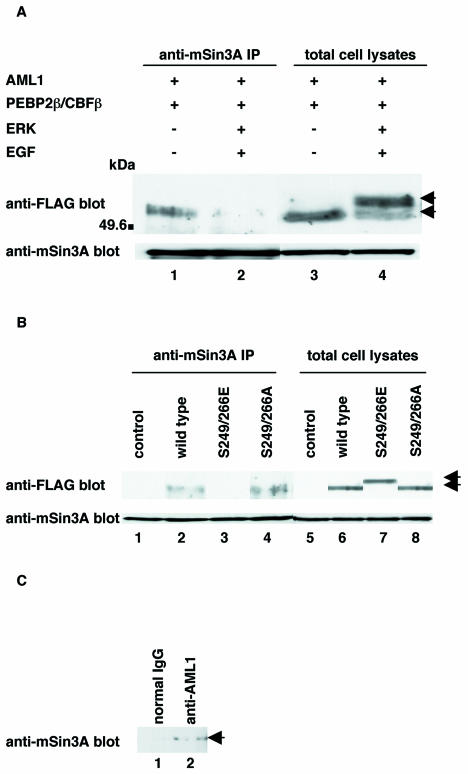
It has been shown that AML1 associates with the mSin3A corepressor (21). Consistent with these observations, we found that FLAG-tagged AML1 is coimmunoprecipitated with mSin3A by the anti-mSin3A antibody when it is overexpressed together with PEBP2β in COS-7 cells (Fig. 1A, lane 1). When COS-7 cells were cotransfected with FLAG-tagged AML1 and PEBP2β plus ERK and stimulated with EGF, we observed shifted bands of AML1 mirroring its phosphorylation (40) (Fig. 1A, lane 4). Significantly, the amount of AML1 coimmunoprecipitated with mSin3A was markedly reduced upon phosphorylation (Fig. 1A, lane 2). The total amount of AML1 was constant regardless of overexpression of ERK combined with EGF stimulation (Fig. 1A, lanes 3 and 4). These results suggest that AML1 dissociates from mSin3A when it is phosphorylated by ERK.
FIG. 1.
Analyses of the interaction between phosphorylated AML1 and mSin3A. (A) COS-7 cells were transfected with pME-FLAG-AML1 and pME-PEBP2β, together with pRc/CMV (lanes 1 and 3) or pCMVMK (lanes 2 and 4), starved in medium containing 0.1% FCS, treated for 5 min with 100 ng of EGF per ml plus 10% FCS (lanes 2 and 4) or left untreated (lanes 1 and 3), and harvested with lysis buffer. The cell lysates were precleared with protein G-Sepharose, mixed with anti-mSin3A antibody, and rotated for 2 h at 4°C. Then, mSin3A was recovered on protein G-Sepharose beads. The washed beads were subjected to SDS-PAGE, followed by Western blotting with anti-FLAG or anti-mSin3A antibody. Expression of each protein was monitored by using 60 μg of total cell lysates. +, present; −, absent; IP, immunoprecipitate; arrows, phosphorylated and unphosphorylated AML1. (B) COS-7 cells were transfected with pME18S, pME-FLAG-AML1, pME-FLAG-S249/266E, or S249/266A with pME-PEBP2β and harvested with lysis buffer. The cell lysates were precleared with protein G-Sepharose, mixed with anti-mSin3A antibody, and rotated for 2 h at 4°C. Then, mSin3A was recovered on protein G-Sepharose beads. The washed beads were subjected to SDS-PAGE, followed by Western blotting with the anti-FLAG or anti-mSin3A antibody. Expression of each protein was monitored by using 60 μg of total cell lysates. Arrows indicate wild-type and mutant AML1. (C) HEL cells were harvested with lysis buffer, and the cell lysates were precleared with protein G-Sepharose. The same amount of cell lysates was mixed with anti-AML1 antibody (Ab-1) or rabbit polyclonal IgG and rotated for 12 h; this was followed by recovery of AML1 on protein G-Sepharose beads. The washed beads were subjected to SDS-PAGE and Western blotting with anti-mSin3A antibody. Arrow, AML1.
The previous study showed that glutamic acid mimics the negative charge of phosphate and that some glutamic acid mutants of serine act like a phosphorylated form of the cognate wild-type proteins (11, 22). To confirm that ERK-mediated phosphorylation can abrogate the ability of AML1 to interact with mSin3A, we generated the mutant of AML1 that mimics phosphorylated AML1, in which serines 249 and 266 of AML1 were changed to glutamic acids (S249/266E). SDS-PAGE and Western blotting revealed that S249/266E showed a mobility shift similar to that of phosphorylated AML1 when expressed in COS-7 cells (Fig. 1B, lane 7). As shown in Fig. 1B, we found by using a coimmunoprecipitation assay that S249/266E had completely lost the ability to interact with mSin3A, although wild-type AML1 was readily coimmunoprecipitated with mSin3A. It had already been shown that the mutant in which serines 249 and 266 were changed to alanines (S249/266A) is not phosphorylated even upon ERK activation (40). In contrast to S249/266E, S249/266A was coimmunoprecipitated with mSin3A as efficiently as wild-type AML1 (Fig. 1B, lane 4). In these experiments, the amounts of wild-type AML1, S249/266E, and S249/266A were almost the same when cotransfected with PEBP2β, which is known to protect AML1 from proteolytic degradation (Fig. 1B, lanes 6 to 8). These results suggest that phosphorylation disrupts the ability of AML1 to heterodimerize with mSin3A.
To confirm that AML1 and mSin3A form a complex in hematopoietic cells, we investigated the interactions between endogenous AML1 and mSin3A in HEL cells, a human erythroleukemia-derived cell line. Endogenous mSin3A is not immunoprecipitated by normal rabbit polyclonal IgG in HEL cells (Fig. 1C, lane 1). On the other hand, the precipitate by the anti-AML1 antibody (Ab-1) contains endogenous mSin3A (Fig. 1C, lane 2). In the previous study, the anti-AML1 antibody (Ab-1) was shown to immunoprecipitate endogenous AML1 in HEL cells (21). These results indicate that mSin3A binds to AML1 in hematopoietic cells.
Transcriptional activity of unphosphorylated AML1 is recovered by addition of HDAC inhibitor.
In a previous study, it was shown that ERK-dependent phoshphorylation of AML1 can potentiate its transcriptional ability (40). However, the identities of regulatory proteins that repress the transcriptional activity of unphosphorylated AML1 have not been well characterized. Given the interaction between AML1 and mSin3A, it might be that mSin3A plays an important role in the repression of AML1 transcriptional activity. Histone deacetylases (HDACs) that are recruited by mSin3A mediate transcriptional repression by rendering the nearby chromatin inaccessible to transcriptional activators through deacetylation of histone proteins (32, 42). Trichostatin A (TSA), a specific HDAC inhibitor, has been shown to relieve transcriptional repression by AML1, suggesting a role for mSin3A-HDAC in the repressor activity of AML1 (21). Therefore, we investigated whether unphosphorylated AML1 can be derepressed when mSin3A-HDAC is inhibited by TSA in a transcriptional-response assay.
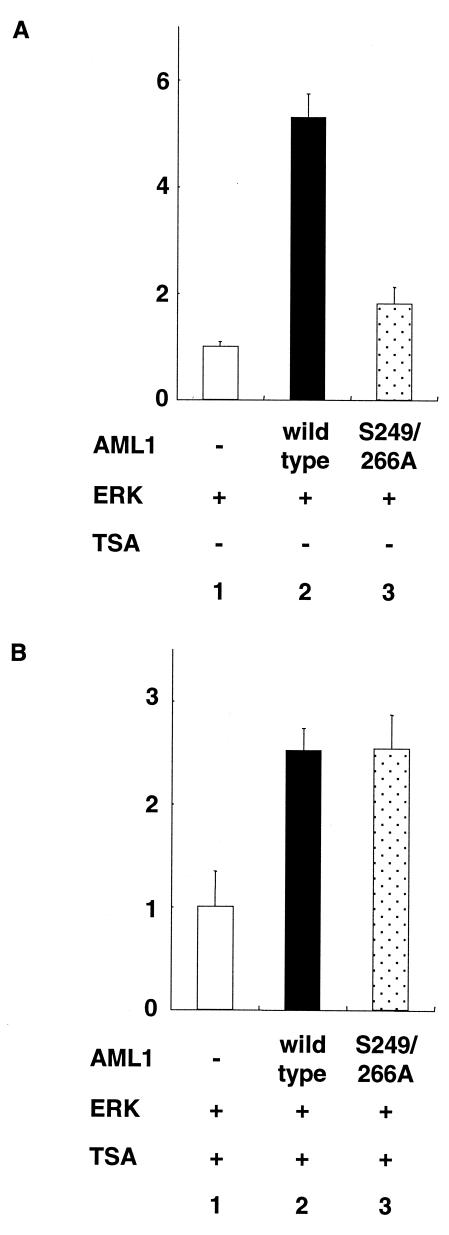
When AML1 and ERK were cotransfected into P19 cells with the Tww-tk-Luc reporter that can be activated by AML1, we observed a fivefold induction of transcriptional activity compared to overexpression of ERK alone (Fig. 2A, lane 2). On the other hand, when S249/266A was coexpressed with ERK, only 1.5-fold induction of the transcriptional activity was observed (Fig. 2A, lane 3), which is consistent with our previous report. Upon treatment with TSA, both wild-type AML1 and S249/266A cotransfected with ERK induced 2.5-fold transcriptional activity compared to overexpression of ERK alone, and the reduced transcriptional activity of S249/266A in the absence of TSA was restored to a level similar to that of wild-type AML1 (Fig. 2B, lanes 2 and 3). These results suggest that the transcriptional activity of unphosphorylated AML1 is suppressed by the interaction with mSin3A-HDAC.
FIG. 2.
Transcriptional response assays of AML1 and its unphosphorylated mutant in the presence of TSA. P19 cells were cotransfected with 500 ng of Tww-tk-Luc, 200 ng of pCMVMK, and 20 ng of pME18S AML1, −), pME-AML1 (AML1, wild type), or pME-S249/266A as indicated with (B) or without (A) treatment with TSA. The cells were cultured for 30 to 36 h after transfection and harvested. The means and standard deviations of the luciferase activities of AML1 and S249/266A in two independent transfections are shown. We used the mean of the luciferase activities of the expression vector (pME18S) in two independent transfections as the control. Similar results were obtained in four additional independent transfections.
Intranuclear targeting of AML1 is induced by its phosphorylation.
The previous study showed that AML1 is localized in the nuclear matrix, a subnuclear organization, and that a nuclear-matrix-targeting sequence has been identified within its carboxyl-terminal domain (47). Furthermore, it was shown that mSin3A is also located in the nuclear matrix, accompanied by HDACs, and represses transcription there (43). Given that the physical interaction between AML1 and mSin3A is disrupted by phosphorylation of AML1, we investigated whether targeting of AML1 to the nuclear matrix could be affected upon phosphorylation.
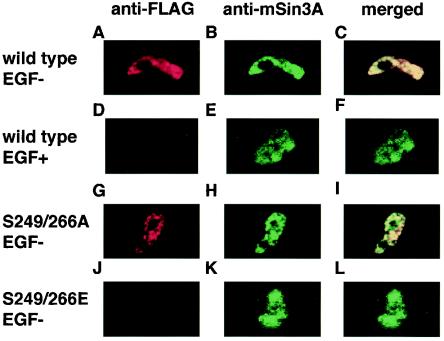
FLAG-tagged AML1, PEBP2β, and ERK were coexpressed in COS-7 cells, and the cells were cultured with or without EGF. They were stained with the anti-FLAG and anti-mSin3A antibodies. Both AML1 and endogenous mSin3A were localized predominantly in the nucleus regardless of EGF stimulation (data not shown). The AML1 mutants, S249/266A and S249/266E, are also located in the nucleus (data not shown). Next, we analyzed the cells in in situ nuclear-matrix preparations (46, 47). Staining the nuclear matrix with anti-FLAG and anti-mSin3A antibodies revealed that AML1 and mSin3A were colocalized in the nuclear matrix without EGF stimulation (Fig. 3A to C). When we stimulated the transfected cells with EGF, mSin3A was detected in the nuclear matrix (Fig. 3E). Under these conditions, however, AML1 was absent from the nuclear matrix, where no colocalization signal of AML1 and mSin3A was detected (Fig. 3D and F). Subsequently, we investigated the intranuclear localization of S249/266A and S249/266E. As shown in Fig. 3, S249/266A and mSin3A were colocalized in the nuclear matrix (Fig. 3G to I), whereas S249/266E was neither detected in the nuclear matrix nor colocalized with mSin3A (Fig. 3J to L). These results indicate that AML1 and mSin3A are colocalized in the nuclear matrix when AML1 is not phosphorylated and that AML1 translocates from the nuclear matrix upon phosphorylation.
FIG. 3.
Intranuclear translocation of AML1 induced by its phosphorylation. (A to F) COS-7 cells were transfected with FLAG-tagged wild-type AML1, together with ERK and PEBP2β, and incubated in the absence (A to C) or the presence (D to F) of EGF. The cells were treated with in situ nuclear-matrix preparations, and immunofluorescent staining was performed using the anti-FLAG antibody (A and D), the anti-mSin3A antibody (B and E), or both (C and F). (G to L) COS-7 cells were cotransfected with FLAG-tagged S249/266A (G to I) or S249/266E (J to L) with PEBP2β. The cells were treated with in situ nuclear matrix preparations, and immunofluorescent staining was performed using the anti-FLAG antibody (G and J), the anti-mSin3A antibody (H and K), or both (I and L).
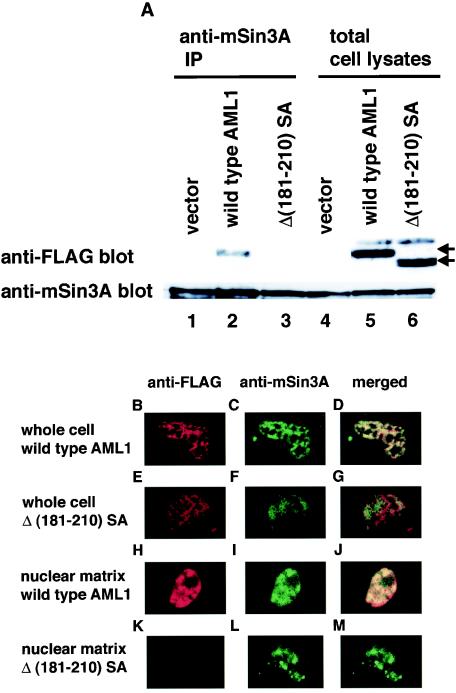
A specific domain that interacts with mSin3A has been mapped to residues 181 to 210 within AML1 (21). To manifest a correlation between mSin3A binding and nuclear localization of AML1, we tested the intranuclear distribution of the AML1 deletion mutant that lacks the domain interacting with mSin3A. To rule out the possibility of phosphorylation-induced release of mSin3A from the mutant, we changed serines 249 and 266 to alanines in AML1Δ(181-210), which generates the double mutant, Δ(181-210)SA. We introduced FLAG-tagged Δ(181-210)SA, together with PEBP2β, into COS-7 cells and assessed the interaction with mSin3A by a coimmunoprecipitation assay. The expression levels of wild-type AML1 and Δ(181-210)SA were almost the same when they were overexpressed with PEBP2β (Fig. 4A, lanes 5 and 6). Consistent with the previous report, Δ(181-210)SA was hardly coimmunoprecipitated with mSin3A by the anti-mSin3A antibody, in contrast to wild-type AML1 (Fig. 4A, lanes 2 and 3). Next, we confirmed the intranuclear localization of Δ(181-210)SA. FLAG-tagged wild-type AML1 or Δ(181-210) SA was introduced into COS-7 cells, and the cells were stained with the anti-FLAG and anti-mSin3A antibodies. Both forms of AML1 were visualized exclusively in the nucleus by whole-cell staining (Fig. 4B and E). Wild-type AML1 was consistently colocalized with mSin3A, whereas Δ(181-210)SA showed altered nuclear distribution, which does not coincide with that of mSin3A (Fig. 4D and G). In in situ nuclear matrix preparations, wild-type AML1 and mSin3A were colocalized in the nuclear matrix (Fig. 4H to J). In contrast, Δ(181-210)SA was not detected in the nuclear matrix, where mSin3A was invariably located (Fig. 4K to M). The data we obtained indicate that targeting of AML1 to the nuclear matrix is dependent on the interaction with mSin3A, which suggests that AML1 is tethered to the nuclear matrix by mSin3A. These results are consistent with our findings that phosphorylation-induced release from mSin3A translocates AML1 from the nuclear matrix.
FIG. 4.
The AML1 mutant lacking the interaction domain with mSin3A does not colocalize with mSin3A in the nuclear matrix. (A) COS-7 cells were cotransfected with pME18S, pME-FLAG-AML1, or pME-FLAG-Δ(181-210)SA with pME-PEBP2β and harvested. The cell lysates were precleared with protein G-Sepharose, mixed with anti-mSin3A antibody, and rotated for 2 h at 4°C. Then, mSin3A was recovered on protein G-Sepharose beads. The washed beads were subjected to SDS-PAGE, followed by Western blotting with anti-FLAG or anti-mSin3A antibody. Expression of each protein was monitored by using 60 μg of total cell lysates. Arrows indicate the wild type and mutant of AML1. (B to M) COS-7 cells were cotransfected with FLAG-tagged wild-type AML1 (B to D and H to J) or Δ(181-210)SA (E to G and K to M) with PEBP2β. The cells were treated with whole-cell (B to G) or in situ nuclear-matrix (H to M) preparations, and immunofluorescent staining was performed using the anti-FLAG antibody (B, E, H, and K), the anti-mSin3A antibody (C, F, I, and L), or both (D, G, J, and M).
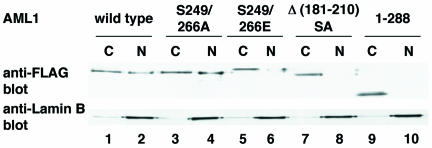
To substantiate our finding that the intranuclear targeting of AML1 is induced by its phosphorylation, we performed a biochemical-fractionation assay (Fig. 5). Wild-type AML1 and S249/266A are detected in the fractions of chromatin and the nuclear matrix, whereas S249/266E is present exclusively in the chromatin fraction. These findings are consistent with the results showing that AML1 translocates from the nuclear matrix upon phosphorylation in in situ immunofluorescence analyses. Furthermore, Δ(181-210)SA is also undetectable in the nuclear-matrix fraction, again suggesting the requirement for mSin3A interaction in nuclear-matrix targeting of AML1
FIG. 5.
Biochemical-fractionation assay of wild-type AML1 and AML1 mutants in COS-7 cells. COS-7 cells were transfected with pME-FLAG-AML1 (lanes 1 and 2), pME-FLAG-S249/266A (lanes 3 and 4), pME-FLAG-S249/266E (lanes 5 and 6), pME-FLAG-Δ(181-210)SA (lanes 7 and 8), or pME-FLAG-AML1 1-288 (lanes 9 and 10). Extract of the chromatin (15%) (lanes 1, 3, 5, 7, and 9) or 5% extract of the nuclear-matrix (lanes 2, 4, 6, 8, and 10) fraction was loaded in each lane. Western blot analyses were performed using the anti-FLAG and the anti-lamin B1 antibodies. C, chromatin; N, nuclear matrix.
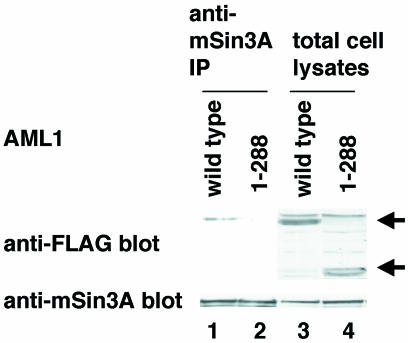
Next, we investigated whether a mutation in the nuclear-matrix attachment site of AML1 affects interaction with mSin3A. It was shown that the carboxy terminus of AML1 contains a segment essential for association with the nuclear matrix (47). Therefore, we generated a carboxy-terminal deletion mutant, AML1 1-288, and overexpressed it in COS-7 cells. In biochemical-fractionation assays, AML1 1-288 is shown to be released from the nuclear matrix (Fig. 5, lane 10). As shown in Fig. 6, AML1 1-288 has lost the ability to interact with mSin3A. These results suggest that the nuclear sublocalization of AML1 plays an important role in interaction with mSin3A.
FIG. 6.
Effect of a mutation in the nuclear-matrix attachment site of AML1 on interaction with mSin3A. COS-7 cells were transfected with pME-FLAG-AML1 (lane 1) or pME-FLAG-AML1 1-288 (lane 2) and harvested. The cell lysates were precleared with protein G-Sepharose, mixed with anti-mSin3A antibody, and rotated for 2 h at 4°C. Then, mSin3A was recovered on protein G-Sepharose beads. The washed beads were subjected to SDS-PAGE, followed by Western blotting with anti-FLAG or anti-mSin3A antibody. Expression of each protein was confirmed by using 30 μg of total lysates. Arrows, wild-type and mutant AML1.
Stability of phosphorylated AML1 proteins in COS-7 cells.
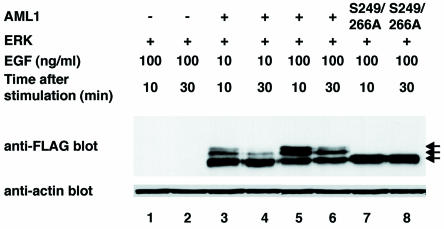
AML1 is continuously subjected to proteolytic degradation mediated by the ubiquitin-proteasome pathway, which is blocked by heterodimerization with PEBP2β (7). mSin3A is also known to block proteasome-mediated degradation of proteins, such as p53 (49). Given the phosphorylation-dependent interaction of AML1 with mSin3A, we examined the stability of phosphorylated AML1 to elucidate a potential role of mSin3A in proteasome-mediated degradation of AML1. COS-7 cells were transfected with FLAG-tagged wild-type AML1 or S249/266A, together with ERK, and then treated for 5 min with graded concentrations of EGF and harvested 10 or 30 min after the removal of EGF. In these experiments, we did not cotransfect PEBP2β into COS-7 cells to eliminate the effect of PEBP2β overexpression on protection of AML1 (7). The cells were cultured in the presence of phosphatase inhibitors to protect AML1 from dephosphorylation. The cell lysates were subjected to SDS-PAGE and Western blotting with the anti-FLAG antibody. As shown in Fig. 7, slowly migrating bands of wild-type AML1, which correspond to phosphorylated forms, were observed 10 min after the removal of EGF in an EGF dose-dependent manner (lanes 3 and 5). After 30 min, the amounts of unphosphorylated AML1 remained unchanged (lanes 4 and 6). Furthermore, S249/266A showed constant levels of expression, regardless of the incubation periods after EGF removal (lanes 7 and 8). These results indicate that unphosphorylated AML1 is stable upon EGF stimulation. In contrast, expression of the phosphorylated forms was markedly diminished at both concentrations of EGF, even in the presence of phosphatase inhibitors (lanes 4 and 6). These data suggest that ERK-dependent phosphorylation promotes proteasome-mediated degradation of AML1.
FIG. 7.
Phosphorylated AML1 is degraded in a time-dependent manner in COS-7 cells. COS-7 cells were cotransfected with pME18S (lanes 1 and 2), pME-FLAG-AML1 (lanes 3 to 6), or pME-FLAG-S249/266A (lanes 7 and 8), together with pCMVMK; starved in medium containing 0.1% FCS; and treated for 5 min with 10 (lanes 3 and 4) or 100 (lanes 1, 2, and 5 to 8) ng of EGF per ml plus 10% FCS in the presence of 50 mM sodium fluoride as a phosphatase inhibitor. The cells were harvested after 10 (lanes 1, 3, 5, and 7) or 30 (lanes 2, 4, 6, and 8) min of EGF stimulation, and 30 μg of total cell lysates was subjected to SDS-PAGE, followed by Western blotting with the anti-FLAG or antiactin antibody. +, present; −, absent; arrows, phosphorylated and unphosphorylated AML1.
Analyses of the stability of AML1 in PEBP2β-deficient fibroblasts.
To determine more explicitly a role for phosphorylation-dependent mSin3A interaction in the degradation of AML1, we established fibroblasts from PEBP2β-deficient embryos, which allowed us to eliminate the effect of endogenous PEBP2β on the protection of AML1 (7).
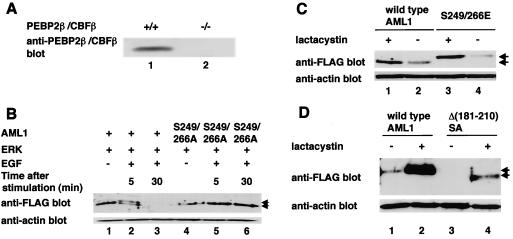
In embryonic fibroblasts from normal mice, we detected endogenous PEBP2β (Fig. 8A, lane 1). On the other hand, PEBP2β was not detected in PEBP2β-deficient fibroblasts (Fig. 8A, lane 2). First, we examined whether AML1 would undergo ERK-mediated phosphorylation and subsequent proteolytic degradation in these cells. We cotransfected wild-type AML1 and ERK into PEBP2β-deficient fibroblasts and stimulated the cells with EGF for 5 min. The cell lysates were analyzed by Western blotting. After 5 min of EGF stimulation, both phosphorylated and unphosphorylated AML1 were observed by Western blotting (Fig. 8B, lane 2). However, when we harvested the transfected cells after 30 min of EGF stimulation, there was a marked reduction in the amount of AML1 such that both forms of AML1 were barely detected (Fig. 8B, lane 3). When S249/266A was transfected into these cells together with ERK, followed by stimulation with EGF, neither the mobility shift nor the decrease in the amount of S249/266A was observed, regardless of EGF stimulation (Fig. 8B, lanes 4 to 6). These results suggest that AML1 is phosphorylated upon the activation of ERK in PEBP2β-deficient fibroblasts and that phosphorylated AML1 is degraded with the lapse of time after EGF stimulation.
FIG. 8.
Phosphorylated AML1 and the AML1 mutants lacking interaction with mSin3A are degraded by proteasome in PEBP2β-deficient fibroblasts. (A) Embryonic fibroblasts from normal mice (lane 1) or PEBP2β-deficient mice (lane 2) were lysed, and 30 μg of cell lysates were subjected to SDS-PAGE and Western blotting with anti-PEBP2β antibody. (B) PEBP2β-deficient fibroblasts were cotransfected with (+) pME-FLAG-AML1 (lanes 1 to 3) or pME-FLAG-S249/266A (lanes 4 to 6), together with pCMVMK. The cells were starved in medium containing 0.1% FCS overnight and treated for 5 min with 100 ng of EGF per ml plus 10% FCS (lanes 2, 3, 5, and 6) or left untreated (lanes 1 and 4). They were harvested, and 30 μg of cell lysates was subjected to SDS-PAGE and Western blotting with anti-FLAG or anti-actin antibody. The EGF-treated cells were harvested after 5 (lanes 2 and 5) or 30 (lanes 3 and 6) min of EGF stimulation. Arrows, phosphorylated and unphosphorylated AML1. (C) PEBP2β-deficient fibroblasts were transfected with pME-FLAG-AML1 or pME-FLAG-S249/266E. The cells were treated with 10 μM lactacystin (lanes 1 and 3) or left untreated (lanes 2 and 4) overnight and harvested; 30 μg of cell lysates was subjected to SDS-PAGE and Western blotting with anti-FLAG or anti-actin antibody. (D) PEBP2β-deficient fibroblasts were transfected with pME-FLAG-AML1 or pME-FLAG-Δ(181-210)SA. The cells were treated with 10 μM lactacystin (lanes 2 and 4) or left untreated (lanes 1 and 3) overnight and harvested; 30 μg of cell lysates was subjected to SDS-PAGE and Western blotting with anti-FLAG or anti-actin antibody. Arrows in panels C and D indicate wild-type and mutant AML1.
Next, we tested the stability of the AML1 mutant that mimics the phosphorylated form in these cells. Wild-type AML1 and S249/266E were expressed at equivalent levels in PEBP2β-deficient fibroblasts when transfected in the presence of lactacystin, which strongly inhibits proteasome-mediated protein degradation (Fig. 8C, lanes 1 and 3). It is expected that AML1 is liable to undergo degradation in PEBP2β-deficient cells without proteolysis inhibition. In support of this, expression of wild-type AML1 and S249/266E noticeably decreased without lactacystin treatment (Fig. 8C, lanes 2 and 4). Furthermore, degradation induced by lactacystin removal was more prominent in S249/266E than in wild-type AML1, which was not distinct in COS-7 cells (Fig. 8C, lanes 2 and 4). These results suggest that phosphorylation-dependent degradation of AML1 is facilitated in PEBP2β-deficient cells.
Recapitulating phosphorylation-dependent degradation of AML1 in PEBP2β-deficient cells, we subsequently tested whether mSin3A can protect AML1 from degradation in these cells. We transfected wild-type AML1 or Δ(181-210)SA into the cells without EGF stimulation. As shown in Fig. 8D, expression of Δ(181-210)SA was much lower than that of wild-type AML1 regardless of lactacystin treatment in PEBP2β-deficient fibroblasts. These results suggest that mSin3A plays a key role in protecting AML1 from proteasome-mediated degradation and provide us with a potential mechanism for phosphorylation-induced degradation of AML1.
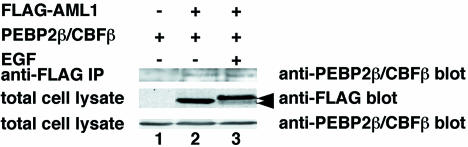
To address the relative contributions of PEBP2β to the stability of AML1 when AML1 is phosphorylated, we investigated whether the interaction between AML1 and PEBP2β would change when AML1 was phosphorylated. We transfected FLAG-tagged AML1 together with ERK and PEBP2β into COS-7 cells. When we stimulated the cells with EGF, we observed the shifted band of AML1 corresponding to the phosphorylated AML1 (Fig. 9, lane 3). When we immunoprecipitated FLAG-tagged AML1 with the anti-FLAG antibody, constant amounts of PEBP2β were coimmunoprecipitated regardless of EGF stimulation (Fig. 9, lanes 2 and 3). These results indicate that the interaction between AML1 and PEBP2β does not change when AML1 is phosphorylated and that the degradation of phosphorylated AML1 is not due to loss of interaction with PEBP2β. These results argue that association with mSin3A, in addition to PEBP2β, is required for AML1 to exist stably in cells and that loss of interaction with mSin3A plays an important role in the degradation of phosphorylated AML1.
FIG. 9.
Analyses of the interaction between phosphorylated AML1 and PEBP2β. COS-7 cells were transfected with pME18S (lane 1) or pME-FLAG-AML1 (lanes 2 and 3), together with pCMV-MK and pME-PEBP2β. The cells were treated for 5 min with EGF (lane 3) or left untreated (lanes 1 and 2) and harvested with lysis buffer. The cell lysates were precleared with protein G-Sepharose, mixed with anti-FLAG antibody, and rotated for 2 h at 4°C. Then, FLAG-AML1 was recovered on protein G-Sepharose beads. The washed beads were subjected to SDS-PAGE, followed by Western blotting with anti-PEBP2β antibody. Expression of each protein was confirmed by using 60 μg of total cell lysates. Arrows, phosphorylated and unphosphorylated AML1.
Transcriptional activities of S249/266E and Δ(181-210)SA mutants of AML1.
We found that S249/266E and Δ(181-210)SA are dissociated from the corepressor mSin3A. These results suggest that the transcriptional activities of these mutants are higher than those of wild-type AML1. Therefore, we tested the transcriptional activities of these mutants in transcriptional-response assays.
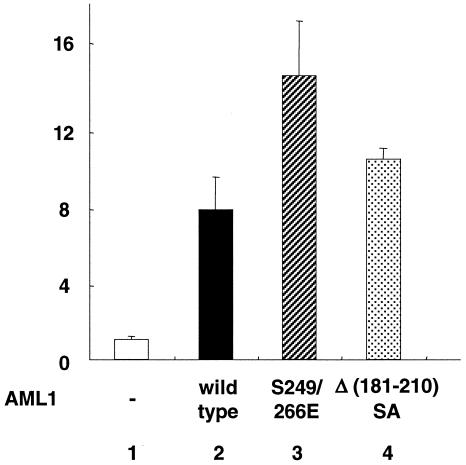
When we cotransfected wild-type AML1, S249/266E, or Δ(181-210)SA into P19 cells in the presence of 10% FCS with the Tww-tk-Luc reporter, there was no significant difference between the transcriptional activities of wild-type AML1 and those of the mutants (data not shown). We showed the loss of stability of S249/266E and Δ(181-210)SA (Fig. 8C and D), and these results may explain the decrease in the transcriptional activities of the mutants. Furthermore, when transfected into COS-7 cells in the presence of 10% FCS, a significant proportion of wild-type AML1 is phosphorylated without EGF stimulation (data not shown). We therefore cotransfected wild-type AML1, S249/266E, or Δ(181-210)SA into COS-7 cells with Tww-tk-Luc reporter and created the cells with 0.1% FCS to eliminate the effect of FCS. In these experiments, S249/266E and Δ(181-210)SA showed higher transcriptional activities than wild-type AML1 (Fig. 10, lanes 3 and 4). These results are consistent with our findings that the mutants lose interaction with the corepressor mSin3A. Compared with S249/266E, Δ(181-210)SA showed reduced transcriptional activities. As shown in Fig. 8C and D, the expression of Δ(181-210)SA is lower than that of wild-type AML1 regardless of lactacystin treatment, although wild-type AML1 and S249/266E were expressed at equivalent levels in the presence of lactacystin. These results suggest that the stability of Δ(181-210)SA is lower than that of S249/266E, and this may explain the reduction in the transcriptional activity of Δ(181-210)SA.
FIG. 10.
Transcriptional activities of S249/266E and Δ(181-210)SA mutants of AML1. COS-7 cells were cotransfected with 500 ng of Tww-tk-Luc and 200 ng of pME18S (AML1, −), pME-FLAG-AML1 AML1, wild type), pME-FLAG-S249/266E, or pME-FLAG-Δ(181-210)SA as indicated. The cells were cultured in DMEM containing 0.1% FCS for 12 h after transfection and harvested. The means and standard deviations of the luciferase activities of AML1 and the mutants in two independent transfections are shown. We used the mean of the luciferase activities of the expression vector (pME18S) in two independent transfections as the control.
Transcriptional activity of AML1 following EGF stimulation.
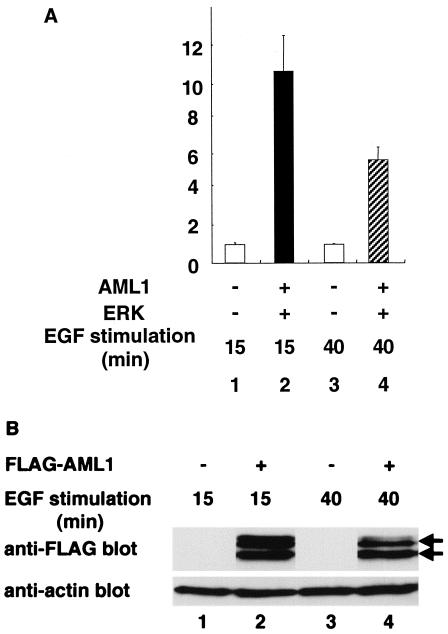
In the present study, we demonstrated that phosphorylated AML1 is prone to proteasome-mediated degradation. Thus, AML1 should exhibit decreased activity in transcriptional assays upon prolonged EGF stimulation. To confirm this, we investigated the change in transcriptional activity of AML1 following EGF stimulation. When we overexpressed AML1 and ERK with the Tww-tk-Luc reporter in COS-7 cells and harvested the cells after stimulation with 100 ng of EGF per ml for 15 min, we observed 10-fold induction of transcriptional activity (Fig. 11A, lane 2). On the other hand, we observed only fivefold induction of transcriptional activity when we harvested the cells after 40 min of stimulation by EGF (Fig. 11A, lane 4). After 15 min of stimulation by EGF, we observed both phosphorylated (Fig. 11B, lane 2) and unphosphorylated (Fig. 11B, lane 2) forms of AML1 in Western blotting. After 40 min of stimulation by EGF, however, we observed a marked reduction in the amount of phosphorylated AML1 (Fig. 11B, lane 4). These results support the idea that degradation may cause decreased transcriptional activity of phosphorylated AML1 with prolonged EGF stimulation.
FIG. 11.
Transcriptional activity of AML1 following EGF stimulation. (A) COS-7 cells were cotransfected with 500 ng of Tww-tk-Luc and 400 ng of pME18S (lanes 1 and 3) or 200 ng of pCMV-MK and pME-FLAG-AML1 (lanes 2 and 4). The cells were cultured in DMEM containing 0.1% FCS for 12 h after transfection. We harvested the cells after incubation in medium containing 100 ng of EGF per ml for 15 (lanes 1 and 2) or 40 (lanes 3 and 4) min. The means and standard deviations of the luciferase activities of AML1 in two independent transfections are shown. We used the mean of luciferase activities of the expression vector (pME18S) in two independent transfections as the control. (B) COS-7 cells were cotransfected with 500 ng of Tww-tk-Luc and 400 ng of pME18S (lanes 1 and 3) or 200 ng of pCMV-MK and pME-FLAG-AML1 (lanes 2 and 4). The cells were cultured in DMEM containing 0.1% FCS for 12 h after transfection. We harvested the cells after incubation in medium containing 100 ng of EGF per ml for 15 (lanes 1 and 2) or 40 (lanes 3 and 4) min; 30 μg of total cell lysates of the transfected cells was subjected to SDS-PAGE, followed by Western blotting with anti-FLAG or antiactin antibody. Arrows, phosphorylated and unphosphorylated AML1.
DISCUSSION
Phosphorylation-dependent regulation of AML1 functions is mediated by the interaction between AML1 and mSin3A.
Transcriptional repression is regulated by several different mechanisms (4, 6). One of them involves the recruitment of corepressor complexes, many of which contain subunits that possess HDACs, to the target genes (32). HDACs act to deacetylate histones and thus convert chromatin into a repressive state (1). In the mSin3A-HDAC complex, mSin3A acts as a linker to transcription factors, such as Mad, p53, TEL, and Ikaros (14, 18, 25). The previous study showed that AML1 also interacts with mSin3A and that this interaction mediates repression of the p21 promoter by AML1 (21). However, it was not shown how the interaction between AML1 and mSin3A is regulated.
We demonstrated that mSin3A binds to AML1 when it is not phosphorylated and that mSin3A is released from phosphorylated AML1 upon activation of ERK. mSin3A is known to recruit class I HDACs (13). When associated with DNA, this complex can deacetylate chromatin and silence transcription. In the previous study, we showed that the transcription of TCRβ induced by AML1 is dependent on phosphorylation of AML1 and that the AML1 mutant cannot be phosphorylated, showing reduced transcriptional activity. Here, we revealed that this reduction is alleviated by treatment with TSA. This suggests that the reduced transcriptional activity of the unphosphorylated AML1 mutant is due to its interaction with mSin3A. We conclude that mSin3A suppresses AML1 transcriptional activity by linking AML1 to HDACs when AML1 is not phosphorylated and that AML1 released from mSin3A upon ERK-induced phosphorylation becomes active as a transcriptional activator. These results provide a novel mechanism by which extracellular stimuli convert inactive forms of transcription factors into active ones.
Intranuclear localization of AML1 is regulated by interaction with mSin3A.
AML1 is a nuclear protein that is localized in the nuclear matrix (47). The in situ immunofluorescence study and the biochemical-fractionation assay demonstrated that wild-type AML1 translocates from the nuclear matrix, where mSin3A is located when it is phosphorylated, supporting our finding that AML1 dissociates itself from mSin3A upon phosphorylation. The deletion mutant of AML1 lacking the interaction domain with mSin3A translocates from the nuclear matrix without phosphorylation, and a mutation in the nuclear-matrix attachment site disrupts the interaction between AML1 and mSin3A. One plausible explanation for this observation is that the interaction of AML1 with mSin3A plays an important role in targeting to the nuclear matrix. This is also supported by our findings that the mutant of AML1 that mimics phosphorylated AML1 lacks its association with mSin3A and that it is released from the nuclear matrix. The previous studies showed that the carboxyl terminus of AML1 is also necessary for targeting to the nuclear matrix (3, 47). From these results, it is suggested that both the mSin3A interaction domain and the carboxyl-terminal domain of AML1 are necessary for its localization in the nuclear matrix.
The importance of the nuclear-matrix targeting in the function of AML1 is not clear. AML1 was shown to colocalize with a subset of hyperphosphorylated RNA polymerase II in the nuclear matrix, which suggests that the association with the nuclear matrix is necessary to support AML1 transcriptional activity (46). In that study, however, only a subset of AML1 was associated with RNA polymerase II in the nuclear matrix whereas a significant amount of AML1 proteins was not. More recently, it was shown that targeting of AML1 to the nuclear matrix is required for the initiation of DNA replication (3). Our data raise the possibility that AML1 works as an active repressor when bound to mSin3A in the nuclear matrix. Once it is phosphorylated, transcriptional repression by AML1 may be abolished. Furthermore, AML1 released from the nuclear matrix may bind to the other regulatory elements in the chromatin and activate gene transcription. Further mechanisms that dictate AML1 activation after mSin3A release remain to be elucidated.
Regulation of the stability of AML1 by phosphorylation.
The previous study reported that heterodimerization with PEBP2β protects AML1 from proteasome-mediated degradation (7). However, our data indicate that phosphorylated AML1 is degraded in a time-dependent manner in COS-7 cells that contain endogenous PEBP2β. These results raise the possibility that some proteins other than PEBP2β may protect unphosphorylated AML1 from degradation and that their loss of interaction may cause degradation of phosphorylated AML1. Our analyses of the AML1 mutant lacking the domain interacting with mSin3A support the ability of mSin3A to protect AML1 from degradation. These results provide a novel regulatory mechanism that governs the stability of AML1.
In PEBP2β-deficient fibroblasts, both phosphorylated and unphosphorylated AML1 were observed by Western blotting after 5 min of EGF stimulation (Fig. 8B, lane 2), and unphosphorylated AML1 was completely degraded after 30 min of EGF stimulation (Fig. 8B, lane 3). It is supposed that unphosphorylated AML1 is phosphorylated and degraded during the 30 min after EGF stimulation. In COS-7 cells, both phosphorylated and unphosphorylated forms of AML1 are observed after 30 min of EGF stimulation (Fig. 7, lanes 4 and 6). This discrepancy is due to the fact that COS-7 cells contain endogenous PEBP2β, which protects AML1 from degradation.
A mechanism for regulation of AML1 activation induced by its phosphorylation.
Recently, it was shown that phosphorylation of the transcription factor Elk-1 in response to ERK activation induces recruitment of the mSin3A-HDAC1 complex and repression of transcription from its target promoters (45). In our study, phosphorylation of AML1 induced the release of AML1 from mSin3A and enhanced AML1 transcriptional activity. Furthermore, phosphorylation regulates the stability of AML1 via interaction with mSin3A, manifested by the fact that phosphorylated AML1 is subjected to proteasome-mediated degradation. Thus, one additional role of corepressor release from AML1 may be to set a ceiling on the degree and duration of AML1 activation in response to cytokine and growth factor stimuli.
In summary, our studies have identified a novel mechanism by which ERK-induced phosphorylation regulates AML1 function. These findings will contribute to elucidation of the leukemogenic mechanisms derived from the dysfunction of AML1.
Acknowledgments
We thank M. Ohki for providing us with AML1 cDNA.
This work was supported in part by grants-in-aid from the Japan Society for the Promotion of Science; the Ministry of Education, Culture, Sports, Science and Technology; and the Ministry of Health, Labor and Welfare.
REFERENCES
- 1.Ayer, D. E. 1999. Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol. 9:193-198. [DOI] [PubMed] [Google Scholar]
- 2.Bruhn, L., A. Munnerlyn, and R. Grosschedl. 1997. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRα enhancer function. Genes Dev. 11:640-653. [DOI] [PubMed] [Google Scholar]
- 3.Chen, L. F., K. Ito, Y. Murakami, and Y. Ito. 1998. The capacity of polyomavirus enhancer binding protein 2αB (AML1/Cbfa2) to stimulate polyomavirus DNA replication is related to its affinity for the nuclear matrix. Mol. Cell. Biol. 18:4165-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowell, I. G. 1994. Repression versus activation in the control of gene transcription. Trends Biochem. Sci. 19:38-42. [DOI] [PubMed] [Google Scholar]
- 5.Golub, T. R., G. F. Barker, S. K. Bohlander, S. W. Hiebert, D. C. Ward, P. Bray-Ward, E. Morgan, S. C. Raimondi, J. D. Rowley, and D. G. Gilliland. 1995. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. USA 92:4917-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herschbach, B. M., and A. D. Johnson. 1993. Transcriptional repression in eukaryotes. Annu. Rev. Cell Biol. 9:479-509. [DOI] [PubMed] [Google Scholar]
- 7.Huang, G., K. Shigesada, K. Ito, H. J. Wee, T. Yokomizo, and Y. Ito. 2001. Dimerization with PEBP2β protects RUNX1/AML1 from ubiquitin-proteasome-mediated degradation. EMBO J. 20:723-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imai, Y., M. Kurokawa, K. Izutsu, A. Hangaishi, K. Maki, S. Ogawa, S. Chiba, K. Mitani, and H. Hirai. 2001. Mutations of the Smad4 gene in acute myelogeneous leukemia and their functional implications in leukemogenesis. Oncogene 20:88-96. [DOI] [PubMed] [Google Scholar]
- 9.Imai, Y., M. Kurokawa, K. Izutsu, A. Hangaishi, K. Takeuchi, K. Maki, S. Ogawa, S. Chiba, K. Mitani, and H. Hirai. 2000. Mutations of the AML1 gene in myelodysplastic syndrome and their functional implications in leukemogenesis. Blood 96:3154-3160. [PubMed] [Google Scholar]
- 10.Imai, Y., M. Kurokawa, K. Tanaka, A. D. Friedman, S. Ogawa, K. Mitani, Y. Yazaki, and H. Hirai. 1998. TLE, the human homolog of Groucho, interacts with AML1 and acts as a repressor of AML1-induced transactivation. Biochem. Biophys. Res. Commun. 252:582-589. [DOI] [PubMed] [Google Scholar]
- 11.Ishida, N., M. Kitagawa, S. Hatakeyama, and K. Nakayama. 2000. Phosphorylation at serine 10, a major phosphorylation site of p27(Kip1), increases its protein stability J. Biol. Chem. 275:25146-25154. [DOI] [PubMed] [Google Scholar]
- 12.Kitabayashi, I., A. Yokoyama, K. Shimizu, and M. Ohki. 1998. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 17:2994-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knoepfler, P. S., and R. N. Eisenman. 1999. Sin meets NuRD and other tails of repression. Cell 99:447-450. [DOI] [PubMed] [Google Scholar]
- 14.Koipally, J., A. Renold, J. Kim, and K. Georgopoulos. 1999. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 18:3090-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunkel, T. A., J. D. Roberts, and R. A. Zakour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 16.Kurokawa, M., K. Mitani, K. Irie, T. Matsuyama, T. Takahashi, S. Chiba, Y. Yazaki, K. Matsumoto, and H. Hirai. 1998. The oncoprotein Evi-1 represses TGF-β signalling by inhibiting Smad3. Nature 394:92-96. [DOI] [PubMed] [Google Scholar]
- 17.Kurokawa, M., T. Tanaka, K. Tanaka, S. Ogawa, K. Mitani, Y. Yazaki, and H. Hirai. 1996. Overexpression of the AML1 proto-oncoprotein in NIH3T3 cells leads to neoplastic transformation depending on the DNA-binding and transactivational potencies. Oncogene 12:883-892. [PubMed] [Google Scholar]
- 18.Laherty, C. D., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, and R. N. Eisenman. 1997. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89:349-356. [DOI] [PubMed] [Google Scholar]
- 19.Levanon, D., R. E. Goldstein, Y. Bernstein, H. Tang, D. Goldenberg, S. Stifani, Z. Paroush, and Y. Groner. 1998. Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl. Acad. Sci. USA 95:11590-11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lutterbach, B., and S. W. Hiebert. 2000. Role of the transcription factor AML-1 in acute leukemia and hematopoietic differentiation. Gene 245:223-235. [DOI] [PubMed] [Google Scholar]
- 21.Lutterbach, B., J. J. Westendorf, B. Linggi, S. Isaac, E. Seto, and S. W. Hiebert. 2000. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem. 275:651-656. [DOI] [PubMed] [Google Scholar]
- 22.Maciejewski, P. M., F. C. Peterson, P. J. Anderson, and C. L. Brooks. 1995. Mutation of serine 90 to glutamic acid mimics phosphorylation of bovine prolactin. J. Biol. Chem. 270:27661-27665. [DOI] [PubMed] [Google Scholar]
- 23.Mitani, K., S. Ogawa, T. Tanaka, H. Miyoshi, M. Kurokawa, H. Mano, Y. Yazaki, M. Ohki, and H. Hirai. 1994. Generation of the AML1-EVI-1 fusion gene in the t(3;21)(q26;q22) causes blastic crisis in chronic myelocytic leukemia. EMBO J. 13:504-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyoshi, H., K. Shimizu, T. Kozu, N. Maseki, Y. Kaneko, and M. Ohki. 1991. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc. Natl. Acad. Sci. USA 88:10431-10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy, M., J. Ahn, K. K. Walker, W. H. Hoffman, R. M. Evans, A. J. Levine, and D. L. George. 1999. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3A. Genes Dev. 13:2490-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niki, M., H. Okada, H. Takano, J. Kuno, K. Tani, H. Hibino, S. Asano, Y. Ito, M. Satake, and T. Noda. 1997. Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcription factor, polyomavirus enhancer binding protein 2/core binding factor. Proc. Natl. Acad. Sci. USA 94:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nuchprayoon, I., S. Meyers, L. M. Scott, J. Suzow, S. Hiebert, and A. D. Friedman. 1994. PEBP2/CBF, the murine homolog of the human myeloid AML1 and PEBP2β/CBFβ proto-oncoproteins, regulates the murine myeloperoxidase and neutrophil elastase genes in immature myeloid cells. Mol. Cell. Biol. 14:5558-5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogawa, E., M. Inuzuka, M. Maruyama, M. Satake, M. Naito-Fujimoto, Y. Ito, and K. Shigesada. 1993. Molecular cloning and characterization of PEBP2β, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2α. Virology 194:314-331. [DOI] [PubMed] [Google Scholar]
- 29.Ohki, M. 1993. Molecular basis of the t(8;21) translocation in acute myeloid leukaemia. Semin. Cancer Biol. 4:369-375. [PubMed] [Google Scholar]
- 30.Okuda, T., J. van Deursen, S. W. Hiebert, G. Grosveld, and J. R. Downing. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell 84:321-330. [DOI] [PubMed] [Google Scholar]
- 31.Osato, M., N. Asou, E. Abdalla, K. Hoshino, H. Yamasaki, T. Okubo, H. Suzushima, K. Takatsuki, T. Kanno, K. Shigesada, and Y. Ito. 1999. Biallelic and heterozygous point mutations in the runt domain of the AML1/PEBP2αB gene associated with myeloblastic leukemias. Blood 93:1817-1824. [PubMed] [Google Scholar]
- 32.Pazin, M. J., and J. T. Kadonaga. 1997. What's up and down with histone deacetylation and transcription? Cell 89:325-328. [DOI] [PubMed] [Google Scholar]
- 33.Preudhomme, C., D. Warot-Loze, C. Roumier, N. Grardel-Duflos, R. Garand, J. L. Lai, N. Dastugue, E. Macintyre, C. Denis, F. Bauters, J. P. Kerckaert, A. Cosson, and P. Fenaux. 2000. High incidence of biallelic point mutations in the Runt domain of the AML1/PEBP2αB gene in M0 acute myeloid leukemia and in myeloid malignancies with acquired trisomy 21. Blood 96:2862-2869. [PubMed] [Google Scholar]
- 34.Romana, S. P., M. Mauchauffe, M. Le Coniat, I. Chumakov, D. Le Paslier, R. Berger, and O. A. Bernard. 1995. The t(12;21) of acute lymphoblastic leukemia results in a tel-AML1 gene fusion. Blood 85:3662-3670. [PubMed] [Google Scholar]
- 35.Song, W. J., M. G. Sullivan, R. D. Legare, S. Hutchings, X. Tan, D. Kufrin, J. Ratajczak, I. C. Resende, C. Haworth, R. Hock, M. Loh, C. Felix, D. C. Roy, L. Busque, D. Kurnit, C. Willman, A. M. Gewirtz, N. A. Speck, J. H. Bushweller, F. P. Li, K. Gardiner, M. Poncz, J. M. Maris, and D. G. Gilliland. 1999. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat. Genet. 23:166-175. [DOI] [PubMed] [Google Scholar]
- 36.Sun, W., B. J. Graves, and N. A. Speck. 1995. Transactivation of the Moloney murine leukemia virus and T-cell receptor β-chain enhancers by cbf and ets requires intact binding sites for both proteins. J. Virol. 69:4941-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi, A., M. Satake, Y. Yamaguchi-Iwai, S. C. Bae, J. Lu, M. Maruyama, Y. W. Zhang, H. Oka, N. Arai, and K. Arai. 1995. Positive and negative regulation of granulocyte-macrophage colony-stimulating factor promoter activity by AML1-related transcription factor, PEBP2. Blood 86:607-616. [PubMed] [Google Scholar]
- 38.Takakura, N., T. Watanabe, S. Suenobu, Y. Yamada, T. Noda, Y. Ito, M. Satake, and T. Suda. 2000. A role for hematopoietic stem cells in promoting angiogenesis. Cell 102:199-209. [DOI] [PubMed] [Google Scholar]
- 39.Tanaka, K., T. Tanaka, M. Kurokawa, Y. Imai, S. Ogawa, K. Mitani, Y. Yazaki, and H. Hirai. 1998. The AML1/ETO(MTG8) and AML1/Evi-1 leukemia-associated chimeric oncoproteins accumulate PEBP2β (CBFβ) in the nucleus more efficiently than wild-type AML1. Blood 91:1688-1699. [PubMed] [Google Scholar]
- 40.Tanaka, T., M. Kurokawa, K. Ueki, K. Tanaka, Y. Imai, K. Mitani, K. Okazaki, N. Sagata, Y. Yazaki, Y. Shibata, T. Kadowaki, and H. Hirai. 1996. The extracellular signal-regulated kinase pathway phosphorylates AML1, an acute myeloid leukemia gene product, and potentially regulates its transactivation ability. Mol. Cell. Biol. 16:3967-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka, T., K. Tanaka, S. Ogawa, M. Kurokawa, K. Mitani, J. Nishida, Y. Shibata, Y. Yazaki, and H. Hirai. 1995. An acute myeloid leukemia gene, AML1, regulates hemopoietic myeloid cell differentiation and transcriptional activation antagonistically by two alternative spliced forms. EMBO J. 14:341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taunton, J., C. A. Hassig, and S. L. Schreiber. 1996. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272:408-411. [DOI] [PubMed] [Google Scholar]
- 43.Wood, J. D., F. C. Nucifora, Jr., K. Duan, C. Zhang, J. Wang, Y. Kim, G. Schilling, N. Sacchi, J. M. Liu, and C. A. Ross. 2000. Atrophin-1, the dentato-rubral and pallido-luysian atrophy gene product, interacts with ETO/MTG8 in the nuclear matrix and represses transcription. J. Cell Biol. 150:939-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yagi, R., L. F. Chen, K. Shigesada, Y. Murakami, and Y. Ito. 1999. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 18:2551-2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, S. H., E. Vickers, A. Brehm, T. Kouzarides, and A. D. Sharrocks. 2001. Temporal recruitment of the mSin3A-histone deacetylase corepressor complex to the ETS domain transcription factor Elk-1. Mol. Cell. Biol. 21:2802-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng, C., S. McNeil, S. Pockwinse, J. Nickerson, L. Shopland, J. B. Lawrence, S. Penman, S. Hiebert, J. B. Lian, A. J. van Wijnen, J. L. Stein, and G. S. Stein. 1998. Intranuclear targeting of AML/CBFα regulatory factors to nuclear matrix-associated transcriptional domains. Proc. Natl. Acad. Sci. USA 95:1585-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng, C., A. J. van Wijnen, J. L. Stein, S. Meyers, W. Sun, L. Shopland, J. B. Lawrence, S. Penman, J. B. Lian, G. S. Stein, and S. W. Hiebert. 1997. Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBF-α transcription factors. Proc. Natl. Acad. Sci. USA 94:6746-6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, D. E., K. Fujioka, C. J. Hetherington, L. H. Shapiro, H. M. Chen, A. T. Look, and D. G. Tenen. 1994. Identification of a region which directs the monocytic activity of the colony-stimulating factor 1 (macrophage colony-stimulating factor) receptor promoter and binds PEBP2/CBF (AML1). Mol. Cell. Biol. 14:8085-8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zilfou, J. T., W. H. Hoffman, M. Sank, D. L. George, and M. Murphy. 2001. The corepressor mSin3A interacts with the proline-rich domain of p53 and protects p53 from proteasome-mediated degradation. Mol. Cell. Biol. 21:3974-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]