Abstract
Background
Previously we showed that BdcA controls Escherichia coli biofilm dispersal by binding the ubiquitous bacterial signal cyclic diguanylate (c-di-GMP); upon reducing the concentration of c-di-GMP, the cell shifts to the planktonic state by increasing motility, decreasing aggregation, and decreasing production of biofilm adhesins.
Findings
Here we report that BdcA also increases biofilm dispersal in other Gram-negative bacteria including Pseudomonas aeruginosa, Pseudomonas fluorescens, and Rhizobium meliloti. BdcA binds c-di-GMP in these strains and thereby reduces the effective c-di-GMP concentrations as demonstrated by increases in swimming motility and swarming motility as well as by a reduction in extracellular polysaccharide production. We also develop a method to displace existing biofilms by adding BdcA via conjugation from E. coli in mixed-species biofilms.
Conclusion
Since BdcA shows the ability to control biofilm dispersal in diverse bacteria, BdcA has the potential to be used as a tool to disperse biofilms for engineering and medical applications.
Background
Bacteria prefer to grow as biofilms [1]; i.e., as aggregates of bacteria that form usually at interfaces. Cells in biofilms are protected and held together by the extracellular matrix that includes polysaccharide, lipids, protein, and DNA [2]. Biofilms are notoriously difficult to remove [3,4] and are related to 80% of all human infections [5]. In addition, biofilm formation leads to significant energy losses in heat exchange equipment [6] and causes operational problems in the oil industry via metal corrosion, flow reduction, contamination, and reservoir plugging [7]. Conversely, biofilms are becoming increasingly important for chemical transformations; for example, biofilms lead to better productivity for biofuels such as ethanol [8], and biofilm communities form on electrodes surfaces to produce power for microbial fuel cells [9]. Also, for biorefineries, it will be important to be able to remove biofilms from fixed film reactors so that diverse products may be synthesized [10]. Hence, effective strategies to remove biofilms for medical applications as well as to control biofilm formation for engineering applications are important. Biofilm development is a dynamic process that includes movement to the surface [11], initial reversible attachment, irreversible attachment, formation of small aggregates, maturation of the biofilm, and biofilm dispersal [12]. Biofilm dispersal is controlled by various factors, including external forces and the physiological changes of the bacteria themselves [4].
Previously we discovered the biofilm dispersal protein BdcA in Escherichia coli, which works by binding to the intracellular signal cyclic diguanylate (c-di-GMP) but does not act as a phosphodiesterase [13]. Reducing c-di-GMP concentrations increases motility and extracellular DNA production and decreases extracellular polysaccharide production (EPS), cell length, and aggregation which leads to biofilm dispersal [13]. We also performed protein engineering on this protein and successfully abolished biofilm formation via single amino acid replacement at E50Q [13]. Hence biofilm dispersal can be achieved by simply by producing BdcA in E. coli. However, it has not been demonstrated that this protein is functional and can cause dispersal of biofilms in other bacteria.
Since BdcA works through c-di-GMP [13], and c-di-GMP exists in almost all bacteria [10] and regulates biofilm formation by controlling cell motility in Gram-negative bacteria [14], we hypothesized that BdcA may also work in most Gram-negative bacteria. Hence, here we constructed a broad-host plasmid to produce BdcA (pMMB206-BdcA) and tested the effect of BdcA production on biofilm dispersal for the opportunistic pathogen Pseudomonas aeruginosa (responsible for chronic cystic fibrosis infections [15,16]), the biocontrol agent Pseudomonas fluorescens (utilized for protecting plants from fungi infection [17,18]), as well as the nitrogen-fixing bacterium Rhizobium meliloti [19,20]. For all three bacteria, we find that BdcA is an effective tool for dispersing biofilms.
Methods
Bacterial strains, media, and growth conditions
The strains and plasmids used in this study are listed in Table 1. Expression of bdcA was induced by 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) (Sigma, St. Louis, MO). Luria-Bertani (LB) [21] was used for all the experiments. Chloramphenicol (50 μg/mL for R. meliloti and 250 μg/mL for Pseudomonas sp.) was used for strains harboring pMMB206 and pMMB206-BdcA. Carbenicillin (50 μg/mL) was used for strains harboring pP25-gfp. 37°C was used for all P. aeruginosa tests, and 30°C was used for P. fluorescens and R. meliloti tests.
Table 1.
Strains and plasmids used in this study
| Strain/Plasmid | Genotype | Source |
|---|---|---|
| Strain | ||
| E. coli BW25113 | lacIq rrnBT14 ΔlacZWJ16 hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | [34] |
| E. coli DH5α | luxS supE44 ΔlacU169(Φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | [21,35] |
| E. coli DH5α/pMMB206/pRK2013 | E. coli harboring pMMB206 and pRK2013 | This study |
| E. coli DH5α/pMMB206-BdcA/pRK2013 | E. coli harboring pMMB206-BdcA and pRK2013 | This study |
| P. aeruginosa PA14 | P. aeruginosa wild-type strain | [36] |
| P. fluorescens | P. fluorescens wild-type strain | R. Frazee |
| R. meliloti 102F34 | R. meliloti wild-type strain | M. Sadowski |
| PA14/pMMB206 | P. aeruginosa wild-type harboring pMMB206 | This study |
| PA14/pMMB206-BdcA | P. aeruginosa wild-type harboring pMMB206-BdcA | This study |
| PA14/pMMB206/pP25-gfp | P. aeruginosa wild-type harboring pMMB206 and pP25-gfp | This study |
| PA14/pMMB206-BdcA/pP25-gfp | P. aeruginosa wild-type harboring pMMB206-BdcA and pP25-gfp | This study |
| P. fluorescens/pMMB206 | P. fluorescens wild-type harboring pMMB206 | This study |
| P. fluorescens/pMMB206-BdcA | P. fluorescens wild-type harboring pMMB206-BdcA | This study |
| R. meliloti 102F34/pMMB206 | R. meliloti wild-type harboring pMMB206 | This study |
| R. meliloti 102F34/pMMB206-BdcA | R. meliloti wild-type harboring pMMB206-BdcA | This study |
| R. meliloti 102F34/pMMB206/pP25-gfp | R. meliloti wild-type harboring pMMB206 and pP25-gfp | This study |
| R. meliloti 102F34/pMMB206-BdcA/pP25-gfp | R. meliloti wild-type harboring the pMMB206-BdcA and pP25-gfp | This study |
| Plasmid | ||
| pMMB206 | Cmr, Mob+, lacIq, broad-host range plasmid | [23] |
| pMMB206-BdcA | Cmr; pMMB206 with Ptac-lacUV5::bdcA | this work |
| pRK2013 | Kmr; Tra+ Mob+ (RK2) Km::Tn7 ColEl origin, helper plasmid for mobilization | M. Bagdasarian, [37] |
| pP25-gfp | Carr; constitutive green fluorescent protein production | [27] |
Cmr, Carr, and Kmr, denote chloramphenicol, carbenicillin, and kanamycin resistance, respectively.
Plasmid construction
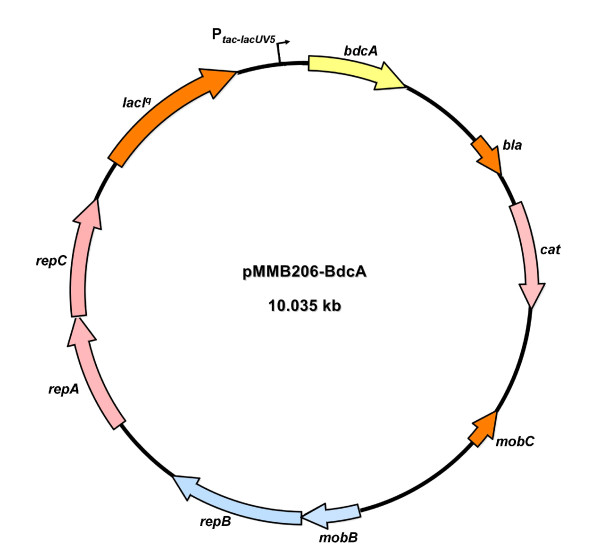
The bdcA gene was amplified from E. coli BW25113 using bdcAc f-primer (5'-CGCGCAGGATCCCACCATCACCACCATCATGGCGCTTTTACAGGTAAGACAGTTC-3') and bdcAc-r primer (5'-GATGTGGAGTCTGCTGAGCTGCAGTTATGCGCCAAACGCGCCATCAATG-3'), and the PCR product was cleaned with the QIAquick PCR purification kit (QIAGEN, Valencia, CA). The bdcA PCR product and the pMMB206 vector were both digested with BamHI (New England Biolabs, Beverly, MA) and PstI (New England Biolabs) overnight at 37°C. The vector was treated with alkaline phosphatase (New England Biolabs), and ligation was performed with T4 DNA ligase (New England Biolabs) overnight at 16°C. The ligation product was electroporated into E. coli DH5α, and white colonies after blue/white screening were selected and sequenced with sequencing primers bdcAseq f-primer (5'-ACACTTTATGCTTCCGGCTCGTATG-3') and bdcAseq r-primer (5'-GCGATTAAGTTGGGTAACGCCAG-3'). The correctly constructed pMMB206-BdcA (Figure 1) was introduced into the Rhizobium strain via electroporation and into the Pseudomonas strains via conjugation as previously described [22]. pMMB206-BdcA is a low-copy-number plasmid, and the expression of bdcA is under the control of the tandem tac-lacUV5 promoter [23].
Figure 1.
Plasmid map for pMMB206-BdcA. The bdcA gene is shown in yellow.
Static biofilm assay using crystal violet
Biofilm formation for the individual single strains was assayed in 96-well polystyrene plates using 0.1% crystal violet staining (Corning Costar, Cambridge, MA) as described previously [24] with small modifications. Briefly, each well was inoculated with overnight cultures at an initial turbidity at 600 nm of 0.05 and grown without shaking for 24 h with chloramphenicol, and then 0.1 mM IPTG was added to the culture to induce bdcA. For P. aeruginosa, the mixture was shaken at 150 rpm for 1 min and then incubated for 2 h (26 h total) to establish the extent of mature biofilm formation. Biofilm dispersal was determined after 19 h of IPTG induction (43 h total). Comparison of these two values gave the percentage of biofilm dispersal. Similarly, for P. fluorescens and R. meliloti, biofilm formation was determined and compared after 2 h and 21.5 h of IPTG induction. At least two independent cultures were used for each strain. To remove growth effects, biofilm formation was normalized by dividing total biofilm by bacterial growth in the bulk liquid.
For BdcA dispersal in dual-species biofilms of E. coli/R. meliloti and E. coli/P. aeruginosa formed in 96-well plates, pMMB206-BdcA was mobilized using E. coli DH5α/pMMB206-BdcA/pRK2013 by adding 300 μL of turbidity 0.05 of these washed cells to the R. meliloti and P. aeruginosa biofilms formed for 24 h. After 4 h, 0.1 mM IPTG was added to produce BdcA, and cells were grown for an additional 26 h. To approximate the conjugation efficiency, biofilm cells were taken from the air/liquid biofilm (top) of each well in the 96-well plates at the end of the experiment and plated on LB, LB + rifampicin, and LB + rifampicin + chloramphenicol selective plates. For rifampicin, 20 μg/mL inhibits E. coli, R. meliloti is resistant to 20 μg/mL, and P. aeruginosa is resistant to 100 μg/mL; chloramphenicol was used to select for cells harboring pMMB206 or pMMB206-BdcA. The conjugation efficiency of R. meliloti and P. aeruginosa was calculated by comparing the number of recipient cells harboring pMMB206 or pMMB206-BdcA with the total number of recipient cells for each species. In addition, five random colonies were selected from LB plates with 100 μg/mL rifampicin and 250 μg/mL chloramphenicol (selective plates for P. aeruginosa/pMMB206-BdcA) and another five colonies were selected from LB plates with 20 μg/mL rifampicin and 50 μg/mL chloramphenicol (selective plates for R. meliloti/pMMB206-BdcA) to verify the presence of the plasmid and to identify the host. The presence of the plasmid in recipient cells was confirmed via PCR of the bdcA gene with the bdcAc f-primer and the bdcAc r-primer, and the bacterial species of the recipient cells was confirmed via sequencing of the 16S rRNA genes (primers are the 1492R-primer 5'-GGTTACCTTGTTACGACTT-3' and the 27F-primer 5'-AGAGTTTGATCCTGGCTCAG-3' [25]).
Similarly, for BdcA dispersal in triple-species biofilms of E. coli, R. meliloti, and P. aeruginosa, pMMB206-BdcA was mobilized using E. coli DH5α/pMMB206-BdcA/pRK2013 by adding 300 μL of turbidity 0.05 of these cells to existing double-species biofilms formed by R. meliloti and P. aeruginosa for 24 h. After another 11 h, 0.1 mM IPTG was added to produce BdcA, and cells were grown for an additional 24 h. To determine the percentage of each of the three strains in the biofilms, cells were taken from the air/liquid biofilm and from the liquid/bottom biofilm and diluted in LB for enumeration. The biofilm composition was determined based on the difference in rifampicin resistance of these three bacteria: 20 μg/mL inhibits E. coli, 100 μg/mL inhibits R. meliloti, and P. aeruginosa is resistant to 100 μg/mL.
Flow cell biofilms and image analysis
The flow cell experiments were performed as previously described [26]. pP25-gfp [27] was used to produce the green fluorescent protein (GFP) for imaging each strain. The flow cells were inoculated with cultures at an initial turbidity at 600 nm of 0.05 at a flow rate of 10 mL/min for 2 h, then fresh LB medium with chloramphenicol and carbenicillin was added at 10 mL/min. IPTG (0.1 mM) was added to each flow cell system after 24 h, and images were taken 48 h and 96 h after IPTG addition for P. aeruginosa and 43 h and 91 h after IPTG addition for R. meliloti. Biofilm images from nine random positions were visualized with IMARIS confocal software (Bitplane, Zurich, Switzerland) and analyzed by COMSTAT confocal software as previously described [26]. Two independent cultures were used for each strain; i.e., eight flow cell experiments were conducted.
Swarming motility, swimming motility, and EPS assays
The swarming motility of P. aeruginosa and P. fluorescens was assayed with BM-2 plates [22], while the swarming motility for R. meliloti was measured with 0.6% Eiken agar (Eiken Chemical Co., Tokyo, Japan) plates in LB medium with 0.5% glucose. Cells grown to a turbidity at 600 nm of ~1.0 were used for inoculation, and 0.1 mM IPTG was applied for each strain to induce the expression of bdcA. The swarming halo was measured after 15 h incubation for P. aeruginosa and R. meliloti and after 20 h incubation for P. fluorescens. Two independent cultures were used for each strain and at least three plates were used for each independent culture.
Swimming motility was performed as previously described [28]. Overnight cultures were used to inoculate the plates (1% tryptone, 0.25% NaCl and 0.3% agar), and the swimming halo was measured after 11 h for P. aeruginosa, after 20 h for P. fluorescens, and after 15 h for R. meliloti. Two independent cultures were used for each strain, and at least three plates were used for each independent culture.
The amount of total EPS was determined as described previously [29]. Briefly, 1 mL cell cultures grown in M9-0.4% mannitol medium were collected after 48 h and boiled in water for 10 min. The supernatants were then used for an anthrone-H2SO4 assay to determine EPS concentrations. This assay was performed with two independent cultures.
Results
BdcA plasmid does not affect growth
In the absence of production of BdcA, all three pairs of bacteria grew at nearly the same rate (0.5860 ± 0.0003/h for P. aeruginosa/pMMB206 vs. 0.571 ± 0.009/h for P. aeruginosa/pMMB206-BdcA at 37°C; 0.512 ± 0.005/h for P. fluorescens/pMMB206 vs. 0.506 ± 0.002/h for P. fluorescens/pMMB206-BdcA at 30°C; and 0.511 ± 0.0008/h for R. meliloti/pMMB206 vs. 0.51 ± 0.01/h for R. meliloti/pMMB206-BdcA at 30°C) so the presence of bdcA on the plasmid does not affect growth. Hence, the bacteria are expected to form similar amounts of biofilm prior to production of BdcA for biofilm dispersal.
BdcA increases swarming and swimming motility and decreases EPS production
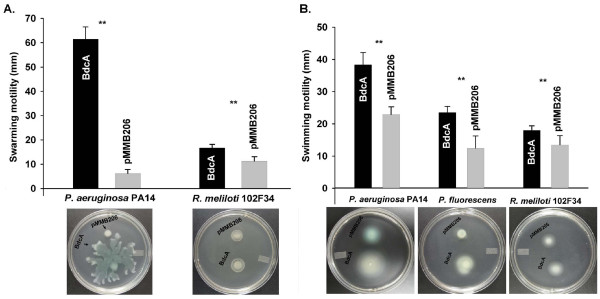
To investigate the impact of BdcA production on cell physiology, we assayed cell motility and EPS production since these phenotypes are controlled by c-di-GMP [13]. Production of BdcA increased both the swarming of P. aeruginosa and R. meliloti (P. fluorescens did not swarm under these conditions) (Figure 2A) as well as increased the swimming of P. aeruginosa, P. fluorescens, and R. meliloti (Figure 2B). Furthermore, EPS production was reduced in all three strains (1.3 ± 0.3-fold for P. aeruginosa, 1.8 ± 0.2-fold for P. fluorescens, and 1.4 ± 0.1-fold for R. meliloti) upon BdcA production. These phenotypes are consistent with reduced c-di-GMP concentrations in these cells in the presence of BdcA [13,22].
Figure 2.
BdcA increases swarming and swimming motility. (A) Swarming motility of P. aeruginosa PA14 and R. meliloti 102F34 with BdcA production. The P. aeruginosa BM-2 plates were incubated at 37°C with 250 μg/mL chloramphenicol for 15 h, and the R. meliloti Eiken agar plates were incubated at 30°C with 50 μg/mL chloramphenicol for 15 h. (B) Swimming motility of P. aeruginosa, P. fluorescens, and R. meliloti 102F34. The P. aeruginosa plates were incubated at 37°C with 250 μg/mL chloramphenicol for 11 h, the P. fluorescens plates were incubated at 30°C with 250 μg/mL chloramphenicol for 20 h, and the R. meliloti plates were incubated at 30°C with 50 μg/mL chloramphenicol for 15 h. Black bars show the motility for strains expressing bdcA via pMMB206-BdcA and the gray bars indicate the control strains with pMMB206. For each strain, 0.1 mM IPTG was added to induce bdcA expression. ** indicates statistical significant difference as determined by a Student's t-test (p < 0.05). Error bars indicate the standard deviation of two independent cultures.
BdcA increases biofilm dispersal in microtitre plates
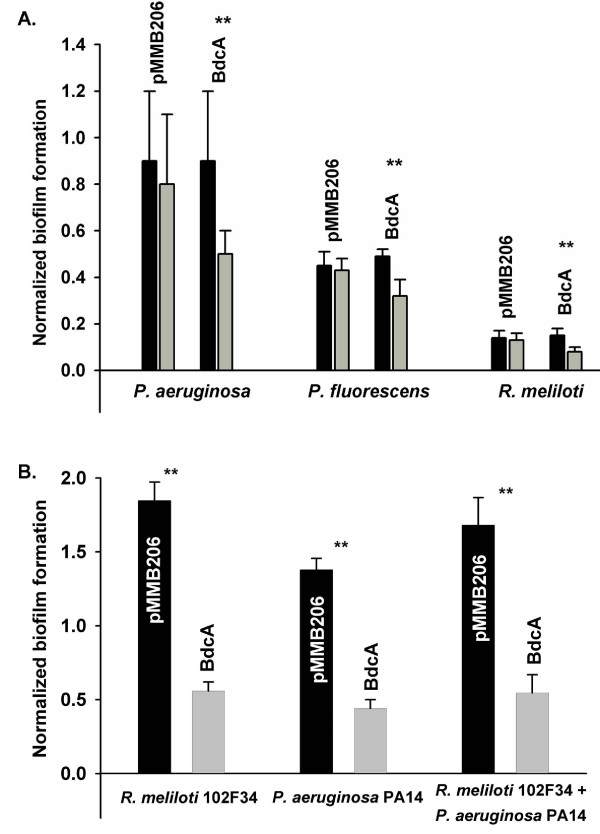
Static biofilm dispersal tests showed that bdcA produced in trans caused 40% biofilm dispersal while the control strain with an empty vector had 5 to 10% biofilm dispersal under the same conditions in P. aeruginosa, P. fluorescens, and R. meliloti (Figure 3A). Critically, similar biofilm formation occurred prior to dispersal for all three strains; hence, production of BdcA only changed the dispersal stage of the biofilm (Figure 3A). Therefore, BdcA is active in vivo in strains other than E. coli, and BdcA can remove existing biofilms.
Figure 3.
BdcA disperses biofilms in static biofilm tests with 96-well plates. (A) Strains as in the Figure 2 caption. Black bars indicate mature biofilm formation (2 h after IPTG induction for each strain), and the gray bars show biofilm formation after dispersal (19 h after IPTG induction for P. aeruginosa/pMMB206-BdcA and 21.5 h after IPTG induction for P. fluorescens/pMMB206-BdcA and R. meliloti/pMMB206-BdcA). For all three strains, 0.1 mM IPTG was added after 24 h incubation to induce bdcA expression. The P. aeruginosa biofilm was formed at 37°C in LB with 250 μg/mL chloramphenicol, the P. fluorescens biofilm was formed at 30°C in LB with 250 μg/mL chloramphenicol, and the R. meliloti biofilm was formed at 30°C in LB with 50 μg/mL chloramphenicol. pMMB206 indicates cells with this plasmid, and BdcA indicates cells containing pMMB206-BdcA. (B) For the dual-species biofilms, R. meliloti (30°C) and P. aeruginosa (37°C) biofilms were formed in LB for 24 h then E. coli DH5α/pMMB206-BdcA/pRK2013 was added. The mixed biofilm was developed for 4 h, then IPTG was added for 26 h to produce BdcA. For the triple species biofilm, the R. meliloti and P. aeruginosa biofilm was formed in LB for 24 h at 30°C, then E. coli DH5α/pMMB206-BdcA/pRK2013 was added. The mixed biofilm was developed for 11 h, then IPTG was added for another 24 h to produce BdcA. Normalized biofilm is indicated by dividing biofilm formation by cell turbidity. pMMB206 (black bars) indicates cells with this plasmid, and BdcA (gray bars) indicates cells containing pMMB206-BdcA. ** indicates statistical significant difference as determined by a Student's t-test (p < 0.05). Error bars indicate the standard deviation of two independent cultures.
BdcA increases biofilm dispersal in flow cells
To corroborate the static 96 well results, we tested biofilm dispersal with the more rigorous flow cell assay by continuously adding fresh medium to the biofilms with P. aeruginosa and R. meliloti. Critically, for both bacteria, biofilms were formed to the same extent prior to BdcA causing dispersal (Table 2). For example, for P. aeruginosa, after 72 h, there was slightly more biofilm biomass for the strain producing BdcA vs. the empty plasmid control and similar biofilm thickness and surface coverage (Table 2). After 67 h, nearly identical biofilm parameters were obtained with R. meliloti.
Table 2.
Flow cell statistical analysis of biofilm formation via COMSTAT
| Time | Biomass (μm3/μm2) |
Surface coverage (%) |
Average thickness (μm) |
Roughness | Biomass (μm3/μm2) |
Surface coverage (%) |
Average thickness (μm) |
Roughness |
|---|---|---|---|---|---|---|---|---|
| P. aeruginosa/pMMB206/pP25-gfp | P. aeruginosa/pMMB206-BdcA/pP25-gfp | |||||||
| 72 h | 3 ± 1 | 36 ± 7 | 6 ± 3 | 0.5 ± 0.3 | 4 ± 1 | 37 ± 9 | 8 ± 3 | 0.5 ± 0.2 |
| 120 h | 3.1 ± 0.9 | 37 ± 8 | 7 ± 3 | 0.5 ± 0.2 | 0.2 ± 0.1 | 13 ± 4 | 0.5 ± 0.4 | 1.6 ± 0.2 |
| R. meliloti /pMMB206/pP25-gfp | R. meliloti /pMMB206-BdcA/pP25-gfp | |||||||
| 67 h | 3 ± 1 | 34 ± 8 | 6 ± 2 | 0.6 ± 0.3 | 3 ± 1 | 40 ± 10 | 6 ± 2 | 0.7 ± 0.2 |
| 115 h | 2.8 ± 0.7 | 30 ± 6 | 6 ± 2 | 0.5 ± 0.1 | 0.4 ± 0.3 | 17 ± 5 | 0.9 ± 0.7 | 1.3 ± 0.3 |
For P. aeruginosa, cultures were grown in LB with 250 μg/mL chloramphenicol and 50 μg/mL carbenicillin for 24 h, then bdcA expression was induced by adding 0.1 mM IPTG. Data were collected at 72 h (48 h after IPTG addition) and 120 h (96 h after IPTG addition). For R. meliloti, cultures were grown in LB with 50 μg/mL chloramphenicol and 50 μg/mL carbenicillin for 24 h, then bdcA expression was induced by adding 0.1 mM IPTG. Data were collected at 67 h (43 h after IPTG addition) and 115 h (91 h after IPTG addition). Data are the average of two independent cultures for each strain.
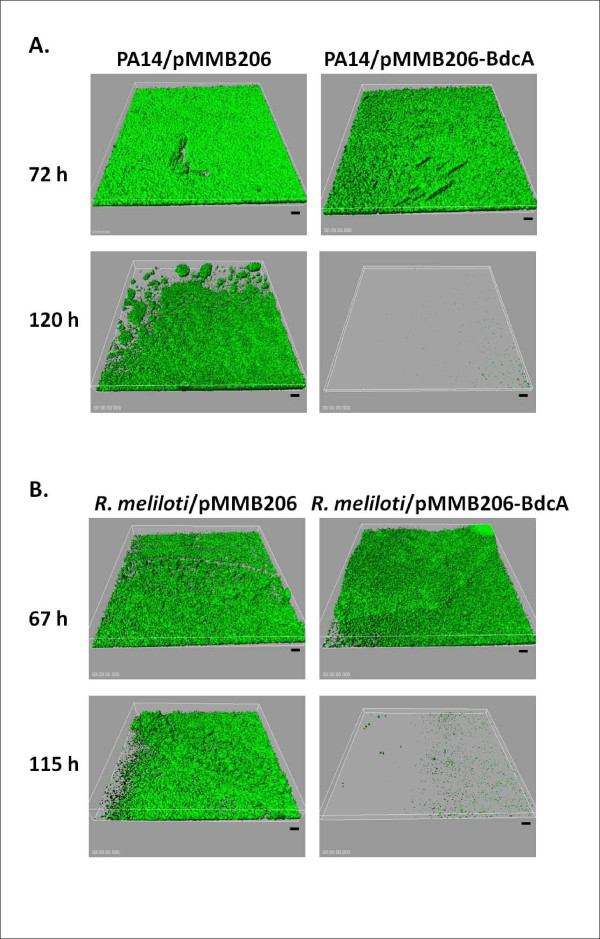
Production of BdcA caused a 7-fold increase in biofilm dispersal (based on biomass) for R. meliloti and 15-fold increased biofilm dispersal in P. aeruginosa (Table 2 and Figure 4). For both strains, the biofilms were almost completely removed for the cells producing BdcA while the empty plasmid control maintained a robust biofilm (Figure 4). Hence, BdcA is extremely effective in causing biofilm dispersal in Gram-negative bacteria other than E. coli.
Figure 4.
BdcA disperses P. aeruginosa and R. meliloti biofilms in flow cell experiments. Representative IMARIS images of P. aeruginosa/pMMB206-BdcA/pP25-gfp and P. aeruginosa/pMMB206/pP25-gfp flow cell biofilm formation after 72 h and 120 h of incubation with LB medium at 37°C (A). IMARIS images of R. meliloti/pMMB206-BdcA/pP25-gfp and R. meliloti/pMMB206/pP25-gfp flow cell biofilm formation after 67 h and 115 h of incubation with LB medium at 30°C (B). After forming biofilms in LB for 24 h, IPTG (0.1 mM) was added to induce BdcA production from pMMB206-BdcA. Each strain has pP25-gfp for producing GFP to visualize the biofilms, and carbenicillin (50 μg/mL) was added to retain pP25-gfp. Chloramphenicol (250 μg/mL for P. aeruginosa and 50 μg/mL for R. meliloti) was used to retain the pMMB206-based plasmids. Scale bars represent 10 μm.
BdcA causes dispersal in mixed-species biofilms
To demonstrate that BdcA may be added to a mixed-species biofilm via conjugation, 96-well static biofilm cultures were used to form biofilms of R. meliloti and P. aeruginosa. After 24 h, BdcA was added to these biofilms by conjugating pMMB206-BdcA from E. coli DH5α/pMMB206-BdcA/pRK2013; E. coli DH5α/pMMB206/pRK2013 was used as the negative control which conjugates but does not provide BdcA. Upon producing BdcA for 26 h, total biofilm formation was reduced by 3.3 ± 0.6 for R. meliloti and 3.1 ± 0.6 for P. aeruginosa (Figure 3B). For these experiments, the efficiency of transferring pMMB206-BdcA to R. meliloti and P. aeruginosa was over 76% and over 80% respectively, probably due to vast excess of donor E. coli cells relative to the recipient cells of the very small biofilm when donor E. coli was added. After conjugation, we checked for the presence of the correct plasmid in recipient cells via PCR for the bdcA gene as well as identified each bacterial species via sequencing for 16S rRNA genes to confirm that the selective plating method was accurate. For both tests, all (10) cells from the selective antibiotic plates were found to have the bdcA plasmid (766 bp), and these cells were all determined to be either P. aeruginosa or R. meliloti, respectively.
In addition, we used the same strategy to treat an existing biofilm formed by both R. meliloti and P. aeruginosa and obtained a 3.1 ± 0.8-fold biofilm reduction based on BdcA production after conjugation from E. coli strain (Figure 3B). Hence, BdcA was able to disperse a multi-species biofilm that consisted of approximately 40% P. aeruginosa PA14, 23% R. meliloti, and 37% E. coli (the composition of control biofilm without BdcA was similar at 37% P. aeruginosa PA14, 26% R. meliloti, and 37% E. coli). Therefore, BdcA may be introduced into mixed biofilms via conjugation and used to reduce biofilm formation.
Discussion
Given the near universal nature of c-di-GMP as a biofilm formation and dispersal signal, we investigated whether BdcA from E. coli would be effective in removing biofilms from various bacteria. To accomplish this goal, the broad host plasmid pMMB206 was used to express bdcA in different organisms including P. aeruginosa, P. fluorescens, and R. meliloti. As we had hoped, BdcA production increased swarming and swimming motility and reduced EPS production; these phenotypes indicate a reduced c-di-GMP concentration in these strains. The reduced c-di-GMP concentrations via BdcA proved extremely effective in dispersing biofilms including that of the opportunistic pathogen P. aeruginosa. This is the first report of reducing c-di-GMP via heterologous protein production to cause biofilm dispersal in these three strains. This is an important result in that it is a significant challenge to remove biofilms [30,31] since cells in biofilms are cemented in place by the secreted polymer matrix consisting of polysaccharide, protein, DNA, and lipids [2]; this matrix of the biofilm colony makes most biofilms difficult or impossible to eradicate [4].
BdcA directly reduces bacterial c-di-GMP concentrations [13], and c-di-GMP is a signal that controls physiology of diverse bacteria [32]. In R. meliloti, c-di-GMP controls growth, motility, EPS production, and even the interactions between bacteria and host plants [33]. In P. aeruginosa, c-di-GMP controls various cell activities including biofilm formation, EPS production, and aggregation [22]. Hence, BdcA may control biofilm dispersal in as many organisms as c-di-GMP functions as a second messenger. In addition, bdcA shows high sequence conservation with other species (including Pseudomonas and Rhizobium) and is well conserved in many bacteria [13]. Hence, BdcA may be used for dispersal in many strains. As a preliminary proof of this principle, our results demonstrate the feasibility of introducing BdcA into existing non-E. coli biofilms (formed by P. aeruginosa and R. meliloti individually, as well as by mixed biofilms of these two bacteria) via conjugation of a broad-host-range plasmid) to reduce total biofilm formation. Therefore, in this study, we successfully disperse biofilms of disparate strains simply by producing BdcA. Hence, through this genetic approach, a single protein may be used to remove biofilms rather than by using traditional chemical or physical means.
List of abbreviations
c-di-GMP: cyclic diguanylate; IPTG: isopropyl-β-D-thiogalactopyranoside; LB: Luria-Bertani; GFP: green fluorescent protein.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
TKW and QM designed the study and wrote the paper. QM and GZ performed the experiments and analyzed the results.
Contributor Information
Qun Ma, Email: qun.ma328@gmail.com.
Guishan Zhang, Email: gszhang86@gmail.com.
Thomas K Wood, Email: Thomas.Wood@chemail.tamu.edu.
Acknowledgements
This work was supported by the National Institutes of Health (R01 GM089999). T.W. is the T. Michael O'Connor Endowed Professor at Texas A & M University.
References
- Heilmann C, Götz F. In: Bacterial Signaling. Krämer R, Jung K, editor. Weinheim: Wiley-Blackwell; 2010. Cell-cell communication and biofilm formation in Gram-positive bacteria. [Google Scholar]
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Walker JT, Bradshaw DJ, Bennett AM, Fulford MR, Martin MV, Marsh PD. Microbial biofilm formation and contamination of dental-unit water systems in general dental practice. Appl Environ Microbiol. 2000;66:3363–3367. doi: 10.1128/AEM.66.8.3363-3367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan JB. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res. 2010;89:205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Richkard AH, Davies DG. Biofilms and biocomplexity. Microb. 2007;2:347–353. [Google Scholar]
- Characklis WG, Nevimons MJ, Picologlou BF. Influence of fouling biofilms on heat transfer. Heat Transfer Eng. 1981;3:23–27. doi: 10.1080/01457638108939572. [DOI] [Google Scholar]
- Schwermer CU, Lavik G, Abed RM, Dunsmore B, Ferdelman TG, Stoodley P, Gieseke A, de Beer D. Impact of nitrate on the structure and function of bacterial biofilm communities in pipelines used for injection of seawater into oil fields. Appl Environ Microbiol. 2008;74:2841–2851. doi: 10.1128/AEM.02027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche B, Li XZ, Hauer B, Schmid A, Buehler K. Microbial biofilms: a concept for industrial catalysis? Trends Biotechnol. 2009;27:636–643. doi: 10.1016/j.tibtech.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Kiely PD, Call DF, Yates MD, Regan JM, Logan BE. Anodic biofilms in microbial fuel cells harbor low numbers of higher-power-producing bacteria than abundant genera. Appl Microbiol Biotechnol. 2010;88:371–380. doi: 10.1007/s00253-010-2757-2. [DOI] [PubMed] [Google Scholar]
- Wood TK, Hong SH, Ma Q. Engineering biofilm formation and dispersal. Trends Biotechnol. 2011;29:87–94. doi: 10.1016/j.tibtech.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TK, González Barrios AF, Herzberg M, Lee J. Motility influences biofilm architecture in Escherichia coli. Appl Microbiol Biotechnol. 2006;72:361–367. doi: 10.1007/s00253-005-0263-8. [DOI] [PubMed] [Google Scholar]
- Van Houdt R, Michiels CW. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res Microbiol. 2005;156:626–633. doi: 10.1016/j.resmic.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Ma Q, Yang Z, Pu M, Peti W, Wood TK. Engineering a novel c-di-GMP-binding protein for biofilm dispersal. Environ Microbiol. 2011;13:631–642. doi: 10.1111/j.1462-2920.2010.02368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev. 2009;73:310–347. doi: 10.1128/MMBR.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiman L, Siegel J. Infection control in cystic fibrosis. Clin Microbiol Rev. 2004;17:57–71. doi: 10.1128/CMR.17.1.57-71.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas D, Keel C. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Annu Rev Phytopathol. 2003;41:117–153. doi: 10.1146/annurev.phyto.41.052002.095656. [DOI] [PubMed] [Google Scholar]
- Girlanda M, Perotto S, Moenne-Loccoz Y, Bergero R, Lazzari A, Defago G, Bonfante P, Luppi AM. Impact of biocontrol Pseudomonas fluorescens CHA0 and a genetically modified derivative on the diversity of culturable fungi in the cucumber rhizosphere. Appl Environ Microbiol. 2001;67:1851–1864. doi: 10.1128/AEM.67.4.1851-1864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto WW, Nixon BT, Ronson CW, Ausubel FM. Identification and characterization of the Rhizobium meliloti ntrC gene: R. meliloti has separate regulatory pathways for activation of nitrogen fixation genes in free-living and symbiotic cells. J Bacteriol. 1987;169:1423–1432. doi: 10.1128/jb.169.4.1423-1432.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eardly BD, Materon LA, Smith NH, Johnson DA, Rumbaugh MD, Selander RK. Genetic structure of natural populations of the nitrogen-fixing bacterium Rhizobium meliloti. Appl Environ Microbiol. 1990;56:187–194. doi: 10.1128/aem.56.1.187-194.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Ueda A, Wood TK. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885) PLoS Pathog. 2009;5:e1000483. doi: 10.1371/journal.ppat.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales VM, Backman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-X. [DOI] [PubMed] [Google Scholar]
- Fletcher M. The effects of culture concentration and age, time, and temperature on bacterial attachment to polystyrene. Can J Microbiol. 1977;23:1–6. doi: 10.1139/m77-001. [DOI] [Google Scholar]
- Kellogg CA, Galkiewicz JP. Cross-kingdom amplification using bacteria-specific primers: complications for studies of coral microbial ecology. Appl Environ Microbiol. 2008;74:7828–7831. doi: 10.1128/AEM.01303-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Ma Q, Wood TK. The R1 conjugative plasmid increases Escherichia coli biofilm formation through an envelope stress response. Appl Environ Microbiol. 2008;74:2690–2699. doi: 10.1128/AEM.02809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, Kulasekara B, Rietsch A, Boyd D, Smith RS, Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Sperandio V, Torres AG, Kaper JB. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol Microbiol. 2002;43:809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- Zhang XS, García-Contreras R, Wood TK. Escherichia coli transcription factor YncC (McbR) regulates colanic acid and biofilm formation by repressing expression of periplasmic protein YbiM (McbA) ISME J. 2008;2:615–631. doi: 10.1038/ismej.2008.24. [DOI] [PubMed] [Google Scholar]
- del Pozo JL, Patel R. The challenge of treating biofilm-associated bacterial infections. Clin Pharmacol Ther. 2007;82:204–209. doi: 10.1038/sj.clpt.6100247. [DOI] [PubMed] [Google Scholar]
- Bardouniotis E, Huddleston W, Ceri H, Olson ME. Characterization of biofilm growth and biocide susceptibility testing of Mycobacterium phlei using the MBEC™ assay system. FEMS Microbiol Lett. 2001;203:263–267. doi: 10.1111/j.1574-6968.2001.tb10851.x. [DOI] [PubMed] [Google Scholar]
- D'Argenio DA, Miller SI. Cyclic di-GMP as a bacterial second messenger. Microbiology. 2004;150:2497–2502. doi: 10.1099/mic.0.27099-0. [DOI] [PubMed] [Google Scholar]
- Wang Y, Xu J, Chen A, Zhu J, Yu G, Xu L, Luo L. GGDEF and EAL proteins play different roles in the control of Sinorhizobium meliloti growth, motility, exopolysaccharide production, and competitive nodulation on host alfalfa. Acta Biochim Biophys Sin. 2010;42:410–417. doi: 10.1093/abbs/gmq034. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci USA. 2006;103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]