Abstract
Background
Cellular constituents such as proteins, DNA, and RNA form a complex web of interactions that regulate biochemical homeostasis and determine the dynamic cellular response to external stimuli. It follows that detailed understanding of these patterns is critical for the assessment of fundamental processes in cell biology and pathology. Representation and analysis of cellular constituents through network principles is a promising and popular analytical avenue towards a deeper understanding of molecular mechanisms in a system-wide context.
Findings
We present Functional Genomics Assistant (FUGA) - an extensible and portable MATLAB toolbox for the inference of biological relationships, graph topology analysis, random network simulation, network clustering, and functional enrichment statistics. In contrast to conventional differential expression analysis of individual genes, FUGA offers a framework for the study of system-wide properties of biological networks and highlights putative molecular targets using concepts of systems biology.
Conclusion
FUGA offers a simple and customizable framework for network analysis in a variety of systems biology applications. It is freely available for individual or academic use at http://code.google.com/p/fuga.
Background
Advances in high throughput data collection and analysis have shown that a discrete biological function can only rarely be attributed to individual molecules [1]. Instead, complex cellular activities can be achieved through a system of interactions between macromolecules such as proteins, DNA, and RNA. Quantitative understanding of these patterns is critical for an in-depth characterization of fundamental principles in cellular biology and pathology.
Network biology is an emerging area of scientific interest aiming at the elucidation of the dynamic structure and pleiotropic function of genes and their products in cellular networks in a systematic and unbiased fashion. The treatment of biological data as graphs, where typically nodes signify cellular entities (e.g. genes, proteins, metabolites) and connections (edges) denote the corresponding functional or physical interactions, is a promising representation in molecular systems biology. Successful applications of network biology have led, for instance, to the classification of breast cancer at the interactome level [2], the characterization of signaling pathways in gastric cancer [3] and the delineation of fundamental organizational principles in metabolic networks [4]. In addition, integration of gene expression and network topological properties have led to more efficient methods for novel biomarker identification in critical conditions such as heart failure [5] and cancer [2].
Analysis of topological features and dynamic properties of cellular networks remains a highly active area of research. Indeed, several tools have been developed to address the need for system-wide analysis (NeAT [6], Systems Biology Toolbox [7]) or visualization (Cytoscape [8], BioLayout [9]). Nonetheless, the adoption of such approaches within the wider biomedical community has been rather limited, possibly due to challenges posed by the integration of experimental (e.g. microarray) and computational (e.g. databases) platforms [10]. Therefore, it is desirable that network-driven pipelines become more accessible to end users, thus facilitating the transformation of information hidden in multi-dimensional datasets into useful hints for the discovery of biomarkers and therapeutic targets.
Implementation
We have developed a system called Functional Genomics Assistant (FUGA), a MATLAB toolbox for the inference of cellular networks, graph topology analysis, random network simulation, network clustering, and functional enrichment (shown schematically in Figure 1). The toolbox is easily customizable and scalable to networks with thousands of nodes and millions of edges. Additionally, FUGA can integrate high throughput datasets into a unified framework, within which other applications can be embedded. Our objective in designing FUGA has been to simplify network analysis concepts and techniques for end users, by providing intuitive MATLAB functions for complex system exploration and network analysis.
Figure 1.
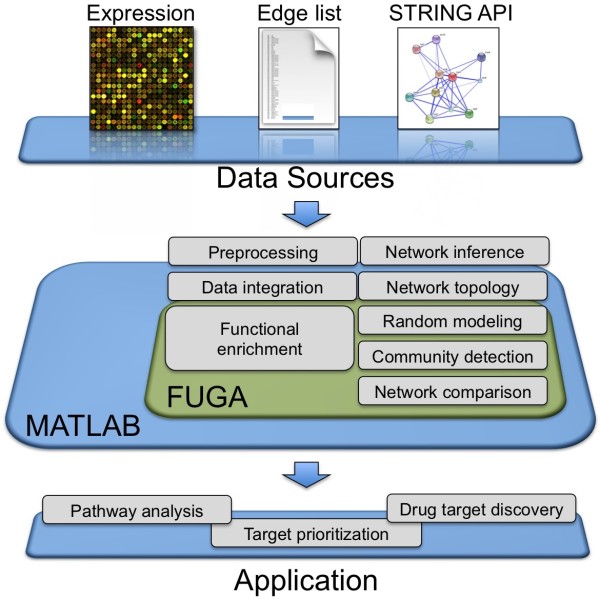
An overview of the most prominent FUGA features. FUGA is used for gene regulatory network inference, random modeling, network topology analysis, clustering, and network comparison. It is also possible to import biological networks from the STRING web-based database. Integration of biomedical data such as gene expression with interactome information may facilitate molecular pathway analysis, network-based target prioritization, and drug target discovery.
MATLAB was the language of choice for the development of FUGA, due to its matrix- and vector-based architecture, simple syntax, and powerful graphics. The code is designed to run on Unix or Mac machines; advanced users may compile sources for other platforms. Overall, FUGA is seamlessly integrated into MATLAB, so as to permit extensive analytical and visualization operations.
Results and Discussion
Recently, FUGA functionality was applied on experimentally derived datasets to define a population-based miRNA signature of type 2 diabetes [11], elucidate the expression patterns of iron regulatory protein 2 (IRP2) [12], and characterize genome-wide expression patterns in physiological cardiac hypertrophy [13]. The current FUGA release contains 137 functions and the main operating features are described briefly below.
Network reconstruction
Biological networks can be inferred with FUGA from computationally or experimentally derived datasets (e.g. BLAST similarity matrices, microarray data) using any form of similarity measure (e.g. Pearson Correlation Coefficient or PCC) to define pairs of nodes. It is also possible to import network edge lists from a text file or download interactomes of interest from the STRING database of known or predicted protein-protein interactions [14] via a web API. In the future releases, FUGA will support a wider range of public interaction databases. Network graphs are typically undirected and may be weighted. Network-specific information, such as node labels and attributes are stored as MATLAB objects for further access.
Global and local network topology
Complex interactions between constituents that regulate phenotypic diversity necessitate the study of a biological system in the context of the entire interactome, rather than just the over- or under- expression of individual entities. FUGA provides access to global network parameters such as shortest path, diameter, or link density in the interaction network. Topological features such as node betweenness and clustering coefficients can also be computed by accessing the Markov clustering (MCL) toolset [15] through the FUGA interface. To compare against random effects in large networks with hundreds of nodes and thousands of edges from high-throughput experiments, FUGA can construct random networks using link rewiring and the Erdos-Rényi model, or it can build a random modular graph, as reported elsewhere [16]. For comparing two or more networks, FUGA implements simple network similarity estimations using the Jaccard index and handles boolean set operations such as network union, intersection, and difference.
Network clustering
Because functionally related genes or proteins tend to co-localize in network vicinity, FUGA offers to identify the modular network structure via the MCL [15] or SPICi protocols [17]. Additionally, FUGA implements a greedy algorithm for community detection that uses network modularity as a measure of community structure [18], as well as several functions for spectral graph partitioning [19]. Such unsupervised algorithms are well adapted to large biological networks and may uncover previously undetected interactions.
Biological interpretation
By default, FUGA works with the ENSEMBL database [20] to annotate network nodes using all major function classification schemes (e.g. Gene Ontology [21] [GO] Biological Process [BP], Molecular Function [MF], Cellular Component [CC], GOSlim, or Reactome pathway terms). In addition, it is possible to define a custom annotation schema such as gene-disease associations. Cluster enrichment is performed by calculating the hypergeometric probability between inter- and intra- cluster gene counts assigned to a priori-defined terms. To facilitate biological discovery, nodes can be explored by merit of their topologies and subsequently linked to databases such as ENTREZ, GeneCards, or UniProt. Similar topological analyses have been previously used to uncover high-quality therapeutic targets in psoriasis [22] and identify putative cancer-associated genes [23], for instance. We illustrate biological discovery with FUGA through an example (see below).
Visualization through Cytoscape
FUGA provides direct access to the Cytoscape graph visualization software. Networks and node attributes, including clusters and topologies, can also be exported to a text file for subsequent exploration with other, user-defined network visualization software.
Extensibility
MATLAB's interactive environment allows flexible and simple addition of new functionalities to FUGA by expanding the existing framework. For example, additional network statistics or clustering algorithms can be implemented using MATLAB matrix operations and the existing FUGA network object architecture.
Comparison with other toolboxes
Similar toolboxes have been developed in MATLAB for systems biology related analysis. These include MATLABBGL [24] and the Brain Connectivity Toolbox (BCT) [25], as well as the bioinformatics toolbox developed by Mathworks. MATLABBGL uses the Boost Graph library to efficiently analyze large sparse graph structures. The BCT package is designed to quantify centrality and structure of brain networks. The bioinformatics toolbox from Mathworks has several functions for graph analysis (e.g. connected components, shortest paths), but its functionality is limited, as it may not scale up well for larger genome-wide networks. A number of non-MATLAB tools are available for network visualization and analysis, including NATbox [26] and NeAT [27]. Each tool has a distinct set of features which are highlighted in Table 1. The FUGA toolbox provides a broader range of graph theory functions and integrates expression analysis, functional annotation, and network visualization. The current FUGA release 2.9.4 contains 137 functions. As such, it provides an important contribution to network biology applications and related biomedical data analyses.
Table 1.
Comparison of network analysis tools.
| Tool | FUGA | BIT | BGL | BCT | NATbox | igraph | NeAT | NWB | IN | GG |
|---|---|---|---|---|---|---|---|---|---|---|
| Interface | MATLAB | MATLAB | MATLAB | MATLAB | R | C/R/Python | Web-based | Java | Java | Java |
| User-defined networks | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Curated pathway/network content: API | ✓ | ✓ | ✓ | ✓ | ||||||
| Computational network reconstruction | ✓ | ✓ | ✓ | ✓ | ||||||
| Statistical network analysis | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ |
| Biological enrichment/annotation | ✓ | ✓ | ✓ | ✓ | ✓ | |||||
| Expression analysis | ✓ | ✓ | ✓ | ✓ | ||||||
BIT = MATLAB Bioinformatics Toolbox, BGL = MATLABBGL, BCT = Brain Connectivity Toolbox, NWB = Network Workbench, IN = Ingenuity Pathways Analysis, GG = GeneGo.
Example
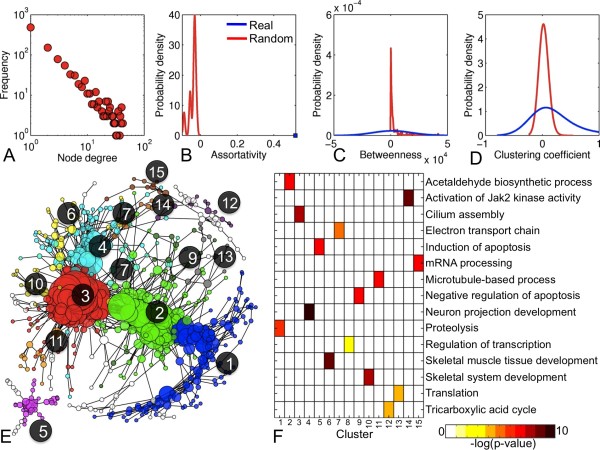
We illustrate the functionality of FUGA by interrogating time-course transcriptional profiles of failing mouse hearts (ArrayExpress: E-MEXP-105 [28]). First, PCCs for all possible gene pairs across all phenotypes were computed and gene pairs with absolute PCC≥0.90 were retained and visualized as an un-weighted, undirected network. The network contained 1018 genes and 2324 links. Average node degree was 4.6, network diameter was 21, and graph architecture was determined to be scale free (Figure 2A). Topological features of the network (assortativity, betweenness, clustering coefficients) were non-random (Figure 2B-D). Then, the greedy method for community detection [18] was used to identify modules of co-expressed genes (Figure 2E). The network and its attributes were visualized with Cytoscape and biological enrichment was performed to identify disease-specific over-represented GO-BP terms (Figure 2F). The above analyses were executed in under 5 minutes in the MATLAB command line prompt on 2.53 GHz Intel Core 2 Duo machine with 4 GB RAM.
Figure 2.
Example analysis of cardiac hypertrophy using FUGA. A-D) Comparison of topological network properties in the gene co-expression network and 10 random networks generated using the Maslov-Sneppen rewiring model [16]. E) Cytoscape visualization of the gene co-expression network in cardiac hypertrophy. Each node represents a gene and links represent a co-expression. Node colors reflect gene cluster assignment (numerical labels) as determined by the Louvain algorithm and node sizes are proportional to node degrees. The largest 15 gene clusters are visualized. F) Enrichment of each network cluster for over-represented Gene Ontology Biological Process terms.
Conclusion
FUGA is an extensible and versatile framework for network analysis in a variety of systems biology applications. While currently FUGA implements the most widely used graph theoretic approaches, future plans include the development of novel centrality and clustering algorithms. We also aim to integrate additional biological annotation repositories to facilitate analysis of networks derived from heterogeneous biological experiments. The FUGA project is an ongoing effort that should facilitate the dissemination of graph theoretic approaches across the wider biomedical community.
Availability and requirements
Project name: FUGA
Project web page: http://code.google.com/p/fuga/
Operating System: Mac OS X/Linux/Windows
Programming language: MATLAB 7.8.0 or higher/C/C++
Other requirements: None
License: GPL
Any restrictions on use by non-academics: License needed
Authors' contributions
ID created the software, and drafted the manuscript. CAO edited the manuscript and advised on software design. AMS coordinated the project design and edited the manuscript. ST coordinated the project design and wrote the manuscript. All authors read and approved the manuscript.
Contributor Information
Ignat Drozdov, Email: ignat.drozdov@kcl.ac.uk.
Christos A Ouzounis, Email: ouzounis@certh.gr.
Ajay M Shah, Email: ajay.shah@kcl.ac.uk.
Sophia Tsoka, Email: sophia.tsoka@kcl.ac.uk.
Acknowledgements
We thank members of the King's College London Centre for Bioinformatics (KCBI), and in particular Chrysanthi Ainali, for ideas, test cases, and feedback.
This project was supported by British Heart Foundation grant RE/08/003 and the Leducq Foundation. Early stages of this work have been supported in part by the Network of Excellence ENFIN (contract number LSHG-CT-2005-518254) funded by the European Commission.
References
- Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5(2):101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal A, Guo DL, Hoshida Y, Yuen ST, Chu KM, So S, Boussioutas A, Chen X, Bowtell D, Aburatani H. et al. Topological and functional discovery in a gene coexpression meta-network of gastric cancer. Cancer Res. 2006;66(1):232–241. doi: 10.1158/0008-5472.CAN-05-2232. [DOI] [PubMed] [Google Scholar]
- Guimera R, Nunes Amaral LA. Functional cartography of complex metabolic networks. Nature. 2005;433(7028):895–900. doi: 10.1038/nature03288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuaje F, Devaux Y, Wagner DR. Coordinated modular functionality and prognostic potential of a heart failure biomarker-driven interaction network. BMC Syst Biol. 2010;4:60. doi: 10.1186/1752-0509-4-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohee S, Faust K, Lima-Mendez G, Vanderstocken G, van Helden J. Network Analysis Tools: from biological networks to clusters and pathways. Nat Protoc. 2008;3(10):1616–1629. doi: 10.1038/nprot.2008.100. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Jirstrand M. Systems Biology Toolbox for MATLAB: a computational platform for research in systems biology. Bioinformatics. 2006;22(4):514–515. doi: 10.1093/bioinformatics/bti799. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theocharidis A, van Dongen S, Enright AJ, Freeman TC. Network visualization and analysis of gene expression data using BioLayout Express(3D) Nat Protoc. 2009;4(10):1535–1550. doi: 10.1038/nprot.2009.177. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Angulo AM, Hennessy BT, Mills GB. Future of personalized medicine in oncology: a systems biology approach. J Clin Oncol. 2010;28(16):2777–2783. doi: 10.1200/JCO.2009.27.0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E. et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107(6):810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- Maffettone C, Chen G, Drozdov I, Ouzounis C, Pantopoulos K. Tumorigenic properties of iron regulatory protein 2 (IRP2) mediated by its specific 73-amino acids insert. PLoS One. 2010;5(4):e10163. doi: 10.1371/journal.pone.0010163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdov I, Tsoka S, Ouzounis CA, Shah AM. Genome-wide expression patterns in physiological cardiac hypertrophy. BMC Genomics. 2010;11:557. doi: 10.1186/1471-2164-11-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009. pp. D412–416. [DOI] [PMC free article] [PubMed]
- Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30(7):1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296(5569):910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- Jiang P, Singh M. SPICi: a fast clustering algorithm for large biological networks. Bioinformatics. 2010;26(8):1105–1111. doi: 10.1093/bioinformatics/btq078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech. 2008. p. P10008.
- Chen D, Burleigh GJ, Fernandez-Baca D. Spectral partitioning of phylogenetic data sets based on compatibility. Syst Biol. 2007;56(4):623–632. doi: 10.1080/10635150701499571. [DOI] [PubMed] [Google Scholar]
- Birney E, Andrews TD, Bevan P, Caccamo M, Chen Y, Clarke L, Coates G, Cuff J, Curwen V, Cutts T. et al. An overview of Ensembl. Genome Res. 2004;14(5):925–928. doi: 10.1101/gr.1860604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dezso Z, Nikolsky Y, Nikolskaya T, Miller J, Cherba D, Webb C, Bugrim A. Identifying disease-specific genes based on their topological significance in protein networks. BMC Syst Biol. 2009;3:36. doi: 10.1186/1752-0509-3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic T, Memisevic V, Ganesan AK, Przulj N. Systems-level cancer gene identification from protein interaction network topology applied to melanogenesis-related functional genomics data. J R Soc Interface. 2010;7(44):423–437. doi: 10.1098/rsif.2009.0192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MatlabBGL. http://www.stanford.edu/~dgleich/programs/matlab_bgl/
- Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 2010;52(3):1059–1069. doi: 10.1016/j.neuroimage.2009.10.003. [DOI] [PubMed] [Google Scholar]
- Chavan SS, Bauer MA, Scutari M, Nagarajan R. NATbox: a network analysis toolbox in R. BMC Bioinformatics. 2009;10(Suppl 11):S14. doi: 10.1186/1471-2105-10-S11-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohee S, Faust K, Lima-Mendez G, Vanderstocken G, van Helden J. Network Analysis Tools: from biological networks to clusters and pathways. Nature protocols. 2008;3(10):1616–1629. doi: 10.1038/nprot.2008.100. [DOI] [PubMed] [Google Scholar]
- Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Holloway E, ArrayExpress update--an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011. pp. D1002–1004. [DOI] [PMC free article] [PubMed]