Abstract
The ets domain transcriptional repressor ERF is an effector of the receptor tyrosine kinase/Ras/Erk pathway, which, it has been suggested, is regulated by subcellular localization as a result of Erk-dependent phosphorylation and is capable of suppressing cell proliferation and ras-induced tumorigenicity. Here, we analyze the effect of ERF phosphorylation on nuclear import and export, the timing of its phosphorylation and dephosphorylation in relation to its subcellular location, Erk activity, and the requirements for ERF-induced cell cycle arrest. Our findings indicate that ERF continuously shuttles between the nucleus and the cytoplasm and that both phosphorylation and dephosphorylation of ERF occur within the nucleus. While nuclear import is not affected by phosphorylation, ERF nuclear export and cytoplasmic release require multisite phosphorylation and dephosphorylation. ERF export is CRM1 dependent, although ERF does not have a detectable nuclear export signal. ERF phosphorylation and export correlate with the levels of nuclear Erk activity. The cell cycle arrest induced by nonphosphorylated ERF requires the wild-type retinoblastoma protein and can be suppressed by overexpression of cyclin. These data suggest that ERF may be a very sensitive and constant sensor of Erk activity that can affect cell cycle progression through G1, providing another link between the Ras/Erk pathway and cellular proliferation.
Subcellular compartmentalization is one of the ways to regulate protein function, especially proteins mediating immediate-early responses in signal transduction pathways (20). Nucleocytoplasmic shuttling usually does not require new protein synthesis and is a common regulatory mechanism for many of these proteins. Since the original report on NF-κB by Baeuerle and Baltimore (6), several transcription factors that trigger immediate-early responses have been found to be regulated in this way, such as NF-AT (11), p53 (22), Smads (61, 61), Yan (41, 56), and ERF (32), as well as signaling kinases such as Erk2 (19) and MAPKAP-K2 (37). However, regulated nucleocytoplasmic transport has been reported for a variety of proteins, such as cyclins (3, 27), tumor suppressors (16), circadian regulators (58), and even junction proteins (26, 55).
There are several mechanisms involved in regulated nucleocytoplasmic transport (for a review see reference 28). The first is structural changes triggered by posttranslational modifications that can result in the masking or unmasking of the nuclear localization signal (NLS) and/or the nuclear export signal (NES) (23, 25, 35, 44, 53, 54). A second mechanism involves signal-dependent sequestration of a protein in or release from a compartment (45, 55). A third mechanism, signal-dependent association with carrier molecules that can transport a protein in or out through the nuclear pores, is common in proteins that lack specific import and export signals (8, 47, 61). In most cases described, these mechanisms serve as on-off switches, allowing the protein to change compartment as a result of the triggering event. In many cases independent mechanisms, which may involve new protein synthesis, are responsible for reversing this process action or destroying the shuttling protein at its final destination (4). However, continuous bidirectional shuttling has been described quite recently (46, 61).
The Ras/Erk signaling pathway is a major mitogenic pathway that links extracellular signals with cell proliferation and differentiation (10, 13, 39, 43, 51, 59). Several ets domain transcription factors in mammalian cells (Ets1, Ets2, Elk1, Sap1, Net, and ERF) are thought to be effectors of this pathway, regulated via ERK phosphorylation, which controls their ability to regulate transcription (12, 14, 17, 33, 42, 52, 62; for review see references 36 and 63). The Ras/Erk signaling pathway is also known to activate a number of immediate-early response proteins and genes by several different mechanisms, generally involving phosphorylation (28). However, very few of the proteins affected by the Ras/Erk pathway are known to be regulated by nucleocytoplasmic shuttling. The only reported examples are Erk itself (1, 2), the ets domain transcription factor ERF (32, 34) in mammalian cells, and the ets domain transcription factor Yan in Drosophila melanogaster (41, 56).
ERF is a ubiquitously expressed ets domain transcriptional repressor with tumor suppressor activity that is regulated by the Ras/Erk signaling pathway (52). We have shown previously that ERF is phosphorylated in vitro and in vivo by Erks at multiple sites. This phosphorylation determines its cytoplasmic localization and inactivation. ERF is dephosphorylated and nuclear in serum-starved cells and, upon serum induction, is found phosphorylated in the cytoplasm. This process is totally dependent on Erk activity and is bidirectional. The ERF protein with its phosphorylation sites mutated to alanine localizes in the nucleus, can arrest fibroblasts at the G0/G1 phase of the cell cycle, and can suppress Ras-induced tumorigenicity, suggesting that ERF is a true effector of Ras (32). In this paper, we analyze the mechanism by which phosphorylation regulates the subcellular localization and the involvement of nuclear ERF in cell cycle arrest. Our data suggest that phosphorylated ERF shuttles continuously between the nucleus and the cytoplasm. Both phosphorylation and dephosphorylation of ERF seem to occur in the nucleus and determine its localization. Whereas ERF nuclear import does not appear to be affected by phosphorylation, ERF nuclear export is dependent on extensive phosphorylation and is a CRM1-dependent process. ERF does not have an identifiable NES, but it has two extended areas required for effective nuclear export. The phosphorylation of ERF and its export from the nucleus closely follow the appearance of active Erk in the nucleus and may serve as a continuous monitor of Erk-mediated receptor tyrosine kinase signaling. The monitoring of nuclear Erk activity by ERF is consistent with the mitogenic effects of Erk activation and the ability of nuclear ERF to arrest the cell cycle in G1. Our data suggest that ERF-induced arrest requires the wild-type retinoblastoma (Rb) protein and can be suppressed by the overexpression of cyclin. Thus, ERF could be a very sensitive monitor of Erk activity that can affect cell cycle progression in an Rb-dependent manner, providing another link between the Ras/Erk pathway and this fundamental cellular process.
MATERIALS AND METHODS
DNA constructs.
The ERF mutations and the relevant green fluorescent protein (GFP)-ERF expression vectors used were described previously (32, 52). Briefly, the mutations to alanine or glutamic acid at positions T148, S161, S246, S251, T271, T357, and T526, referred to as positions 1 through 7, respectively, were generated by PCR and verified by sequencing. GFP-ERF fusions were generated by the in frame insertion of the appropriate restriction fragment of the ERF human cDNA into the pEGFP-C1 vector (Clontech). A 30-residue synthetic oligonucleotide encoding amino acids 480 to 489 of ERF was inserted into the pEGFP-C1 vector to test its possible NLS function. Carboxyl-terminal deletion constructs had 2 to 7 additional amino acids encoded by the vector linker sequence. In frame fusion was verified by sequencing and/or protein production by immunoblotting (see below).
Cell culture and transfection.
Ref-1 cells, Ref-9 cells (Ref-1 cells overexpressing wild-type ERF), Saos2 cells, primary mouse fibroblasts (MEFs) prepared from day 12.5 mouse embryos, and MEFs carrying a homozygous deletion of the Rb gene (kindly provided by C. Sardet, Institut de Génétique Moléculaire de Montpellier, Montpellier, France) were cultured in Dulbecco's modified Eagle medium (Life Technologies) supplemented with 10% fetal bovine serum (Life Technologies) and penicillin-streptomycin (Life Technologies) at 37°C in 8% CO2. Ref-1, Ref-9, and Saos2 cells were transfected with Lipofectamine (Life Technologies) and MEFs were transfected with FuGENE (Roche) according to the respective company protocols and analyzed 24 h after transfection. Transfected cells were fixed with 4% paraformaldehyde and analyzed by microscopy for GFP autofluorescence. Ref-mut7A cell lines overexpressing the mutant ERF mut7A, carrying an alanine-to-threonine mutation at position 526, were generated by their ability to express a cotransfected neomycin resistance gene and to proliferate in the presence of 400 μg of Geneticin sulfate (Life Technologies)/ml. Positive cell lines were scored for their ability to overexpress ERF mut7A, as detected by the anti-ERF S17S antibody (52) but not the antibody specific for the ERF protein phosphorylated at threonine 526 (P7; see below).
Antibodies.
The anti-GFP rabbit polyclonal antibody was purchased from Clontech and used according to the company specifications. The S17S and M15C rabbit polyclonal anti-ERF antibodies were described previously (52). Phospho-specific antibodies P3-4 and P7 were generated after sheep immunization with the GPEPLS(p)PFPVS(p)PLAG (P3-4) and AGGPLT(p)PRRVSS (P7) peptides, respectively, and purified by affinity chromatography. The specificity of the P3-4 and P7 antibodies was analyzed by both indirect immunofluorescence and immunoblotting. The corresponding phosphopeptide, but not the corresponding nonphosphorylated peptide, effectively blocked the antibody signal. Signal intensities were proportional to ERF levels, as determined by using the S17S and M15C anti-ERF antibodies. P3-4 and P7 antibodies did not recognize the ERF proteins when serines 246 and 251 or threonine 526, respectively, was mutated to alanine. In addition, both antibodies recognized ERF proteins produced in bacteria or a cell-free system only after in vitro phosphorylation by Erk2 kinase. Both serines 246 and 251 are phosphorylated in vivo and in vitro; however, it is not clear at this point if the P3-4 antibody recognizes ERF molecules phosphorylated at both or one of the two phospho-serines. The rabbit polyclonal anti-Erk antibody was purchased from New England Biolabs, and the mouse monoclonal phospho-specific anti-Erk MAPK-YT antibody was purchased from Sigma. The anti-CRM1 rabbit polyclonal antibody was kindly provided by E. Paraskeua (Zentrum für Molekulare Biologie der Universität Heidelberg, Heidelberg, Germany). Leptomycin B (LMB) was a generous gift from B. Wolff-Winiski, Novartis Forschungsinstitut, Vienna, Austria.
Cell staining.
GFP and GFP-ERF fusions were detected and scored after transient transfection of the respective plasmids, as described previously (32). The ERF protein was detected in methanol-acetone-fixed cells by the S17S antibody at a 1:100 dilution in 50 mM Tris-HCl-138 mM NaCl-2.7 mM KCl plus 3% bovine serum albumin (BSA) and was visualized with a biotin-labeled goat anti-rabbit antibody at a 1:200 dilution and streptavidin-fluorescein isothiocyanate (Jackson ImmunoResearch) at a 1:500 dilution in the same buffer. The phosphorylated ERF protein was detected under the same conditions with the P3-4 sheep polyclonal anti-ERF antibody at 1:50 dilution and visualized with a biotin-labeled donkey anti-sheep antibody at a 1:100 dilution (Jackson ImmunoResearch) and streptavidin-fluorescein isothiocyanate at a 1:500 dilution (Jackson ImmunoResearch). For the double ERF/phospho-ERF staining, cells were fixed in methanol-acetone and blocked with 5% normal donkey serum in 10 mM phosphate buffer (pH 7.4)-138 mM NaCl-2.7 mM KCl-0.05% Tween 20 (PBST). They were then incubated with both the P3-4 and S17S antibodies at 1:25 and 1:100 dilutions, respectively, in PBST-1% BSA, followed by incubation with a biotinylated donkey anti-sheep antibody diluted 1:100 in PBST-1% BSA. The cells were then blocked with 5% normal goat serum in PBST and incubated with a rhodamine-labeled goat anti-rabbit antibody and streptavidin-fluorescein isothiocyanate at 1:100 and 1:500 dilutions, respectively, in PBST-1% BSA. The secondary antibodies from Jackson ImmunoResearch (ML; multilabeling grade) were absorbed against proteins from other species. The specificity of the signal was evaluated by omitting one primary or secondary antibody at a time to assure no cross-reactivity. The anti-phospho-Erk mouse monoclonal antibody MAPK-YT (Sigma) was used on 4% formaldehyde-fixed cells at 1:50 dilution in PBST-1% BSA. The monomeric cyanine nucleic acid stain TO-PRO-3 (Molecular Probes) was used at 1 μg/ml. Fluorescence microscopy was performed on a Zeiss Axiovert fitted with a 100× oil submersion objective. For confocal microscopy, samples were scanned on the x-y plane with a Leica TCS NT microscope with a 63× oil submersion objective or a Bio-Rad Radiance 2100 fitted on a Zeiss AxioskopeII+ microscope with a 40× objective. Fluorescein was excited by laser light at 488 nm, rhodamine was excited by laser light at 543 nm, and TO-PRO-3 was excited by laser light at 637 nm. Beam power and channel gains were adjusted to minimize cross-detection, while for multicolor detection samples were scanned with lambda (laser) strobing. Images were electronically merged with the LCS, version 2.5, Leica software or the Laser Sharp 2000 Bio-Rad software and saved as TIFF files. Representative single optical sections were assembled with GraphicConverter, version 4.0.9 (Lemke software), and Canvas, version 7 (Deneba software). Colocalization images were analyzed first with the confocal microscope software and then reconstructed from individual scans with GraphicConverter or Photoshop, version 6 (Adobe software).
Immunoblotting and immunoprecipitation.
Cell extracts (usually 10 μg of total protein) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and proteins were transferred onto nitrocellulose membranes (Schleicher & Schuell) and detected by immunoblotting as described previously (32, 52). Immunoprecipitations and immunodetections of ERF complexes were performed in 10 mM Tris-HCl-100 mM NaCl-0.1% Triton X-100 (TNT buffer) with 1 mg of either the M15C, the P3-4, or the P7 anti-ERF polyclonal antibody for every 100 mg of cell extract. The extracts were incubated overnight at 4°C with the antibodies. Sepharose-protein A or Sepharose-protein G beads (Pharmacia Biotech) were added for 1 h at 4°C to absorb the antibodies, and the beads were washed four times with 100 volumes of TNT buffer. Bound proteins were resolved by SDS-PAGE and transferred onto nitrocellulose membranes. CRM1 was detected with the rabbit polyclonal anti-CRM1 antibody at 1:1,000 dilution in 50 mM Tris (pH 8.0)-150 mM NaCl-0.05% Tween 20 (TBST) with 1% nonfat dry milk. The anti-ERF S17S antibody was used in the same buffer at a dilution of 1:2,000. P3-4 and P7 polyclonal sheep antibodies were used at 1:2,000 and 1:1,000 dilutions, respectively, in the same buffer. Goat anti-rabbit and donkey anti-sheep horseradish peroxidase-conjugated antibodies (Jackson ImmunoResearch) were used as secondary antibodies at a 1:5,000 dilution in TBST. Western blots were visualized by chemiluminescence using the ECL reagents (Amersham Pharmacia Biotech) and exposure to X-ray film.
BrdU incorporation.
Monitoring of the newly synthesized DNA in cells was performed as described previously (32). Briefly cells were transfected with a CD4-expressing plasmid as a marker and the plasmids indicated in the legend to Fig. 6 and labeled with 50 mM bromodeoxyuridine (BrdU) 8 h before harvest. Twenty-four and 48 h after transfection the cells were fixed and stained with an anti-BrdU mouse monoclonal antibody (Sigma) and analyzed by fluorescence microscopy. CD4-positive cells were detected with a fluorescein isothiocyanate-conjugated anti-CD4 mouse monoclonal antibody (Immunotech).
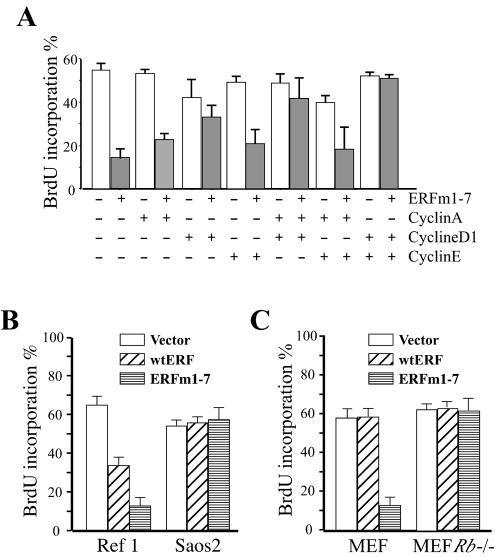
FIG. 6.
Nuclear ERF arrests cell proliferation in an Rb-dependent manner. (A) Ref-1 cells were cotransfected in the presence (gray bars) and absence (white bars) of ERF mut1-7A with expression plasmids encoding cyclin A, cyclin D1, and cyclin E as indicated and a plasmid encoding CD4 as a marker for the transfected cells. Twenty-four hours after transfection, cells were exposed to BrdU for 8 h, and the percentage of transfected cells that proceed to DNA replication during this 8-h window was monitored by indirect immunofluorescence with an anti-BrdU antibody. A minimum of 50 transfected cells were scored for each combination in three independent experiments. (B) Ref-1 cells and Saos2 cells were transfected with an empty vector, wild-type ERF (wtERF), or ERF mut1-7A (ERFm1-7) and a plasmid encoding CD4 to detect transfected cells. DNA synthesis was monitored as for panel A. A minimum of 50 transfected cells were scored in three independent experiments (C) Primary MEF cultures and MEF cultures from mice currying a homozygous deletion of the Rb gene (Rb−/− MEFs) were transfected with an empty vector, wtERF, or ERF mut1-7A and a plasmid encoding CD4 to detect transfected cells. DNA synthesis was monitored as for panel A. A minimum of 50 transfected cells were scored in three independent experiments.
RESULTS
Nucleocytoplasmic shuttling of ERF.
We have previously shown that subcellular positioning of ERF is phosphorylation dependent. Phosphorylated ERF is found almost exclusively in the cytoplasm, while the nuclear protein is nonphosphorylated (32). However, it was unclear whether phosphorylation blocks nuclear import or facilitates nuclear export. To address the possible role of phosphorylation in nuclear import, we used ERF-GFP fusion proteins carrying phosphorylation-mimicking glutamic acid mutations at positions S246, S251, and T526 (mut347E), which are known to be phosphorylated in vivo by Erks. We also used proteins carrying the T526E mutation alone (mut7E) since T526 is a major site phosphorylated by Erk after serum induction, even in cells that have high basal levels of Erk activity in the absence of serum, such as HeLa cells (52). Wild-type and mutated GFP-ERF fusions were introduced into Ref-1 cells by transient transfection. The wild-type ERF and ERFs with glutamic acid mutations enter the nucleus with identical kinetics after serum starvation (Fig. 1A). This was also true for the ERF proteins carrying alanine mutations at one or more of the seven phosphorylation sites (not shown), suggesting that nuclear import of ERF may be independent of its phosphorylation state and that under exponential growth the phosphorylated ERF protein may enter the nucleus and be rapidly reexported. Thus the observed localization under exponential growth conditions is probably due to the balance of the import and export kinetics.
FIG. 1.
ERF nuclear import is a phosphorylation-independent process. (A) Ref cells were transfected with plasmids expressing the indicated GFP-ERF fusion proteins. The localization of the fusion proteins under the indicated growth conditions was determined by measuring the GFP fluorescence under UV light. A minimum of 100 transfected cells were counted for each plasmid and growth condition, in at least three independent experiments. The mutations are as follows: 7A, T526A; 7E, T526E; 347E, S246E-S251E-T526E. (B) The localization of ERF in exponentially growing Ref-1 cells after 30 and 60 min of treatment with 300 mM LMB was determined by indirect immunofluorescence. The S17S antibody was used to determine total ERF protein (top), and the P3-4 phospho-specific antibody was used to determine the localization of ERF protein phosphorylated at S246 and S251 (middle). The localization of the activated Erks under the same conditions was determined with the phospho-specific anti-Erk mouse monoclonal antibody MAPK-YT, as a control (bottom). (C) The total amount of the phosphorylated ERF protein under the conditions described for panel B was determined by immunoblotting using the phospho-specific anti-ERF antibodies P3-4 and P7.
To examine the possibility of continuous dynamic shuttling of ERF and to verify the ability of phosphorylated ERF to enter the nucleus, we used LMB, a specific inhibitor of CRM1 (18, 30, 31), to block CRM1-dependent nuclear export. To determine the phosphorylation status of the ERF protein and the phosphorylation events that may be important for its regulation, we developed two specific antibodies that recognize the phosphorylated forms of ERF (see Materials and Methods). The P3-4 antibody recognizes ERF proteins phosphorylated at serine 246 and/or serine 251 (positions 3 and 4), while the P7 antibody recognizes ERF phosphorylated at threonine 526 (position 7).
Treatment of exponentially growing cells with LMB resulted in the nuclear accumulation of ERF. After 30 min of treatment with 300 nM LMB, ERF could be detected both in the cytoplasm and the nucleus, while complete nuclear localization of the protein could be observed 1 h after treatment with LMB. The nuclear export of ERF after serum induction was also blocked by LMB (not shown), suggesting that nuclear export follows the same CRM1-mediated pathway during continuous shuttling and serum-induced export. Determination of the subcellular localization of ERF by indirect immunofluorescence with both the S17S and the P3-4 anti-ERF antibodies yielded identical patterns (Fig. 1B). In addition, all the ERFs with glutamic acid and alanine mutations accumulate in the nucleus with comparable kinetics, and they are all nuclear after 60 min of LMB treatment (not shown). These data indicate that ERF nuclear entry is unaffected by its phosphorylation status. Activated Erk also accumulated in the nucleus after LMB treatment with similar kinetics (Fig. 1B), consistent with previous results (2). These data suggest that the cytoplasmic localization of ERF in exponentially growing cells is a dynamic process and that the observed distribution is probably due to the much higher affinity of phosphorylated ERF for the export machinery than for the import machinery.
To minimize the possibility that the overall levels of ERF phosphorylation and nuclear accumulation change as a result of treatment with LMB, we examined the extent of phosphorylation of ERF at either positions S246 and S251 or position T526 by Western blotting. Our data indicate that treatment with LMB did not affect the level of ERF phosphorylation, as determined with the phospho-specific antibodies (Fig. 1C), suggesting that in exponentially growing cells it is the phosphorylated form of ERF that constantly shuttles between the nucleus and the cytoplasm.
ERF is phosphorylated in the nucleus prior to its exit.
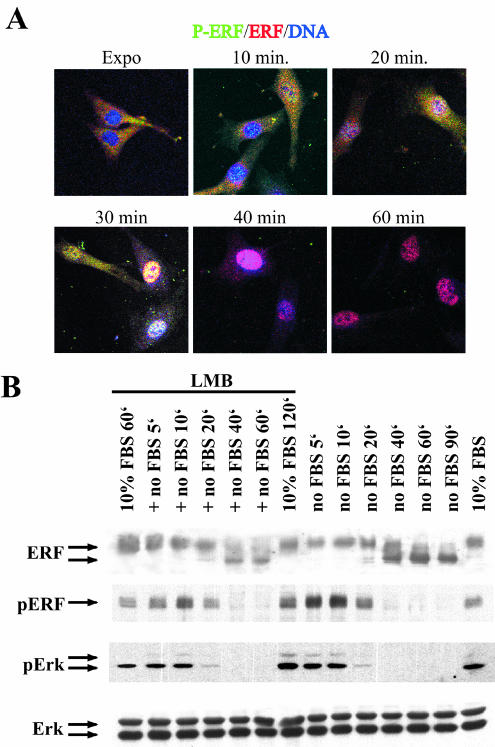
We have shown previously that the multisite phosphorylation of ERF mediates its subcellular localization. To further our understanding of this process, we analyzed the timing and the cellular compartment where phosphorylation may take place, both by confocal microscopy and biochemical fractionation. We utilized the specific antibodies (see Materials and Methods) that can recognize ERF only when it is phosphorylated at positions S246 and S251 (P3-4) or position T526 (P7) to correlate phosphorylation status with subcellular localization. We first analyzed ERF localization during serum deprivation and serum stimulation by indirect immunofluorescence utilizing the P3-4 and the S17S anti-ERF antibodies (Fig. 2A). Under normal growth conditions, phosphorylated ERF is almost exclusively cytoplasmic and only minimal nuclear staining is observed. Two hours after serum withdrawal no staining was evident with the phospho-specific antibody, and the ERF protein was nuclear, as is evident by the purple color of the nuclei (Fig. 2A) (32). Two minutes after serum addition, ERF phosphorylated at positions S246 and S251 can be detected in the nucleus, as shown by the white spots in the nuclei of some cells. Four minutes after serum stimulation phosphorylated ERF can be detected in the nuclei of all the cells, while 7 min after serum stimulation phosphorylated ERF can be detected in the cytoplasm of some cells, as shown by the yellow color in the cytoplasm. Ten minutes after serum stimulation the majority of the ERF is phosphorylated and cytoplasmic, and the export process is completed 15 min after serum stimulation (Fig. 2A, left). The timing of ERF phosphorylation and export closely follows the activation of Erk and its localization to the nucleus (Fig. 2A, right). These data suggest that, in response to Erk activation, ERF is phosphorylated within the nucleus and is exported as a phospho-protein into the cytoplasm.
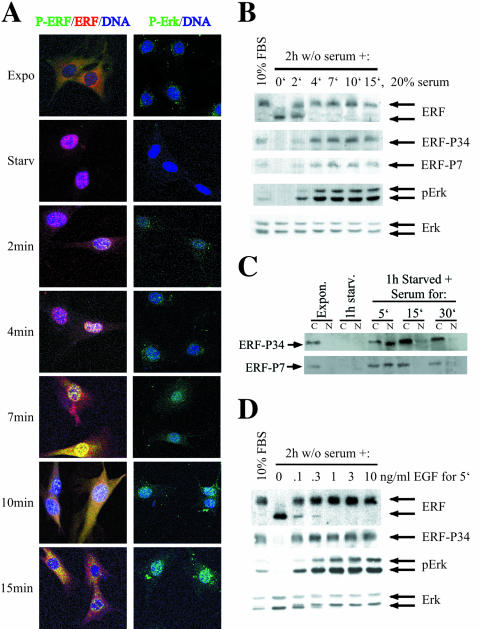
FIG. 2.
ERF is phosphorylated in the nucleus. (A) Ref cells were subjected to serum arrest for 2 h and consequent serum stimulation for the indicated times. The localization of the phosphorylated ERF was determined by indirect immunofluorescence using the P3-4 phospho-specific anti-ERF antibody (green), and that of total ERF was determined with the S17S anti-ERF antibody (red). Nuclei ware stained blue by TO-PRO-3 (left). At the same time points, activated Erks were detected with the phospho-specific anti-Erk monoclonal antibody MAPK-YT (green). Nuclei ware stained blue by TO-PRO-3 (right). Red and blue color colocalization shows as magenta, green and blue colocalization shows as cyan, green and red colocalization shows as yellow, and colocalization of all three shows as white. (B) Under the same conditions as in panel A, total cell extract was analyzed by immunoblotting for total and phosphorylated ERF and Erks, as indicated. The S17S, P3-4, P7, MAPK-YT, and anti-Erk specific polyclonal antibodies were used to determine total ERF, ERF phosphorylated at S246 and S251, ERF phosphorylated at T526, phosphorylated Erks, and total Erks, respectively. FBS, fetal bovine serum. (C) Ref cells were deprived of serum for 1 h and then induced with serum. At the indicated times cells were harvested and separated into nuclear and cytoplasmic fractions. The amount of the phosphorylated ERF protein in each fraction was determined by immunoblotting. Both the P3-4 (top) and the P7 (bottom) phospho-specific anti-ERF antibodies were used. (D) Ref cells were serum arrested for 2 h and then treated with the indicated amounts of epidermal growth factor for 5 min. Total cell extracts were analyzed for total and phosphorylated ERF and Erks, as for panel B.
To better quantify phosphorylation during this process, we determined the levels of ERF, phospho-ERF, Erk, and phospho-Erk by immunoblotting. The total ERF and Erk protein levels remain constant during this process, and the level of ERF phosphorylation closely parallels the Erk activation level (Fig. 2B). Interestingly, 2 min after serum stimulation the mobility of the phosphorylated ERF protein appears to be somewhat faster than at the other time points, suggesting that distinct phosphorylation steps may be involved during nuclear export.
The phosphorylation-induced nuclear export was further confirmed by biochemical subcellular fractionation (Fig. 2C). During exponential growth, phosphorylated ERF can be found only in the cytoplasmic fraction, while in the absence of serum no phosphorylated protein can be detected in either fraction (Fig. 2C) and the protein is exclusively nuclear (Fig. 2A) (32). Five minutes after the addition of serum phosphorylated ERF is found primarily in the nucleus but also in the cytoplasm. ERF phosphorylated at positions S246 and S251 exhibits a somewhat slower mobility in the cytoplasmic fraction than the nuclear protein, suggesting that additional modifications may occur after or during nuclear export (Fig. 2C). At the same time point ERF phosphorylated at position T526 is distributed almost equally between the nucleus and the cytoplasm and has the same mobility (Fig. 2C, bottom). This is consistent with a distinct role for the phosphorylation at T526 during export (see below). Fifteen minutes after serum addition phosphorylated ERF is almost exclusively cytoplasmic (Fig. 2C).
Utilizing specific inhibitors of the activation of MEK1, the protein kinase that activates Erk, we have previously shown (32) that ERF phosphorylation and its cytoplasmic localization and serum-induced nuclear export are totally dependent on Erk activity. To verify the Erk dependence of ERF phosphorylation, we induced the activation of Erk in serum-arrested cells with increasing amounts of epidermal growth factor (EGF). Within 5 min of EGF addition Erk became activated and the ERF was phosphorylated (Fig. 2D) and, as with serum, phosphorylation of ERF led to nuclear export (not shown). Even very low concentrations of EGF (0.1 to 0.3 ng/ml) inducing partial activation of Erk were sufficient to induce ERF phosphorylation (Fig. 2D). This is consistent with the phosphorylation status of ERF in exponentially growing cells where only a low level of activation of Erk can be observed, characteristic of many established cell lines. Thus, our data unambiguously suggest that, in response to Erk activation, nuclear ERF is first phosphorylated in the nucleus and then exported to the cytoplasm.
ERF phosphorylation at position 526 has a distinct role in export.
We have shown that multisite phosphorylation of ERF is required for nuclear export. To determine the possible effect of the phosphorylation of the different sites on the export process, we first used GFP-ERF fusions carrying either glutamic acid or alanine mutations at the putative phosphorylation sites and analyzed their localization. The wild type and all the mutated forms of ERF introduced into Ref-1 cells exhibited a predominantly nuclear localization 1 h after serum deprivation or LMB treatment (not shown), suggesting that the mutations do not affect nuclear import. However, when serum-deprived cells were stimulated for 30 min with serum, the subcellular distribution of each mutated form was different (Fig. 3A). Wild-type ERF was exported into the cytoplasm efficiently and had a distribution identical to that observed under steady-state conditions. Similarly, both the 3-7A and the 1-7A mutated forms (see the Fig. 3 legend for the nomenclature of the mutations) remained mostly nuclear under both conditions, with 1-7A form exhibiting an almost exclusively nuclear localization. This indicates that multisite phosphorylation is required for effective exit. The 1-5A form exhibited partially impaired nuclear exit after serum induction but was more cytoplasmic at this stage compared to its steady-state distribution. This suggests that phosphorylation at the remaining phosphorylation sites was able to support effective nuclear export under serum induction conditions. The 7A mutation had a marginal effect on export under serum induction conditions but a strong effect on steady-state distribution, suggesting that phosphorylation on this site may not by itself be critical for export (Fig. 3A and B). In addition, both the ERF 347E and 7E mutations impaired or delayed export and had no effect on steady-state distribution, suggesting that a charged amino acid at position 526 delays export (Fig. 3A and B). This is consistent with the efficient export of the 7A mutant form. However, phosphorylation at position 526 must be required at some stage of the shuttling process, since the alanine mutation at this position affects the steady-state distribution of the ERF protein.
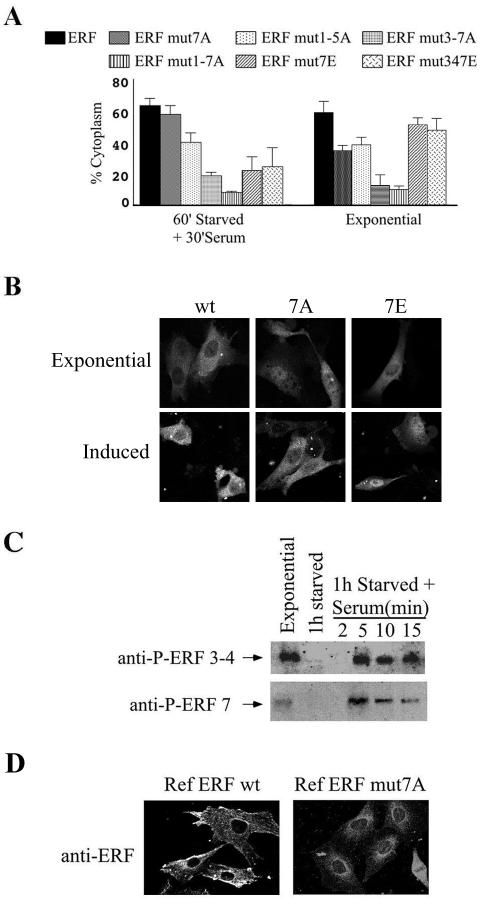
FIG. 3.
Phosphorylation of threonine 526 plays a distinct role in nuclear export. (A) The localization of the indicated GFP-ERF fusion proteins after transfection of the corresponding plasmids into Ref-1 cells was determined as described for Fig. 1A under exponential growth and after 60 min of serum withdrawal followed by 30 min of serum stimulation. A minimum of 100 transfected cells were counted for each plasmid and growth condition in at least three independent experiments. The mutations are as follows: 7A, T526A; 1-5A, T148A-S161A-S246A-S251A-T271A; 3-7A, S246A-S251A-T271A-T357A-T526A; 1-7A, T148A-S161A-S246A-S251A-T271A-T357A-T526A; 7E, T526E; 347E, S246E-S251E-T526E. (B) Localization of the wild-type (wt) and T526A (7A) and T526E (7E) mutated forms of the ERF-GFP fusions during exponential growth and 15 min after serum induction. (C) Ref cells were subjected to serum withdrawal and stimulation. At the indicated time points the levels of the total phosphorylated ERF protein were determined by immunoblotting using the P3-4 (top) and the P7 (bottom) anti-Erf phospho-specific antibodies to determine phosphorylation at serines 246 and 251 and T526, respectively. (D) The localization of ERF was determined by indirect immunofluorescence using the S17S anti-ERF antibody in Ref-1-derived cell lines overexpressing comparable levels of wt ERF (left) or ERF carrying the T526A mutation (right).
We have shown previously (52) that T526 is the major phosphorylation site after serum induction in HeLa cells that have high basal Erk activity, indicating that this site may be preferentially dephosphorylated in vivo. Using the P3-4 and P7 phospho-specific anti-ERF antibodies we compared the total amounts of ERF protein phosphorylated at these positions during exponential growth, serum starvation, and serum induction (Fig. 3C). Five minutes after serum induction, the time at which maximum phosphorylation can be observed with each antibody, was used as a reference point. It is evident that phosphorylation at position 526 is decreased not only during exponential growth but also 10 and 15 min after serum induction, suggesting that both phosphorylation and dephosphorylation of threonine 526 may be required during the export process. This would be consistent with the export kinetics data for the alanine and glutamic acid mutations in this position (Fig. 3A and B) and could explain the mobility differences observed during serum induction (Fig. 2B and C).
To further analyze the role of T526 phosphorylation in ERF subcellular localization, we generated Ref-1 cell lines that express ERF with the threonine-to-alanine mutation at position 526. In these cells, we compared the localization of ERF with the T526A mutation to that of wild-type ERF by indirect immunofluorescence. To minimize the contribution of endogenous ERF and account for possible effects of overexpression, we used for comparison Ref-1 cells overexpressing wild-type ERF and adjusted the antibody conditions so that the endogenous ERF would not be detectable. The T526A mutant ERF could be detected both in the cytoplasm and in the nuclei of these cells, in contrast to wild-type ERF, which was cytoplasmic. The mutated ERF however, exhibited a characteristic perinuclear staining, which included the nuclear membrane (Fig. 3D), indicating that the protein accumulated on the cytoplasmic side of the nuclear membrane and was not distributed throughout the cytoplasm, like the wild-type ERF protein.
These data suggest that phosphorylation at threonine 526 probably occurs at a later point during export, facilitating the release of the protein in the cytoplasm. At the same time, as suggested by the glutamic acid mutation, the loading of ERF on the cargo protein in the nucleus could be inhibited by the phosphorylation of this site.
ERF has no autonomous NES and two NLSs.
ERF is actively exported from the nucleus via a CRM1-dependent mechanism both during exponential growth and after serum induction, as indicated by the effect of LMB on its export (Fig. 1B). However, no previously described NESs can be detected within the ERF primary sequence (18, 24, 38). In contrast, the NLS within the ets DNA-binding domain described for other ets proteins (9) is also conserved in ERF. To determine regions of ERF responsible for nuclear export and import, we utilized GFP-ERF hybrids that contained various fragments of ERF. These fusions were transfected into Ref-1 cells, and their subcellular localization was determined during exponential cell growth, serum arrest, and serum induction and in the presence of LMB (Fig. 4A). Under these conditions we could determine effects on dynamic equilibrium, import, export, and active transport versus diffusion, respectively. Our data indicate that two distinct regions on ERF harbor an NLS, one within the ets DNA-binding domain, also described for other ets proteins, and an additional one within the carboxyl-terminal repression domain. Indeed the ERF protein sequence between amino acids 480 and 489 has all the characteristics of a basic amino acid NLS (Fig. 4A and B). When this 10-amino-acid region was fused to GFP, its nuclear localization was increased to levels seen for the 472-to-548 fragment (see below). The NLS within the ets domain appears to be more potent, since its elimination leads to only partial nuclear localization in the absence of serum (Fig. 4A, fragment consisting of amino acids 100 to 548 [fragment 100-548] versus the full-length protein). However, the NLS within the repression domain can drive nuclear localization of the GFP fusion effectively in the absence of other ERF sequences (Fig. 4A, fragment 472-548), suggesting that is indeed a functional NLS.
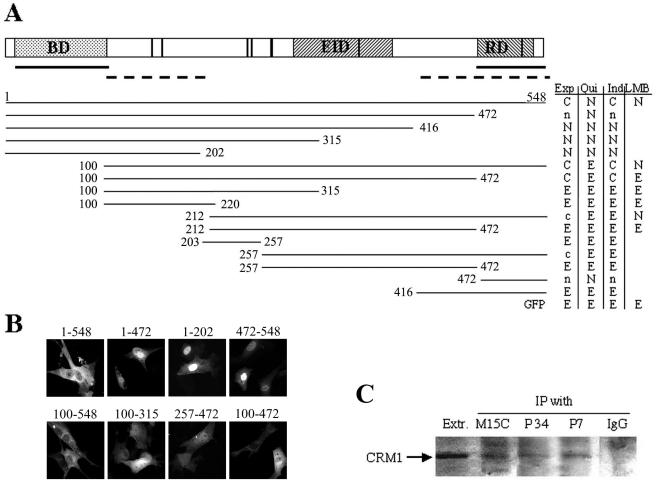
FIG. 4.
ERF has two NLSs but no autonomous NES to mediate CRM1-dependent export. (A) Plasmids encoding GFP-ERF hybrids were transfected into Ref-1 cells, and the protein localization was determined by measuring the GFP fluorescence. The localization was determined during exponential growth 24 to 28 h after transfection (Exp column), after 3 h of serum deprivation (Qui column), after 3 h of serum deprivation and 30 min of serum stimulation (Ind column), and after 60 min of LMB treatment in the presence of serum (LMB column). A minimum of 100 transfected cells were counted for each plasmid and growth condition in at least three independent experiments, and the localization of the hybrids is indicated as follows: C, >60% cytoplasmic localization; c, 30 to 60% cytoplasmic localization; N, >60% nuclear localization; n, 30 to 60% nuclear localization; E, ubiquitous distribution. A diagram of ERF is presented at the top. BD, ets DNA-binding domain; EID, Erk interaction domain; RD, repressor domain. The vertical lines indicate the seven optimal putative Erk phosphorylation sites. Solid lines below the diagram, NLS-containing regions; dashed lines, regions contributing to nuclear export. The regions of ERF fused to the C terminus of GFP are indicated by lines below. The end points of the ERF segments are indicated by the corresponding amino acid number of the human ERF protein. (B) Representative images of the localization of GFP-ERF deletion mutations during exponential growth. (C) ERF was immunoprecipitated (IP) from cellular extracts of exponentially growing cells with the M15C, P3-4, or P7 anti-ERF specific antibodies or with control serum (IgG lane). The presence of CRM1 in the immunoprecipitated complexes and the original extract (Extr. lane) was detected by immunoblotting using an anti-CRM1 specific antibody.
In contrast to what was found for the NLS, we were unable to determine a single region within the ERF protein capable of serving as a NES. At least two extended areas are required for nuclear export. One harbors the ERF repressor domain at the carboxyl terminus of the protein and extends from amino acid 418 to the end of the protein. Elimination of the last 76 amino acids (Fig. 4A, fragment 1-472) leads to partial nuclear localization in exponentially growing cells, which is stronger after the elimination of 56 additional residues (Fig. 4A, fragment 1-416). The contribution of this region to nuclear export is also evident in a comparison of the 100-472 and 100-315 hybrids. The 416-to-548 region by itself has a ubiquitous subcellular distribution (Fig. 4, fragment 416-548), in contrast to the nuclear localization of the last 76 residues (Fig. 4A, fragment 472-548). The other region required for effective nuclear export lies between residues 100 and 212. Thus, the 212-548 fusion has a weaker cytoplasmic distribution than the 100-548 fusion and is not exported effectively after serum induction. These differences are more evident when the last 76 residues are eliminated (Fig. 4A, fragment 100-472 versus 212-472). However, this region by itself is incapable of serving as a NES (Fig. 4A, fragment 100-220). These data indicate that multiple regions on ERF are required for efficient nuclear export. This is consistent with our findings regarding the role of multisite phosphorylations in nuclear export (32) and suggests a mechanism involving structural changes in ERF that allow interaction with an adaptor/carrier protein linking ERF with the CRM1 export machinery.
However, given the lack of recognizable NES signals within the ERF sequence, the CRM1 block by LMB may have either a direct or an indirect effect on ERF localization. To address this question, we investigated the possible physical association of the two proteins. We used three different anti-ERF antibodies to immunoprecipitate ERF from exponentially growing cells and determined the presence of CRM1 in these complexes using a CRM1-specific antibody. All three anti-ERF antibodies, namely, M15C, an antibody against the N terminus of ERF (52), and the P3-4 and P7 antibodies were able to immunoprecipitate ERF (not shown). In all cases we were able to detect the presence of CRM1 in the ERF-immunoprecipitated complexes (Fig. 4C). Interestingly, complexes immunoprecipitated with the antibody against the ERF protein phosphorylated at T526 contained amounts of CRM1 comparable to those contained by the complexes precipitated with the P3-4 or the amino terminus-specific M15C antibody. Considering the lower abundance of the ERF molecules phosphorylated at T526 in exponentially growing cells (Fig. 3B), this could be an additional indication that phosphorylation at this position is directly involved in the export process.
These data further indicate that ERF may associate with CRM1-containing complexes and may be exported from the nucleus via a CRM1-mediated mechanism. However, we were unable to detect a direct interaction between bacterially expressed ERF and CRM1 proteins, regardless of the phosphorylation state of ERF (not shown). This suggests that the interaction of CRM1 with ERF may be mediated by other proteins, consistent with the absence of a NES motif within the ERF protein sequence, the extensive regions on ERF required for effective export, and the multisite phosphorylations that precede ERF nuclear export in vivo.
ERF is a sensor of nuclear Erk activity.
Our data suggest that nuclear phosphorylation is the critical step regulating CRM1-mediated ERF nuclear export. How this process is reversed was unclear. To elucidate steps in this process, we studied the dephosphorylation and localization of ERF during serum arrest. As early as 10 min after serum withdrawal phosphorylated ERF protein can be found in the nucleus, while 20 min after serum withdrawal all cells contain phosphorylated ERF protein. Thirty minutes after serum withdrawal most of the ERF protein is nuclear but still phosphorylated, while after 40 min most of the cells contain nonphosphorylated nuclear ERF and after 60 min the nuclear translocation and dephosphorylation process is completed (Fig. 5A). Within the 30-min period after serum deprivation, we did not observe any major distribution differences between total and phosphorylated ERF. These data further suggest that the phosphorylated protein can enter the nucleus effectively, where it may eventually be dephosphorylated.
FIG. 5.
ERF is dephosphorylated in the nucleus as a consequence of decreased nuclear Erk activity. (A) Ref cells were serum arrested and at the indicated time points after serum deprivation the localization of total (red) and phosphorylated (green) ERF was determined by indirect immunofluorescence as for Fig. 2A. Nuclei ware stained blue by TO-PRO-3. Red and blue color colocalization shows as magenta color, green and blue colocalization shows as cyan, green and red colocalization shows as yellow, and colocalization of all three shows as white. (B) Ref cells were serum arrested in the presence (+ no FBS lanes) or absence (no FBS lanes) of LMB. For the LMB treatment cells were grown in complete media for 1 h in the presence of LMB (10% FBS 60′ lane) and then were treated with LMB-containing serum-free media for the indicated times. Cells were also treated for 2 h with LMB in complete media (10% FBS 120′ lane) to account for any possible effects of the prolonged LMB treatment. At the indicated time points total cellular extracts were analyzed for ERF levels with the S17S antibody (top panel), phosphorylated ERF (pERF) levels with the P3-4 antibody (second panel), phosphorylated Erk1 and -2 levels with an anti-phospho-Erk antibody (third panel), and Erk1 and -2 levels with an anti-Erk antibody (fourth panel).
To test whether nuclear dephosphorylation of ERF is possible, we took advantage of the nuclear accumulation of phosphorylated ERF in the presence of LMB. LMB treatment of exponentially growing cells leads to nuclear accumulation of both the phosphorylated ERF protein and activated Erks (Fig. 1B). In the presence of LMB and the absence of serum, ERF and Erks were both dephosphorylated while remaining nuclear (not shown), indicating that ERF can be dephosphorylated within the nucleus.
To better quantify the kinetics of ERF dephosphorylation, we analyzed the level of phosphorylation after serum withdrawal and in the absence and presence of LMB (Fig. 5B). Under both conditions, the dephosphorylation of ERF follows identical kinetics. Bands of faster mobility, suggestive of ERF dephosphorylation, can be detected with an ERF-specific antibody 20 min after serum withdrawal, while after 40 min only faster-moving molecules can be detected (Fig. 5B, top panel). The dephosphorylation of ERF was confirmed by the initial small decrease (20 min) and later absence (40 min) of specific bands detected with the phospho-ERF-specific antibody, P3-4 (Fig. 5B, second panel). The similar dephosphorylation kinetics in the absence of serum of the ERF that is blocked in the nucleus and the ERF that can still shuttle through the nuclear pores suggest that ERF dephosphorylation may indeed take place in the nucleus.
ERF phosphorylation is very sensitive to the level of Erk activity (Fig. 2). We therefore monitored the Erk activity level during serum withdrawal using the phospho-specific anti-Erk antibody. Both in the absence and presence of LMB, Erk dephosphorylation and inactivation slightly preceded ERF dephosphorylation. Twenty minutes after serum withdrawal there was a significant decrease in the abundance of the phosphorylated Erk, which was absent at later time points (Fig. 5B, third panel), while there was no detectable change in the overall Erk level (Fig. 5B, fourth panel).
Taken together, these data suggest that the balance between a nuclear phosphatase that dephosphorylates ERF and nuclear Erk activity may determine the phosphorylation status of ERF. Thus, it appears that ERF acts as a constant quantitative sensor of nuclear Erk activity, providing a link between signal duration and strength on the one hand and transcriptional and biological outcomes on the other.
Cell cycle arrest by nuclear ERF is Rb dependent.
A biological outcome in which ERF has been implicated is cell proliferation. We have shown previously that the mutated forms of ERF exhibiting predominantly nuclear localization can arrest cells at the G0/G1 phase of the cell cycle, as determined by DNA content and new DNA synthesis (32). However, it was unclear if the cell cycle is blocked at G0 or early or late G1, or if cell cycle machinery components were directly involved in ERF-mediated inhibition. To address these questions, we overexpressed different cyclins, alone or in combination, in the presence or absence of mutated ERF that localizes in the nucleus (mut1-7) and arrests cellular proliferation (32). Overexpression of the S-phase cyclin A had no effect on ERF-mediated arrest. The same was true for the G1/S-phase cyclin E. However, addition of cyclin D1 significantly decreased ERF-mediated arrest (Fig. 6A), suggesting a G1 arrest. Coexpression of cyclin D1 and cyclin E eliminated ERF-mediated cell cycle arrest (Fig. 6A), suggesting a possible late G1 arrest by ERF. Coexpression of cyclins D1 and A had an effect similar to that of cyclin D1 alone, while coexpression of cyclins A and E had no effect on ERF-mediated arrest. These data indicate that overexpression of ERF may block cells at late G1, around the G1 restriction point. However, cyclin overexpression can affect different components of the cell machinery. Thus, to test the possible effect of ERF at late G1, we utilized the Saos2 cells that do not have a functional Rb gene. We have previously shown (32) that Ref-1 cells are partially inhibited by the overexpression of wild-type ERF and are blocked by overexpression of mutated ERF. The Saos2 cells were not affected by the overexpression of either the wild type or the nuclear form of ERF (Fig. 6B), suggesting that a functional G1 restriction point may be required for ERF-mediated arrest. To eliminate the possible contribution of other mutations that frequently accumulate in established cell lines, we tested both the ability of ERF to arrest the cell cycle and the role of a functional G1 restriction point in this arrest in primary MEFs. Wild-type and mutated ERFs were introduced into MEFs from wild-type and isogenic mice carrying a homozygous mutation of the Rb gene. Normal MEFs were arrested by the overexpression of the mutated ERF that localizes in the nucleus, a condition which mimics the state where Erk is inactive, while Rb−/− MEFs were unaffected (Fig. 6C). Wild-type ERF has no effect in either cell type. These data suggest that the nuclear localization of ERF can arrest cell cycle progression in fibroblasts in an Rb-dependent manner, indicating that ERF arrests cells in late G1, probably by interfering with the levels of specific cell cycle machinery components. Thus, ERF provides a link between the level of active Erk and cell cycle progression through the G1 restriction point.
DISCUSSION
The Ras/Erk signaling pathway regulates multiple cellular functions. A number of immediately-early response effectors that are regulated by Erk have been identified, including transcription factors, notably those of the ets family. However, for none of these effectors is it clear how the strength and the duration of the signal are integrated in their regulation. Here we present evidence that ERF, an Erk-regulated transcriptional repressor, can sense signal strength and duration via continuous phosphorylation-dependent nucleocytoplasmic shuttling, affecting the progress through G1 in an Rb-dependent manner.
ERF import.
ERF is a stable protein with a half life of more than 24 h (our unpublished observation) with a constant transcription rate in most cell types and tissues tested. The only known form of regulation is Erk-dependent phosphorylation, which alters the subcellular localization of the protein (32). Nuclear accumulation following LMB treatment of the cells strongly suggests that the subcellular localization of ERF is a dynamic process, the result of equilibrium between the import and export rates. The nuclear import of ERF does not appear to be regulated by phosphorylation. Phosphorylated ERF can be readily detected in the nucleus following CRM1 blockade by LMB and during nuclear accumulation induced by serum withdrawal. Phosphorylation-mimicking glutamic acid mutations do not affect nuclear import. Blocking nuclear import with carbonyl cyanide m-chlorophenyl-hydrazone does not affect the subcellular distribution of either cytoplasmic or nuclear mutated forms of ERF (not shown). We have shown previously that substituting the NLS present in the ets domain of ERF is not sufficient to alter the subcellular localization of chimeric ERF proteins (5), arguing against a possible phosphorylation-dependent NLS-masking mechanism. Also, the presence of two independent NLSs in ERF argues against this possibility. However, is not inconceivable that, under certain conditions in some cell types, masking of either or both NLSs might be a mechanism affecting the rate of nuclear import. Another possibility is a transient dephosphorylation of ERF during its entry into the nucleus. However, we were not able to detect such a form by confocal microscopy or by immunoblotting in proliferating cells during serum starvation or LMB-induced nuclear accumulation. Finally, our export data indicate that phosphorylated ERF can pass through nuclear pores very effectively. Thus, phosphorylation-dependent nuclear import does not appear to regulate ERF localization.
ERF export.
In contrast to the rate of import, the rate of nuclear export of ERF is totally dependent on its state of phosphorylation. We have shown previously that phosphorylation-blocking mutated forms have an increasing nuclear accumulation and that multiple phosphorylations are required for effective nuclear export (32). Analysis of the export kinetics during serum stimulation suggests that there are two distinct ERF phosphorylation events in the nucleus. First, all sites except T526 are phosphorylated, probably resulting in the loading of ERF on the export machinery, and immediately after T526 is phosphorylated, allowing the effective release of the protein into the cytoplasm. The abundance T526-phosphorylated ERF molecules associated with CRM1-containing complexes suggests that phosphorylation at this position is directly involved in the export process. The delayed export of the T526E form indicates that a negative charge at this position may block loading onto the export machinery, while the perinuclear localization of the T526A form indicates a delayed release from the export machinery to the cytoplasm. This delayed release may also contribute to the increased nuclear accumulation observed under steady-state conditions (exponential growth).
It is not clear whether T526 dephosphorylation occurs in the cytoplasm or the nucleus. The decreased levels of T526-phosphorylated ERF protein observed 10 to 15 min after serum stimulation, when almost all the protein is cytoplasmic, suggest that ERF dephosphorylation occurs in the cytoplasm. Also, the level of the T526 phosphorylated ERF protein at steady state, which is estimated to be 10 to 20% of the total protein, supports this hypothesis. However, the T526E mutation does not affect nuclear import, and T526-phosphorylated ERF can be found in the nucleus after blockade of CRM1. Although rephosphorylation of ERF in the nucleus cannot be excluded under these conditions, ERF can be dephosphorylated in the nucleus after blockade of CRM1 and serum withdrawal, suggesting that nuclear dephosphorylation of T526 is also possible. Thus, it is likely that T526 dephosphorylation can occur in both compartments, ensuring the efficient export of the ERF protein under exponential-growth conditions.
ERF does not contain a CRM1-dependent leucine-rich (18) or bipartite NES (38), a CRM1-independent hnRNP K nuclear shuttling element (15, 24), or any single region capable of serving as an autonomous NES. However, the block of ERF nuclear export by LMB strongly suggests CRM1-mediated export. Indeed CRM1 can be detected in ERF complexes immunoprecipitated from cell extracts, but no interaction between these proteins has yet been detected in vitro. This suggests that interaction in vivo may require another protein. Such adaptor proteins have been found in many exported proteins, including the Drosophila ets domain repressor Yan (56) and Erk itself (1, 2). It has been shown that the nuclear export of several members of the Forkhead family of transcription factors is facilitated by their interaction of the phosphorylated proteins with 14-3-3s (40, 48; for recent review see reference 7). 14-3-3s proteins can associate with ERF in vitro (T. Dubois and A. Aitken, University of Edinburgh, Edinburgh, United Kingdom, unpublished findings). However, the 14-3-3ζ that we tested interacted in vitro equally well with phosphorylated and nonphosphorylated ERFs. Moreover, 14-3-3 proteins cannot be found in ERF-containing complexes after low-stringency immunoprecipitation and detection with a pan-14-3-3 antibody, nor can we detect any ERF-specific bands using a far-Western approach with labeled 14-3-3s. Furthermore, overexpression of wild-type or NES-deficient mutant 14-3-3ζ did not affect the localization of ERF, its binding to DNA, or its ability to function as a transcriptional repressor (our unpublished data). Thus we have no evidence to support a role for 14-3-3s in the nuclear export of ERF.
Erk1 and Erk2 associate strongly with ERF, shuttle between the nucleus and the cytoplasm, and could therefore also serve as adaptors that could also facilitate the distinct phosphorylation of T526. However, the interaction between ERF and Erk is phosphorylation independent (unpublished data) and thus cannot explain the nuclear localization of the phosphorylation-deficient mutant ERFs. In addition, the Erk interaction site is distinct from the two regions required for ERF export, (32; our unpublished data), arguing that the Erks may not be the adaptor proteins responsible for ERF export. Finally, the localization of ERF and Erk during serum induction does not support this hypothesis. We were not able to detect any protein that can serve as an export adaptor in repetitive yeast two-hybrid screenings or in low-stringency pull-downs and immunoprecipitations (our unpublished data), suggesting that a multimeric adaptor may be required for ERF export.
It is conceivable that the phosphorylation of ERF may also release it from a nuclear anchor, thereby allowing its nuclear export. It has been shown that phosphorylation does not affect DNA binding (52) and that substitution of the ERF DNA-binding domain does not affect nuclear shuttling (5; our unpublished data). We could not detect any DNA-binding-independent association with either histones or chromatin (unpublished observations). It is possible though that a nuclear protein masks the NESs. This hypothesis is consistent with the multisite phosphorylation required for ERF export. However, the lack of an autonomous NES region on ERF and its inability to associate directly with exportin suggest that such a mechanism would be additional to the phosphorylation-dependent loading onto the export machinery.
Erk monitoring and growth arrest.
We have shown previously that ERF phosphorylation and localization are totally dependent on Erk activity and that nuclear ERF can arrest the cell cycle at the G0/G1 phase (32). ERF appears to be a highly specific target for the Erk subfamily of the mitogen-activated protein kinases (MAPK). It harbors specific determinants for its interaction with Erks that are not utilized by other MAPK subfamilies (our unpublished data). Our data suggest that ERF phosphorylation mirrors the level of Erk activity and, more specifically, the level of nuclear Erk activity. The constant overall level of ERF within cells, its regulation via subcellular repositioning, and its continuous nucleocytoplasmic shuttling imply that ERF serves as a continuous monitor of nuclear Erk activity that modulates transcription in response to Erk activation and inactivation. A similar mechanism and role have only been recently suggested for Smads in the transforming growth factor β signaling pathway (60, 61).
Although the Ras/Erk pathway is considered to be a major mitogenic pathway, there is little information regarding the mechanisms via which Erks affect cell cycle machinery components. It has been suggested that the balance between cyclin D1 and p21 inhibitors is a major regulatory step (for a review see reference 49), while up-regulation of cyclinD1 and down-regulation of p27 constitute another proposed mechanism (21). Either of these would be consistent with Rb-dependent cell cycle activation by Erks (13). Our data indicate that loss of Rb or overexpression of cyclins could abolish the cell cycle arrest induced by the overexpression of the inactive-Erk-mimicking, phosphorylation-deficient mutated form of ERF. This suggests that ERF could be one of the factors mediating Ras/Erk signaling to the cell cycle machinery. This role and mode of function of ERF would be consistent with the function of the recently identified ERF homologue METS (29), which can repress cell cycle progression during macrophage terminal differentiation in a ras- and Rb-dependent manner. It is not clear which transcription factors are involved in the regulation of cyclin D1, p21, and p27. ERF as a transcriptional repressor could directly affect cyclin D1 down-regulation in the absence of Erk activity, indirectly activate p21/p27, or affect other cell cycle components required for cell cycle progression, such as cyclin-dependent kinases. ERF could also affect other genes that are involved in cell cycle regulation and that harbor ets-binding regulatory sites, such as myc (50) and p53 (57).
In conclusion our data support the hypothesis that ERF through its Erk phosphorylation-regulated nuclear export and continuous shuttling may be a link between the Ras/Erk signaling pathway and the cell cycle components regulating progression through G1 (Fig. 7).
FIG. 7.
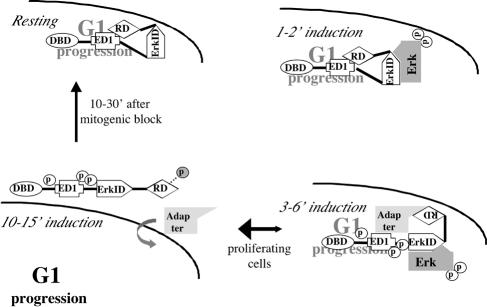
A model for the regulation and function of ERF during Erk-mediated mitogenic stimulation. (Upper left) In the absence of Erk activity ERF is nuclear and represses the transcription of genes required for the progression through G1. (Upper right) Two to 3 min after mitogenic stimulation the activated Erk kinase translocates into the nucleus and phosphorylates ERF. (Lower right) This results in conformational changes that allow the interaction of ERF with an adaptor protein, which leads to its nuclear export. (Lower left) After the loading of ERF onto the export machinery, ERF is further and transiently phosphorylated at position T526 (grey phosphorus) in order to be released into the cytoplasm, allowing for the transcription of genes required for progression through G1. Upon the elimination of nuclear Erk activity the shuttling ERF molecules are retained in the nucleus due to their dephosphorylation (vertical arrow). In proliferating cells the export rate of ERF is high due to the Erk activity, resulting in its observed cytoplasmic localization (bidirectional horizontal arrow). The indicated domains of ERF are as follows: DBD, ets DNA-binding domain; ED1, export domain 1; ErkID, Erk interaction domain; RD, repressor domain (also contains part of export domain 2). G1 progression, transcription of genes involved in cell cycle progression through G1.
Acknowledgments
We thank Valérie Lefevbre for help with the localization experiments, Kaliopi Borboudaki for technical assistance, Katerina Mpilitou for help with the antibody work, Meropi Athanasiou (BRP-SAIC, Frederick, Md.) for immunohistochemistry reagents, Jane Leitch (Division of Signal Transduction Therapy, School of Life Sciences, University of Dundee, Dundee, Scotland) for the production of the anti-phospho-ERF antibodies, Thierry Dubois (University of Edinburgh) and others in the Aitken laboratory for the 14-3-3 reagents, and Pantelis Hatzis for help with the manuscript.
This work was supported by the EU grants ERBFMRXCT 96-0041 and BIO4CT975133 (to G.M.) and by the UK Medical Research Council and The Royal Society (to P.C.).
REFERENCES
- 1.Adachi, M., M. Fukuda, and E. Nishida. 2000. Nuclear export of MAP kinase (ERK) involves a MAP kinase kinase (MEK)-dependent active transport mechanism. J. Cell Biol. 148:849-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi, M., M. Fukuda, and E. Nishida. 1999. Two co-existing mechanisms for nuclear import of MAP kinase: passive diffusion of a monomer and active transport of a dimer. EMBO J. 18:5347-5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alt, J. R., A. B. Gladden, and J. A. Diehl. 2002. p21(Cip1) promotes cyclin D1 nuclear accumulation via direct inhibition of nuclear export. J. Biol. Chem. 277:8517-8523. [DOI] [PubMed] [Google Scholar]
- 4.Argentini, M., N. Barboule, and B. Wasylyk. 2001. The contribution of the acidic domain of MDM2 to p53 and MDM2 stability. Oncogene 20:1267-1275. [DOI] [PubMed] [Google Scholar]
- 5.Athanasiou, M., L. LeGallic, D. K. Watson, D. G. Blair, and G. Mavrothalassitis. 2000. Suppression of the Ewing's sarcoma phenotype by FLI1/ERF repressor hybrids. Cancer Gene Ther. 7:1188-1195. [DOI] [PubMed] [Google Scholar]
- 6.Baeuerle, P. A., and D. Baltimore. 1988. Activation of DNA-binding activity in an apparently cytoplasmic precursor of the NF-κB transcription factor. Cell 53:211-217. [DOI] [PubMed] [Google Scholar]
- 7.Birkenkamp, K. U., and P. J. Coffer. 2003. Regulation of cell survival and proliferation by the FOXO (Forkhead box, class O) subfamily of Forkhead transcription factors. Biochem. Soc. Trans. 31:292-297. [DOI] [PubMed] [Google Scholar]
- 8.Black, B. E., J. M. Holaska, F. Rastinejad, and B. M. Paschal. 2001. DNA binding domains in diverse nuclear receptors function as nuclear export signals. Curr. Biol. 11:1749-1758. [DOI] [PubMed] [Google Scholar]
- 9.Boulukos, K. E., P. Pognonec, B. Rabault, A. Begue, and J. Ghysdael. 1989. Definition of an Ets1 protein domain required for nuclear localization in cells and DNA-binding activity in vitro. Mol. Cell. Biol. 9:5718-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, F., L. S. Steelman, J. G. Shelton, J. T. Lee, P. M. Navolanic, W. L. Blalock, R. Franklin, and J. A. McCubrey. 2003. Regulation of cell cycle progression and apoptosis by the Ras/Raf/MEK/ERK pathway. Int. J. Oncol. 22:469-480. [PubMed] [Google Scholar]
- 11.Chow, C. W., M. Rincon, J. Cavanagh, M. Dickens, and R. J. Davis. 1997. Nuclear accumulation of NFAT4 opposed by the JNK signal transduction pathway. Science 278:1638-1641. [DOI] [PubMed] [Google Scholar]
- 12.Cruzalegui, F. H., E. Cano, and R. Treisman. 1999. ERK activation induces phosphorylation of Elk-1 at multiple S/T-P motifs to high stoichiometry. Oncogene 18:7948-7957. [DOI] [PubMed] [Google Scholar]
- 13.D'Abaco, G. M., S. Hooper, H. Paterson, and C. J. Marshall. 2002. Loss of Rb overrides the requirement for ERK activity for cell proliferation. J. Cell Sci. 115:4607-4616. [DOI] [PubMed] [Google Scholar]
- 14.Ducret, C., S. M. Maira, A. Dierich, and B. Wasylyk. 1999. The net repressor is regulated by nuclear export in response to anisomycin, UV, and heat shock. Mol. Cell. Biol. 19:7076-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eleftheriou, A., M. Yoshida, and B. R. Henderson. 2001. Nuclear export of human beta-catenin can occur independent of CRM1 and the adenomatous polyposis coli tumor suppressor. J. Biol. Chem. 276:25883-25888. [DOI] [PubMed] [Google Scholar]
- 16.Fabbro, M., and B. R. Henderson. 2003. Regulation of tumor suppressors by nuclear-cytoplasmic shuttling. Exp. Cell Res. 282:59-69. [DOI] [PubMed] [Google Scholar]
- 17.Frost, J. A., H. Steen, P. Shapiro, T. Lewis, N. Ahn, P. E. Shaw, and M. H. Cobb. 1997. Cross-cascade activation of ERKs and ternary complex factors by Rho family proteins. EMBO J. 16:6426-6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390:308-311. [DOI] [PubMed] [Google Scholar]
- 19.Furuno, T., N. Hirashima, S. Onizawa, N. Sagiya, and M. Nakanishi. 2001. Nuclear shuttling of mitogen-activated protein (MAP) kinase (extracellular signal-regulated kinase (ERK) 2) was dynamically controlled by MAP/ERK kinase after antigen stimulation in RBL-2H3 cells. J. Immunol. 166:4416-4421. [DOI] [PubMed] [Google Scholar]
- 20.Gama-Carvalho, M., and M. Carmo-Fonseca. 2001. The rules and roles of nucleocytoplasmic shuttling proteins. FEBS Lett. 498:157-163. [DOI] [PubMed] [Google Scholar]
- 21.Greulich, H., and R. L. Erikson. 1998. An analysis of Mek1 signaling in cell proliferation and transformation. J. Biol. Chem. 273:13280-13288. [DOI] [PubMed] [Google Scholar]
- 22.Gu, J., L. Nie, D. Wiederschain, and Z. M. Yuan. 2001. Identification of p53 sequence elements that are required for MDM2-mediated nuclear export. Mol. Cell. Biol. 21:8533-8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harbers, M., T. Nomura, S. Ohno, and S. Ishii. 2001. Intracellular localization of the Ret finger protein depends on a functional nuclear export signal and protein kinase C activation. J. Biol. Chem. 276:48596-48607. [DOI] [PubMed] [Google Scholar]
- 24.Henderson, B. R., and A. Eleftheriou. 2000. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 256:213-224. [DOI] [PubMed] [Google Scholar]
- 25.Humbert-Lan, G., and T. Pieler. 1999. Regulation of DNA binding activity and nuclear transport of B-Myb in Xenopus oocytes. J. Biol. Chem. 274:10293-10300. [DOI] [PubMed] [Google Scholar]
- 26.Islas, S., J. Vega, L. Ponce, and L. Gonzalez-Mariscal. 2002. Nuclear localization of the tight junction protein ZO-2 in epithelial cells. Exp. Cell Res. 274:138-148. [DOI] [PubMed] [Google Scholar]
- 27.Jackman, M., Y. Kubota, N. den Elzen, A. Hagting, and J. Pines. 2002. Cyclin A- and cyclin E-Cdk complexes shuttle between the nucleus and the cytoplasm. Mol. Biol. Cell 13:1030-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jans, D. A., C. Y. Xiao, and M. H. Lam. 2000. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays 22:532-544. [DOI] [PubMed] [Google Scholar]
- 29.Klappacher, G. W., V. V. Lunyak, D. B. Sykes, D. Sawka-Verhelle, J. Sage, G. Brard, S. D. Ngo, D. Gangadharan, T. Jacks, M. P. Kamps, D. W. Rose, M. G. Rosenfeld, and C. K. Glass. 2002. An induced Ets repressor complex regulates growth arrest during terminal macrophage differentiation. Cell 109:169-180. [DOI] [PubMed] [Google Scholar]
- 30.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96:9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kudo, N., B. Wolff, T. Sekimoto, E. P. Schreiner, Y. Yoneda, M. Yanagida, S. Horinouchi, and M. Yoshida. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242:540-547. [DOI] [PubMed] [Google Scholar]
- 32.Le Gallic, L., D. Sgouras, G. Beal, Jr., and G. Mavrothalassitis. 1999. Transcriptional repressor ERF is a Ras/mitogen-activated protein kinase target that regulates cellular proliferation. Mol. Cell. Biol. 19:4121-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, Q. J., S. H. Yang, Y. Maeda, F. M. Sladek, A. D. Sharrocks, and M. Martins-Green. 2003. MAP kinase phosphorylation-dependent activation of Elk-1 leads to activation of the co-activator p300. EMBO J. 22:281-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu, J. C., R. E. Baker, C. Sun, V. C. Sundmark, and H. P. Elsholtz. 2002. Activation of Go-coupled dopamine D2 receptors inhibits ERK1/ERK2 in pituitary cells. A key step in the transcriptional suppression of the prolactin gene. J. Biol. Chem. 277:35819-35825. [DOI] [PubMed] [Google Scholar]
- 35.Malek, S., Y. Chen, T. Huxford, and G. Ghosh. 2001. IκBβ, but not IκBα, functions as a classical cytoplasmic inhibitor of NF-κB dimers by masking both NF-κB nuclear localization sequences in resting cells. J. Biol. Chem. 276:45225-45235. [DOI] [PubMed] [Google Scholar]
- 36.Mavrothalassitis, G., and J. Ghysdael. 2000. Proteins of the ETS family with transcriptional repressor activity. Oncogene 19:6524-6532. [DOI] [PubMed] [Google Scholar]
- 37.Meng, W., L. L. Swenson, M. J. Fitzgibbon, K. Hayakawa, E. Ter Haar, A. E. Behrens, J. R. Fulghum, and J. A. Lippke. 2002. Structure of mitogen-activated protein kinase-activated protein (MAPKAP) kinase 2 suggests a bifunctional switch that couples kinase activation with nuclear export. J. Biol. Chem. 277:37401-37405. [DOI] [PubMed] [Google Scholar]
- 38.Michael, W. M., P. S. Eder, and G. Dreyfuss. 1997. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 16:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mittnacht, S., H. Paterson, M. F. Olson, and C. J. Marshall. 1997. Ras signalling is required for inactivation of the tumour suppressor pRb cell-cycle control protein. Curr. Biol. 7:219-221. [DOI] [PubMed] [Google Scholar]
- 40.Muslin, A. J., and H. Xing. 2000. 14-3-3 proteins: regulation of subcellular localization by molecular interference. Cell. Signal. 12:703-709. [DOI] [PubMed] [Google Scholar]
- 41.O'Neill, E. M., I. Rebay, R. Tjian, and G. M. Rubin. 1994. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78:137-147. [DOI] [PubMed] [Google Scholar]
- 42.Paumelle, R., D. Tulasne, Z. Kherrouche, S. Plaza, C. Leroy, S. Reveneau, B. Vandenbunder, V. Fafeur, and D. Tulashe. 2002. Hepatocyte growth factor/scatter factor activates the ETS1 transcription factor by a RAS-RAF-MEK-ERK signaling pathway. Oncogene 21:2309-2319. [DOI] [PubMed] [Google Scholar]
- 43.Peeper, D. S., T. M. Upton, M. H. Ladha, E. Neuman, J. Zalvide, R. Bernards, J. A. DeCaprio, and M. E. Ewen. 1997. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature 386:177-181. [DOI] [PubMed] [Google Scholar]
- 44.Polizotto, R. S., and M. S. Cyert. 2001. Calcineurin-dependent nuclear import of the transcription factor Crz1p requires Nmd5p. J. Cell Biol. 154:951-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prigent, M., I. Barlat, H. Langen, and C. Dargemont. 2000. IκBα and IκBα/NF-κB complexes are retained in the cytoplasm through interaction with a novel partner, RasGAP SH3-binding protein 2. J. Biol. Chem. 275:36441-36449. [DOI] [PubMed] [Google Scholar]
- 46.Rehberg, S., P. Lischka, G. Glaser, T. Stamminger, M. Wegner, and O. Rosorius. 2002. Sox10 is an active nucleocytoplasmic shuttle protein, and shuttling is crucial for Sox10-mediated transactivation. Mol. Cell. Biol. 22:5826-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rena, G., Y. L. Woods, A. R. Prescott, M. Peggie, T. G. Unterman, M. R. Williams, and P. Cohen. 2002. Two novel phosphorylation sites on FKHR that are critical for its nuclear exclusion. EMBO J. 21:2263-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rittinger, K., J. Budman, J. Xu, S. Volinia, L. C. Cantley, S. J. Smerdon, S. J. Gamblin, and M. B. Yaffe. 1999. Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell 4:153-166. [DOI] [PubMed] [Google Scholar]
- 49.Roovers, K., and R. K. Assoian. 2000. Integrating the MAP kinase signal into the G1 phase cell cycle machinery. Bioessays 22:818-826. [DOI] [PubMed] [Google Scholar]
- 50.Roussel, M. F., J. N. Davis, J. L. Cleveland, J. Ghysdael, and S. W. Hiebert. 1994. Dual control of myc expression through a single DNA binding site targeted by ets family proteins and E2F-1. Oncogene 9:405-415. [PubMed] [Google Scholar]
- 51.Sage, J., G. J. Mulligan, L. D. Attardi, A. Miller, S. Chen, B. Williams, E. Theodorou, and T. Jacks. 2000. Targeted disruption of the three Rb-related genes leads to loss of G1 control and immortalization. Genes Dev. 14:3037-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sgouras, D. N., M. A. Athanasiou, G. J. Beal, Jr., R. J. Fisher, D. G. Blair, and G. J. Mavrothalassitis. 1995. ERF: an ETS domain protein with strong transcriptional repressor activity, can suppress ets-associated tumorigenesis and is regulated by phosphorylation during cell cycle and mitogenic stimulation. EMBO J. 14:4781-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stommel, J. M., N. D. Marchenko, G. S. Jimenez, U. M. Moll, T. J. Hope, and G. M. Wahl. 1999. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 18:1660-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tickenbrock, L., J. Cramer, I. R. Vetter, and O. Muller. 2002. The coiled coil region (amino acids 129-250) of the tumor suppressor protein adenomatous polyposis coli (APC). Its structure and its interaction with chromosome maintenance region 1 (Crm-1). J. Biol. Chem. 277:32332-32338. [DOI] [PubMed] [Google Scholar]
- 55.Tolwinski, N. S., and E. Wieschaus. 2001. Armadillo nuclear import is regulated by cytoplasmic anchor Axin and nuclear anchor dTCF/Pan. Development 128:2107-2117. [DOI] [PubMed] [Google Scholar]
- 56.Tootle, T. L., P. S. Lee, and I. Rebay. 2003. CRM1-mediated nuclear export and regulated activity of the receptor tyrosine kinase antagonist YAN require specific interactions with MAE. Development 130:845-857. [DOI] [PubMed] [Google Scholar]
- 57.Venanzoni, M. C., L. R. Robinson, D. R. Hodge, I. Kola, and A. Seth. 1996. ETS1 and ETS2 in p53 regulation: spatial separation of ETS binding sites (EBS) modulate protein: DNA interaction. Oncogene 12:1199-1204. [PubMed] [Google Scholar]
- 58.Vielhaber, E. L., D. Duricka, K. S. Ullman, and D. M. Virshup. 2001. Nuclear export of mammalian PERIOD proteins. J. Biol. Chem. 276:45921-45927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weinstein-Oppenheimer, C. R., W. L. Blalock, L. S. Steelman, F. Chang, and J. A. McCubrey. 2000. The Raf signal transduction cascade as a target for chemotherapeutic intervention in growth factor-responsive tumors. Pharmacol. Ther. 88:229-279. [DOI] [PubMed] [Google Scholar]
- 60.Xiao, Z., N. Watson, C. Rodriguez, and H. F. Lodish. 2001. Nucleocytoplasmic shuttling of Smad1 conferred by its nuclear localization and nuclear export signals. J. Biol. Chem. 276:39404-39410. [DOI] [PubMed] [Google Scholar]
- 61.Xu, L., Y. Kang, S. Col, and J. Massague. 2002. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFβ signaling complexes in the cytoplasm and nucleus. Mol. Cell 10:271-282. [DOI] [PubMed] [Google Scholar]
- 62.Yang, S. H., E. Vickers, A. Brehm, T. Kouzarides, and A. D. Sharrocks. 2001. Temporal recruitment of the mSin3A-histone deacetylase corepressor complex to the ETS domain transcription factor Elk-1. Mol. Cell. Biol. 21:2802-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yordy, J. S., and R. C. Muise-Helmericks. 2000. Signal transduction and the Ets family of transcription factors. Oncogene 19:6503-6513. [DOI] [PubMed] [Google Scholar]