Abstract
KLF7, a member of the Krüppel-like transcription factor family, is believed to regulate neurogenesis and cell cycle progression. Here, a yeast two-hybrid screen for KLF7 cofactors in the developing nervous system identified a novel 140-kDa protein named MoKA, for modulator of KLF7 activity. Interaction between MoKA and KLF7 was confirmed by the in vitro glutathione S-transferase pull-down assay and by coimmunoprecipitation of the proteins overexpressed in mammalian cells. Functional assays documented that MoKA is a KLF7 coactivator, and in situ hybridizations identified the developing nervous system and the adult testes as two sites of MoKA and Klf7 coexpression. Chromatin immunoprecipitation experiments demonstrated KLF7 binding to the p21WAF1/Cip1 gene while transient transfection assays documented KLF7 stimulation of the p21WAF1/Cip1 proximal promoter. Additional tests revealed that distinct structural motifs of MoKA direct interaction with KLF7 and shuttling between the nucleus and cytoplasm of asynchronously cycling cells. Altogether, our results strongly suggest that MoKA and KLF7 interact functionally to regulate gene expression during cell differentiation and identify the cell cycle regulator p21WAF1/Cip1 as one of the targeted genes.
Developmental programs rely on the dynamic interplay between extrinsic signals and intrinsic factors that gradually restrict the potential of progenitor cells with the acquisition of stage- and tissue-specific profiles of gene expression (21). Transcriptional regulators play a critical role in this process by modulating gene activity through binding to specific DNA sequences, alone or in combination with other nuclear proteins. Krüppel-like factors (KLFs) have recently emerged as critical contributors to vertebrate development (1, 3, 6, 13). Mammalian KLFs and the related group of Sp1-like proteins comprise 20 distinct transcription factors characterized by three highly homologous, C-terminally located zinc fingers of the C2H2 type that bind to similar Sp1 sites (GC-rich sequences and related GT or CACCC boxes) on DNA (3, 13). Structure-function considerations further segregate KLFs into four phylogenetically distinct groups (1). KLFs stimulate and/or repress transcription of a large variety of genes, such as those encoding differentiation products, cytoskeletal proteins, cell cycle regulators, cell surface receptors, soluble growth factors, extracellular matrix components, and KLFs themselves (3, 6).
Gene-targeted deletions in mice have documented the involvement of KLFs in cell growth, proliferation, and differentiation (6). Examples include KLF1 control of erythroid cell proliferation and β-globin gene cluster activity (5, 24, 28); KLF2 contribution to lung formation, blood vessel stabilization, and c-Myc-dependent T-cell quiescence (4, 16, 36); KLF4 involvement in terminal differentiation of dermal and intestinal epithelia (14, 31); and KLF5 participation in cardiovascular remodeling (33). Additionally, a screen for mutations in prostate cancer has indicated that KLF6 is a tumor suppressor gene product that is normally implicated in inhibiting cell proliferation (23). KLF-like gene products have been identified in Xenopus and zebra fish as well, where they are believed to control erythroid cell differentiation, blood vessel formation, and epidermal development (12, 25). Finally, a Drosophila melanogaster orthologue of the mammalian KLF6/KLF7 group has been recently shown to be a critical determinant of fly development (7).
Indirect lines of evidence suggest an important role of KLF7 in cell differentiation. First, KLF7 overexpression in cultured fibroblasts and neuroblastoma cells leads to accumulation of the cdk inhibitor p21 protein and growth arrest (17). Second, expression of the mouse Klf7 gene is restricted to postmitotic neuroprogenitor cells of the embryonic and neonatal nervous systems (17, 18). Lastly, loss of Klf7 activity in mice is associated with a neurodeficient phenotype and postnatal death (unpublished data). KLF7 may also have additional functions in the adult organism. Klf7 activity is in fact maintained at high levels in a few neuronal subtypes of the central and peripheral nervous systems and, less prominently, in several nonneural tissues (17, 18). Cell context and stage-specific mechanisms have been invoked as potential means to control the postulated functions of KLF7 (17).
Combinatorial interactions of nuclear proteins are one of the mechanisms responsible for functional diversification of transcription factors, including the KLFs (3, 11, 13, 21). We therefore undertook a genetic screen for KLF7 cofactors by using the yeast two-hybrid system and RNA purified from mouse embryonic neural tissues. As result, we report here the identification and characterization of a novel 140-kDa protein that enhances KLF7 transactivating potential. The protein was named MoKA, for modulator of KLF7 activity, and the corresponding gene was shown to be coexpressed with Klf7 in the embryonic nervous system and in the adult testes. We therefore propose that MoKA and KLF7 interact functionally to regulate gene expression during cell differentiation.
MATERIALS AND METHODS
Yeast two-hybrid screen.
Mouse KLF7 coding sequences were PCR amplified and subcloned into pLexA vector (Clontech) by homologous recombination in EGY48 yeast cells (9). The (Δ1-58)KLF7 construct was used as the bait to screen a mouse cDNA expression library generated from 20 μg of poly(A)+ RNA purified from the brain and spinal cord of embryonic day 12.5 (E12.5) to E13.5 embryos. The cDNAs were linked to specific adaptors, PCR amplified, and subcloned into the PJG4-5 expression vector by homologous recombination in EGY48 yeast cells (Clontech) (9). Yeast screening was performed by using the Matchmaker LexA two-hybrid system (Clontech) according to the manufacturer's instructions. Plasmids from positives clones were purified and cotransformed with (Δ1-58)KLF7 into EGY48 yeast cells to identify candidates that specifically interact with the bait. Growth on selective medium lacking His, Trp, and Leu and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining of yeast colonies grown on selective medium lacking His, Ura, and Trp indicated positive interactions between the bait and prey (30).
Cloning and recombinant expression of MoKA.
The original KLF7-interacting clone 154A was used as a probe to screen ∼106 phage recombinants from an E13.5 mouse embryonic cDNA library (Clontech). A computer-aided BLAST search for expressed sequence tags (EST) in the mouse database was employed to identify overlapping EST clones. The full-length coding sequence of MoKA has been submitted to GenBank under accession number AY267463. Expression of full-length MoKA was pursued in bacteria and mammalian cells with pET15-b and pCMV-Tag 1 vectors, respectively. Sizes of recombinantly expressed proteins were estimated after sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) fractionation of cell lysates by Coomassie brilliant blue staining and Western blot analysis.
In vitro GST pull-down assay.
The sequence of the 154A clone was PCR amplified and subcloned into the BamHI and NotI sites of the bacterial expression vector pGEX-5X-1 (Pharmacia Biotech). [35S]methionine-labeled KLF7 was produced in vitro by using the TNT coupled transcription-translation system (Promega) according to the manufacturer's protocol. Bacterial protein extracts containing glutathione S-transferase (GST) or the GST fusion product were mixed with labeled KLF7 in binding buffer containing 50 mM Tris-HCl, 100 mM NaCl, 10 mM MgCl2, and 1 mM dithiothreitol (pH 7.9) and gently rocked at 4°C for 1 h. Proteins were purified with glutathione-conjugated agarose beads (Pharmacia Biotech), washed three times in binding buffer containing 0.04% NP-40, divided into three aliquots, and fractioned by SDS-PAGE. One gel was stained with Coomassie brilliant blue, one was Western blotted against anti-GST antibodies, and the last one was Enhance-treated (NEN) and dried before autoradiography.
In vivo coimmunoprecipitation assay.
The full-length coding sequence of MoKA was PCR amplified and subcloned into the blunt-ended BamHI site of mammalian expression vector pCMV-Tag1 (Stratagene). The resulting MoKA-Flag construct was used as a substrate to generate a Flag-tagged deletion mutant (Δ116-472)MoKA by using conveniently located restriction endonuclease sites. Mammalian vectors expressing c-Myc-tagged KLF7 or KLF4 were kindly provided by Lei Lei and Vincent Yang, respectively (18, 32). MoKA-Flag was transfected together with either KLF7-Myc or KLF4 expression vectors into COS7 cells with Lipofectamine 2000 (Invitrogen). Fusion proteins were purified with monoclonal anti-Flag (Sigma), anti-c-Myc (Santa Cruz), or anti-KLF4 antibodies (a gift from V. Yang); the same antibodies were used to detect Flag-tagged MoKA Myc-tagged KLF7 or KLF4. Horseradish peroxidase-conjugated anti-immunoglobulin G antisera (Amersham-Pharmacia) were used as secondary antibodies for Western blot analysis.
Luciferase reporter assays.
Functional assays employed reporter gene constructs harboring the 2.4-kb and 225-bp upstream sequences of the p21WAF1/Cip1 gene [p21(−2400)-Luc and p21(−225)-Luc], or the CACCC binding sites of the β globin gene (pC1G3TKCAT) (2, 8, 19); these reporter plasmids were kindly provided by Tsai Wang and James Bieker, respectively. The pC1G3TKCAT construct was further modified so to generate (CAC)4TK-Luc, a luciferase reporter gene construct driven by four copies of the CAC site linked to the minimal thymidine kinase (TK) promoter. Reporter gene constructs were transfected into COS7, NIH 3T3, or NB-OK1 cells (17, 20) together with the Myc-tagged KLF7 expression vector or both Myc-tagged KLF7 and Flag-tagged MoKA expression vectors in the amounts indicated in the respective figure legends. Luciferase activities were evaluated by using a commercial kit (Promega), and transfection efficiencies were normalized against constitutive expression of Renilla luciferase by the pRL-TK vector (30 ng/transfection). Each functional assay was performed at least three times in duplicate, and the statistical value of the data was evaluated by the Mann-Whitney U test.
ChIP assay.
Chromatin immunoprecipitation (ChIP) was performed by using a commercial kit (Upstate Biotech, Lake Placid, N.Y.) according to the manufacturer's protocol. All solutions contained 1 mM phenylmethylsulfonyl fluoride, 1 mg of aprotinin/ml, and 1 mg of pepstatin/ml. 293T cells were transfected with 20 μg of Myc-tagged KLF7 expression vector or the control Myc-tagged plasmid. After 24 h, cells were formaldehyde cross-linked for 12 min at 37°C prior to ChIP. The cross-linking was quenched with 125 mM glycine for 5 min at room temperature. After three washes in phosphate-buffered saline, cell pellets were resuspended in SDS lysis buffer, incubated for 15 min on ice, and sonicated three times for 12 s each time with an ultrasonic liquid processor Sonicator 3000 at power setting 2 and 100% duty cycle (Misonix, Farmingdale, N.Y.). Supernatants were isolated by centrifugation, diluted with ChIP dilution buffer, and incubated overnight at 4°C with and/or without anti-c-Myc (Santa Cruz). Immune complexes were recovered by the addition of 60 μl of salmon sperm DNA-protein A-agarose followed by incubation at 4°C for 3 h. Beads were washed with low- and high-salt buffers first and then with LiCl buffer and Tris-EDTA buffer. The immune complexes were then eluted by incubation with 500 μl of elution buffer (1% SDS, 100 mM NaHCO3, and 10 mM dithiothreitol) for 15 min at room temperature. To solubilize the chromatin, eluted samples were adjusted to 0.2 M NaCl, incubated overnight at 65°C, and treated with proteinase K for 1 h at 45°C. DNA was phenol-chloroform purified, isopropanol precipitated, and dissolved in 25 μl of water. PCR was carried out for 35 cycles with 5 μl of the sample DNA solution and 10% dimethyl sulfoxide; amplified products were detected by standard 2% agarose gel electrophoresis.
Ubiquitination assay.
NIH 3T3 cells were transfected with KLF7-Myc alone or together with MoKA-Flag as described above. After 24 h, cell extracts were immunoprecipitated with anti-c-Myc (Santa Cruz) in cell lysis buffer containing 50 mM Tris-HCl (pH 7.4), 0.1% NP-40, 0.3% sodium deoxycholate, 150 mM NaCl, 1 mM EGTA, 1mM phenylmethylsulfonyl fluoride, 1-mg/ml (each) aprotinin, leupeptin, and pepstatin, 1 mM Na3VO4, and 1 mM NaF. The immune complexes were resolved by SDS-10% PAGE and Western blotted with anti-c-Myc (Santa Cruz) and/or with anti-ubiquitin antibodies (Upstate). Before probing with anti-ubiquitin antibodies, the membranes were subjected to high-temperature antigen retrieval, and ubiquitin-GST fusion protein (Calbiochem) was used as a positive control for Western blot analysis. Proteins were visualized with horseradish peroxidase-conjugated anti-immunoglobulin G antisera (Amersham-Pharmacia) and a chemiluminescent detection system. For the affinity chromatography, the ubiquitinated protein enrichment kit (Calbiochem) was used according to the manufacturer's protocol and 10 ng of ubiquitin-GST fusion protein (Calbiochem) was used as a positive control for chromatography. Western blot analysis was performed as described above.
RNA hybridizations.
In situ hybridizations on whole-mount embryos or tissue sections were carried out by using a MoKA probe that spans from nucleotides 1500 to 4250 according to the published protocol (17). Northern analyses were performed by using commercially available blots (Clontech) and the full-length MoKA cDNA.
MoKA subcellular localization.
The full-length sequence of MoKA was PCR amplified and subcloned into the SalI and BamHI sites of the mammalian expression vector pEGFP-C3 (Clontech) to create MoKA-GFP. The same PCR-based strategy was employed to generate (Δ350-1193)MoKA-GFP and (Δ1-472)MoKA-GFP. Amplified PCR products were also subcloned into the NotI and BamHI sites or into the BamHI site of pCMV-Tag 1 vector to create (Δ350-1193)MoKA-Flag and (Δ1-472)MoKA-Flag. All constructs were transfected into COS7, NIH 3T3, or NB-OK1 cells as described previously (17, 20); 24 h after transfection, cells were fixed with 3.7% paraformaldehyde for 20 min. Prior to immunostaining, cells expressing Flag or GAL4 fusion products were permeabilized with three 10-min changes of phosphate-buffered saline containing 0.1% Triton X-100. Monoclonal antibodies against Flag or the DNA-binding domain of GAL4 (Clontech) were used for immunofluorescence detection of the respective fusion proteins together with Alexa 488 (Jackson). Subcellular localization of fusion products was monitored with a confocal scanning microscope (Leica TCS-SP).
RESULTS
Isolation of a KLF7 interacting protein.
Expression of Klf7 in the developing nervous system of the mouse reaches its maximum at about E13.5 (17). A yeast two-hybrid screen was therefore employed to identify potential KLF7 cofactors in neural tissues from E12.5 to E13.5 mouse embryos. Toward this end, a Klf7 cDNA without the sequence encoding the first 58 amino acids was fused to the DNA-binding domain of LexA and used as the bait to screen a brain and spinal cord cDNA expression library. The deleted sequence includes most of the transactivation domain of KLF7 (20) and was omitted from the bait because of unacceptably high background, even in the absence of the prey (data not shown).
About 105 independent cDNA transformants were screened for the ability to stimulate expression of LexA-responsive Leu2 and LacZ reporter genes only in the presence of galactose. The screen yielded a single 160-bp-long cDNA-positive clone (154A) containing a potential open reading frame (ORF) that codes for a novel peptide sequence. Additional tests with yeast cells documented specificity by showing the inability of the 154A peptide to interact with LexA alone, an unrelated protein, or a shorter (Δ1-119) KLF7 protein (Fig. 1A). The last result implicitly located the site of interaction with the 154A peptide within the KLF7 segment that resides between amino acids 59 and 119 and which includes an evolutionarily conserved leucine zipper sequence (17, 18, 20). A GST pull-down assay further documented positive in vitro interaction between full-length KLF7 and 154A, in addition to showing the inability of GST alone to pull down the radiolabeled KLF7 protein (Fig. 1B).
FIG. 1.
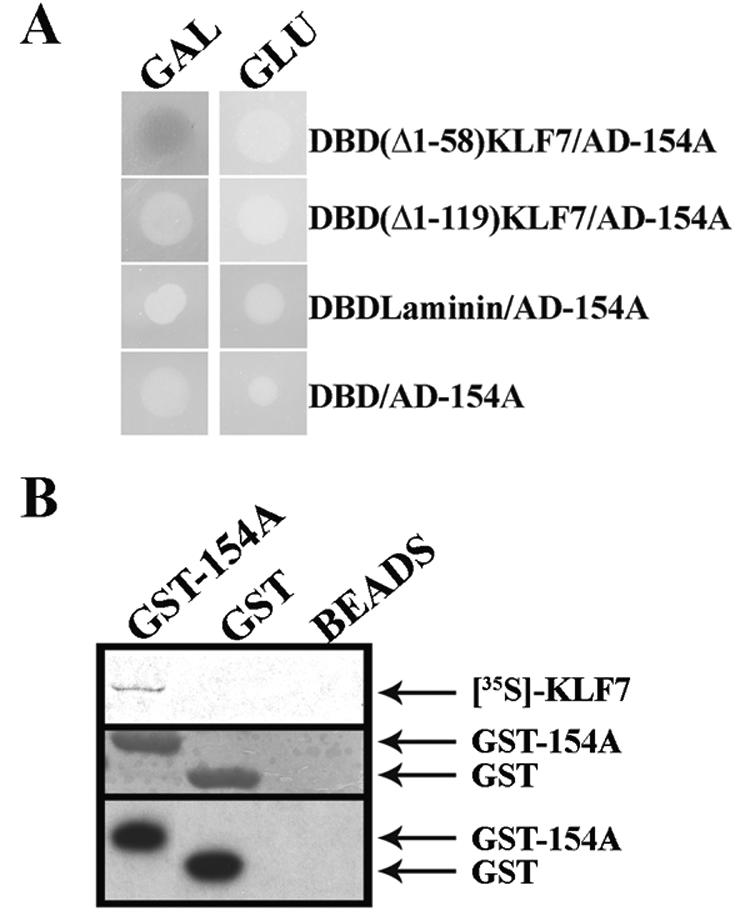
Interaction between KLF7 and the 154A peptide. (A) β-Galactosidase-induced expression of yeast cells grown on selective medium in the presence of galactose (GAL) or glucose (GLU) and cotransformed with clone 154A and various bait constructs. (B) GST pull-down assay between radiolabeled KLF7 and 154A-GST fusion protein. Samples were fractionated by SDS-PAGE and visualized by autoradiography (upper panel), Coomassie blue staining (middle panel), and Western analysis with anti-GST antibody (lower panel).
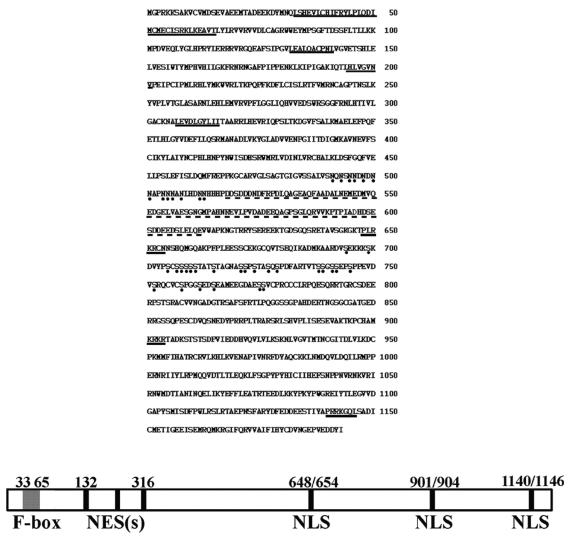
Next, the 154A sequence was used to probe an E13.5 whole-embryo cDNA library constructed in the λgt11 phage vector. The screen yielded a 1.1-kb clone (154B) that extended the ORF of 154A 867 nucleotides farther 3′ and 123 nucleotides farther 5′ to include the putative start site of translation. A search for the 154B sequence in the mouse database of EST identified an overlapping clone (BG174008) that extended the ORF 2,533 nucleotides farther 3′ to include the termination codon. The assembled ORF of the phage and EST clones codes for a 1,193-amino-acid-long polypeptide with a predicted molecular mass of ∼140 kDa (Fig. 2). Electrophoretic fractionation of the protein recombinantly expressed in bacteria confirmed this size prediction (Fig. 3A). Western analysis of lysates from COS7 cells overexpressing the full-length Flag-tagged cDNA identified a major fusion product larger than 140 kDa, in addition to a minorly expressed species of the same size (Fig. 3B). We interpreted this last result as evidence that the 140-kDa protein may undergo posttranslational modifications.
FIG. 2.
MoKA primary structure. The deduced amino acid sequences of the combined mouse clones coding for full-length MoKA are shown. Structural features included in the schematic below are underlined, whereas others discussed in the text are highlighted by dotted lines or single dots. A schematic representation of the MoKA protein is depicted below.
FIG. 3.
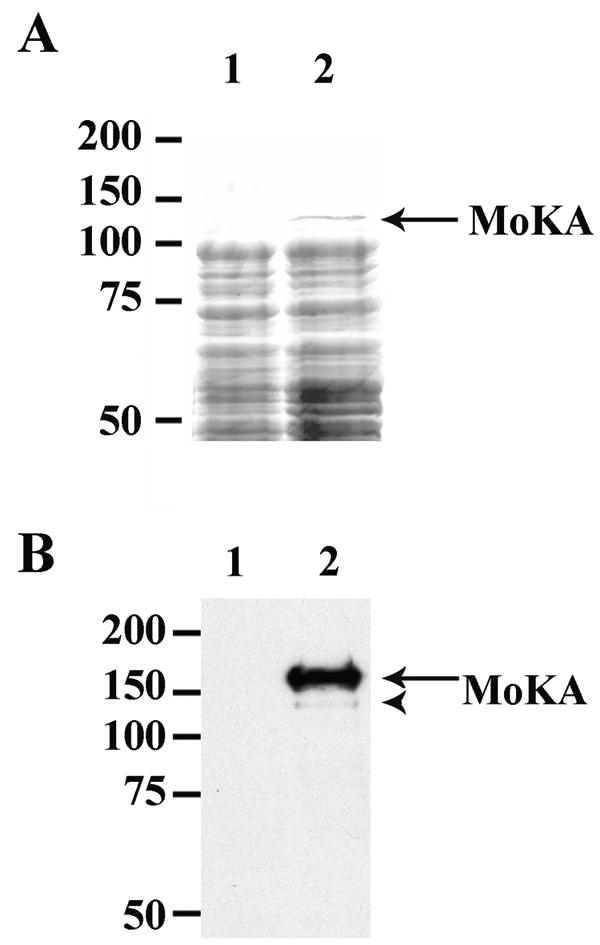
Recombinant MoKA expression. (A) Coomassie blue staining of uninduced (lane 1) and induced (lane 2) bacterial cells expressing MoKA (arrow). (B) Western analysis with anti-Flag antibody of COS7 cells mock-transfected (lane 1) or transfected with Flag-tagged MoKA expression vector (lane 2). The arrowhead and arrow highlight the normal and slower-migrating 140-kDa proteins, respectively. Protein marker sizes (in kilodaltons) are shown on the left in both panels.
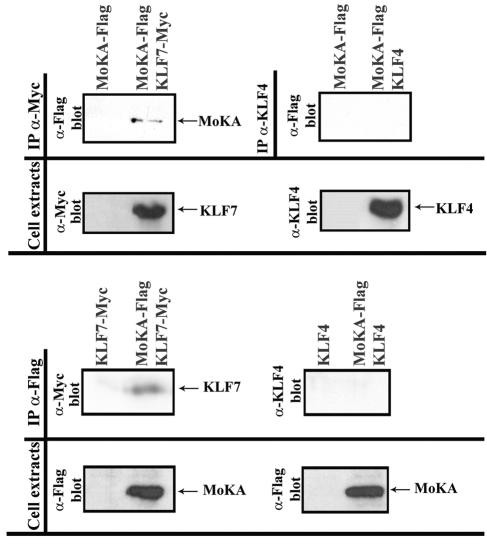
COS7 cells do not produce detectable amounts of transcripts for either KLF7 or the 140-kDa protein and were cotransfected with plasmids expressing the Flag-tagged 140 kDa protein and Myc-tagged KLF7. Lysates from transfected cells were immunoprecipitated with anti-Flag or anti-Myc antisera, and the resulting immunoprecipitates were analyzed by Western blots with anti-Myc or anti-Flag antibodies, respectively. Coimmunoprecipitation of MoKA and KLF7 was observed in both samples, demonstrating that the two proteins interact in vivo as well (Fig. 4). Importantly, there was no immunoprecipitate when the 140-kDa protein was coexpressed with KLF4 (Fig. 4). Based on these data and the functional evidence described below, the 140-kDa protein was named MoKA for modulator of KLF7 activity.
FIG. 4.
Association of MoKA and KLF7 in mammalian cells. Western analyses with indicated antibodies (α) of immunoprecipitates (IP) and cell extracts from COS7 cultures cotransfected with 12 μg of plasmids expressing Flag-tagged MoKA and Myc-tagged KLF7 or KLF4.
MoKA is a novel protein.
A BLAST search failed to identify significant sequence homologies between MoKA and known vertebrate and invertebrate proteins; it also revealed that MoKA is a single copy gene located in human chromosome 5q32 and mouse chromosome 18E1-E2. A PROSITE-led analysis identified several structural motifs potentially involved in MoKA activity (Fig. 2). They include an F-box within the interacting segment of 154A (amino acids 33 to 65) which was originally identified in proteins that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex (34); three putative nuclear localization signals (NLSs) in the C-terminal half of the protein, between residues 648 and 654 (PLRKRCN), 901 and 904 (KRKR), and 1140 and 1146 (PRRKGQL); and three potential nuclear export signals (NESs) in the N-terminal half of the protein, between amino acids 132 and 141 (LEALQACPNL), 194 and 201 (LHLVGVNV), and 307 and 316 (LEVDLGYLII) (Fig. 2). Additional structural features of MoKA include a highly acidic sequence (pI, 3.66) between residues 518 and 612 as well as asparagine- and serine-rich stretches between amino acids 491 and 513 and 694 and 776, respectively (Fig. 2).
MoKA enhances KLF7 activity.
We have previously shown that the zinc finger domain of KLF7 binds in vitro specifically to the CACCC element of the β-globin promoter (20). Functional interaction between KLF7 and MoKA was therefore assessed in transiently transfected COS7 cells with an artificial luciferase reporter gene construct in which the basal TK promoter lies downstream of four copies of the CACCC element [(CAC4)TK-Luc] (2). The same TK promoter without the CACCC elements (TK-Luc) was used as a negative control. The results of the cell transfections showed that KLF7 stimulates transcription of the (CAC)4TK-Luc reporter gene and that the stimulation is further enhanced by coexpression of MoKA (Fig. 5A). They also documented that MoKA activity requires binding of KLF7 to the CACCC elements (Fig. 5A). Incidentally, expression of the KLF7 plasmid was not increased by MoKA; moreover, identical results were obtained with NIH 3T3 fibroblasts and the neuroblastoma line NB-OK1 (data not shown). These in vitro data therefore established that MoKA acts as a positive modulator of KLF7 activity.
FIG. 5.
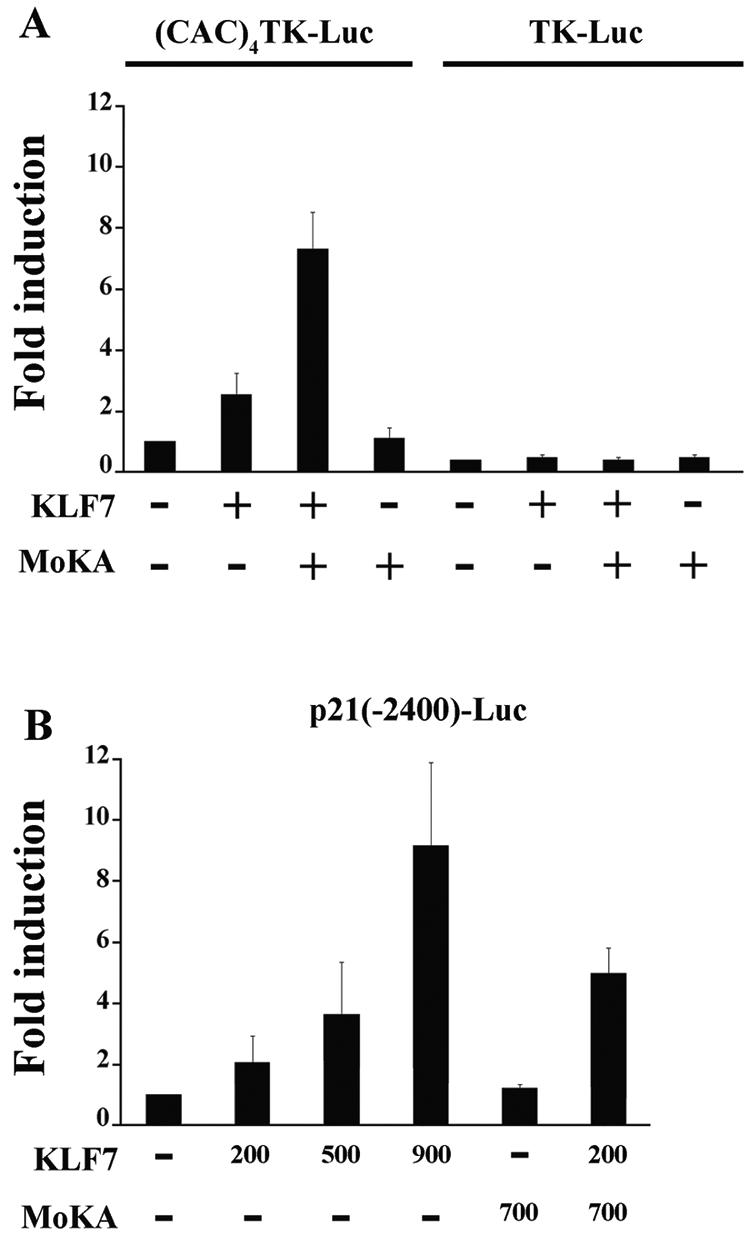
MoKA superstimulates KLF7 activity. (A) COS7 cells were cotransfected with 100 ng of (CAC)4TK-Luc or TK-Luc reporter plasmid and with 200 ng of the KLF7 expression plasmid alone and a twofold molar excess of the MoKA expression plasmid. (B) COS7 cells were cotransfected with the p21(−2400)-Luc reporter construct and the indicated amounts (in nanograms) of the KLF7 plasmid alone or together with the MoKA plasmid. In all experiments, the total amount of transfected DNA was adjusted to the same level in each transfection. +, present; −, absent.
MoKA and Klf7 gene expression overlap.
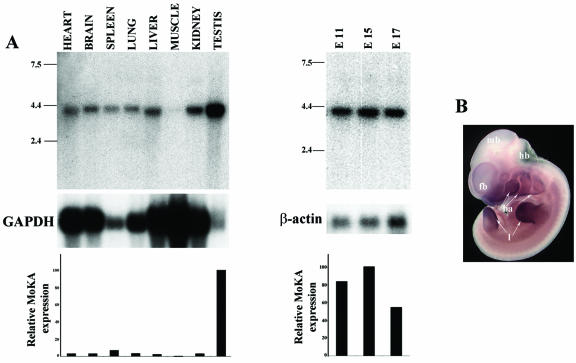
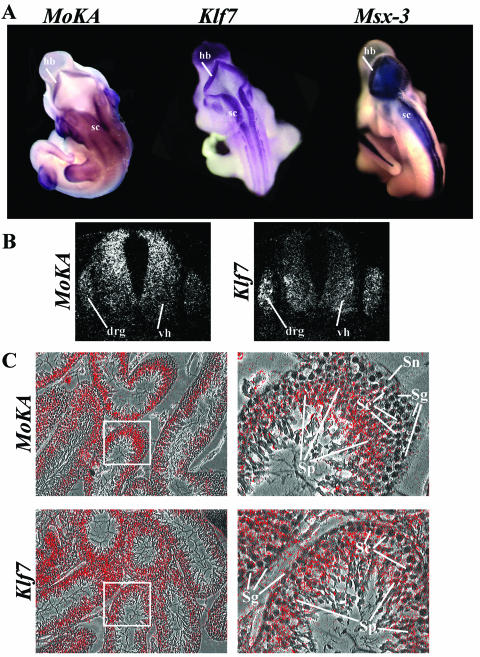
To provide supporting in vivo evidence for the above conclusion, expression of the MoKA gene was examined in the developing embryo and adult mouse. Northern analyses showed high levels of MoKA transcripts throughout embryogenesis and low levels in all adult tissues examined except the testes (Fig. 6A). Whole-mount in situ hybridization of E10.5 embryos revealed that the MoKA gene is actively transcribed in the developing brain, branchial arches, and limbs (Fig. 6B). A comparative analysis of whole-mount in situ hybridizations showed similar profiles of MoKA, Klf7, and Msx3 gene expression in the developing brain and spinal cord (Fig. 7A) (17, 18, 35). In situ hybridizations of cross sections of the spinal cord confirmed coexpression of MoKA and Klf7 in the dorsal root ganglia and the ventral horn of the neural tube (Fig. 7B). Similarly, cross sections of 2-month-old mouse testes hybridized in situ to MoKA and Klf7 probes showed high levels of both transcripts in postmeiotic spermatids (Fig. 7C). Altogether, these results strongly suggested that MoKA and KLF7 may functionally interact in specific cell lineages of the developing and adult mouse in addition to identifying unique sites of MoKA expression.
FIG. 6.
MoKA spatiotemporal expression pattern. (A) Northern blot hybridizations of RNA from embryonic stage mice (E11 to E17) normalized against actin (right) and of RNA from adult tissues normalized against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (left). Relative intensities of the hybridizing bands were quantified with the public domain NIH Image program (http://rsb.info.nih.gov/nih-image/). Numbers to the left of the blots indicate sizes (in kilobases) of RNA markers. (B) Whole-mount in situ hybridization of an E10.5 mouse embryo to antisense MoKA with indicated gene expression in the forebrain (fb), midbrain (mb), hindbrain (hb), branchial arches (ba), and forming limbs (l).
FIG. 7.
MoKA and KLF7 coexpressing tissues. (A) Comparative whole-mount hybridizations of E10.5 embryos to MoKA, Klf7, and Msx3 antisense probes showing coexpression of the three genes in the hindbrain (hb) and spinal cord (sc). (B) In situ hybridizations to antisense probes for MoKA (left panel) and Klf7 (right panel) of cross sections of the spinal cord at the thoracic level from an E12.5 mouse embryo. MoKA and Klf7 are coexpressed in the forming dorsal root ganglia (drg) and in the ventral horns (vh) of the neural tube. (C) In situ hybridizations of tissue sections from 2-month-old mice to antisense probes for MoKA (upper panels) or Klf7 (lower panels). The panels on the right are high-magnification views of the boxed regions shown on the left, with highlighted postmeiotic spermatids (Sp), spermatogonia (Sg), Sertoli cells (Sn), and spermatocytes (Sc). In all hybridizations, sense probes yielded no signal above the background level (data not shown).
KLF7 stimulates p21WAF1/Cip1 transcription.
It was previously reported that KLF7 promotes p21 accumulation and growth arrest of transfected cells, and thus, it was argued that one of its roles may be to regulate cell cycle progression during differentiation (17). This hypothesis was further investigated here by using two separate approaches. The first approach employed KLF7 and MoKA expression plasmids and the natural promoter sequence of the p21WAF1/Cip1 gene in cell transfection assays. The second approach examined KLF7 binding to the endogenous p21WAF1/Cip1 promoter sequence with the ChIP technique.
Transient transfection of COS7 cells with increasing amounts of the KLF7 plasmid stimulated p21(−2400)-Luc expression in a dose-dependent manner up to ninefold (Fig. 5B). Consistent with the results observed with the artificial (CAC)4TK-Luc reporter gene, cotransfection of MoKA alone did not stimulate p21(−2400)-Luc expression (Fig. 5B). On the other hand, coexpression of MoKA and KLF7 plasmids resulted in a 2.5-fold increase in p21(−2400)-Luc stimulation compared with KLF7 alone (Fig. 5B). These in vitro data therefore provided indirect evidence that KLF7 can potentially stimulate p21WAF1/Cip1 transcription; moreover, they documented ability of MoKA to superstimulate KLF7 activity in the context of a natural promoter sequence as well.
The proximal promoter of the p21WAF1/Cip1 gene contains several Sp1 sites that are the targets of distinct regulatory pathways (10). Binding of KLF7 to the proximal promoter (nucleotides −150 to +1) of the endogenous p21WAF1/Cip1 gene was assessed by ChIP with DNA purified from 293T cells transiently transfected with the Myc-tagged KLF7 expression vector. The results of the ChIP analysis showed that transiently expressed Myc-tagged KLF7 binds specifically to the proximal promoter of the endogenous p21WAF1/Cip1 gene (Fig. 8). Consistent with this finding, functional promoter assays showed the ability of Myc-tagged KLF7 to stimulate the activity of p21(−225)-Luc, albeit to a lesser extent than the p21(−2400)-Luc construct (Fig. 8). Different stimulation of the two reporter plasmids may indicate multiple binding sites of KLF7 in the −2.4-kb promoter or synergistic interaction between KLF7 and nuclear factors binding farther upstream of the −225 position. This point notwithstanding, the analyses conclusively demonstrated the potential of KLF7 to modulate expression of the cell cycle regulator p21WAF1/Cip1 gene.
FIG. 8.
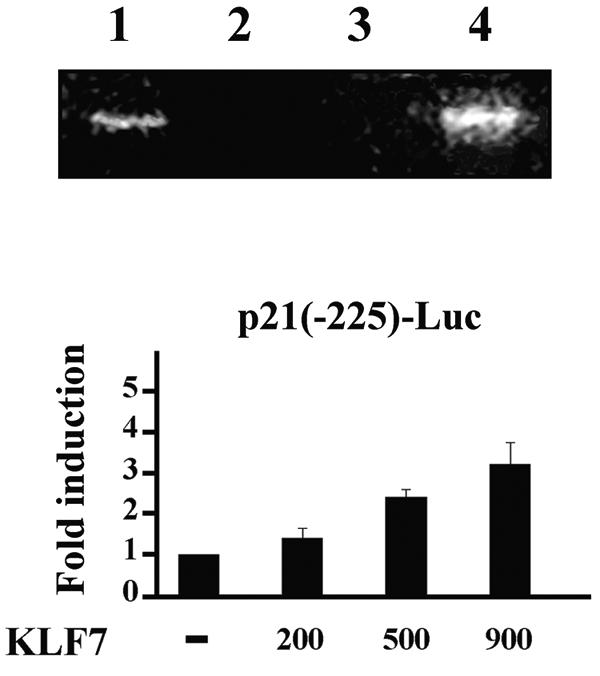
KLF7 binds in vivo to p21WAF1/CP1. At the top is a ChIP analysis of chromatin from mock-transfected cells before (lane 1) and after (lane 2) immunoprecipitation with anti-Myc antibody and from Myc-tagged KLF7-transfected cells following immunoprecipitation with (lane 4) and without (lane 3) anti-cMyc antibody. At the bottom the reporter gene activity of the p21(−225)-Luc construct cotransfected with the indicated amounts (in nanograms) of the KLF7 expression plasmid is shown. −, absent.
MoKA is a shuttling protein.
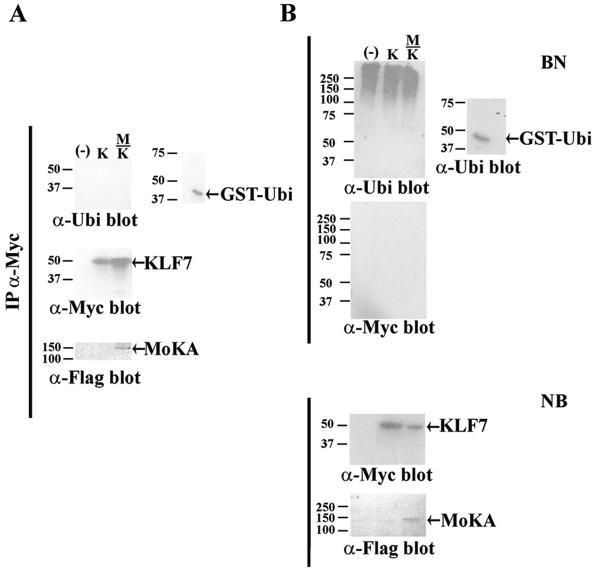
The next set of experiments was designed to characterize structure-function features of the MoKA protein. First, we investigated whether or not the F-box-mediated interaction between MoKA and KLF7 might target the transcription factor to the ubiquitination machinery (34). Myc-tagged KLF7 was therefore transiently expressed in NIH 3T3 cells, alone or together with Flag-tagged MoKA, and immunoprecipitated with anti-Myc antibody. The immunoprecipitates were then fractionated by SDS-PAGE and Western blotted against the anti-ubiquitin antibody. No ubiquitinated product was detected in the cell transfected with KLF7 alone or in the cell transfected with KLF7 and MoKA (Fig. 9A). We also performed the converse experiment, whereby extracts from cells overexpressing Myc-tagged KLF7 alone or together with Flag-tagged MoKA were incubated with polyubiquitin affinity beads. Once again, the results yielded no evidence that MoKA interaction promotes KLF7 ubiquitination (Fig. 9B).
FIG. 9.
MoKA-KLF7 interaction and ubiquitination. (A) Western analyses with indicated antibodies (α) of anti-Myc immunoprecipitates (IP) from NIH 3T3 cells cotransfected with 12 μg of plasmids expressing Myc-tagged KLF7 (K), Flag-tagged MoKA (M), or neither (−). The autoradiograph at the bottom is of the membrane above stripped and probed against the anti-Flag antibody. Also shown on the right side is a Western blot of the ubiquitin-GST fusion protein with the anti-ubiquitin antibody. (B) Western analyses of cell lysates from indicated transfections that did bind (BN) or did not bind (NB) to the polyubiquitin affinity matrix. Binding of the ubiquitin-GST fusion protein to the polyubiquitin affinity matrix is shown as a positive control. Numbers to the left of the panels indicate sizes (in kilodaltons) of protein markers.
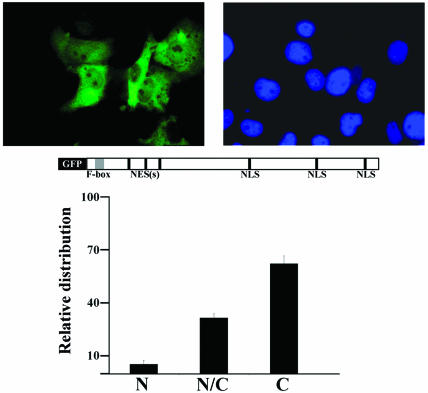
Next, we examined the subcellular localization of MoKA by expressing the full-length protein fused to the green fluorescent protein (GFP) in unsynchronized and exponentially growing COS7 cells. Autofluorescence analysis revealed that the MoKA fusion product localizes to both the nuclear and cytosolic compartments of transfected cells (Fig. 10). To be precise, counting ∼5 × 103 transfected cells showed that an average of 62% of them display MoKA-GFP only in the cytoplasm, 6% display it only in the nucleus, and 32% display it in both compartments (Fig. 10). Identical results were obtained with NIH 3T3 fibroblasts or with a MoKA-Flag fusion protein (data not shown).
FIG. 10.
MoKA subcellular localization. At the top the immunofluorescence localization of full-length GFP-tagged MoKA in transfected COS7 cells is shown on the left, with 4′,6′-diamidino-2-phenylindole staining shown on the right. At the bottom the relative distribution of transfected MoKA-GFP in cells within a two-well chamber slide totaling 4 cm2 is shown. Bars indicate standard errors. N, nuclear; N/C, nuclear and cytoplasmic; C, cytoplasmic.
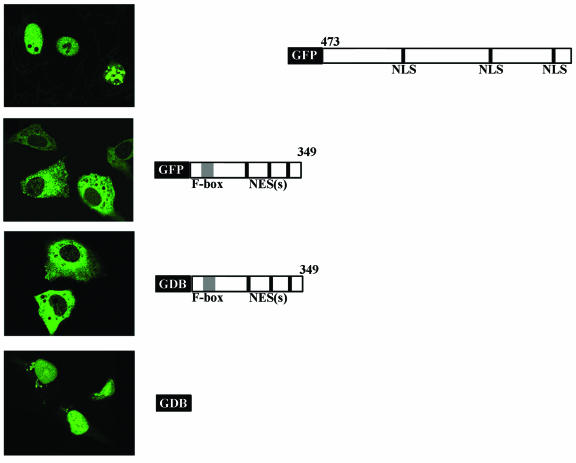
MoKA contains putative NLSs and NESs that may be responsible for the diversified distribution of the protein in asynchronously cycling cells. To dissociate the potential contribution of these two kinds of localization signals, mutant versions of MoKA-GFP that lack either the sequence around the NLSs or the NESs were transiently expressed in COS7 cells. Confocal microscopy revealed that the NES-containing (Δ350-1193)MoKA-GFP fusion product and the NLS-containing (Δ1-472)MoKA-GFP fusion product localized, respectively, to the cytoplasmic and nuclear compartments of transfected cells (Fig. 11). The three potential NESs of MoKA do not adhere to the canonical sequence that has been established for those signals (37). To estimate the relative strength of the noncanonical NESs, the mutant (Δ350-1193)MoKA was fused with the DNA-binding domain of GAL4 (GDB) and transiently expressed in COS7 cells. In spite of the strong NLS of GAL4, the chimeric (Δ350-1193)MoKA-GDB product still localized to the cytoplasm in >95% of the cells (Fig. 11). These results therefore confirmed that the canonical NLSs and the noncanonical NESs are responsible for the subcellular distribution of MoKA molecules.
FIG. 11.
Mapping of MoKA subcellular localization signals. (Top to bottom) COS7 cells were transfected with (Δ1-472)MoKA-GFP, (Δ350-1193)MoKA-GFP, (Δ350-1193)MoKA-GDB, and GDB alone. Subcellular localizations of fluorescent or immunolabeled fusion proteins were monitored by confocal microscopy.
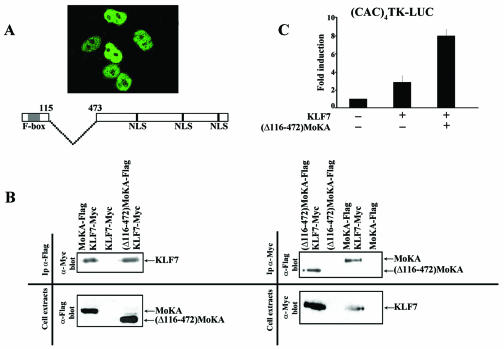
The above conclusion was corroborated by additional experiments in which an internally deleted Flag-tagged MoKA protein that lacks the NESs but retains the F-box and the NLSs [(Δ116-472)MoKA-Flag] was coexpressed in COS7 cells together with the Myc-tagged KLF7 plasmid and the (CAC)4 TK-Luc reporter construct. The results of the cell transfections showed that the (Δ116-472)MoKA-Flag fusion product localizes exclusively to the nucleus, coimmunoprecipitates with KLF7-Myc, and superstimulates KLF7 activation of the reporter gene (Fig. 12). Nuclear localization of the internally deleted MoKA protein confirmed the exclusive role of the NES-containing region in directing cytoplasmic localization of the full-length molecule. Conversely, it confirmed that other MoKA sequences are responsible for nuclear transport and interaction with and superactivation of KLF7.
FIG. 12.
Mapping of MoKA functional domains. (A) The Flag-tagged MoKA deletion is depicted below the image of COS7 cells transfected with 400 ng of the fusion product and stained with anti-Flag antibody. (B) Western analyses with indicated antibodies (α) of immunoprecipitates (IP) and cell extracts from COS7 cultures cotransfected with 12 μg of Flag-tagged full-length MoKA or deletion constructs of MoKA and Myc-tagged KLF7. (C) Luciferase activity of COS7 cells transfected with 100 ng of (CAC)4TK-Luc reporter gene together with 200 ng of KLF7 plasmid and a twofold molar excess of the (Δ116-472)MoKA-Flag plasmid. +, present; −, absent.
DISCUSSION
A large body of work has implicated Sp1 sites in the transcriptional control of both housekeeping and tissue-specific genes (3, 13). Indeed, the Sp/KLF superfamily of nuclear proteins is believed to constitute a transcriptional network that integrates disparate intracellular and extracellular signals to fine-tune gene expression during cell proliferation, growth, and differentiation (3). Binding affinities, promoter composition, cellular context, posttranslational modifications, and interacting cofactors are the major contributors to functional specificity of individual Sp/KLF proteins (3, 6, 13). With respect to cofactors, protein-protein interactions have been described both among Sp/KLF superfamily members and between them and other nuclear factors (3, 6, 13). An illustrative example of the latter is the dual role of CBP or p300 in superactivating KLF1 and promoting its association with the SWI-SNF chromatin-remodeling complex (38). To begin to elucidate the mechanisms and factors underlying KLF7 activity, we performed a genetic screen for interacting partners and identified a novel gene product that was named MoKA. Three independent sets of experimental evidence support the notion that MoKA is involved in positively modulating KLF7 action in vivo. First, a yeast two-hybrid screen, a GST pull-down assay, and protein copurification from mammalian cells all demonstrated physical interaction between MoKA and KLF7. Second, transient transfections and reporter gene assays correlated the protein-protein interaction with MoKA superactivation of KLF7. Third, in situ hybridizations documented the overlapping Klf7 and MoKA patterns of gene expression in embryonic and adult tissues. Functional assays also implicated KLF7 and MoKA in the transcriptional control of the p21WAF1/Cip1 gene in addition to identifying structural motifs responsible for different MoKA functions. They include interaction with KLF7 via an F-box motif and intracellular shuttling by canonical NLSs and noncanonical NESs.
The F-box is a 50-residue-long protein-protein interaction motif that was originally described in components of the SFC ubiquitin-ligase complex and, more recently, in proteins involved in several other cellular activities (15, 27). The F-box is usually found in the N-terminal portion of proteins and is often coupled at the C terminus with other protein-protein interaction motifs, most commonly leucine-rich and WD repeats (15). Homotypic interactions at the N termini link F-box-containing proteins to the core ubiquitin-ligase complex, whereas heterotypic interactions at the C termini bind them to phosphorylated substrates destined for degradation (34). We have excluded that MoKA interaction may target KLF7 to the ubiquitination machinery by two complementary sets of experiments. It is also important that MoKA diverges from prototypical F-box-containing proteins in two major ways. First, the MoKA-KLF7 interaction is heterotypic in that the latter protein contains no F-box motif. Indeed, our results suggest that a leucine zipper motif in KLF7 is the most likely candidate to mediate the interaction with the F-box of MoKA. Second, MoKA does not contain any of the coupled protein-protein interaction motifs thus far described in canonical F-box proteins. This last observation does not obviously exclude the possibility that structural motifs other than leucine-rich sequences and WD repeats may enable MoKA to interact in the nuclear and/or cytoplasmic compartments with different proteins, including members of the Sp/KLF superfamily. For example, MoKA may regulate the activity of KLF7 and other transcription factors through multiple protein-protein interactions that ultimately direct their intracellular distribution. These events may be also connected with MoKA subcellular redistribution during the cell cycle, as preliminary data with synchronized cells seem to suggest. The availability of anti-MoKA antibodies will enable us to test this and other hypotheses.
The ability of MoKA to shuttle between the nucleus and cytoplasm of asynchronously growing cells is under the control of NLSs and NESs located in the C-and N-terminal thirds of the protein, respectively. Although the NES consensus sequence was originally defined as a leucine-rich motif of the type LX1-3 LX2-3 LXL (where X can be any amino acid), recent evidence indicates that other hydrophobic residues can substitute for the second to fourth leucines in functional NESs (22, 29, 37). The NESs of MoKA display the same substitutions and therefore adhere to the noncanonical sequence; nonetheless, they can effectively compete with the canonical NLS of GAL4. The presence of both NESs and NLSs, as well as the distribution of the transfected protein in both nucleus and cytoplasm, imply that other factors are involved in controlling MoKA shuttling. Posttranslational modifications could conceivably be one of such modulating factors. For example, serine dephosphorylation has been shown to promote transcription factor NFAT1 relocation from the cytoplasm to the nucleus as the result of a conformational switch that masks the NES while concomitantly exposing the NLSs (26). A similar mechanism may be operating in MoKA where a serine-rich sequence lies within the two NLSs. Indeed, a computer-aided search for canonical phosphorylation sites suggests that a number of these residues could be potentially targeted. Indeed, the larger size of MoKA overexpressed in COS7 cells than in bacterial cells is a strong indication of posttranslational modifications. Another possibility is that binding of KLF7 in close proximity to the NESs may trigger nuclear accumulation of the protein. Preliminary experiments do not, however, support this last model, in that comparable patterns of MoKA subcellular distribution have been seen, irrespective of whether the protein is overexpressed alone or together with KLF7. This preliminary finding is also consistent with the idea that functional interaction between MoKA and KLF7 is not limited by the concentration of the former protein in the nucleus. This idea is based on the finding that the internally deleted protein without NESs, which localizes exclusively to the nucleus, superstimulates KLF7 activity to nearly the same extent as the full-length MoKA, which is distributed in both the nucleus and the cytoplasm (Fig. 5A, 10, and 12A and C). Work in progress is examining the mechanism of superstimulation of KLF7 activity and the biological significance of MoKA intracellular shuttling.
Four lines of evidence support a functional connection between KLF7 and MoKA and the cell cycle. First, overexpression of KLF7 in stably transfected cells results in p21 accumulation and growth arrest (17). Second, KLF7 stimulates p21WAF1/Cip1 promoter activity by itself and together with MoKA. Third, KLF7 binds to the proximal promoter region of the endogenous p21WAF1/Cip1 gene. Lastly, Klf7 and MoKA are coexpressed in cells that are in the process of exiting the cell cycle. In the testes, overlapping expression of MoKA and Klf7 in postmeiotic spermatids coincides with sperm maturation and/or chromatin remodeling. In the embryonic nervous system, MoKA and Klf7 are coexpressed in postmitotic neuroprogenitors, such as those in the ventral horn of the neural tube. Like Klf7, MoKA is actively transcribed in other tissues and at other developmental stages as well indicating independent involvement of the two proteins in other regulatory pathways. Along these lines, MoKA is actively transcribed in both postmitotic motor neurons (together with Klf7) and proliferating progenitors (without Klf7) of the spinal cord. Our current effort is aimed at deciphering structure-function relationships between MoKA and KLF7 in vitro and in genetically targeted mice as well as in defining their involvement in cell cycle progression and p21 regulation.
Acknowledgments
We thank J. Bieker, L. Carta, G. Karsenty, L. Lei, E. Johnson, T. Pietropaolo, M. Walsh, T. Wang, V. Yang, A. Zervos, and W. Zhang for advice and reagents and K. Johnson for typing the manuscript.
This work was supported by grants form the National Institutes of Health (AR38648), the New York Spinal Cord Injury Foundation, and the St. Giles Foundation.
REFERENCES
- 1.Bieker, J. J. 2001. Kruppel-like factors: three fingers in many pies. J. Biol. Chem. 276:34355-34358. [DOI] [PubMed] [Google Scholar]
- 2.Bieker, J. J., and C. M. Southwood. 1995. The erythroid Kruppel-like factor transactivation domain is a critical component for cell-specific inducibility of a beta-globin promoter. Mol. Cell. Biol. 15:852-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, A. R., J. D. Black, and J. Azizkhan-Clifford. 2001. Sp1 and Kruppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell Physiol. 188:143-160. [DOI] [PubMed] [Google Scholar]
- 4.Buckley, A. F., C. T. Kuo, and J. M. Leiden. 2001. Transcription factor LKLF is sufficient to program T cell quiescence via a c-Myc-dependent pathway. Nat. Immunol. 2:698-704. [DOI] [PubMed] [Google Scholar]
- 5.Coghill, E., S. Eccleston, V. Fox, L. Cerruti, C. Brown, J. Cunningham, S. Jane, and A. Perkins. 2001. Erythroid Kruppel-like factor (EKLF) coordinates erythroid cell proliferation and hemoglobinization in cell lines derived from EKLF null mice. Blood 97:1861-1868. [DOI] [PubMed] [Google Scholar]
- 6.Dang, D. T., J. Pevsner, and V. W. Yang. 2000. The biology of the mammalian Kruppel-like family of transcription factors. Int. J. Biochem. Cell Biol. 32:1103-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeGraeve, F., S. Smaldone, F. Laub, M. Mlodzik, M. Bhat, and F. Ramirez. 2003. Identification of the Drosophila progenitor of mammalian Krüppel-like factors 6 and 7 and a determinant of fly development. Gene 314:55-62. [DOI] [PubMed] [Google Scholar]
- 8.Donze, D., T. M. Townes, and J. J. Bieker. 1995. Role of erythroid Kruppel-like factor in human γ- to β-globin gene switching. J. Biol. Chem. 270:1955-1959. [DOI] [PubMed] [Google Scholar]
- 9.Fusco, C., E. Guidotti, and A. S. Zervos. 1999. In vivo construction of cDNA libraries for use in the yeast two-hybrid system. Yeast 15:715-720. [DOI] [PubMed] [Google Scholar]
- 10.Gartel, A. L., and A. L. Tyner. 1999. Transcriptional regulation of the p21(WAF1/CIP1) gene. Exp. Cell Res. 246:280-289. [DOI] [PubMed] [Google Scholar]
- 11.Gillemans, N., R. Tewari, F. Lindeboom, R. Rottier, T. de Wit, M. Wijgerde, F. Grosveld, and S. Philipsen. 1998. Altered DNA-binding specificity mutants of EKLF and Sp1 show that EKLF is an activator of the β-globin locus control region in vivo. Genes Dev. 12:2863-2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huber, T. L., A. C. Perkins, A. E. Deconinck, F. Y. Chan, P. E. Mead, and L. I. Zon. 2001. Neptune, a Kruppel-like transcription factor that participates in primitive erythropoiesis in Xenopus. Curr. Biol. 11:1456-1461. [DOI] [PubMed] [Google Scholar]
- 13.Kaczynski, J., T. Cook, and R. Urrutia. 2003. Sp1- and Kruppel-like transcription factors. Genome Biol. 4:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katz, J. P., N. Perreault, B. G. Goldstein, C. S. Lee, P. A. Labosky, V. W. Yang, and K. H. Kaestner. 2002. The zinc-finger transcription factor Klf4 is required for terminal differentiation of goblet cells in the colon. Development 129:2619-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kipreos, E. T., and M. Pagano. 2000. The F-box protein family. Genome Biol. 1:3002.1-3002.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo, C. T., M. L. Veselits, K. P. Barton, M. M. Lu, C. Clendenin, and J. M. Leiden. 1997. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 11:2996-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laub, F., R. Aldabe, V. Friedrich, Jr., S. Ohnishi, T. Yoshida, and F. Ramirez. 2001. Developmental expression of mouse Kruppel-like transcription factor KLF7 suggests a potential role in neurogenesis. Dev. Biol. 233:305-318. [DOI] [PubMed] [Google Scholar]
- 18.Lei, L., L. Ma, S. Nef, T. Thai, and L. F. Parada. 2001. mKlf7, a potential transcriptional regulator of TrkA nerve growth factor receptor expression in sensory and sympathetic neurons. Development 128:1147-1158. [DOI] [PubMed] [Google Scholar]
- 19.Lu, S., M. Liu, D. E. Epner, S. Y. Tsai, and M. J. Tsai. 1999. Androgen regulation of the cyclin-dependent kinase inhibitor p21 gene through an androgen response element in the proximal promoter. Mol. Endocrinol. 13:376-384. [DOI] [PubMed] [Google Scholar]
- 20.Matsumoto, N., F. Laub, R. Aldabe, W. Zhang, F. Ramirez, T. Yoshida, and M. Terada. 1998. Cloning the cDNA for a new human zinc finger protein defines a group of closely related Kruppel-like transcription factors. J. Biol. Chem. 273:28229-28237. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell, P. J., and R. Tjian. 1989. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science 245:371-378. [DOI] [PubMed] [Google Scholar]
- 22.Mowen, K., and M. David. 2000. Regulation of STAT1 nuclear export by Jak1. Mol. Cell. Biol. 20:7273-7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Narla, G., K. E. Heath, H. L. Reeves, D. Li, L. E. Giono, A. C. Kimmelman, M. J. Glucksman, J. Narla, F. J. Eng, A. M. Chan, A. C. Ferrari, J. A. Martignetti, and S. L. Friedman. 2001. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 294:2563-2566. [DOI] [PubMed] [Google Scholar]
- 24.Nuez, B., D. Michalovich, A. Bygrave, R. Ploemacher, and F. Grosveld. 1995. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375:316-318. [DOI] [PubMed] [Google Scholar]
- 25.Oates, A. C., S. J. Pratt, B. Vail, Y. Yan, R. K. Ho, S. L. Johnson, J. H. Postlethwait, and L. I. Zon. 2001. The zebrafish klf gene family. Blood 98:1792-1801. [DOI] [PubMed] [Google Scholar]
- 26.Okamura, H., J. Aramburu, C. Garcia-Rodriguez, J. P. Viola, A. Raghavan, M. Tahiliani, X. Zhang, J. Qin, P. G. Hogan, and A. Rao. 2000. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol. Cell 6:539-550. [DOI] [PubMed] [Google Scholar]
- 27.Patton, E. E., A. R. Willems, and M. Tyers. 1998. Combinatorial control in ubiquitin-dependent proteolysis: don't Skp the F-box hypothesis. Trends Genet. 14:236-243. [DOI] [PubMed] [Google Scholar]
- 28.Perkins, A. C., A. H. Sharpe, and S. H. Orkin. 1995. Lethal β-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375:318-322. [DOI] [PubMed] [Google Scholar]
- 29.Roth, J., M. Dobbelstein, D. A. Freedman, T. Shenk, and A. J. Levine. 1998. Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J. 17:554-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato, T., M. Hanada, S. Bodrug, S. Irie, N. Iwama, L. H. Boise, C. B. Thompson, E. Golemis, L. Fong, H. Wang, and J. C. Reed. 1994. Interactions among members of the Bcl-2 protein family analyzed with a yeast two-hybrid system. Proc. Natl. Acad. Sci. USA 91:9238-9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Segre, J. A., C. Bauer, and E. Fuchs. 1999. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 22:356-360. [DOI] [PubMed] [Google Scholar]
- 32.Shields, J. M., R. J. Christy, and V. W. Yang. 1996. Identification and characterization of a gene encoding a gut-enriched Kruppel-like factor expressed during growth arrest. J. Biol. Chem. 271:20009-20017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shindo, T., I. Manabe, Y. Fukushima, K. Tobe, K. Aizawa, S. Miyamoto, K. Kawai-Kowase, N. Moriyama, Y. Imai, H. Kawakami, H. Nishimatsu, T. Ishikawa, T. Suzuki, H. Morita, K. Maemura, M. Sata, Y. Hirata, M. Komukai, H. Kagechika, T. Kadowaki, M. Kurabayashi, and R. Nagai. 2002. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat. Med. 8:856-863. [DOI] [PubMed] [Google Scholar]
- 34.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 35.Wang, W., X. Chen, H. Xu, and T. Lufkin. 1996. Msx3: a novel murine homologue of the Drosophila msh homeobox gene restricted to the dorsal embryonic central nervous system. Mech. Dev. 58:1203-1215. [DOI] [PubMed] [Google Scholar]
- 36.Wani, M. A., S. E. Wert, and J. B. Lingrel. 1999. Lung Kruppel-like factor, a zinc finger transcription factor, is essential for normal lung development. J. Biol. Chem. 274:21180-21185. [DOI] [PubMed] [Google Scholar]
- 37.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]
- 38.Zhang, W., S. Kadam, B. M. Emerson, and J. J. Bieker. 2001. Site-specific acetylation by p300 or CREB binding protein regulates erythroid Kruppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol. Cell. Biol. 21:2413-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]