Abstract
Genetic studies using a set of overlapping deletions centered at the piebald locus on distal mouse chromosome 14 have defined a genomic region associated with respiratory distress and lethality at birth. We have isolated and characterized the candidate gene Phr1 that is located within the respiratory distress critical genomic interval. Phr1 is the ortholog of the human Protein Associated with Myc as well as Drosophila highwire and Caenorhabditis elegans regulator of presynaptic morphology 1. Phr1 is expressed in the embryonic and postnatal nervous system. In mice lacking Phr1, the phrenic nerve failed to completely innervate the diaphragm. In addition, nerve terminal morphology was severely disrupted, comparable with the synaptic defects seen in the Drosophila hiw and C. elegans rpm-1 mutants. Although intercostal muscles were completely innervated, they also showed dysmorphic nerve terminals. In addition, sensory neuron terminals in the diaphragm were abnormal. The neuromuscular junctions showed excessive sprouting of nerve terminals, consistent with inadequate presynaptic stimulation of the muscle. On the basis of the abnormal neuronal morphology seen in mice, Drosophila, and C. elegans, we propose that Phr1 plays a conserved role in synaptic development and is a candidate gene for respiratory distress and ventilatory disorders that arise from defective neuronal control of breathing.
Establishment of a regular breathing pattern is a critical physiological response that accompanies birth. Effective respiration is dependent on the development and maturation of the lungs and airways, the respiratory muscles, and the central and peripheral neural pathways that control their function (11). Genetic disorders that disrupt the development of any of these respiratory components can result in congenital central hypoventilation and respiratory distress syndromes (3, 8).
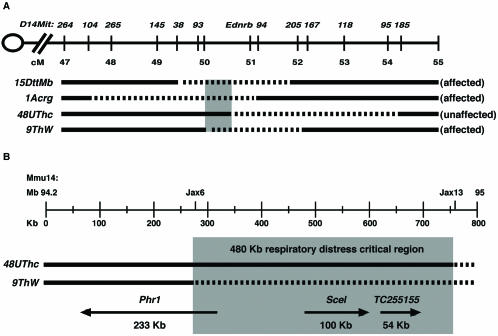
We have been using mice from the piebald deletion complex, a panel of nested megabase-scale chromosomal deficiencies, to identify genes that are essential for mouse development. Our studies have demonstrated that mice that are homozygous for the ∼1.75-Mb Ednrbs-15DttMb piebald deletion (hereafter, piebald deletions are abbreviated by using allele names [e.g., Ednrbs-15DttMb is abbreviated 15DttMb]) die of respiratory distress at birth and show a hunched posture indicating a lack of muscle tone, both of which are consistent with impaired motor function (15). Complementation mapping in genetic crosses with selected piebald deletion mice has focused the search for candidate genes responsible for this phenotype to a 480-kb critical region (17).
The evolutionarily conserved gene Phr1 (for PAM, highwire, rpm-1) maps within the respiratory distress critical region (16). Phr1 was first identified as the human gene PAM (encoding Protein Associated with Myc; present designation, KIAA0916) in a screen to isolate proteins that bind to the proto-oncogene c-myc (9). PAM encodes a large, 4,641-amino-acid (aa) protein that contains several conserved domains and protein interaction motifs. Genes homologous to Phr1 were also identified as highwire (hiw) in Drosophila (22) and as regulator of presynaptic morphology 1 (rpm-1) in Caenorhabditis elegans (19, 26). Genetic and functional studies with these organisms indicate a role for hiw and rpm-1 in regulating presynaptic differentiation and activity (5).
In the present study we have isolated and characterized the mouse Phr1 gene. The Phr1 gene covers a 233-kb region positioned at ∼94 Mb on mouse chromosome 14 within the piebald deletion complex, a region syntenic with human chromosome 13q22, which contains the human PAM gene (16). Conservation of the coding sequence and genomic location indicates that Phr1 is the mouse ortholog of human PAM, and conservation of sequence as well as function, as described below, indicates that it is also an ortholog of hiw and rpm-1.
In rodents Phr1 is expressed in several regions of the developing postnatal and adult brain (25). Our studies confirm and extend these results to show that Phr1 is expressed in several regions of the central nervous system (CNS) and peripheral nervous system (PNS) during development, consistent with a role for Phr1 in synapse formation in mammals.
The piebald deletion respiratory distress mutants fail to inflate their lungs following birth. This is a condition observed with mutations that impair motor function, such as agrin mutations (1). Given that the minimal genomic region deletes the Phr1 gene, we examined the innervation of the respiratory muscles of these mice. We observed a striking dysmorphology of motor neuron terminals in the intercostal muscles and of both sensory and motor neuron endings in the diaphragm. In addition, the phrenic nerve contained fewer axons, and innervation of the diaphragm was incomplete. Our results provide evidence that Phr1 plays an evolutionarily conserved role in synapse formation in mammals and is necessary for motor function and respiration at birth.
MATERIALS AND METHODS
Mice.
The 15DttMb, 9ThW, 48UThc, 1Acrg piebald deletion stock mice were maintained as deletion heterozygotes on a C57BL/6J genetic background as previously described (17). Embryos were collected by Caesarian section from timed matings of intercrosses between deletion stock mice, where detection of the vaginal plug was defined as embryonic day 0.5 (E0.5). Genotypes were determined by PCR, using the simple sequence length polymorphism markers D14Mit265 and the absence of the sequence-tagged site marker Jax22 (16, 17).
Cloning and analysis of the mouse Phr1 cDNA.
The full-length Phr1 coding sequence was isolated as a contig of five overlapping cDNA clones. Reverse transcription reactions were performed with 1 μg of total RNA from C57BL/6J E10.5 embryos. Long-range PCR was performed according to the recommendations of the manufacturer (Qiagen). Individual fragments were PCR amplified by using gene-specific primers (Table 1). PCR products were cloned into either the pCRII vector or the pCR-XL-TOPO vector for sequencing (Invitrogen). Overlapping sequence reads were assembled by using Sequencher 4.1, and the multiple sequence alignments were generated with a local version of ClustalW, version 1.8. The multiple sequence alignments were visualized and edited by using GeneDoc (http://www.psc.edu/biomed/genedoc).
TABLE 1.
PCR primers used to isolate Phr1 cDNA
| Primer pair | Product size (bp) | Sequence
|
|
|---|---|---|---|
| Forward | Reverse | ||
| Phr1.F1.2-Phr1.R1.1 | 2,641 | GCAGCCGCCACCATCTCTTCC | GCTGAGCTGGATGGGATCTGAG |
| Phr1.F2-Phr1.R2 | 3,000 | GTTGCAAGGCATGTGCAAGAG | GCTATGTCACTGGGTGAATCG |
| Phr1.F3-Phr1.R3 | 3,792 | GGAGGTGTTGGTTGATGATAGTG | CCTTCATCCACACTCTTCAAGTC |
| Phr1.F4-Phr1.R4 | 2,204 | GTGAGGTAGTTGAAGTCTGTAC | CATCATAAGGTCAGAGATGGTC |
| Phr1.F5-Phr1.R5 | 3,652 | CTTGCCGTGTGTTTGCAATGG | CATCCTTCTCAGTCCATGAGC |
In situ hybridization.
Embryos were dissected in cold phosphate-buffered saline (PBS), fixed overnight in 4% paraformaldehyde-PBS at 4°C, washed twice in PBST (PBS plus 0.1% Tween 20), dehydrated through a methanol-PBST series, and stored at −20°C in 100% methanol. Whole-mount in situ hybridization was performed as previously described (24). The digoxigenin-labeled antisense riboprobes Phr1 (nucleotides 8021 to 9542 of the coding sequence) and TC255155 (nucleotides 153 to 764) were used in this study.
The same probes were also used for hybridization to cryostat sections. Hybridization was performed with fresh, frozen tissue according to published protocols (20). Signals were visualized either with alkaline phosphatase-conjugated antidigoxigenin (Roche) followed by colorimetric nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate development or with peroxidase-conjugated antidigoxigenin followed by fluorescence detection (NEN/PE TSA-plus system), both according to the manufacturers' protocols, with the exception that the antidigoxigenin antibodies (Roche) were diluted 1:2,000.
Immunohistochemistry and histology.
For whole-mount nerve and muscle staining, tissue was dissected and fixed for 2 h in cold 2% paraformaldehyde-PBS. Staining was performed as described previously (6). The following antibodies were used: rabbit anti-neurofilament 200 (Sigma), rabbit antisynaptophysin (Zymed), Alexa 488 goat anti-rabbit (Molecular Probes), and rhodamine-conjugated alpha-bungarotoxin (Molecular Probes). Samples were examined on a Nikon fluorescence microscope, and images were collected with a SPOT 9000 camera or a Leica confocal microscope. For histological analysis, tissue was fixed by immersion in Bouin's fixative and embedded in paraffin for sectioning. Sections were stained by standard hematoxylin-eosin procedures. Four E18.5 Acrg/9ThW animals were examined by serial sectioning through the cervical spinal cord and were compared to wild-type littermates.
Nucleotide sequence accession number.
The sequence reported in this paper has been deposited in the GenBank database (accession no. AY325887).
RESULTS
Phr1 maps within the respiratory distress critical region of the piebald deletion complex.
Respiratory distress resulting in lethality at birth was first observed in mice homozygous for the ∼1.75-Mb 15DttMb deletion. In previous studies we used mice carrying additional piebald deletions in complementation crosses to refine the respiratory distress interval (17). The 1Acrg deletion chromosome provides DNA covering a distal portion of the 15DttMb deletion yet fails to rescue the defect. The 9ThW deletion chromosome provides additional proximal DNA and also fails to rescue the phenotype. Finally, the 48UThc deletion chromosome complements a larger proximal portion of the 15DttMb deletion than 9ThW deletion chromosome and is able to rescue the lethal breathing defect. These studies define the critical region as a 480-kb interval delimited by the proximal breakpoints of the 9ThW and 48UThc deletions (Fig. 1).
FIG. 1.
Functional map of the respiratory distress critical region. (A) Summary of complementation studies showing the original respiratory distress-defining 15DttMb deletion along with the nonrescuing 1Acrg and 9ThW deletions and the rescuing 48UThc deletion. The deleted portion of each chromosome is depicted as a dashed line, and the critical functional interval is shown in grey. Deletions are shown relative to mouse chromosome 14 from 47 to 55 cM on the Mouse Genome Database genetic map. D14Mit genetic markers used for molecular mapping studies and the piebald (Ednrb) locus are positioned along the chromosome. (B) The 480-kb respiratory distress critical region, in grey, is defined by the 48UThc and 9ThW proximal deletion breakpoints. The deletions were defined by the Jax6 and Jax13 sequence-tagged site markers. The positions and sizes of the Phr1, Scel, and TC255155 transcripts based on alignments with the genomic sequence are shown below the deletion chromosomes. Arrows indicate the direction of transcription.
In this study we have focused on the characterization of the candidate gene Phr1. The 15DttMb deletion removes the entire Phr1 gene. The proximal breakpoint of the nonrescuing 9ThW deletion maps within Phr1, removing the promoter region and the first two exons at the 5′ end of the gene. Reverse transcription-PCR (RT-PCR) and in situ hybridization studies indicate that Phr1 is not expressed in 9ThW/15DttMb compound deletion heterozygous mice (data not shown). The proximal breakpoint of the 48UThc deletion maps ∼452 kb distal to the 5′ end of the Phr1 gene (Fig. 1). RT-PCR and in situ hybridization studies indicate that Phr1 is expressed in 48UThc/15DttMb compound deletion heterozygous mice (data not shown). Thus, the loss of Phr1 expression correlates with the respiratory distress phenotype.
Distinct and evolutionarily conserved features of the PHR1 protein.
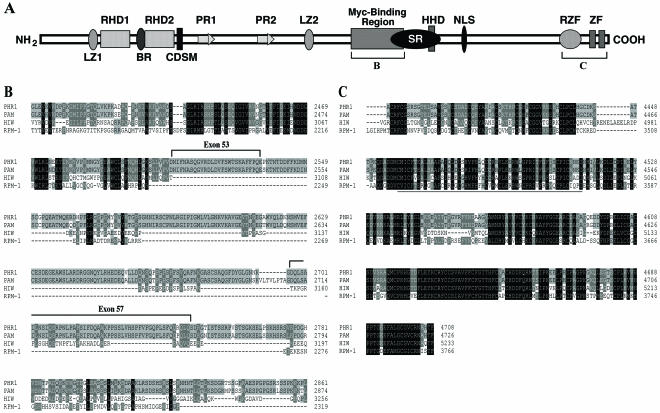
To further characterize the mouse Phr1 gene, we performed RT-PCR to isolate overlapping clones covering 14,498 bp of the Phr1 cDNA. The complete open reading frame (ORF) encodes a large, 4,708-aa PHR1 protein with a predicted mass of 517 kDa. Sequence alignment showed 96% identity with human Pam across the entire ORF. The PHR1 protein shares several conserved domains with Pam, HIW, and RPM-1 (Fig. 2). These included the N-terminal RCC1 (regulator of chromatin condensation 1) homology domains, the two PHR1 (for Pam, HIW, RPM-1) signature repeats, leucine zipper motifs, and the highly conserved cysteine-rich C-terminal region containing the ubiquitin ligase activity-associated ring zinc finger domain and the C2H2-type zinc finger motifs (Fig. 2C). Although other proteins in the mammalian genome share many of these motifs, there are no clear homologs of Phr1 that would suggest that it is part of a gene family. Interestingly, the mammalian orthologs contained conserved motifs that are absent in the invertebrate proteins. The HIW and RPM-1 proteins lack the putative nuclear localization signal and a portion of the ∼300-aa region associated with Myc binding activity (9). This region shares 87% identity between the mouse PHR1 and human Pam proteins.
FIG. 2.
Structure and conservation of PHR1. (A) Schematic representation of PHR1. The following conserved motifs were identified: LZ1 and LZ2, potential leucine zippers; RHD1 and RHD2, RCC1 homology domains; PR1 and PR2, Pam repeats; CDSM, cell division sequence motif; HHD, histone homology domain; SR, serine-rich region; NLS, nuclear localization signal; RZF, ring Zn finger domain; and ZF, potential C2H2-type zinc finger motifs. The brackets indicate the regions displayed in the protein alignments in panels B and C. (B) The Myc binding domain is conserved between PHR1 and Pam, with only the first 100 residues conserved in HIW and RPM-1. Exons 53 and 57 encode novel amino acids. (C) The C-terminal region is highly conserved between PHR1, Pam, HIW, and RPM-1. PHR1 shares 97, 65, and 55% identity across this region with Pam, HIW, and RPM-1, respectively. The ring Zn finger domain is underlined.
The PHR1 ORF that we identified encodes a protein that is 68 aa larger than the originally described human Pam protein. Alignment of the Phr1 cDNA with the mouse genomic sequence revealed 85 exons covering a ∼233-kb genomic segment. The additional 68 aa correspond to alternative transcripts containing two additional exons within the Myc binding domain (Fig. 2A and B). These exons are also present in the human genomic sequence, and human expressed sequence tags (ESTs) with (BG391609 and AU117955) and without (BE815141 and AU141436) these exons are represented in the GenBank EST database. Using RT-PCR, we have also confirmed mouse transcripts with and without these exons (data not shown). Thus, the PHR1 and Pam proteins have acquired sequences associated with the Myc interaction domain during evolution, and the region harboring the Myc interaction domain is differentially expressed through alternative splicing.
Phr1 is expressed in the developing CNS and PNS.
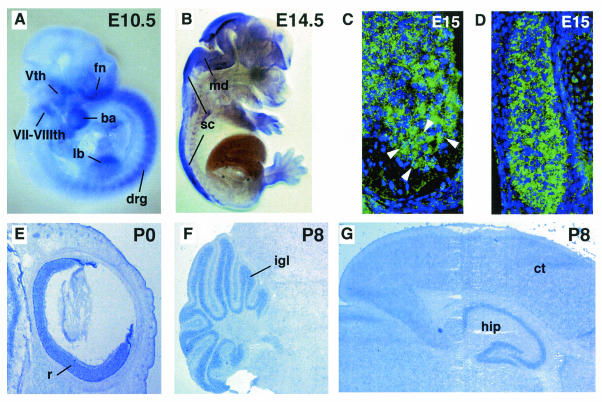
Phr1 transcripts are detected by RT-PCR in developing embryos from E8.5 through E18.5 of development. We performed in situ hybridization studies and observed a dynamic tissue-specific developmental expression profile for Phr1 (Fig. 3). Phr1 is expressed prominently in peripheral neurons of the dorsal root ganglia (DRG) and cranial ganglia at midgestation (E10.5) and during later stages of development. Craniofacial tissues and the limb buds also express Phr1 in embryos at E10.5 (Fig. 3A). We observed expression in the mid- and hindbrain regions of the CNS in E11.5 to E12.5 embryos, and as development progresses, Phr1 transcripts are detected along the length of the spinal cord (Fig. 3B). In the E15 spinal cord, populations of cells across the dorsoventral axis express Phr1, including the motor neuron pools of the developing cervical and thoracic spinal cord, containing cells that project axons that innervate the diaphragm and intercostal muscles (Fig. 3C). Motor neurons in the ventral horn of the spinal cord express Phr1 strongly. Sensory neuron cell bodies throughout the DRG also express Phr1 at a uniformly robust level (Fig. 3D). Interestingly, Phr1 was also detected by RT-PCR from skeletal muscle and cultured C2C12 cells (data not shown), suggesting that it may have a role in both tissues for reciprocal signaling during synaptogenesis, as suggested for hiw in Drosophila.
FIG. 3.
Phr1 is expressed in the developing nervous system. (A) Whole-mount in situ hybridization of E10.5 embryo showing Phr1 expression in peripheral neurons of the craniofacial ganglia and in the DRG flanking the spinal cord. Expression is also detected in the developing craniofacial region and forelimb bud. fn, fronto-nasal process; Vth, trigeminal ganglion; VII-VIIIth, facial and auditory ganglia; ba, first branchial arch; lb, forelimb bud. (B) Sagittal view at E14.5, showing expression throughout the mid- and hindbrain region and along the length of the developing spinal cord. md, medulla; sc, spinal cord. Transverse sections at E15.5 showed expression in cells across the dorsoventral axis of the spinal cord and in the DRG flanking the spinal cord. (C) The motor neurons of the ventral horn are robustly positive for Phr1 expression (arrowheads) (ventral is down). Motor neurons were identified based on their anatomical location and large nuclei with prominent nucleoli. (D) The sensory neuron cell bodies in the DRG are also uniformly positive for Phr1 expression. (E) Sagittal section of eye at P0, demonstrating Phr1 expression in retinal neurons. r, retina. (F) Sagittal section at P8, showing expression in the granule cell layer of the cerebellum. igl, internal granule cell layer. (G) Sagittal section at P8, showing expression in the hippocampus and throughout the cerebral cortex. ct, cerebral cortex; hip, hippocampus. In panels E, F, and G, anterior is to the right.
We also documented Phr1 expression in the brain during postnatal development. Expression was particularly strong in the cerebellum, hippocampus, and retina (Fig. 3E, F, and G). Lower levels of expression were observed throughout the cerebral cortex. As previously reported, we observed that levels of Phr1 expression in the postnatal brain increased over the first 2 weeks of development and persisted into adulthood (25). Our results indicate that the level and tissue distribution of Phr1 transcripts are high and actively regulated during pre- and postnatal CNS and PNS development, consistent with a role in synapse formation and function.
The other two genes that fall within the critical interval for respiratory distress are the sciellin gene, which is implicated in the formation of cornified epithelium, and TC255155, an uncharacterized EST. Both of these genes are detectable at birth (postnatal day 0 [P0]) in lung by RT-PCR; however, Northern blotting from P8 tissue reveals that TC255155 is detectable only in the brain and only with a very long exposure (data not shown). Given the low level of expression and the lack of evidence for an evolutionarily conserved role in the nervous system for these genes, we have focused on Phr1 for subsequent studies. However, both TC255155 and sciellin remain formal candidates for contributing to the phenotypic defects seen in the compound-deficiency mice.
Defective innervation and synapse formation in respiratory distress piebald deletion mutant mice.
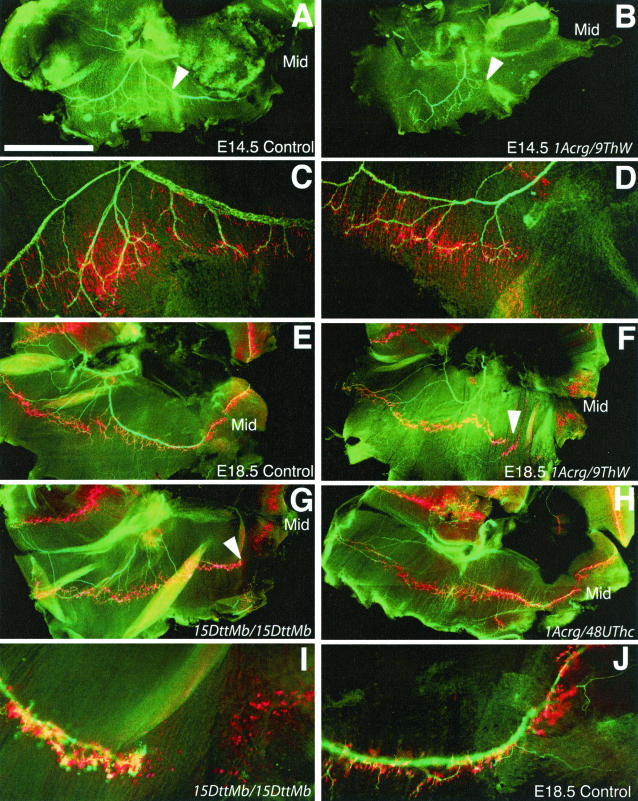
Based on the lethal breathing defect and the role of the Drosophila and C. elegans homologs of Phr1 in synaptic development, we examined the innervation of the diaphragm and intercostal muscles in piebald deletion mutant mice. Normally, axons of the phrenic nerve exit the cervical spinal cord and contact the developing diaphragm at E12. The nerve and muscle develop together, and by E13 the nerve forms three main branches, one extending dorsally to the crus, one innervating the dorsal portion of the diaphragm, and one innervating the ventral portion of the diaphragm (8). Whole-mount staining of nerves and acetylcholine receptors (AChRs) revealed that motor innervation of the diaphragm was incomplete even at early stages in 9ThW/1Acrg mice, the minimal deficiency combination that results in respiratory distress (Fig. 4A to D). While the entire diaphragm is contacted by axons of the phrenic nerve in wild-type mice at E14.5, axons consistently failed to reach the ventral-most portion of the diaphragm in the mutants. The pattern of AChR staining in the ventral muscle was diffuse and not tightly restricted to the endplate band of the muscle, consistent with muscle fibers that have never been innervated. This defect persisted, with only minor improvement in the elaboration of the nerve by E18.5 (Fig. 4E and F). Therefore, the lack of innervation reflects an early-stage developmental defect and does not appear to be due to a loss of synaptic connections or a retraction of phrenic axons.
FIG. 4.
Innervation of the diaphragm by the phrenic nerve. (A) In control mice at E14.5, the axons of the phrenic nerve, stained green with antibodies against neurofilament and synaptophysin, have reached the diaphragm, branched, and extended both dorsally and ventrally to the full extent of the muscle. (B) In 9ThW/1Acrg littermate mice at E14.5, the phrenic nerve is less robust and fails to reach large portions of the diaphragm. In particular, the ventral portion of the muscle is not innervated. (C and D) Higher-magnification views of the regions indicated in panels A and B shows that the aneural muscle in 1Acrg/9ThW mice has a pattern of postsynaptic differentiation that is consistent with muscle that has never beeninnervated. (E) By E18.5, control mice have a similar but more elaborate pattern of motor innervation, with the nerves terminating on plaques of AChRs, stained red with rhodamine-conjugated α-bungarotoxin, on the muscle fibers. (F) In 9ThW/1Acrg mice at E18.5, the ventral diaphragm is still not innervated and the pattern of aneural muscle is similar to that at earlier ages. The defects in innervation of the diaphragm correspond to deficiency combinations that cause lethality due to respiratory distress. (G) Mice homozygous for 15DttMb show defects in the phrenic nerve that are very similar to those seen in 9ThW/1Acrg mice. (H) However, 48UThc/1Acrg mice are viable postnatally and show no defects in the diaphragm at E18.5. (I and J) Higher-magnification views of the 15DttMb diaphragm and a control diaphragm in the ventral affected region. In all cases, the right side of the diaphragm is shown, dorsal is left, ventral is right, Mid indicates the ventral midline, and the arrowheads indicate regions of incomplete innervation. Bars, 1 mm (A and B) and 1.6 mm (E to H).
The defective innervation of the diaphragm corresponds to the respiratory distress genetic interval and Phr1 expression. Mice homozygous for the 15DttMb deletion show the same deficit in innervation seen in the 9ThW/1Acrg mutants (Fig. 4G). However, 48UThc/1Acrg deletion mice, which preserve Phr1 expression, are viable, establish a normal breathing pattern, and do not show a defect in innervation at E18.5 (Fig. 4H).
Neuronal morphology is disrupted in mice with deficiencies which remove Phr1.
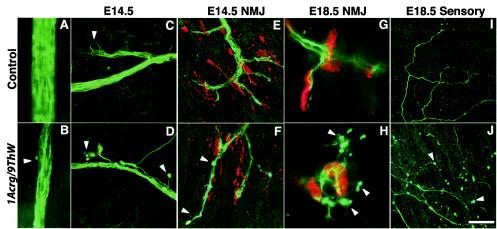
Soon after the nerve contacts the muscle, neuromuscular junctions form and are stabilized in response to agrin signaling from the motor axon (12). Embryonically, the nerve has wispy terminals that cover a plaque of receptors on the muscle. Although the final structure of the neuromuscular junction is not complete until weeks postnatally, it is a functional cholinergic synapse soon after it forms. In 9ThW/1Acrg mice the phrenic nerve was markedly decreased in diameter at all ages examined, consisting of many fewer axons than in control animals (Fig. 5A and B). At E14.5, the ingrowing nerve also had abnormally shaped endings with large varicosities at the ends of the axons (Fig. 5C and D). The neuromuscular junctions that formed at E14.5 were only modestly malformed, with a greater degree of terminal overshooting and some varicose endings (Fig. 5E and F). At E15.5, the neuromuscular junctions appeared fairly normal, suggesting that signaling by proteins such as agrin is intact in these mice (data not shown). By E18.5 the neuromuscular junction morphology was more clearly disrupted, with a striking amount of nerve terminal sprouting beyond the postsynaptic receptor plaque, consistent with inadequate neurotransmitter release and subsequent lack of muscle stimulation (Fig. 5G and H). The E18.5 terminal sprouts also had varicose endings, resembling the abnormalities seen in the ingrowing nerve. The sensory neurons in the diaphragm also showed defects at E18.5 (Fig. 5I and J). Their axons were of much smaller caliber than controls and appeared to be either increasingly branched or defasiculated. Like the motor axons, the sensory fibers also had large varicosities near their terminals. Thus, more than one population of neurons in the diaphragm is affected. Neuromuscular junctions in the intercostal muscles showed a nerve terminal dysmorphology similar to that of those in the diaphragm, indicating that there is a general defect in motor axons, even though the intercostal muscles appeared to be completely innervated (data not shown).
FIG. 5.
Motor and sensory neuronal morphology. (A and B) The phrenic nerve contains markedly fewer axons in 9ThW/1Acrg mice than in littermate control mice. Furthermore, at E14.5, ingrowing axons show striking abnormalities in terminal morphology. (C) Control mice have tightly fascilulated axons and no visible terminal morphological defects (arrowhead). (D) 9ThW/1Acrg mice have stray axons and large varicosities at the termini (arrowheads). (E) At E14.5, neuromuscular junctions (NMJ) have formed in control mice, with nerve terminals (green) overlying AChR-rich plaques (red) on the muscle fibers. At this age, a large number of aneural receptor plaques are still evident. (F) In E14.5 9ThW/1Acrg mice, the neuromuscular junction morphology is mildly perturbed, with some overgrowth of terminals beyond the receptor plaques (lower arrowhead) and some varicose terminals (upper arrowhead). These defects are less pronounced by E15.5 (not shown) (G) In E18.5 control mice, the nerves are more robust and the receptor plaques are more condensed than at earlier stages. (H) 9ThW/1Acrg mice show similar maturation of the neuromuscular junction but also show striking sprouting of the nerve terminals beyond the receptor plaques. The terminal sprouts have varicose endings (arrowheads) similar to those seen on the ingrowing nerve at E14. (I) By E18.5, sensory neurons have also projected processes into the diaphragm from the lateral edges. These endings have a characteristic arborization pattern. (J) In 9ThW/1Acrg mice, the sensory axons are present but are of much finer caliber, have numerous varicosities, and are either more elaborately branched or less tightly fasiculated than in control mice. Bars, 36 μm (A and B), 40 μm (C to F, I, and J), and 11 μm (G and H).
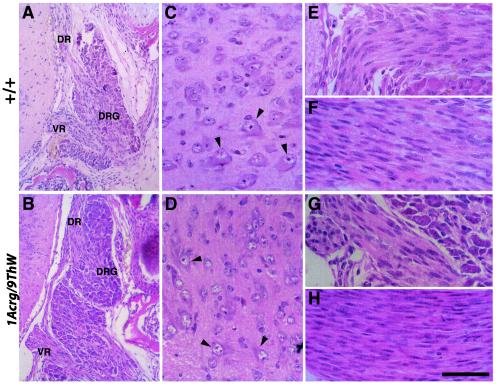
The morphology defects seen in the sensory and motor axons were most easily observed in the terminals. A histological examination at E18.5 of the sensory cell bodies of the DRG and motor cell bodies in the ventral horn of the spinal cord did not reveal any gross abnormalities (Fig. 6A to D). The structures and cells were of comparable size in 1Acrg/9ThW and wild-type control littermates. The motor neurons had large eosinophilic cytoplasm and a prominent nucleolus. The nerves themselves were also qualitatively similar in the two genotypes (Fig. 6E to H). In the mouse, the ventral root motor axons exit the spinal cord and pass through the DRG cell bodies before joining the mixed sensory and motor spinal nerve distally. Both the ventral roots and the spinal nerves of the 1Acrg/9ThW mice were of normal size and morphology, with axons that are not yet myelinated intermingled with a large number of elongated Schwann cell nuclei. No dying neurons or dystrophic axons were seen, suggesting that the defects seen in 1Acrg/9ThW peripheral sensory and motor innervation may be restricted to terminal processes and that there is not a striking increase or decrease in cell number.
FIG. 6.
Histological examination of E18.5 spinal cords. (A) The cell bodies of sensory neurons are located in the DRG. These bipolar neurons project dorsal root axons into the dorsal horn of the spinal cord. The ventral root motor axons exit the spinal cord, passing through the DRG. (B) The same anatomy is observed in 1Acrg/9ThW mutant mice. Neither pathology nor differences in cell size or number were observed in the mutants. (C) In the ventral horn of the spinal cord, motor neurons are identifiable by their large eosinophilc cytoplasm and large nuclei with prominent nucleoli (arrowheads). (D) The ventral horn of 1Acrg/9ThW mice also showed motor neurons in the proper anatomical position with normal morphology. Similarly, the axons themselves showed no pathology. Ventral roots (E and G) contain motor axons as well as elongated Schwann cell nuclei. The spinal nerves (F and H) contain both sensory afferent and motor efferent axons as well as Schwann cells. No myelination is under way at this age. DR, dorsal root; VR, ventral root. Bars, 144 μm (A and B) and 48 μm (C to H).
DISCUSSION
In this study we have focused on a set of piebald deletions that are associated with respiratory distress in newborn mice. Our studies reveal innervation and synaptic defects in the diaphragm and intercostal muscles that control breathing in the piebald deletion mutants that die of respiratory failure. We have also identified and characterized the candidate gene Phr1, which encodes an evolutionarily conserved protein that is required for normal synapse development in Drosophila and C. elegans.
The respiratory distress critical genomic interval is defined by the proximal breakpoints of the 9ThW and 48UThc piebald deletions. Computational and experimental analyses reveal three genes within this interval, i.e., Phr1, the sciellin gene (Scel), and a novel transcript, TC255155 (16). Scel encodes a precursor of the cornified envelope of keratinizing tissues and is expressed in the upper cell layers of the epithelium during development (2). TC255155 is broadly expressed at very low levels in several tissues, including the nervous system (data not shown). Both genes are detectable by RT-PCR in lung as well. Although Scel and TC255155 are formally candidates, we have focused on the characterization of Phr1 as the most promising candidate gene in the region to play an important role in neuronal development and respiratory function. This is based on the robust neuronal expression of Phr1 and its homology with genes known to affect synaptic morphology.
Phr1 is expressed in several regions of the developing PNS and CNS, including the ventral motor neurons at the cervical and thoracic levels of the spinal cord that project axons that innervate the diaphragm and intercostal respiratory muscles. Phr1 expression postnatally includes regions associated with long-term changes in synaptic activity, and Pam has been shown to inhibit adenylyl cyclase activity, suggesting an involvement in learning and memory (21, 25). Similarly, autoantibodies against Pam have been implicated in some forms of schizophrenia (23). Our data demonstrating abnormalities in synapse organization in Phr1-deficient mice are consistent with a proposed role for Phr1 in the regulation of synaptic differentiation and homeostasis. Our results also suggest that loss of Phr1 function is lethal at birth. Therefore, in vivo studies to investigate a role for Phr1 in postnatal processes such as learning and memory will require a conditional knockout approach.
PHR1 is an evolutionarily conserved protein that contains several interactive motifs and domains associated with diverse activities. This structure suggests that PHR1 potentially performs multiple functions. The RCC1-like domains, the putative histone binding protein homologous domains, and the nuclear localization signal are consistent with studies that have localized Pam in the nucleus (9). These data and the interaction with the transcription factor Myc are consistent with PHR1 activity in the nucleus. In addition, the interaction with Myc suggests a potential role in biological processes such as cell proliferation. Preliminary experiments using bromodeoxyuridine labeling to examine proliferation and migration in the retina and external granule cell layer of the cerebellum, both of which are sites of high Phr1 expression, did not reveal an obvious role for Phr1 in proliferation during CNS development (data not shown).
The PHR1 protein also contains a cluster of Zn finger domains in the carboxy-terminal region of the protein, including a ring Zn finger motif proposed to mediate ubiquitin ligase activity. Genetic studies with Drosophila using mutations of hiw, encoding a proposed ubiquitin ligase, and the deubiquinating enzyme fat facets have provided a link between the ubiquitin pathway and synaptic development (5, 10). The PHR1 ring Zn finger domain is highly conserved between HIW, RPM-1, and Pam. Our genetic studies with the mouse suggest that this conserved domain is important for an evolutionarily conserved role for Phr1 in synaptic development, making Phr1 a true ortholog of hiw and rpm-1.
Interestingly, a portion of the Myc interactive domain is absent in the invertebrate proteins HIW and RPM-1. Our studies confirm that the mouse and human proteins both contain the complete Myc binding region and that this region is subject to alternative splicing. We have also determined that this region of Phr1 is represented in the Fugu genome (data not shown). Therefore, it appears that domain acquisition has occurred during the evolution of the Phr1 gene. Thus far, our genetic studies have not revealed a function for Phr1 associated with unique activities potentially acquired through the addition of the Myc interactive domain. Rather, our results provide evidence for a conserved function in synapse development consistent with the ubiquitin activity-associated ring Zn finger domain that is highly conserved between invertebrates and vertebrates. Additional studies are needed to address the function of this large, multidomain protein, given that PHR1 is likely to perform both conserved and potentially unique and modular activities in mammals.
The neurodevelopmental phenotype of the piebald deletion mice that lack Phr1 is characteristic of both a defect in the outgrowth of the phrenic nerve and impaired synaptic activity at the neuromuscular junction. The expression of Phr1 in muscle cells also indicates that a function in retrograde signaling cannot be ruled out. In mice lacking Phr1, the phrenic nerve is incompletely formed, containing fewer axons even at early stages and the diaphragm is incompletely innervated in a very stereotyped pattern. Furthermore, the mice show limited spontaneous motility, are not able to breathe effectively, and have sprouting at motor terminals at E18.5, consistent with ineffective synaptic transmission at the neuromuscular junction. However, they did show some mobility, making the phenotype less severe than that in mice with a complete lack of functional neuromuscular junctions (4, 6, 13). The Drosophila hiw mutant phenotype shows a marked decrease but not a complete loss of presynaptic release. The quantal content of evoked synaptic transmission is decreased by approximately 75% (22). Similarly, rpm-1 null mutations in C. elegans result in a temperature-sensitive reduction in synaptic transmission but not in a complete elimination (19, 26).
The synaptic dysmorphology observed is also consistent with the Drosophila and C. elegans phenotypes. The piebald deletion mutants exhibit both a motor neuron defect and a novel sensory neuron dysmorphology. The similarity of the defects in the sensory and motor axons that we have observed in the piebald mutant mice and the expression pattern suggest that Phr1 plays a role in the establishment of nerve terminal morphology and activity for multiple neuronal cell types in the developing nervous system. In Drosophila, hiw mutants have an overgrowth of motor terminals at the neuromuscular junctions. In C. elegans, rpm-1 mutants have disrupted morphology in GABAergic motor terminals. The defects that we have observed in the deficient mice are consistent with these phenotypes. However, the sensory as well as the motor axons are affected, suggesting an even greater involvement of Phr1 in neuronal differentiation.
As in Drosophila, the synaptic overgrowth at the NMJ is proposed to reflect that PHR1 regulates the balance of factors that are important for synaptic function. The partial innervation of the diaphragm and the thinner phrenic nerve that we have observed in mice also raises the possibility that PHR1 regulates factors critical for proper axonal outgrowth (14). Thus, PHR1 activity would be important not only for the proper function of neuronal connections but also for their formation.
We have focused on peripheral neurons; however, the respiratory distress phenotype could also be manifested by defects in the CNS. For instance, the nucleus ambiguus in the brain stem controls unconscious respiratory rhythm. Given the broad expression of Phr1 in the CNS, it seems likely that additional defects in connectivity and neuronal morphology exist and could be exacerbating the phenotype.
The establishment and maintenance of a normal breathing pattern are dependent on central and peripheral neuronal pathways. Deficits in these pathways can lead to hypoxia and death. Synaptic defects that impair the function of these neuronal pathways could contribute a variety of breathing disorders. For instance, congenital central hypoventilation syndrome is often associated with Hirschsprung's disease (7). While Hirschsprung's disease is associated with mutations in the endothelin receptor B (Ednrb, the piebald gene that is the focal point of the deletion complex), studies involving a subset of cases of congenital central hypoventilation syndrome associated with Hirschsprung's disease have not shown mutations in the coding region of the Ednrb gene (18). It is interesting to consider the possibility of a contiguous gene syndrome, where mutations affect additional neighboring genes such as Phr1, which sits 463 kb proximal to Ednrb. Thus, Phr1 is a candidate for additional studies to understand the genetic basis of respiratory distress and related syndromes in humans.
Acknowledgments
We thank Susan Ackerman and Greg Cox for valuable discussions and comments on the manuscript, Greg Martin and Pete Finger for assistance with confocal microscopy, and Jill Wentzell for assistance with the histology.
This work was supported by NIH grants HD36434 and HD41066 (to T.P.O.) and the shared service facilities of The Jackson Laboratory Cancer Center (CORE grant CA34196).
REFERENCES
- 1.Burgess, R. W., Q. T. Nguyen, Y. J. Son, J. W. Lichtman, and J. R. Sanes. 1999. Alternatively spliced isoforms of nerve- and muscle-derived agrin: their roles at the neuromuscular junction. Neuron 23:33-44. [DOI] [PubMed] [Google Scholar]
- 2.Champliaud, M. F., H. P. Baden, M. Koch, W. Jin, R. E. Burgeson, and A. Viel. 2000. Gene characterization of sciellin (SCEL) and protein localization in vertebrate epithelia displaying barrier properties. Genomics 70:264-268. [DOI] [PubMed] [Google Scholar]
- 3.Cole, F. S., A. Hamvas, and L. M. Nogee. 2001. Genetic disorders of neonatal respiratory function. Pediatr. Res. 50:157-162. [DOI] [PubMed] [Google Scholar]
- 4.DeChiara, T. M., D. C. Bowen, D. M. Valenzuela, M. V. Simmons, W. T. Poueymirou, S. Thomas, E. Kinetz, D. L. Compton, E. Rojas, J. S. Park, C. Smith, P. S. DiStefano, D. J. Glass, S. J. Burden, and G. D. Yancopoulos. 1996. The receptor tyrosine kinase MuSK is required for neuromuscular junction formation in vivo. Cell 85:501-512. [DOI] [PubMed] [Google Scholar]
- 5.DiAntonio, A., A. P. Haghighi, S. L. Portman, J. D. Lee, A. M. Amaranto, and C. S. Goodman. 2001. Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature 412:449-452. [DOI] [PubMed] [Google Scholar]
- 6.Gautam, M., P. G. Noakes, L. Moscoso, F. Rupp, R. H. Scheller, J. P. Merlie, and J. R. Sanes. 1996. Defective neuromuscular synaptogenesis in agrin-deficient mutant mice. Cell 85:525-535. [DOI] [PubMed] [Google Scholar]
- 7.Gozal, D., and R. M. Harper. 1999. Novel insights into congenital hypoventilation syndrome. Curr. Opin. Pulm. Med. 5:335-338. [DOI] [PubMed] [Google Scholar]
- 8.Greer, J. J., D. W. Allan, M. Martin-Caraballo, and R. P. Lemke. 1999. An overview of phrenic nerve and diaphragm muscle development in the perinatal rat. J. Appl. Physiol. 86:779-786. [DOI] [PubMed] [Google Scholar]
- 9.Guo, Q., J. Xie, C. V. Dang, E. T. Liu, and J. M. Bishop. 1998. Identification of a large Myc-binding protein that contains RCC1-like repeats. Proc. Natl. Acad. Sci. USA 95:9172-9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hegde, A. N., and A. DiAntonio. 2002. Ubiquitin and the synapse. Nat. Rev. Neurosci. 3:854-861. [DOI] [PubMed] [Google Scholar]
- 11.Katz, D. M., and A. Balkowiec. 1997. New insights into the ontogeny of breathing from genetically engineered mice. Curr. Opin. Pulm. Med. 3:433-439. [DOI] [PubMed] [Google Scholar]
- 12.Lin, W., R. W. Burgess, B. Dominguez, S. L. Pfaff, J. R. Sanes, and K. F. Lee. 2001. Distinct roles of nerve and muscle in postsynaptic differentiation of the neuromuscular synapse. Nature 410:1057-1064. [DOI] [PubMed] [Google Scholar]
- 13.Misgeld, T., R. W. Burgess, R. M. Lewis, J. M. Cunningham, J. W. Lichtman, and J. R. Sanes. 2002. Roles of neurotransmitter in synapse formation: development of neuromuscular junctions lacking choline acetyltransferase. Neuron 36:635-648. [DOI] [PubMed] [Google Scholar]
- 14.Murthy, V., S. Han, R. L. Beauchamp, N. Smith, L. A. Haddad, N. Ito, and V. Ramesh. 14October2003. Pam and its ortholog highwire interact with and may negatively regulate the TSC1-TSC2 complex. J. Biol. Chem. 14559897. [DOI] [PubMed]
- 15.O'Brien, T. P., D. L. Metallinos, H. Chen, M. K. Shin, and S. M. Tilghman. 1996. Complementation mapping of skeletal and central nervous system abnormalities in mice of the piebald deletion complex. Genetics 143:447-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peterson, K. A., B. L. King, A. Hagge-Greenberg, J. J. Roix, C. J. Bult, and T. P. O'Brien. 2002. Functional and comparative genomic analysis of the piebald deletion region of mouse chromosome 14. Genomics 80:172-184. [DOI] [PubMed] [Google Scholar]
- 17.Roix, J. J., A. Hagge-Greenberg, D. M. Bissonnette, S. Rodick, L. B. Russell, and T. P. O'Brien. 2001. Molecular and functional mapping of the piebald deletion complex on mouse chromosome 14. Genetics 157:803-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakai, T., A. Wakizaka, and Y. Nirasawa. 2001. Congenital central hypoventilation syndrome associated with Hirschsprung's disease: mutation analysis of the RET and endothelin-signaling pathways. Eur. J. Pediatr. Surg. 11:335-337. [DOI] [PubMed] [Google Scholar]
- 19.Schaefer, A. M., G. D. Hadwiger, and M. L. Nonet. 2000. rpm-1, a conserved neuronal gene that regulates targeting and synaptogenesis in C. elegans. Neuron 26:345-356. [DOI] [PubMed] [Google Scholar]
- 20.Schaeren-Wiemers, N., and A. Gerfin-Moser. 1993. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry 100:431-440. [DOI] [PubMed] [Google Scholar]
- 21.Scholich, K., S. Pierre, and T. B. Patel. 2001. Protein associated with Myc (PAM) is a potent inhibitor of adenylyl cyclases. J. Biol. Chem. 276:47583-47589. [DOI] [PubMed] [Google Scholar]
- 22.Wan, H. I., A. DiAntonio, R. D. Fetter, K. Bergstrom, R. Strauss, and C. S. Goodman. 2000. Highwire regulates synaptic growth in Drosophila. Neuron 26:313-329. [DOI] [PubMed] [Google Scholar]
- 23.Wang, X. F., D. Wang, W. Zhu, K. K. Delrahim, D. Dolnak, and M. H. Rapaport. 2003. Studies characterizing 60 kda autoantibodies in subjects with schizophrenia. Biol. Psychiatry 53:361-375. [DOI] [PubMed] [Google Scholar]
- 24.Welsh, I. C., and T. P. O'Brien. 2000. Loss of late primitive streak mesoderm and interruption of left-right morphogenesis in the Ednrb(s-1Acrg) mutant mouse. Dev. Biol. 225:151-168. [DOI] [PubMed] [Google Scholar]
- 25.Yang, H., K. Scholich, S. Poser, D. R. Storm, T. B. Patel, and D. Goldowitz. 2002. Developmental expression of PAM (protein associated with MYC) in the rodent brain. Brain Res. Dev. Brain Res. 136:35-42. [DOI] [PubMed] [Google Scholar]
- 26.Zhen, M., X. Huang, B. Bamber, and Y. Jin. 2000. Regulation of presynaptic terminal organization by C. elegans RPM-1, a putative guanine nucleotide exchanger with a RING-H2 finger domain. Neuron 26:331-343. [DOI] [PubMed] [Google Scholar]