Abstract
The inner ear arises from multipotent placodal precursors that are gradually committed to the otic fate and further differentiate into all inner ear cell types, with the exception of a few immigrating neural crest-derived cells. The otocyst plays a pivotal role during inner ear development: otic progenitor cells sub-compartmentalize into non-sensory and prosensory domains, giving rise to individual vestibular and auditory organs and their associated ganglia. The genes and pathways underlying this progressive subdivision and differentiation process are not entirely known. The goal of this study was to identify a comprehensive set of genes expressed in the chicken otocyst using the serial analysis of gene expression (SAGE) method. Our analysis revealed several hundred transcriptional regulators, potential signaling proteins, and receptors. We identified a substantial collection of genes that were previously known in the context of inner ear development, but we also found many new candidate genes, such as SOX4, SOX5, SOX7, SOX8, SOX11, and SOX18, which previously were not known to be expressed in the developing inner ear. Despite its limitation of not being all-inclusive, the generated otocyst SAGE library is a practical bioinformatics tool to study otocyst gene expression and to identify candidate genes for developmental studies.
Electronic supplementary material
The online version of this article (doi:10.1007/s10162-011-0286-z) contains supplementary material, which is available to authorized users.
Keywords: gene array, inner ear development, otic vesicle, SAGE, Sox
Introduction
The otic vesicle, or otocyst, is one of the earliest morphological manifestations of the vertebrate inner ear. It arises by invagination of the otic placode, which is an ectodermal thickening that develops near the developing hindbrain. This process happens in chicken embryos during the second and third days of embryonic development (E2–E3). In intermediate stages, the otic placode has folded inward to form a pouch that is also called otic pit. The otic pit subsequently pinches off from the surface ectoderm into the underlying mesenchyme, resulting in the formation of the otocyst.
It has been hypothesized that axis formation of the developing inner ear already happens during otocyst formation, where signals from the surrounding tissues result in the regionalization of the developing otic placode, pit, and otocyst (reviewed in Fekete 1996; Fekete and Wu 2002). Despite a complex patterning process that is already manifest at the otocyst stage by regionalized expression of specific markers (for a review, see Streit 2007), the otocyst itself is a remarkable structure because it contains all the necessary progenitor cells to form the major cell types of the inner ear. This autonomy was revealed by grafting otocysts into other regions of the developing body (Swanson et al. 1990) as well as by determining that the otic lineage that is specified during otic induction has already reached a largely committed state when the otocyst is formed (Groves and Bronner-Fraser 2000).
Inner ear cell regeneration research has been utilizing the fact that stem cell-derived mammalian otic progenitor cells, defined by the expression of otocyst markers such as PAX2, PAX8, and DLX5, display a certain degree of commitment toward the otic lineage (Li et al. 2003a, b; Oshima et al. 2007, 2010). This was demonstrated by showing the potential of otic progenitors to differentiate into different cell types that express makers indicative of neurons, hair cells, and supporting cells. Based on these findings, it has been hypothesized that stem cell-derived otic progenitors are similar to otocyst cells (Brigande and Heller 2009; Diensthuber et al. 2009).
Here, we present the results of an unbiased interrogation of gene expression in the chicken otocyst. This analysis was spurred by the lack of comprehensive gene expression information at this important stage of development. We decided to utilize serial analysis of gene expression (SAGE), which is a quantitative method that can be used to identify known as well as new genes (Saha et al. 2002; Velculescu et al. 1995). Of the 39,326 seventeen-base pair sequence tags that we found, we evaluated 16,008 unique sequences that resulted in 4,153 unequivocally identified genes. Although our study lacked the sensitivity of more modern high-throughput deep sequencing methods, we consider the results as an important contribution because they provide a comprehensive summary of genes that are expressed at medium and high levels. Our analysis revealed potential signaling proteins and receptors as well as almost 300 transcriptional regulators that are expressed in the chicken otocyst. Some of these regulators have been previously known to play important roles during inner ear development, but we found additional candidate genes, such as several members of the Sox gene family, which have thus far not been evaluated in the context of the developing inner ear.
Methods
Tissue dissection, RNA preparation, and SAGE library preparation
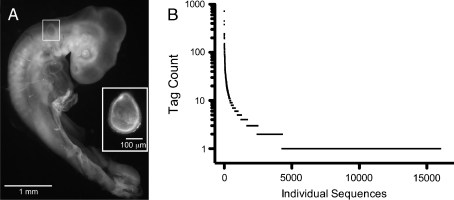
Fertilized eggs (Gallus gallus) of the white leghorn strain (California Golden Eggs, Dinuba, CA) were incubated at 38°C for 72 h on a rocking platform in a humidified egg incubator (Sportsman 1202A, GQF Manufacturing, Savannah, GA). The embryos were removed from the eggs, washed in Hanks’ balanced salt solution (HBSS; Invitrogen, Carlsbad, CA), and collected in HBSS. Staging was conducted according to Hamburger and Hamilton’s (HH) guidelines (Hamburger and Hamilton 1992), and only HH stage 18–19 embryos with clearly identifiable closed otic vesicles and unpigmented eyes were used. Otic vesicles were microdissected using fine forceps and attention was given to remove as much of the surrounding periotic mesenchyme as possible (Fig. 1A). The vesicles were individually inspected and frozen in bulks of 10–20 in liquid nitrogen and stored at −80°C.
FIG. 1.
SAGE library from the chicken otocyst. A HH stage 18–19 chicken embryo. The otocyst is indicated with a white box. Inset, otocyst after dissection. B SAGE tag frequency (tag count) plot for all tags identified.
Total RNA was isolated using a commercial kit (RNeasy Mini Kit, Qiagen, Hilden, Germany). RNA integrity and quality was confirmed by gel electrophoresis and by visual assessment. Five micrograms total RNA, the combined yield of 200 otocysts, was used for SAGE library synthesis (I-SAGE Long kit, Invitrogen) starting with attaching polyA+ RNA to oligo(dT)-paramagnetic beads, reverse transcription, and second strand synthesis. The resulting cDNA was cleaved with NlaIII, divided into two fractions, and bound to two different adapters containing a type IIS restriction nuclease recognition site. The adapters with adjacent 21-bp cDNA pieces were released from the oligo(dT)-magnetic beads using the type IIS restriction endonuclease MmeI. The two pools of released adapter-linked tags were ligated to 130-bp ditags and amplified with 27 PCR cycles with specific primers to each adapter. The adapters were then cleaved with NlaIII, 34-bp ditags were purified from the adapters by polyacrylamide gel electrophoresis, and concatenated. Concatemers were fractioned by size, gel-purified, and then cloned into pZErO-1 provided with the kit.
Quality control was conducted in two steps. First, 20 colonies were picked, plasmid DNA was prepared, and the resulting plasmids were digested with NsiI, which resulted in the release of the individual inserts. We obtained 20 distinct restriction patterns with a mean insert length of 671 bp (±425 bp (SD)). The smallest fragment was 200 bp and the largest was 1,700 bp. In a second step, we sequenced 24 inserts, obtaining 12,025 bp of raw data. These insert lengths slightly exceeded the expected ≈25 SAGE tags per clone predicted by the kit manufacturer. Three thousand eight hundred forty colonies were robotically picked and the plasmid concatemer inserts were directly sequenced (GeneWiz, South Plainfield, NJ).
Modifications from the manufacturer’s protocol included that the gel electrophoreses for the 130- and 34-bp ditags were performed on 10% polyacrylamide gels (Novex TBE Gels, Invitrogen) and that DNA was isolated from polyacrylamide gels with QIAEXII beads (Qiagen). Concatemers were separated on a 2% agarose gel and purified before subcloning with a column-based gel extraction kit (QIAquick, Qiagen).
Data analysis
At the highest stringency settings for DNA sequence quality and tag extraction, 39,326 individual 17-bp tags were extracted from the 3,512 sequencing data files using SAGE2000 analysis software (version 4.5, Invitrogen). Only unequivocal sequences were used for library construction, which resulted in a library about three times smaller than predicted by the quality control samples. A potential reason for this shortfall might be that the sequence quality achieved with direct sequencing was lower than the sequencing results obtained with plasmid DNA, which was used for the quality control clones. For computerized mapping, we appended the NlaIII restriction site (5′-CATG-3′) to the 5′ end of each tag. The resulting 21-bp tags were mapped using 22,290 G. gallus cDNA sequences available through the Ensembl 52 database (www.ensembl.org/info/data/ftp/), 19,307 G. gallus cDNAs available from the RefSeq database (ftp.ncbi.nih.gov), and 33,383 G. gallus sequences from the Unigene database (www.ncbi.nlm.nih.gov/unigene). Mapping was conducted using MAQ software (Li et al. 2008; maq.sourceforge.net) with all parameters set to default allowing for two mismatches in the sequence alignments. Ingenuity Pathway Analysis (IPA) software was accessed via the Stanford University Bioinformatics Resource (cmgm.stanford.edu).
Reverse transcriptase PCR
Chicken otocyst RNA was isolated (Absolutely RNA Miniprep Kit, Stratagene/Agilent Technologies, La Jolla, CA) and treated with RNase-free DNase I (Roche Diagnostics, Mannheim, Germany). The RNA concentration was determined by spectrophotometric analysis using a NanoDrop (Thermo Fisher Scientific, Wilmington, DE). Total RNA extracts were then used for reverse transcription (RT) into cDNA (first strand) using SuperScript III Reverse Transcriptase (Invitrogen) and Oligo(dT)18 primer (Invitrogen) with 350 ng of total RNA per 20 μl reaction. To prevent RNA degradation, 1 μl RiboLock RNase Inhibitor (Fermentas, Thermo Fisher Scientific, Glen Burnie, MD) was also included in each reaction. Control reactions were done without reverse transcriptase.
Oligonucleotide primer pairs were designed for each gene of interest using NCBI Primer-BLAST (www.ncbi.nlm.nih.gov/tools/primer-blast) with NCBI Reference Sequences as template. For each gene tested, at least two primer pairs covering two non-overlapping 300- to 700-bp regions were used to confirm mRNA expression. A full list of primers tested can be found in Electronic supplementary materials (ESM) Table 1. PCR was performed using GoTaq Green Master Mix (Promega, Madison, WI) with 2 μl cDNA template and 1 μl 400 nM each of forward and reverse gene-specific primers. The following cycling conditions were employed: initial denaturation at 94°C (3 min); 30 cycles of denaturation at 94°C (30 s), annealing at 55°C (1 min), and elongation at 72°C (1 min); and a hold at 4°C. Aliquots of PCR products were electrophoresed in a 2.0% agarose gel, stained with SYBR Safe (Invitrogen) in 1X TAE buffer at 120 V for 35 min, and documented using UV transillumination and digital photography (Kodak Gel Logic 200 Imaging System).
In situ hybridization
The T7 promoter sequence (5′-TAATACGACTCACTATAGGG-3′) was added to the 5′ end of the forward or reverse primer for the different Sox2 cDNAs to allow for conversion of the PCR product to sense and antisense cRNA probes for in situ hybridization. Of the PCR product, 500 ng was used to synthesize digoxigenin-labeled antisense probes (DIG RNA Labeling Kit, Roche Diagnostics), which were resuspended in 30 μl RNAse-free water. Embryos were dissected at HH stage 18–19 (E3) and HH stage 26–28 (E5), fixed overnight with 4% paraformaldehyde in phosphate-buffered saline (PBS, pH 7.4), transferred into 30% sucrose in PBS for 24–36 h, and embedded in O.C.T compound (Tissue-Tek). Sections were cut with a cryomicrotome (CM3050 S, Leica), collected on ultrastick slides (precleaned Gold Seal, Rite-on, Micro Slides), dried at 37°C for 45 min, and stored frozen at −70°C. For hybridization, the sections were brought to room temperature and rehydrated in 100 μl diluted probe (1:100) in 50% formamide, 10% dextran sulfate, 1 mg/ml yeast RNA, 1x Denhardt’s solution, 185 mM NaCl, 5.6 mM NaH2PO4, 5 mM Na2HPO4, 5 mM EDTA, and 15 mM Tris at pH 7.5. After coverslipping and overnight incubation at 65°C in a chamber humidified with 50% formamide in 150 mM NaCl, 15 mM trisodium citrate, pH 7 (1× SSC), the coverslips were removed in 5x SSC and the slides washed twice for 30 min each in 50% formamide and 0.1% Triton X-100 in 1x SSC at 65°C. Thereafter, the slides were washed for 15 min in 0.2x SSC and for 15 min in 150 mM NaCl and 100 mM Tris at pH 7.5 at room temperature. For antibody detection, the sections were blocked for 30 min in 0.5% blocking powder (Roche Diagnostics), 10% heat-inactivated goat serum, 100 mM NaCl, 0.1% Triton X-100, and 100 mM Tris at pH 7.5. The slides were then incubated for 2 h at room temperature in a blocking solution pre-incubated for 1 h with alkaline phosphatase-conjugated anti-digoxigenin Fab fragments (1:500, Roche Diagnostics). Unbound Fab fragments were removed by washing twice for 30 min each in 150 m NaCl and 100 mM Tris at pH 7.5. The sections were first incubated in detection buffer (100 mM NaCl, 50 mM MgCl2, 100 mM Tris at pH 9.5) for 10 min. For detection, the sections were then covered with 200 μl of chromogen solution consisting of 20 μl NBT/BCIP stock solution (Roche Diagnostics) and 50 μl Levamisol stock solution (20x concentrate, Invitrogen) in 1 ml detection buffer, coverslipped, and incubated overnight at room temperature in a humidified chamber. Coverslips were removed and color reaction was stopped in 1 mM EDTA and 10 mM Tris at pH 8.1. Slides were embedded in 50% glycerol in PBS and coverslipped. Analysis and photography was conducted on an Axiovert 25 microscope with an AxioCam MRC camera, using AxioVision software (V 4.6.3.0, Zeiss).
Results
SAGE library of the chicken otocyst
Otocysts were dissected from HH stage 18–19 chicken embryos (Fig. 1A), total RNA was extracted, and subjected to a commercial long-SAGE protocol, resulting in a library of concatemerized tags. Individual clones of SAGE concatemers were sequenced, resulting in 39,326 seventeen-base pair tags with tag counts up to 718 for the most abundant tag; 3,292 tags were represented between two and five times, whereas the majority of tags (11,717) were only found once (Fig. 1B). Overall, we identified 16,008 unique sequence tags (ESM Table 2; NCBI Geo DataSet accession no. GSM651351).
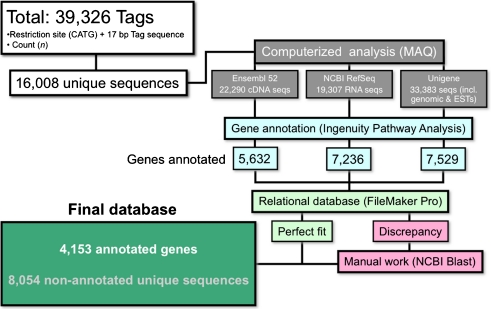
Although the chicken genome has been fully sequenced (The Chicken Genome Consortium 2004), the annotation of chicken genes is far from complete, and consequently, gene annotation needed to be conducted by combining several database resources (Fig. 2). We started by mapping all 16,008 tags consisting of the 4-bp NlaIII restriction site and each individual 17-bp SAGE tag using the MAQ software (Li et al. 2008). Three reference databases were used—Ensembl 52, RefSeq, and Unigene—resulting in 7,026, 8,682, and 13,405 matches, respectively. To compare the matches from the three different databases that were composed of assortments of unofficial and official gene names, we imported the results into the IPA software, which revealed official gene symbols as common identifiers. IPA software recognized 5,632, 7,326, and 7,529 gene identifiers from Ensembl, RefSeq, and Unigene database matches, respectively. In a next step, official gene symbols created in IPA were used to directly compare matches of different databases using a relational database. Only when all three different databases suggested a match for a given gene was the gene accepted for further analysis. In case of discrepancies in matches between different databases or in case of missing matches in some of the databases, we performed manual BLAST searches (blast.ncbi.nlm.nih.gov/Blast.cgi) with each tag. If a match was found, the resulting gene was added for further analysis. In 52 cases, a single tag corresponded to two different genes, and in one case, a single tag corresponded to five different genes. These 53 ambiguous tags (ESM Table 3) were removed from further analysis. After this step, the library consisted of 7,912 gene matches with unique tags. However, many genes were represented in the library with two or more different tags, which could be due to alternative poly-adenylation sites, internal priming, or alternative splicing near the 3′ end of the mRNA. These duplicates were combined, resulting in 4,180 genes that were re-imported into IPA for final analysis. At this final step, IPA recognized 4,153 genes, which in the final annotated SAGE library were associated with the aggregate count number of all the tags of every given gene (ESM Table 4).
FIG. 2.
Flowchart of SAGE tag analysis resulting in a library of 4,153 annotated genes expressed in the chicken otocyst.
Gene annotation reveals abundance of transcriptional regulators
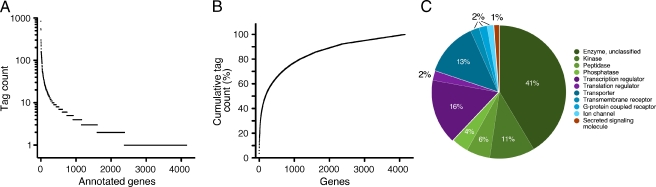
Analysis of the relation between tag count and annotated gene number revealed that 50% of all tags in the otocyst were encoded by only 180 genes, whereas the majority of genes were represented by fewer than 10 tags (Fig. 3A, B). Not unexpectedly, the genes with the highest tag count were housekeeping genes involved in the maintenance of basal cellular functions (Table 1). The IPA database provided unequivocal gene family information for about half of the identified genes. Analysis of all unambiguous gene family annotation identifiers revealed that the majority of otocyst genes encoded unclassified enzymes as well as kinases, phosphatases, and peptidases. The second largest family of genes identified encoded transcriptional regulators, followed by transporters, transmembrane domain-containing receptors, and ion channels (Fig. 3C). Translational regulators, growth factors, cytokines, and other protein families comprised the rest.
FIG. 3.
SAGE library after annotation. A Tag count frequency plot for all 4,153 annotated genes. B Cumulative tag count analysis reveals that 180 of the most abundantly expressed annotated genes represent 50% of all identified tags. C Family representation of all unambiguously identified annotated genes.
TABLE 1.
Genes with the highest expression based on SAGE tag count
| Gene symbol | Count | Family | Gene name |
|---|---|---|---|
| COX1 | 840 | Enzyme | Cytochrome c oxidase I |
| ARIH1 | 560 | Enzyme | Ariadne homolog, ubiquitin-conjugating enzyme E2 binding protein, 1 |
| COX2 | 526 | Enzyme | Cytochrome c oxidase II |
| COX3 | 307 | Enzyme | Cytochrome c oxidase III |
| ATP6 | 298 | Transporter | ATP synthase 6, ATPase subunit 6 |
| NPM1 | 273 | Transcription regulator | Nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
| ND4 | 257 | Enzyme | NADH dehydrogenase, subunit 4 (complex I) |
| GAPDH | 254 | Enzyme | Glyceraldehyde-3-phosphate dehydrogenase |
| RPL13 | 230 | Ribosomal structure | Ribosomal protein L13 |
| RPL10A | 218 | Ribosomal structure | Ribosomal protein L10a |
| RPL4 | 187 | Ribosomal structure | Ribosomal protein L4 |
| TUBA4A | 162 | Cytoskeletal structure | Tubulin, α4a |
| RPL23 | 161 | Ribosomal structure | Ribosomal protein L23 |
| RPS27A | 156 | Ribosomal structure | Ribosomal protein S27a |
| MDK | 154 | Growth factor | Midkine (neurite growth-promoting factor 2) |
| ND5 | 151 | Enzyme | NADH dehydrogenase, subunit 5 (complex I) |
| RPS3 | 146 | Ribosomal structure | Ribosomal protein S3 |
| EEF1A1 | 142 | Translation regulator | Eukaryotic translation elongation factor 1 α1 |
| ATP5B | 139 | Transporter | ATP synthase, H+ transporting, mitochondrial F1 complex, β polypeptide |
| RPS29 | 136 | Ribosomal structure | Ribosomal protein S29 |
| RPS27L | 131 | Ribosomal structure | Ribosomal protein S27-like |
| ACTB | 124 | Cytoskeletal structure | Actin, β |
| RPS15 | 120 | Ribosomal structure | Ribosomal protein S15 |
| TUBB2A | 118 | Cytoskeletal structure | Tubulin, β 2A |
| RPLP1 | 116 | Ribosomal structure | Ribosomal protein, large, P1 |
| RPS3A | 112 | Ribosomal structure | Ribosomal protein S3A |
| RPL21 | 107 | Ribosomal structure | Ribosomal protein L21 |
| RPL36 | 102 | Ribosomal structure | Ribosomal protein L36 |
| RPL35 | 96 | Ribosomal structure | Ribosomal protein L35 |
| HNRNPA3 | 94 | Nucleic acid binding | Heterogeneous nuclear ribonucleoprotein A3 |
Overall, we identified 299 genes that encode transcriptional regulators (ESM Table 5) which can be categorized into transcription factors containing zinc-coordinating DNA-binding domains (11%), helix-loop-helix domains (13%), basic domains (15%), ß-scaffold factors with minor groove contacts (16%), and others (45%). Fifty-one transcriptional regulators were previously known to be expressed in the developing inner ear. Known examples for each respective category are GATA2 and GATA3 (Lillevali et al. 2007) for zinc-coordinating DNA-binding domains, PAX2 and FOXG1 (Herbrand et al. 1998; Li et al. 2004; Pauley et al. 2006) for helix-loop-helix domains, NEUROG1 and NEUROD1 (Liu et al. 2000; Ma et al. 2000) for basic domains, and SOX10 and NOTCH1 (Lewis et al. 1998; Stone and Rubel 1999; Watanabe et al. 2000) for ß-scaffold factors with minor groove contacts. Two hundred forty-eight transcriptional regulators were previously unknown in the context of early inner ear development.
Secreted proteins and transmembrane proteins
Genes that encode growth factors, cytokines, and other secreted proteins are the second group of developmentally interesting otocyst genes (ESM Table 6). Of the 172 genes that we identified in this group, several were previously known in inner ear development and include BMP7, FGF10, FGF19, FRZB, TGFß2, NETRIN1, SLIT1, WNT3, and WNT5A (Abraira et al. 2008; Alsina et al. 2004; Battisti and Fekete 2008; Hollyday et al. 1995; Liu et al. 2008; Oh et al. 1996; Okano et al. 2005; Sanchez-Calderon et al. 2007; Sienknecht and Fekete 2009). Transcripts encoding the secreted signaling protein midkine (MDK) were by far the most abundantly expressed mRNA that we detected. Midkine has been previously reported in the postnatal mouse cochlea, and it has been shown that the protein is involved in regulating the expression of the tectorial membrane component ß-tectorin (Jia et al. 2001; Zou et al. 2006), but early developmental roles in the inner ear have not been reported. Other proteins, such as opticin (OPTC), have previously been shown in the otic vesicle, but their function in inner ear development remains unknown (Frolova et al. 2004). Several genes emerged in our screen as novel candidates for roles in inner ear development, such as olfactomedin-like 2A, 2B, 3 (OLFML2A, OLFML2B, OOLFML3), which belong to a class of proteins implicated in a variety of developmental processes (reviewed by Tomarev and Nakaya 2009), or neudesin (NENF), which may play roles in neuronal differentiation and development (Kimura et al. 2006). We identified various TGFß antagonists such as twisted gastrulation protein homolog 1 (TWSG1) or follistatin-like 3 (FSTL3). Lastly, we identified various secreted proteins of unknown function during development, but with previous implications in cancer or other cell growth- and death-related processes; examples for these proteins are AGR3, CLU, EGFL7, and HDGF.
Our analysis of transmembrane-spanning proteins revealed high transcript expression levels of many tight junction and cell adhesion proteins such as claudin 1 (CLDN1), CLDN3, and CLDN17; integrins α3 (ITGA3) and α6 (ITGA6); integrins ß1, ß2, ß3, and ß5 (ITGB1, ITGB2, ITGB3, ITGB5); neurexin 1 (NRXN1); as well as cell adhesion molecule 1 (CADM1) and epithelial cell adhesion molecule TACSTD1, among others. One of the most abundant genes identified in this category encodes protein tyrosine kinase 7 (PTK7), a protein implicated in the regulation of planar cell polarity, convergent extension, and Wnt signaling (Lu et al. 2004; Puppo et al. 2011; Yen et al. 2009). Another interesting gene in this regard encodes the Ig superfamily protein protogenin (PRTG), which has been shown to play a role in suppressing premature neural differentiation and whose roles in other tissues might similarly be in controlling the timing of transitions between early progenitor state and differentiation (Ito et al. 2011; Wong et al. 2010). Probably the most interesting group of genes that we identified encodes receptors for signaling proteins because they might reveal information about the developmental processes happening in the otocyst. These include genes that encode receptors for ligands that are already known for playing roles in otic development such as FGFR1, FZD1, FZD2, FZD3, FZD4, FZD7, NGFR, and NOTCH1, which have previously been shown to be expressed in the vertebrate otocyst (Adam et al. 1998; Pirvola et al. 2002; Sienknecht and Fekete 2009; Stevens et al. 2003; von Bartheld et al. 1991; Wright and Mansour 2003). BMPR1, BMPR2, LGFR1, SMO1, PTCH1, DISP1, and TGFBR2 are genes that were presumed to be expressed in the otocyst because their ligands, such as BMPs and other TGFß family members, IGF, as well as hedgehog signaling proteins, have been shown to be expressed and active during inner ear development (Bok et al. 2005; Frenz et al. 1991, 1992; Liu et al. 2002; Oh et al. 1996; Riccomagno et al. 2002; Yamashita and Oesterle 1995). Other identified genes include receptors for somatostatin (SSTR1), interleukin 11 (IL11RA), endothelin (EDNRB), and tumor necrosis factors (TNFRSF1A, TNFRSF6B, TNFRSF19) and orphan receptors such as lathrophilin 3 (LPNHN3; Sudhof 2001).
Other potentially interesting transcripts encoded transmembrane proteins involved in cell recognition and adhesion that play roles in axonal guidance and cell migration such as the semaphorins SEMA4B, SEMA5B, SEMA6D, SEMA7A and some components of their receptor complex such as Plexins A1 and B2 (PLXNA1, PLXNB2; Perrot et al. 2002). Additional genes with similar roles include ephrin B1 (EFNB1) and ephrin receptors (EPHA4, EPHA5, EPHB3), netrin G1 and the netrin receptor UNC5B, the Slit receptors ROBO1 and ROBO2, as well as the Slit-like transmembrane protein SLITRK6. The possible roles of some of these genes in axon guidance and cell migration has been discussed in the context of the inner ear (Fekete and Campero 2007; Webber and Raz 2006), and their expression patterns and potential function are the focus of intensive research (Battisti and Fekete 2008; Katayama et al. 2009; Matilainen et al. 2007).
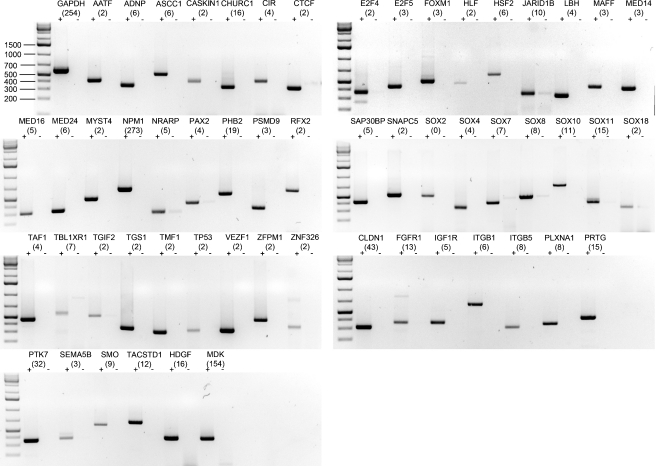
As initial validation of mRNA expression, we conducted RT-PCR on otic vesicle RNA template with oligonucleotide primer pairs specific for selected genes representing transcriptional regulators, signaling proteins, receptors, and other genes of interest (Fig. 4 and Table 2). Transcripts for all selected genes were detectable, and although the RT-PCR was not conducted in quantitative fashion, we observed a general trend where strong amplification products corresponded to genes with higher SAGE tag counts.
FIG. 4.
Shown are agarose gels stained with ethidium bromide to visualize RT-PCR fragments for genes expressed in the chicken otocyst (indicated with “+”). Control reactions without reverse transcription are labeled with “−”. Faint bands in the “−” control lanes are likely the result of residual genomic DNA contamination. The corresponding gene names are listed in Table 2. Predicted PCR product sizes are listed in ESM Table 1. SAGE tag count numbers are indicated in parentheses.
TABLE 2.
Index for gene names shown in Figure 4
| Gene symbol | Gene name | Tag count |
|---|---|---|
| Housekeeping genes | ||
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | 254 |
| Transcriptional regulators | ||
| AATF | Apoptosis antagonizing transcription factor | 2 |
| ADNP | Activity-dependent neuroprotector homeobox | 6 |
| ASCC1 | Activating signal cointegrator 1 complex subunit 1 | 6 |
| CASKIN1 | CASK interacting protein 1 | 2 |
| CHURC1 | Churchill domain containing 1 | 16 |
| CIR | CBF1 interacting corepressor | 4 |
| CTCF | CCCTC-binding factor (zinc finger protein) | 2 |
| E2F4 | E2F transcription factor 4, p107/p130-binding | 2 |
| E2F5 | E2F transcription factor 5, p130-binding | 3 |
| FOXM1 | Forkhead box M1 | 3 |
| HLF | Hepatic leukemia factor | 2 |
| HSF2 | Heat shock transcription factor 2 | 6 |
| JARID1B | Jumonji, AT rich interactive domain 1B | 10 |
| LBH | Limb bud and heart development homolog (mouse) | 4 |
| MAFF | v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) | 3 |
| MED14 | Mediator complex subunit 14 | 3 |
| MED16 | Mediator complex subunit 16 | 5 |
| MED24 | Mediator complex subunit 24 | 6 |
| MYST4 | MYST histone acetyltransferase (monocytic leukemia) 4 | 2 |
| NPM1 | Nucleophosmin (nucleolar phosphoprotein B23, numatrin) | 273 |
| NRARP | NOTCH-regulated ankyrin repeat protein | 5 |
| PAX2 | Paired box 2 | 4 |
| PHB2 | Prohibitin 2 | 19 |
| PSMD9 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 | 3 |
| RFX2 | Regulatory factor X, 2 (influences HLA class II expression) | 2 |
| SAP30BP | SAP30 binding protein | 5 |
| SNAPC5 | Small nuclear RNA activating complex, polypeptide 5, 19 kDa | 2 |
| SOX2 | SRY (sex determining region Y)-box 2 | 0 |
| SOX4 | SRY (sex determining region Y)-box 4 | 4 |
| SOX7 | SRY (sex determining region Y)-box 7 | 7 |
| SOX8 | SRY (sex determining region Y)-box 8 | 8 |
| SOX10 | SRY (sex determining region Y)-box 10 | 11 |
| SOX11 | SRY (sex determining region Y)-box 11 | 15 |
| SOX18 | SRY (sex determining region Y)-box 18 | 2 |
| TAF1 | TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250 kDa | 4 |
| TBL1XR1 | Transducin (beta)-like 1 X-linked receptor 1 | 7 |
| TGIF2 | TGFB-induced factor homeobox 2 | 2 |
| TGS1 | Trimethylguanosine synthase homolog (S. cerevisiae) | 2 |
| TMF1 | TATA element modulatory factor 1 | 2 |
| TP53 | Tumor protein p53 | 15 |
| VEZF1 | Vascular endothelial zinc finger 1 | 2 |
| ZFPM1 | Zinc finger protein, multitype 1 | 2 |
| ZNF326 | Zinc finger protein 326 | 2 |
| Transmembrane proteins | ||
| CLDN1 | Claudin 1 | 43 |
| FGFR1 | Fibroblast growth factor receptor 1 | 13 |
| IGF1R | Insulin-like growth factor 1 receptor | 5 |
| ITGB1 | Integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | 6 |
| ITGB5 | Integrin, beta 5 | 8 |
| PLXNA1 | Plexin A1 | 8 |
| PRTG | Protogenin homolog (Gallus gallus) | 15 |
| PTK7 | PTK7 protein tyrosine kinase 7 | 32 |
| SEMA5B | Sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5B | 3 |
| SMO | Smoothened homolog (Drosophila) | 9 |
| TACSTD1 | Epithelial cell adhesion molecule | 12 |
| Growth factors | ||
| HDGF | Hepatoma-derived growth factor (high-mobility group protein 1-like) | 16 |
| MDK | Midkine (neurite growth-promoting factor 2) | 154 |
Known and novel Sox genes expressed in the otocyst
One of the most strongly represented groups of transcription factors in the chicken otocyst SAGE library were the Sox genes. Previous reports show the expression of SOX1, SOX2, SOX3, SOX6, SOX9, SOX10, and SOX21 in the chicken otocyst, or in the otic vesicle of various species including African clawed frog, zebrafish, and mouse (Barrionuevo et al. 2008; Liu et al. 2003; Neves et al. 2007; Uchikawa et al. 1999; Watanabe et al. 2000). Clearly highlighting the limitation of SAGE, showing that about 40,000 tags are far from exhaustive, is the fact that tags for SOX2, SOX3, and SOX6 were not represented in our SAGE library. Nevertheless, we found six Sox genes that previously were not known to be expressed in the developing inner ear, which include SOX4, SOX5, SOX7, SOX8, SOX11, and SOX18.
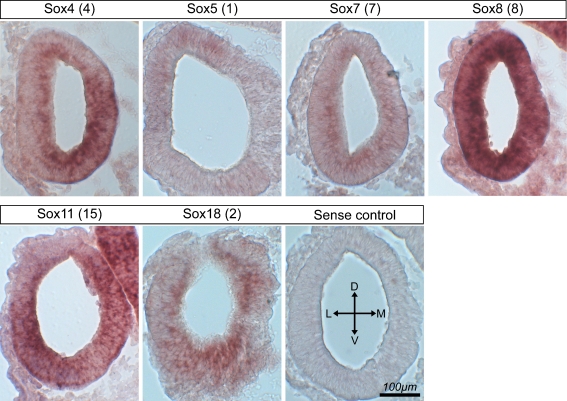
We verified the expression of the Sox genes by RT-PCR (Fig. 4) and by in situ hybridization at the otocyst stage (Fig. 5). SOX8 and SOX11 were abundantly expressed throughout the otocyst and also in the adjacent hindbrain. SOX4 mRNA appeared to be more regionalized with stronger expression in the ventromedial regions of the otocyst, indicative of a potential subsequent expression in the prosensory domains. SOX5 transcripts were weakly detectable throughout the otocyst, with a possible stronger expression in the dorsolateral part of the otocyst. SOX7 and SOX18 mRNA was detectable throughout the otocyst, perhaps with a slightly stronger expression in the ventral portion.
FIG. 5.
In situ hybridization analysis of Sox gene expression in sections of the chicken otocyst. Numbers in parentheses indicate the SAGE tag count for each individual gene. Sense controls were negative for all probes used; a representative control is shown.
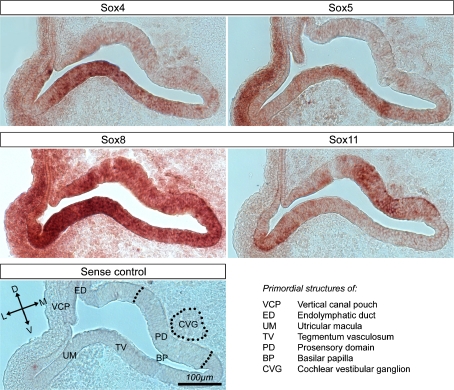
At E5, when the basilar papilla and vestibular compartments of the chicken inner ear are clearly defined and prosensory domains have been formed, only SOX11 appeared to be specifically associated with the prosensory domains of the basilar papilla and vestibular maculae (Fig. 6). SOX7 was no longer detectable, whereas SOX4 and SOX5 transcripts were detectable in the roof of the basilar papilla, presumably in the premordial tegmentum vasculosum. SOX8 expression was strong throughout the inner ear as well as in the adjacent hindbrain (not shown).
FIG. 6.
In situ hybridization analysis of Sox gene expression in cross-sections of the E5 chicken inner ear. Sense controls were negative for all probes used; a representative control is shown.
Discussion
The chicken embryo is one of the major animal models used to study inner ear induction and development. In the past decades, many genes have been found that are expressed by cells of the otocyst, and the specific roles of some of these genes have been elucidated. Nevertheless, no comprehensive study has been conducted on gene expression in the chicken otocyst. We hypothesized that the existing collection of otocyst markers and genes is just the tip of an iceberg, and we consequently decided to investigate, using a high-throughput method, gene expression in this clearly defined transient structure. Unlike the mouse and human genomes, the chicken genome is comparably poorly annotated, which complicated the analysis strategy. We refrained from using gene arrays whose preselected genes are constrained by these shortcomings. Additionally, at the onset of this study, no comprehensive chicken gene arrays were commercially available and next-generation sequencing techniques, likewise, were not yet developed. We decided to employ SAGE, which is a relatively unbiased method, based on sequencing of short tags that are directly adjacent to a NlaIII restriction site in the 3′ region of any given mRNA (Velculescu et al. 1995). The NlaIII recognition sequence is 4-bp long (5′-CATG-3′) and theoretically occurs once in every 256 bp. Using long-SAGE (Saha et al. 2002), which employs 17-mer tags instead of 10-mer tags, which were used in initial SAGE protocols, we were able to utilize a specificity of 421. Indeed, we only found 53 ambiguous tags, which either occur more than once in the transcriptome or were associated with more than one gene as a result of annotation ambiguities.
Our analysis is not based on comparative or subtractive studies, and consequently, many genes identified are widely expressed. Nevertheless, the results of our study do not preclude the use of bioinformatic tools to extract subtractive or otherwise user-defined datasets, and the reader is encouraged to use our dataset as needed. A recent very elegant gene array study focusing on FGF-based otic induction in mouse embryos is an example of the powerful specificity that can be achieved by selecting proper tissues for comparison (Urness et al. 2010). In this specific case, wild-type mouse otic placode tissue was compared with tissue from the prospective otic placode of Fgf3−/−;Fgf10−/− mice in which otic development fails to be initiated. This study revealed several transcriptional regulator genes that depend on FGF-based otic induction, including Hmx2, Hmx3, Foxg1, and Sox9, which we also found in our dataset. Other studies that focused on the identification of otocyst genes used differential display of chicken otocyst RNA against RNA from surrounding tissues (Gong et al. 1997) and on cDNA subtraction of mouse otocyst minus liver cDNA (Powles et al. 2004). The differential display study identified only a small number of unknown genes, and the collection of 280 specific transcripts found in the mouse otocyst cannot be directly compared with our data because the dataset was only partially annotated and has not been deposited in a format usable for in silicio comparison, for example via the NCBI Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/).
An obvious limitation of the SAGE method is the number of tags which results in libraries that are reasonable large, but that are far from exhaustive, particularly when dealing with complex tissues consisting of different cell types. Analysis of our chicken otocyst dataset clearly revealed this limitation. For example, known and easily detectable otocyst genes such as SOX2, PAX8, and FOXI3 (Groves and Bronner-Fraser 2000; Ohyama and Groves 2004; Uchikawa et al. 1999; Wood and Episkopou 1999) were not represented in our library, and 45% of all annotated tags were only represented once. The consequence of this limitation is probably a major reason why the SAGE method appears to be a transient technology that is in the process of being replaced with much more comprehensive and massive parallel next-generation sequencing methods capable of generating datasets of tens of millions of tags with a single run. Likewise, microarray and cross-species comparison methods are becoming increasingly more accessible to study gene expression in avian species and have already been successfully used in recent years (Hawkins et al. 2007).
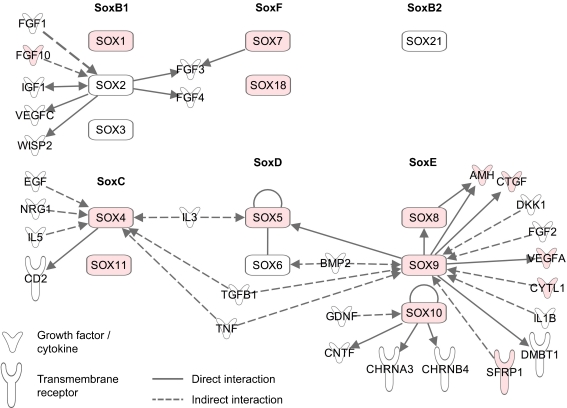
Despite these limitations, we were able to identify a large number of genes that previously were not known in the context of early inner ear development. Particularly transcriptional regulators were well represented in the library, and we were able to validate the expression of SOX4, SOX5, SOX7, SOX8, SOX11, and SOX18, all of which were previously not known to be expressed in the early developing inner ear. Analysis of the interaction networks of these transcription factors revealed some relation to various signaling pathways, including FGF, TGFß/BMP, EGF, and IGF signaling (Fig. 7). Furthermore, we showed for the first time the expression of SoxC group genes (SOX4 and SOX11) as well as SoxF genes (SOX18) in the developing otocyst. We are just beginning to understand the interplay of different signaling pathways and the particular roles of Sox transcription factors in specific developmental processes. Previous studies with mouse Sox genes that are expressed in the otocyst show involvement in placode invagination for Sox9 (Barrionuevo et al. 2008) as well as prosensory specification for Sox2 (Kiernan et al. 2005). The overlapping expression of many Sox genes combined with a potential functional redundancy will make it difficult to study individual Sox gene function in the otocyst because single knockouts might lack a phenotype. Phylogeny, neofunctionalization, and redundancies within the Sox gene family have recently been comprehensively reviewed, highlighting the importance of this group of genes in many developmental processes (Guth and Wegner 2008).
FIG. 7.
IPA software analysis of direct and indirect interactions of inner ear Sox genes with growth factors and transmembrane receptors. Genes present in the annotated chicken otocyst SAGE library are labeled in pink.
Our analysis also revealed many other potentially important genes that have not previously been considered in the context of inner ear development, and some have just recently been investigated. We found a number of secreted proteins that are novel candidates for signaling functions in the developing otocyst. Transmembrane proteins consisted of members of previously known families of proteins that are essentially involved in inner ear development such as receptors for FGFs, BMPs, and WNTs, as well as NOTCH1, among others. Interestingly, we found a relatively large group of proteins belonging to families that have been implicated in axonal guidance and cell migration; some of these proteins have previously been shown to be expressed in the otocyst and other developmental stages of the inner ear (Battisti and Fekete 2008; Matilainen et al. 2007). The expression and function of Slit-like transmembrane protein SLITRK6, for example, has recently been analyzed in mouse inner ear development (Katayama et al. 2009). Slitrk6 is strongly expressed in the prosensory and sensory patches of the auditory and vestibular organs; the innervation density of these organs was reduced or abolished in Slitrk6−/− mice.
In summary, we have used the SAGE method to assemble a list of sequence tags that can be associated with gene expression in the chicken otocyst. Although not all-inclusive, this SAGE library is a practical bioinformatics tool to study otocyst gene expression. For user-defined analyses, the library is available in electronic formats that can be directly queried online such as NCBI GEO, or it can be imported into commercial or public domain bioinformatic software packages such as IPA. We used the Sox gene family as an example to highlight the depth as well as the limitations of the library and to demonstrate that the collection of otocyst SAGE tags is a useful tool for molecular and developmental studies of early inner ear development.
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Oligonucleotide primer pairs used FOR RT-PCR (PDF 75 kb)
List of all 16,008 unique sage tags with individual tag count (XLS 1267 kb)
Ambiguous tags (PDF 64 kb)
Annotated SAGE library using official gene symbols recognized by IPA software. Listed are the gene symbols, gene names, tag count, the predicted location of the encoded protein, the gene family, as well as potential drugs that target the encoded protein (XLS 602 kb)
Transcriptional regulators identified in the annotated chicken otocyst SAGE library. Listed are the gene symbols, gene names, tag count, the predicted location of the encoded protein, potential drugs that target the encoded protein, as well as additional information including world wide web links, and references of publications that mention specific genes in the context of inner ear development (XLS 127 kb)
Secreted proteins identified in the annotated chicken otocyst SAGE library. Listed are the gene symbols, gene names, and tag count (PDF 104 kb)
Acknowledgments
This project was supported by the Sigrid Jusélius Foundation and Instrumentarium Science Foundation (to S.T.S.), a Stanford Dean’s Fellowship, and by fellowship D/06/41764 from the German Academic Exchange Service (to V.S.), as well as grants DC006167, DC010042, and P30 DC010363 from the National Institutes of Health (to S.H.).
Footnotes
Saku T. Sinkkonen and Veronika Starlinger equally contributed to this work.
References
- Abraira VE, Rio T, Tucker AF, Slonimsky J, Keirnes HL, Goodrich LV. Cross-repressive interactions between Lrig3 and netrin 1 shape the architecture of the inner ear. Development. 2008;135:4091–4099. doi: 10.1242/dev.029330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam J, Myat A, Roux I, Eddison M, Henrique D, Ish-Horowicz D, Lewis J. Cell fate choices and the expression of Notch, Delta and Serrate homologues in the chick inner ear: parallels with Drosophila sense-organ development. Development. 1998;125:4645–4654. doi: 10.1242/dev.125.23.4645. [DOI] [PubMed] [Google Scholar]
- Alsina B, Abello G, Ulloa E, Henrique D, Pujades C, Giraldez F. FGF signaling is required for determination of otic neuroblasts in the chick embryo. Dev Biol. 2004;267:119–134. doi: 10.1016/j.ydbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Barrionuevo F, Naumann A, Bagheri-Fam S, Speth V, Taketo MM, Scherer G, Neubuser A. Sox9 is required for invagination of the otic placode in mice. Dev Biol. 2008;317:213–224. doi: 10.1016/j.ydbio.2008.02.011. [DOI] [PubMed] [Google Scholar]
- Battisti AC, Fekete DM. Slits and Robos in the developing chicken inner ear. Dev Dyn. 2008;237:476–484. doi: 10.1002/dvdy.21429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok J, Bronner-Fraser M, Wu DK. Role of the hindbrain in dorsoventral but not anteroposterior axial specification of the inner ear. Development. 2005;132:2115–2124. doi: 10.1242/dev.01796. [DOI] [PubMed] [Google Scholar]
- Brigande JV, Heller S. Quo vadis hair cell regeneration? Nat Neurosci. 2009;12:679–685. doi: 10.1038/nn.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diensthuber M, Oshima K, Heller S. Stem/progenitor cells derived from the cochlear sensory epithelium give rise to spheres with distinct morphologies and features. J Assoc Res Otolaryngol. 2009;10:173–190. doi: 10.1007/s10162-009-0161-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete DM. Cell fate specification in the inner ear. Curr Opin Neurobiol. 1996;6:533–541. doi: 10.1016/S0959-4388(96)80061-4. [DOI] [PubMed] [Google Scholar]
- Fekete DM, Campero AM. Axon guidance in the inner ear. Int J Dev Biol. 2007;51:549–556. doi: 10.1387/ijdb.072341df. [DOI] [PubMed] [Google Scholar]
- Fekete DM, Wu DK. Revisiting cell fate specification in the inner ear. Curr Opin Neurobiol. 2002;12:35–42. doi: 10.1016/S0959-4388(02)00287-8. [DOI] [PubMed] [Google Scholar]
- Frenz DA, Water TR, Galinovic-Schwartz V. Transforming growth factor beta: does it direct otic capsule formation? Ann Otol Rhinol Laryngol. 1991;100:301–307. doi: 10.1177/000348949110000407. [DOI] [PubMed] [Google Scholar]
- Frenz DA, Galinovic-Schwartz V, Liu W, Flanders KC, Water TR. Transforming growth factor beta 1 is an epithelial-derived signal peptide that influences otic capsule formation. Dev Biol. 1992;153:324–336. doi: 10.1016/0012-1606(92)90117-Y. [DOI] [PubMed] [Google Scholar]
- Frolova EI, Fokina VM, Beebe DC. The expression pattern of opticin during chicken embryogenesis. Gene Expr Patterns. 2004;4:335–338. doi: 10.1016/j.modgep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Gong TW, Hegeman AD, Shin JJ, Lindberg KH, Barald KF, Lomax MI. Novel genes expressed in the chick otocyst during development: identification using differential display of RNA. Int J Dev Neurosci. 1997;15:585–594. doi: 10.1016/S0736-5748(96)00113-X. [DOI] [PubMed] [Google Scholar]
- Groves AK, Bronner-Fraser M. Competence, specification and commitment in otic placode induction. Development. 2000;127:3489–3499. doi: 10.1242/dev.127.16.3489. [DOI] [PubMed] [Google Scholar]
- Guth SI, Wegner M. Having it both ways: Sox protein function between conservation and innovation. Cell Mol Life Sci. 2008;65:3000–3018. doi: 10.1007/s00018-008-8138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Hawkins RD, Bashiardes S, Powder KE, Sajan SA, Bhonagiri V, Alvarado DM, Speck J, Warchol ME, Lovett M. Large scale gene expression profiles of regenerating inner ear sensory epithelia. PLoS ONE. 2007;2:e525. doi: 10.1371/journal.pone.0000525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbrand H, Guthrie S, Hadrys T, Hoffmann S, Arnold HH, Rinkwitz-Brandt S, Bober E. Two regulatory genes, cNkx5-1 and cPax2, show different responses to local signals during otic placode and vesicle formation in the chick embryo. Development. 1998;125:645–654. doi: 10.1242/dev.125.4.645. [DOI] [PubMed] [Google Scholar]
- Hollyday M, McMahon JA, McMahon AP. Wnt expression patterns in chick embryo nervous system. Mech Dev. 1995;52:9–25. doi: 10.1016/0925-4773(95)00385-E. [DOI] [PubMed] [Google Scholar]
- Ito K, Nakamura H, Watanabe Y. Protogenin mediates cell adhesion for ingression and re-epithelialization of paraxial mesodermal cells. Dev Biol. 2011;351:13–24. doi: 10.1016/j.ydbio.2010.11.024. [DOI] [PubMed] [Google Scholar]
- Jia XQ, Nakashima T, Kadomatsu K, Muramatsu T. Expression of midkine in the cochlea. Hear Res. 2001;160:10–14. doi: 10.1016/S0378-5955(01)00313-6. [DOI] [PubMed] [Google Scholar]
- Katayama K, Zine A, Ota M, Matsumoto Y, Inoue T, Fritzsch B, Aruga J. Disorganized innervation and neuronal loss in the inner ear of Slitrk6-deficient mice. PLoS ONE. 2009;4:e7786. doi: 10.1371/journal.pone.0007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan AE, Pelling AL, Leung KK, Tang AS, Bell DM, Tease C, Lovell-Badge R, Steel KP, Cheah KS. Sox2 is required for sensory organ development in the mammalian inner ear. Nature. 2005;434:1031–1035. doi: 10.1038/nature03487. [DOI] [PubMed] [Google Scholar]
- Kimura I, Konishi M, Miyake A, Fujimoto M, Itoh N. Neudesin, a secreted factor, promotes neural cell proliferation and neuronal differentiation in mouse neural precursor cells. J Neurosci Res. 2006;83:1415–1424. doi: 10.1002/jnr.20849. [DOI] [PubMed] [Google Scholar]
- Lewis AK, Frantz GD, Carpenter DA, Sauvage FJ, Gao WQ. Distinct expression patterns of notch family receptors and ligands during development of the mammalian inner ear. Mech Dev. 1998;78:159–163. doi: 10.1016/S0925-4773(98)00165-8. [DOI] [PubMed] [Google Scholar]
- Li H, Liu H, Heller S. Pluripotent stem cells from the adult mouse inner ear. Nat Med. 2003;9:1293–1299. doi: 10.1038/nm925. [DOI] [PubMed] [Google Scholar]
- Li H, Roblin G, Liu H, Heller S. Generation of hair cells by stepwise differentiation of embryonic stem cells. Proc Natl Acad Sci U S A. 2003;100:13495–13500. doi: 10.1073/pnas.2334503100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Liu H, Corrales CE, Mutai H, Heller S. Correlation of Pax-2 expression with cell proliferation in the developing chicken inner ear. J Neurobiol. 2004;60:61–70. doi: 10.1002/neu.20013. [DOI] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillevali K, Haugas M, Pituello F, Salminen M. Comparative analysis of Gata3 and Gata2 expression during chicken inner ear development. Dev Dyn. 2007;236:306–313. doi: 10.1002/dvdy.21011. [DOI] [PubMed] [Google Scholar]
- Liu M, Pereira FA, Price SD, Chu MJ, Shope C, Himes D, Eatock RA, Brownell WE, Lysakowski A, Tsai MJ. Essential role of BETA2/NeuroD1 in development of the vestibular and auditory systems. Genes Dev. 2000;14:2839–2854. doi: 10.1101/gad.840500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Li G, Chien JS, Raft S, Zhang H, Chiang C, Frenz DA. Sonic hedgehog regulates otic capsule chondrogenesis and inner ear development in the mouse embryo. Dev Biol. 2002;248:240–250. doi: 10.1006/dbio.2002.0733. [DOI] [PubMed] [Google Scholar]
- Liu D, Chu H, Maves L, Yan YL, Morcos PA, Postlethwait JH, Westerfield M. Fgf3 and Fgf8 dependent and independent transcription factors are required for otic placode specification. Development. 2003;130:2213–2224. doi: 10.1242/dev.00445. [DOI] [PubMed] [Google Scholar]
- Liu W, Li L, Li G, Garritano F, Shanske A, Frenz DA. Coordinated molecular control of otic capsule differentiation: functional role of Wnt5a signaling and opposition by sfrp3 activity. Growth Factors. 2008;26:343–354. doi: 10.1080/08977190802442013. [DOI] [PubMed] [Google Scholar]
- Lu X, Borchers AG, Jolicoeur C, Rayburn H, Baker JC, Tessier-Lavigne M. PTK7/CCK-4 is a novel regulator of planar cell polarity in vertebrates. Nature. 2004;430:93–98. doi: 10.1038/nature02677. [DOI] [PubMed] [Google Scholar]
- Ma Q, Anderson DJ, Fritzsch B. Neurogenin 1 null mutant ears develop fewer, morphologically normal hair cells in smaller sensory epithelia devoid of innervation. J Assoc Res Otolaryngol. 2000;1:129–143. doi: 10.1007/s101620010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilainen T, Haugas M, Kreidberg JA, Salminen M. Analysis of Netrin 1 receptors during inner ear development. Int J Dev Biol. 2007;51:409–413. doi: 10.1387/ijdb.072273tm. [DOI] [PubMed] [Google Scholar]
- Neves J, Kamaid A, Alsina B, Giraldez F. Differential expression of Sox2 and Sox3 in neuronal and sensory progenitors of the developing inner ear of the chick. J Comp Neurol. 2007;503:487–500. doi: 10.1002/cne.21299. [DOI] [PubMed] [Google Scholar]
- Oh SH, Johnson R, Wu DK. Differential expression of bone morphogenetic proteins in the developing vestibular and auditory sensory organs. J Neurosci. 1996;16:6463–6475. doi: 10.1523/JNEUROSCI.16-20-06463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama T, Groves AK. Expression of mouse Foxi class genes in early craniofacial development. Dev Dyn. 2004;231:640–646. doi: 10.1002/dvdy.20160. [DOI] [PubMed] [Google Scholar]
- Okano J, Takigawa T, Seki K, Suzuki S, Shiota K, Ishibashi M. Transforming growth factor beta 2 promotes the formation of the mouse cochleovestibular ganglion in organ culture. Int J Dev Biol. 2005;49:23–31. doi: 10.1387/ijdb.041905jo. [DOI] [PubMed] [Google Scholar]
- Oshima K, Grimm CM, Corrales CE, Senn P, Martinez Monedero R, Geleoc GS, Edge A, Holt JR, Heller S. Differential distribution of stem cells in the auditory and vestibular organs of the inner ear. J Assoc Res Otolaryngol. 2007;8:18–31. doi: 10.1007/s10162-006-0058-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima K, Shin K, Diensthuber M, Peng AW, Ricci AJ, Heller S. Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell. 2010;141:704–716. doi: 10.1016/j.cell.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauley S, Lai E, Fritzsch B. Foxg1 is required for morphogenesis and histogenesis of the mammalian inner ear. Dev Dyn. 2006;235:2470–2482. doi: 10.1002/dvdy.20839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrot V, Vazquez-Prado J, Gutkind JS. Plexin B regulates Rho through the guanine nucleotide exchange factors leukemia-associated Rho GEF (LARG) and PDZ-RhoGEF. J Biol Chem. 2002;277:43115–43120. doi: 10.1074/jbc.M206005200. [DOI] [PubMed] [Google Scholar]
- Pirvola U, Ylikoski J, Trokovic R, Hebert JM, McConnell SK, Partanen J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35:671–680. doi: 10.1016/S0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Powles N, Babbs C, Ficker M, Schimmang T, Maconochie M. Identification and analysis of genes from the mouse otic vesicle and their association with developmental subprocesses through in situ hybridization. Dev Biol. 2004;268:24–38. doi: 10.1016/j.ydbio.2003.11.024. [DOI] [PubMed] [Google Scholar]
- Puppo F, Thome V, Lhoumeau AC, et al. Protein tyrosine kinase 7 has a conserved role in Wnt/beta-catenin canonical signalling. EMBO Rep. 2011;12:43–49. doi: 10.1038/embor.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccomagno MM, Martinu L, Mulheisen M, Wu DK, Epstein DJ. Specification of the mammalian cochlea is dependent on Sonic hedgehog. Genes Dev. 2002;16:2365–2378. doi: 10.1101/gad.1013302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Sparks AB, Rago C, Akmaev V, Wang CJ, Vogelstein B, Kinzler KW, Velculescu VE. Using the transcriptome to annotate the genome. Nat Biotechnol. 2002;20:508–512. doi: 10.1038/nbt0502-508. [DOI] [PubMed] [Google Scholar]
- Sanchez-Calderon H, Francisco-Morcillo J, Martin-Partido G, Hidalgo-Sanchez M. Fgf19 expression patterns in the developing chick inner ear. Gene Expr Patterns. 2007;7:30–38. doi: 10.1016/j.modgep.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Sienknecht UJ, Fekete DM. Mapping of Wnt, frizzled, and Wnt inhibitor gene expression domains in the avian otic primordium. J Comp Neurol. 2009;517:751–764. doi: 10.1002/cne.22169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens CB, Davies AL, Battista S, Lewis JH, Fekete DM. Forced activation of Wnt signaling alters morphogenesis and sensory organ identity in the chicken inner ear. Dev Biol. 2003;261:149–164. doi: 10.1016/S0012-1606(03)00297-5. [DOI] [PubMed] [Google Scholar]
- Stone JS, Rubel EW. Delta1 expression during avian hair cell regeneration. Development. 1999;126:961–973. doi: 10.1242/dev.126.5.961. [DOI] [PubMed] [Google Scholar]
- Streit A. The preplacodal region: an ectodermal domain with multipotential progenitors that contribute to sense organs and cranial sensory ganglia. Int J Dev Biol. 2007;51:447–461. doi: 10.1387/ijdb.072327as. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. alpha-Latrotoxin and its receptors: neurexins and CIRL/latrophilins. Annu Rev Neurosci. 2001;24:933–962. doi: 10.1146/annurev.neuro.24.1.933. [DOI] [PubMed] [Google Scholar]
- Swanson GJ, Howard M, Lewis J. Epithelial autonomy in the development of the inner ear of a bird embryo. Dev Biol. 1990;137:243–257. doi: 10.1016/0012-1606(90)90251-D. [DOI] [PubMed] [Google Scholar]
- Tomarev SI, Nakaya N. Olfactomedin domain-containing proteins: possible mechanisms of action and functions in normal development and pathology. Mol Neurobiol. 2009;40:122–138. doi: 10.1007/s12035-009-8076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchikawa M, Kamachi Y, Kondoh H. Two distinct subgroups of Group B Sox genes for transcriptional activators and repressors: their expression during embryonic organogenesis of the chicken. Mech Dev. 1999;84:103–120. doi: 10.1016/S0925-4773(99)00083-0. [DOI] [PubMed] [Google Scholar]
- Urness LD, Paxton CN, Wang X, Schoenwolf GC, Mansour SL. FGF signaling regulates otic placode induction and refinement by controlling both ectodermal target genes and hindbrain Wnt8a. Dev Biol. 2010;340:595–604. doi: 10.1016/j.ydbio.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Bartheld CS, Patterson SL, Heuer JG, Wheeler EF, Bothwell M, Rubel EW. Expression of nerve growth factor (NGF) receptors in the developing inner ear of chick and rat. Development. 1991;113:455–470. doi: 10.1242/dev.113.2.455. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Takeda K, Katori Y, Ikeda K, Oshima T, Yasumoto K, Saito H, Takasaka T, Shibahara S. Expression of the Sox10 gene during mouse inner ear development. Brain Res Mol Brain Res. 2000;84:141–145. doi: 10.1016/S0169-328X(00)00236-9. [DOI] [PubMed] [Google Scholar]
- Webber A, Raz Y. Axon guidance cues in auditory development. Anat Rec A Discov Mol Cell Evol Biol. 2006;288:390–396. doi: 10.1002/ar.a.20299. [DOI] [PubMed] [Google Scholar]
- Wong YH, Lu AC, Wang YC, Cheng HC, Chang C, Chen PH, Yu JY, Fann MJ. Protogenin defines a transition stage during embryonic neurogenesis and prevents precocious neuronal differentiation. J Neurosci. 2010;30:4428–4439. doi: 10.1523/JNEUROSCI.0473-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood HB, Episkopou V. Comparative expression of the mouse Sox1, Sox2 and Sox3 genes from pre-gastrulation to early somite stages. Mech Dev. 1999;86:197–201. doi: 10.1016/S0925-4773(99)00116-1. [DOI] [PubMed] [Google Scholar]
- Wright TJ, Mansour SL. Fgf3 and Fgf10 are required for mouse otic placode induction. Development. 2003;130:3379–3390. doi: 10.1242/dev.00555. [DOI] [PubMed] [Google Scholar]
- Yamashita H, Oesterle EC. Induction of cell proliferation in mammalian inner-ear sensory epithelia by transforming growth factor alpha and epidermal growth factor. Proc Natl Acad Sci U S A. 1995;92:3152–3155. doi: 10.1073/pnas.92.8.3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen WW, Williams M, Periasamy A, Conaway M, Burdsal C, Keller R, Lu X, Sutherland A. PTK7 is essential for polarized cell motility and convergent extension during mouse gastrulation. Development. 2009;136:2039–2048. doi: 10.1242/dev.030601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou P, Muramatsu H, Sone M, Hayashi H, Nakashima T, Muramatsu T. Mice doubly deficient in the midkine and pleiotrophin genes exhibit deficits in the expression of beta-tectorin gene and in auditory response. Lab Invest. 2006;86:645–653. doi: 10.1038/labinvest.3700428. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
Oligonucleotide primer pairs used FOR RT-PCR (PDF 75 kb)
List of all 16,008 unique sage tags with individual tag count (XLS 1267 kb)
Ambiguous tags (PDF 64 kb)
Annotated SAGE library using official gene symbols recognized by IPA software. Listed are the gene symbols, gene names, tag count, the predicted location of the encoded protein, the gene family, as well as potential drugs that target the encoded protein (XLS 602 kb)
Transcriptional regulators identified in the annotated chicken otocyst SAGE library. Listed are the gene symbols, gene names, tag count, the predicted location of the encoded protein, potential drugs that target the encoded protein, as well as additional information including world wide web links, and references of publications that mention specific genes in the context of inner ear development (XLS 127 kb)
Secreted proteins identified in the annotated chicken otocyst SAGE library. Listed are the gene symbols, gene names, and tag count (PDF 104 kb)