Abstract
In the present study, genotype–phenotype correlations in eight Dutch DFNB8/10 families with compound heterozygous mutations in TMPRSS3 were addressed. We compared the phenotypes of the families by focusing on the mutation data. The compound heterozygous variants in the TMPRSS3 gene in the present families included one novel variant, p.Val199Met, and four previously described pathogenic variants, p.Ala306Thr, p.Thr70fs, p.Ala138Glu, and p.Cys107Xfs. In addition, the p.Ala426Thr variant, which had previously been reported as a possible polymorphism, was found in one family. All affected family members reported progressive bilateral hearing impairment, with variable onset ages and progression rates. In general, the hearing impairment affected the high frequencies first, and sooner or later, depending on the mutation, the low frequencies started to deteriorate, which eventually resulted in a flat audiogram configuration. The ski-slope audiogram configuration is suggestive for the involvement of TMPRSS3. Our data suggest that not only the protein truncating mutation p.T70fs has a severe effect but also the amino acid substitutions p.Ala306Thr and p.Val199Met. A combination of two of these three mutations causes prelingual profound hearing impairment. However, in combination with the p.Ala426Thr or p.Ala138Glu mutations, a milder phenotype with postlingual onset of the hearing impairment is seen. Therefore, the latter mutations are likely to be less detrimental for protein function. Further studies are needed to distinguish possible phenotypic differences between different TMPRSS3 mutations. Evaluation of performance of patients with a cochlear implant indicated that this is a good treatment option for patients with TMPRSS3 mutations as satisfactory speech reception was reached after implantation.
Electronic supplementary material
The online version of this article (doi:10.1007/s10162-011-0282-3) contains supplementary material, which is available to authorized users.
Keywords: DFNB8/10, TMPRSS3 mutations, cochlear implantation, ski-slope audiogram, genotype–phenotype correlations
Introduction
Autosomal recessive nonsyndromic hearing impairment (arNSHI) is the most common type of inherited hearing impairment, accounting for approximately 80% of inherited prelingual hearing impairment. The phenotype associated with nonsyndromic recessive hearing impairment is usually prelingual, non-progressive, and severe to profound. Autosomal recessive inheritance is rare in nonsyndromic hearing impairment with postlingual onset (Petersen and Willems 2006).
Already 72 loci have been described to be involved in arNSHI and 40 causative genes have been indentified thus far, indicating enormous genetic heterogeneity. However, there is little knowledge about the contribution of the different genes to arNSHI in the European population. GJB2 mutations are a frequent cause of arNSHI, as in most populations; however, the relative contribution varies per country. Other genes reported to be relatively important for arNSHI in populations of western European origin are TMC1, OTOF, and CDH23 (Hilgert et al. 2009).
Mutations in the transmembrane protease serine 3 (TMPRSS3, OMIM 605511) gene on chromosome 21q22 are responsible not only for arNSHI with a prelingual onset (DFNB10, OMIM 605316; Bonne-Tamir et al. 1996) but also for postlingual (DFNB8, OMIM 601072; Veske et al. 1996) arNSHI. TMPRSS3 mRNA has been detected in the spiral ganglion, the entire epithelium that supports the cells of the organ of Corti, and in low levels in the stria vascularis, but was not detected in sensory hair cells with in situ hybridization (Guipponi et al. 2002). However, in a more recent study by Guipponi et al. (2008), expression of the TMPRSS3 protein was shown in the inner hair cells of the organ of Corti and in the cell bodies of the spiral ganglion neurons. The function of TMPRSS3 in the auditory pathway is currently poorly understood, but it has been hypothesized that lack of TMPRSS3 activity results in hearing impairment because of an increased sodium concentration in the endolymph by insufficient ENaC activation (Guipponi et al. 2002). However, individuals with pseudohypoaldosteronism type 1, which are homozygous for null alleles of ENaC subunits, demonstrate no hearing impairment (Peters et al. 2006). This indicates that hearing impairment associated with TMPRSS3 mutations cannot be explained by inactive ENaC. Involvement of TMPRSS3 in the proteolytic cleavage of proneurotrophins could play a role (Guipponi et al. 2002). Proneurotrophins function in the development and maintenance of inner ear innervation (Farinas et al. 2001). The serine protease plasmin, which belongs to the same family as TMPRSS3, has been shown to cleave the neurotrophic factor BDNF (Bom et al. 2001; Guipponi et al. 2002).
So far, mutations in TMPRSS3 have mainly been identified in families from Asian and Mediterranean countries, and did not seem to contribute substantially to prelingual arNSHI in the Caucasian population (Wattenhofer et al. 2002). However, in the present study, we demonstrate that mutations in TMPRSS3 are a common cause of progressive arNSHI with a childhood onset in the Dutch population. We characterized the hearing impairment in eight Dutch families with compound heterozygous mutations in TMPRSS3 and describe genotype–phenotype correlations. The present family E was previously described as family 2 in a report on autosomal recessive progressive high-frequency hearing impairment with childhood onset (Cremers et al. 1987).
Methods
Patients
The pedigrees of eight families with arNSHI were constructed (Fig. 1; Cremers et al. 1987). After informed consent had been obtained from the participating family members, audiograms were obtained to establish the hearing impairment phenotype of these families. The study was approved by the medical ethics committee of the Radboud University Nijmegen Medical Centre.
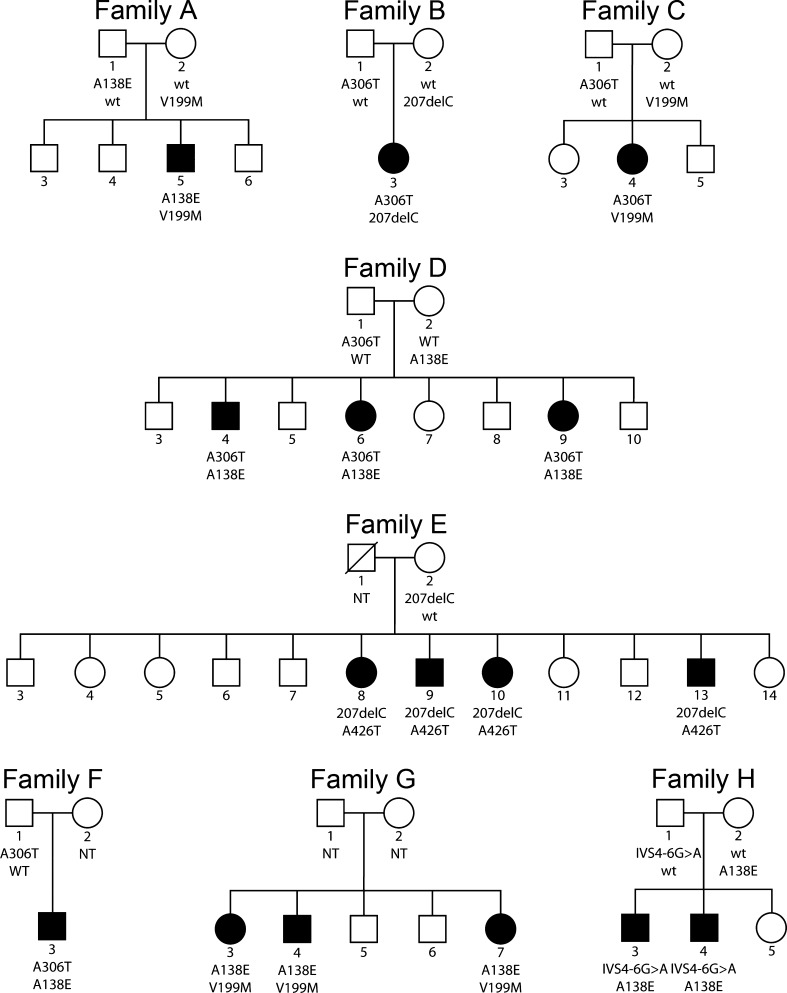
FIG. 1.
Pedigrees of the Dutch families with autosomal recessive hearing impairment and segregation of the TMPRSS3 mutations. Family E is previously described by Cremers et al. (1987). All unaffected sibs were either carrier of one mutant allele or of two wild-type alleles. Square, male; circle, female; open symbol, clinically unaffected; solid symbol, clinically affected; slash, deceased individuals. NT not tested.
Examination of the family members included a medical history guided by a questionnaire, otoscopy, pure tone audiometry, and drawing blood samples for DNA isolation. Some of the affected family members underwent speech audiometry, vestibulo-ocular examination, and high-resolution spiral computed tomography imaging of the temporal bones. Furthermore, concomitant disease, use of medication, and any other possible causes of acquired hearing impairment were ruled out. Previous medical records and audiograms were traced for individual longitudinal analysis. The previously published audiograms of family E were also included (Cremers et al. 1987).
Linkage analysis
Genomic DNA of all participating individuals was extracted from peripheral blood lymphocytes according to standard protocols. Families D and E were selected for linkage analysis. For family D, the unaffected parents and three affected and five unaffected siblings were genotyped using the Illumina 6k single nucleotide polymorphism (SNP) array according to the manufacturer’s protocol. Similarly, for family E, the unaffected mother and four affected and five unaffected siblings were genotyped. Multipoint linkage analysis was performed with GeneHunter version 2.1r5 in the EasyLinkage software package (Hoffmann and Lindner 2005). An autosomal recessive mode of inheritance with a penetrance of 95% and a disease allele frequency of 0.001 were used for limit of detection (LOD) score calculations.
Sequence analysis and prediction of effects of mutations on protein structure
Amplification of all coding exons and flanking intronic sequences by PCR was performed on 40 ng of genomic DNA with Taq DNA polymerase (Roche). Primer sequences and PCR conditions are available in Electronic supplementary material (ESM) Table 1. PCR fragments were purified using NucleoFast 96 PCR plates (Clontech) according to the manufacturer’s protocol. Sequence analysis was performed with the ABI PRISM Big Dye Terminator Cycle Sequencing V3.1 Ready Reaction kit and the ABI PRISM 3730 DNA analyzer (Applied Biosystems).
For segregation analysis of the identified TMPRSS3 mutations in the families, restriction digestion could be performed. Relevant exons were amplified and PCR products were purified as described for sequencing. Digestion of the PCR products with restriction enzymes was performed according to the manufacturer’s protocol, and restriction fragments were analyzed on 2% agarose gels. Primer sequences and restriction enzymes are listed in ESM Table 1. The same approach was used for testing the occurrence of the c.595G>A and c.1276G>A variants in Dutch control individuals. As reference sequence, NM_024022 was used.
The effects of the amino acid substitutions in TMPRSS3 on its structure were analyzed using the Project HOPE web server (http://www.cmbi.ru.nl/hope; Venselaar et al. 2010). The exact 3D structure of TMPRSS3 is unknown; therefore, HOPE built a model based on the homologous structure protein data bank entry 1z8g (Herter et al. 2005). The model was built using an automatic script in the Yasara and WHAT IF Twinset (Vriend 1990; Krieger et al. 2002).
Audiometry and data analysis
Audiometric examination comprised conventional pure tone audiometry in a sound-treated room. Air conduction (AC) thresholds were measured in decibel hearing level (HL) at 0.25, 0.5, 1, 2, 4, and 8 kHz; bone conduction thresholds were measured in decibel HL at 0.5, 1, 2, 4, and 8 kHz. The individual 95th percentile threshold values of presbyacusis in relation to the patient’s sex and age were derived for each frequency using the ISO 7029 method. Individuals were considered affected if the best hearing ear showed thresholds beyond the 95th percentile threshold values for presbyacusis at three frequencies or more. Binaural mean air conduction threshold values were calculated for each frequency.
Individual longitudinal regression analysis of binaural mean air conduction threshold values on age was performed for individual 10 of family E because the audiometric data of this individual were most comprehensive (Cremers et al. 1987). The regression coefficient (slope) was called annual threshold deterioration (ATD), expressed in decibels per year. Progression was significant if the 95% confidence interval of the ATD did not include zero.
Speech audiometry was performed under the aforementioned conditions using standard Dutch monosyllabic consonant–vocal–consonant word lists. The maximum phoneme recognition score (mean percentage correct for both ears) was retained from the monaural performance versus intensity curves. These maximum phoneme recognition scores were analyzed in relation to age and to pure tone average (mean value for both ears) at frequencies of 1, 2, and 4 kHz (PTA1,2,4 kHz). Cross-sectional analysis was performed using linear regression analysis to determine the local average slope, called deterioration rate, in the score-against-age plot and deterioration gradient in the score-against-PTA1,2,4 kHz plot. A previously described group of subjects with only presbyacusis was used as the reference group (De Leenheer et al. 2002).
Vestibulo-ocular examination and data analysis
Nine of 16 affected individuals (A5, B3, C4, E8, E9, E10, E13, and G3) underwent vestibular and ocular motor tests. The test included evaluation of the vestibulo-ocular reflex using electronystagmography with computer analysis and saccadic, smooth pursuit, and optokinetic nystagmus responses. Vestibular stimulation comprised rotatory and caloric tests. Details and normal values have been described previously (Kunst et al. 2000).
Genotype–phenotype correlations
We compared the phenotype of the present families by focusing on the genotype to study whether the mutations differ in severity. The phenotypes of the different families were compared when at least one identical mutation in TMPRSS3 was present. Therefore, the effect of the second mutation on the phenotype in these families could be determined. All family members with the same compound heterozygous mutations were taken together, for example, families A/G and D/F. The threshold values of each family member were compared to the threshold values of every family member of the other family. We compared the thresholds at all frequencies in relation to age.
Evaluation of the effect of CI on phoneme recognition
Nine patients underwent cochlear implantation (CI). Speech audiometry was performed in a quiet environment using standard monosyllabic Dutch word lists after cochlear implantation. Words were presented through a loudspeaker at a fixed distance of 1 m from the patient at a normal conversational level of 70 dB sound pressure level (SPL). Subject responses were scored as the percentage of phonemes correct. As a reference, the test results after 1 year of use in postlingually implanted adults were used. The first reference group (n = 70) was implanted with a CII/HR90K device of Advanced Bionics® and the second reference group (n = 65) with a Nucleus 24RCA of Cochlear®. A mean phoneme score of 64% (SD 23%) was demonstrated in the first reference group, and the second group (n = 65) showed a mean phoneme score of 72% (SD 18%; Verhaegen et al. 2008).
Results
Compound heterozygous TMPRSS3 mutations in Dutch arNSHI patients
Linkage analysis was performed for two arNSHI families, D and E, with progressive hearing impairment. For family D, five regions were found with a maximum LOD score of ~1.82 (ESM Fig. 1). Two of the regions contained a known deafness gene, namely, GRXCR1 on chromosome 4 and TMPRSS3 on chromosome 21. Mutations in GRXCR1 were excluded in a previous study (Schraders et al. 2010). For family E, three regions with suggestive linkage were found with a LOD score of ~2.42. One of those regions harbored TMPRSS3. In both families, the LOD score for the TMPRSS3 region reached the theoretical maximum LOD score.
Thus, mutation analysis was performed for all coding exons and exon–intron boundaries of TMPRSS3. In both families, compound heterozygous sequence variants were detected (Table 1). The variants c.413C>A, c.916G>A, and c.207delC have been described before to be pathogenic in families with arNSHI. The c.1276G>A variant is present in the SNP database (http://www.ncbi.nlm.nih.gov/sites/SNP; rs56264519). However, this SNP has not been validated. We did not identify this variant in 590 Dutch control alleles, and, importantly, the resulting amino acid change Ala426Thr has been shown to affect protein function (Lee et al. 2003). We therefore considered the Ala426Thr substitution to be pathogenic. All four TMPRSS3 mutations co-segregated with the hearing impairment in the families (Fig. 1).
TABLE 1.
Overview of the clinical characteristics and genotypes in the eight families
| Family | Family research no. | Patient | Age of onset | Phenotype | Mutation | Protein change (predicted) | Protein domain |
|---|---|---|---|---|---|---|---|
| A | W06-357 | 5 | 5 years | High-frequency hearing impairment with normal thresholds of the low frequencies until at least 12 years of age | c.413C>G; c.595G>A | p.Ala138Glu; p.Val199Met | SRCR; SRCR |
| B | W06-418 | 3 | Prelingual (2 years) | Down-sloping audiogram configuration with impairment of the low frequencies at a very young age | c.207delC; c.916G>A | p.Thr70fs; p.Ala306Thr | Truncation after TM; serine protease |
| C | W06-445 | 4 | Prelingual (2 years) | Down-sloping audiogram configuration with impairment of the low frequencies at a very young age | c.595G>A; c.916G>A | p.Val199Met; p.Ala306Thr | SRCR; serine protease |
| D | W02-068 | 4 | 7 years | Down-sloping audiogram configuration with deterioration of the low frequencies after 11 years of age | c.413C>A; c.916G>A | p.Ala138Glu; p.Ala306Thr | SRCR; serine protease |
| 6 | 16 years | High-frequency hearing impairment with normal thresholds of the low frequencies until at least 19 years of age | |||||
| 9 | 18 years | ||||||
| E | W07-0050 | 8 | 17 years | Mainly high-frequency hearing impairment with deterioration of the low frequencies after approximately 30 years of age | c.207delC; c.1276G>A | p.Thr70fs; p.Ala426Thr | Truncation after TM; serine protease |
| 9 | 12 years | ||||||
| 10 | 7 years | ||||||
| 13 | 17 years | ||||||
| F | W03-357 | 3 | 10 years | High-frequency hearing impairment with mild decline of the low frequencies at the age of 22 years | c.413C>A; c.916G>A | p.Ala138Glu; p.Ala306Thr | SRCT; serine protease |
| G | W08-1756 | 3 | 15 years | Flat audiogram configuration with thresholds of about 110 dB at 49 years of age | c.413C>A; c.595G>A | p.Ala138Glu; p.Val199Met | SRCR; SRCR |
| 4 | 33 years | Down-sloping audiogram configuration with impairment of the low frequencies starting after 44 years | |||||
| 7 | 21 years | ||||||
| H | W10-1160 | 3 | 5 years | Down-sloping audiogram configuration with deterioration of the low frequencies at 23 years of age | c.413C>A; c.323-6G>Aa | p.Ala138Glu; p.Cys107fs | SRCR; SRCR |
| 5 | 4 years | High-frequency hearing impairment with normal threshold of the low frequencies until at least 16 years of age |
To investigate the contribution of TMPRSS3 mutations in progressive hearing impairment, we screened a panel of 22 unrelated supposed arNSHI patients with progressive hearing impairment; compound heterozygous mutations were found in four of them, belonging to families A, B, C, and F (Fig. 1 and Table 1). The novel variant, c.595G>A, leading to the substitution of a methionine for a valine at position 199 of the protein (p.Val199Met) was present in two patients. This variant was not found in 590 Dutch control alleles. In summary, we identified compound heterozygous TMPRSS3 mutations in 6 of 24 families (25%) with progressive arNSHI.
To evaluate the prevalence of TMPRSS3 mutations in unselected patients with (putative) arNSHI in the Netherlands, we screened a panel of 212 index patients for whom GJB2 mutations had been excluded as the cause of their hearing impairment. Compound heterozygous mutations were detected in two of these index patients (Table 1), which segregated with the hearing impairment in the corresponding families G and H (Fig. 1). Hearing impairment was found to be progressive in these families as well.
Large variation in age of onset
The clinical characteristics and genotypes of the patients are listed in Table 1. For none of the affected family members, there was evidence of other causes of hearing impairment, and otoscopy was normal. All affected individuals reported progressive bilateral hearing impairment. Some individuals had prelingual hearing impairment, whereas other individuals were not aware of being hearing impaired until in their late teens, which indicates postlingual hearing impairment (Table 1).
Imaging of temporal bones
Computed tomography (CT) of the temporal bone was performed in ten individuals (A5, C4, D4, E8, E9, E10, E13, F3, G3, and H3), mainly as part of a preoperative CI selection procedure. Magnetic resonance imaging (MRI) was accomplished in individual B3. The CT scans and MRI showed normal middle and inner ear structures in all cases.
Vestibulo-ocular examination
Although vestibular symptoms were not reported, testing of vestibular function in three affected individuals (A5, E8, and E9) revealed mild hyperreflexia of the velocity step responses with time constants between 28 and 32 s (normal limit, 26 s). Furthermore, individuals G3 and H3 showed significant hyporeflexia of the velocity step responses with time constants between 3 and 9 s for both nystagmus directions. Caloric tests revealed borderline bilateral caloric weakness in individual G3 and normal caloric functioning in individual H3. None of the relative prevalences of the abnormal vestibular findings was above chance level. The tests of vestibular function in four additional affected individuals (B3, C4, E10, and E13) demonstrated no abnormalities. The vestibulo-ocular examination in all nine affected individuals was performed before cochlear implantation. None of the patients reported by Bonne-Tamir et al. (1996) showed signs of vestibular involvement. However, formal vestibular tests were not performed. Information on the vestibular function of the other TMPRRS3 families previously reported were not available.
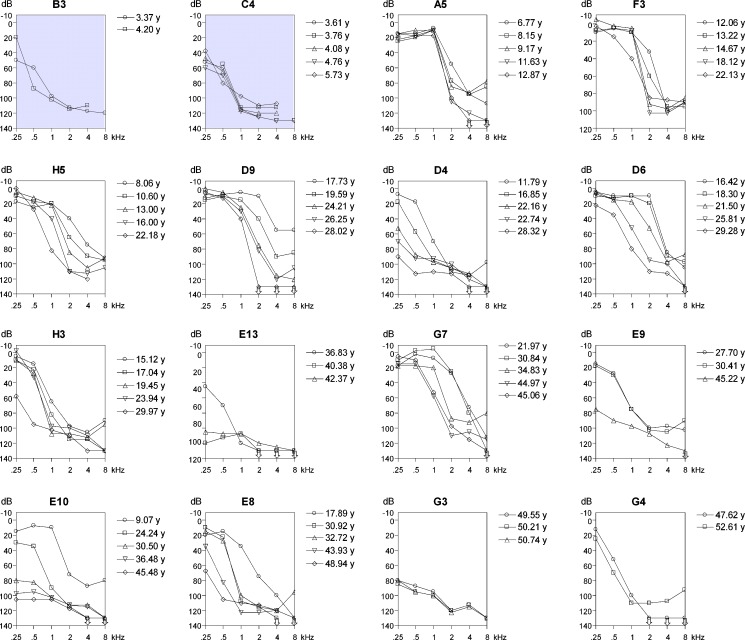
Typical ski-slope audiogram configuration
A representative selection of the available pure tone audiograms of all 16 affected individuals are shown in Figure 2. The thresholds in families B and C (gray audiograms in Fig. 2) are higher than or similar to the thresholds in the other families, at a much younger age. This is in accordance with the reported prelingual hearing impairment in these families. Most frequently, a down-sloping audiogram configuration was observed, indicating high-frequency hearing impairment. Remarkable is the typical ski-slope audiogram configuration for patients with postlingual hearing impairment, as seen in, for example, individuals A5, F3, D6, and H3. However, flatter audiogram configurations, close to residual hearing, were also observed (individuals D4, E8, E9, E10, E13, and G3), but only at ages above 28 years. The families with postlingual hearing impairment initially presented with hearing impairment of the high frequencies which was followed by an increase of the low- and mid-frequency threshold values with advancing age.
FIG. 2.
Selection of binaural mean air conduction threshold values of clinically affected family members at different ages, ordered by age (from top left to bottom right) at last visit (Cremers et al. 1987). Family number and sequence number are above each audiogram. Gray background, relatively poorer thresholds.
The thresholds in individual D4 are exceptionally high at the low and mid-frequencies at ages between 21 and 28 years as compared with the thresholds of individuals D6 and D9. The threshold of individual G3 at these frequencies are also remarkably high in comparison to the thresholds of individuals G4 and G7 at the same ages. So far, there is less variation in thresholds in families E and H at matching ages. However, it is not clear at this point whether the hearing impairment in individual H4 will show the same progression as the hearing impairment of his brother H3 until the age of 29 years.
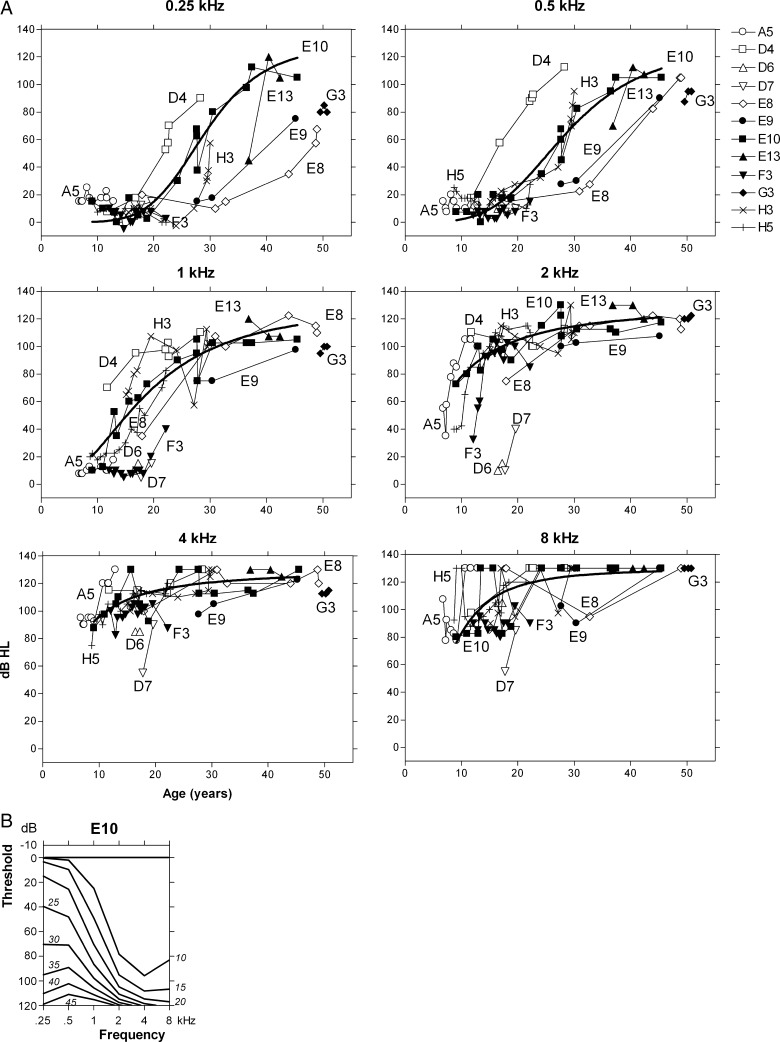
Progressive bilateral hearing impairment at all frequencies
Regression analysis for individual E10 demonstrated a significant deterioration of threshold levels with advancing age for all frequencies. The longitudinal threshold data of individual E10 covered approximately 36 years (from age 9 to 45 years). Figure 3a shows the threshold data for all frequencies separately. A nonlinear regression model of a dose–response curve with a variable slope fitted far better to the data than a linear regression model (comparative data not shown); the fitted curves are included in Figure 3a. The maximum local slope (ATD) was about 6 dB/year at 0.25 kHz at ages between 25 and 35 years. This maximum ATD can also be derived from the age-related typical audiogram (ARTA; Fig. 3b); the threshold at 0.25 kHz increases from about 40 dB at 25 years to about 95 dB at 35 years. Figure 3 illustrates that most of the progression at the higher frequencies, in the absence of a pronounced congenital hearing impairment, already occurred before the age of 10 years. If that indeed was the case in individual E10, the threshold levels at around 9 years of age would suggest an average progression of about 10 dB/year at 2–8 kHz.
FIG. 3.
a Longitudinal individual measurements of individuals A5, D4, D6, D7, E8, E9, E10, E13, F3, G3, H3, and H5 for each frequency separately (different symbols for each individual; Cremers et al. 1987). A dose–response curve with a variable slope could be obtained for the longitudinal data of individual E10 for each frequency (bold line). b ARTA derived from a longitudinal regression analysis of mean AC threshold levels of individual E10. Italics indicate age in years.
Figure 3a also shows all the longitudinal threshold data of individuals A5, D4, D6, D7, E8, E9, E13, F3, G3, H3, and H5 (postlingual DFNB8 patients; see below). The available audiograms of individual A5 were measured between 6 and 13 years of age. The hearing impairment in this individual indeed demonstrated progression only at 2–8 kHz, which is on the order of about 10 dB/year. Figure 3a also demonstrates that the longitudinal data for individual E10 are, more or less, representative for the whole group of DFNB8 patients in the present collection of families. The sigmoidal regression curve for individual E10 takes a fairly median position; however, some of the other individuals are represented by threshold values that indicate major deviations from that position (Fig. 3a).
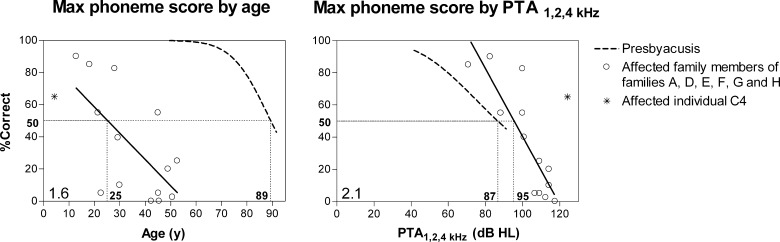
Relatively good speech recognition scores
Figure 4 shows the available single-snapshot measurements of the phoneme scores in the affected members of families A, D, E, F, G, and H. For the more severely affected families B and C (see below), we only had one score for individual C4. Speech recognition in the patients with a TMPRSS3 mutation was severely impaired, even at young ages. The 50% score was attained at the age of 25 years, whereas in the presbyacusis group, this score was attained at 89 years. At approximately 55 years of age, there was no remaining speech recognition. The PTA1,2,4 kHz levels at which speech recognition scores of 50% were attained in the TMPRSS3 and the presbyacusis patients were 95 and 87 dB, respectively. Therefore, the TMPRSS3 patients tended to have somewhat higher scores than the presbyacusis patients at similar PTA1,2,4 kHz levels. The deterioration rate in the score-against-age plot was 1.6% per year, and the deterioration gradient in the score-against-PTA1,2,4 kHz plot was 2.1% per decibel. The presbyacusis group showed a higher deterioration rate (3.3% per year) and a lower deterioration gradient (1.1% per decibel). Compared with matching scores of the other family members, the speech recognition score of individual C4 was observed at a younger age and a higher PTA1,2,4 kHz level (Fig. 4). Unfortunately, speech recognition scores were not reported for previously described families with TMPRSS3 mutations.
FIG. 4.
Single-snapshot measurements of the affected family members of binaural mean phoneme recognition scores against age (left) and against binaural mean pure tone average at 1, 2, and 4 kHz (right; Cremers et al. 1987). The solid regression line covers the cross-sectional analysis. The dotted curve represents presbyacusis and was previously established for patients with presbyacusis. See text for the meaning of the figures and the straight horizontal and vertical lines.
Genotype–phenotype correlations
In the present eight families, four different missense mutations and two frameshift mutations were detected. Four of these mutations were recurrent and present in two to four of the eight families. To study whether the mutations differ in severity, we compared the phenotypes of the families with one identical mutation in TMRPSS3. This allows a comparison of the effect of the second mutation in these families. For example, patients in families B and E have the p.Thr70fs mutation in common (Table 1); therefore, a comparison of the phenotypic effect of the p.Ala306Thr mutation in family B and the p.Ala426Thr mutation in family E is possible. Thresholds in family B at age 3–4 years are poorer than those in family E at ages between 9 and 30 years, but fairly similar to those in family E at age 24–37 years and better than those in family E at age 36–49 years (Fig. 2). This implies that threshold levels similar to those shown by family B in the first decade of life are unlikely to be found in family E before the third or fourth decade of life and that poorer levels are only likely to be found in family E from the fourth to the fifth decade of life onwards. This suggests that the p.Ala426Thr mutation has a milder effect than the p.Ala306Thr mutation. The same procedure was repeated for the other mutations. The p.Ala306Thr mutation has a fairly similar phenotypic effect as p.Val199Met (families A/G versus D/F) and p.Cys107fs (families D/F versus family H), whereas p.Val199Met has a fairly similar effect as p.Thr70fs (family B versus C) and p.Cys107fs (families A/G versus family H): this suggests that also p.Ala306Thr and p.Thr70fs must have fairly similar phenotypic effects. Both p.Ala426Thr and p.Ala138Glu have milder phenotypic effects than p.Ala306Thr (family B versus E and family C versus families A/G, respectively); this suggests that p.Ala426Thr and p.Ala138Glu have fairly similar phenotypic effects. Furthermore, p.Ala138Glu has a milder effect than p.Val199Met (family C versus families D/F) and also a milder effect than p.Thr70fs (family B versus families D/F). The thresholds of individuals D4 and G3 were higher compared with the thresholds of their sibs, and therefore individuals D4 and G3 were excluded from the genotype–phenotype assessment. Nevertheless, the classification of mutations did not change when individuals D4 and G3 were included. In line with these considerations and the levels of the thresholds in the various families, we suggest that the mutations in TMPRSS3 can be classified in mild and severe mutations. Our classification of TMPRSS3 mutations is in accordance with the two types of hearing impairment phenotypes:
DFNB10: a severe congenital or early childhood type with prelingual hearing impairment (families B and C) caused by the presence of two severe mutations
DFNB8: a later-onset progressive but initially milder type with postlingual hearing impairment (families A/G, D/F, and E) caused by the presence of one mild and one severe mutation
Predicted effects of amino acid substitutions on TMPRSS3 structure
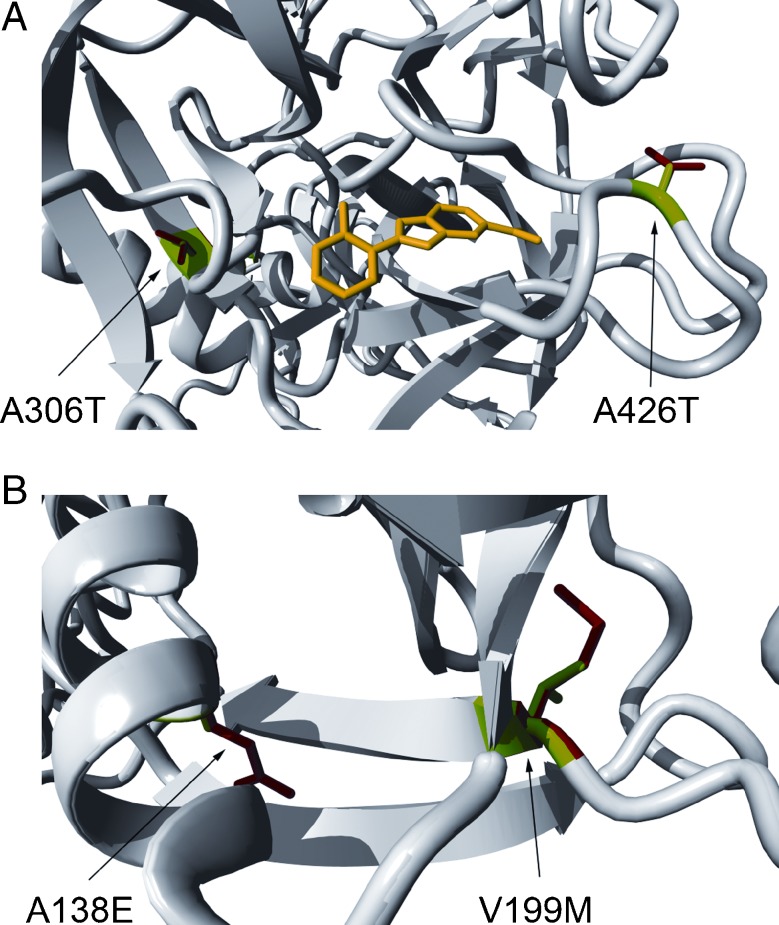
Genotype–phenotype correlations in the present study suggest that the amino acid substitutions p.Ala138Glu and p.Ala426Thr have a less severe effect as compared with p.Val199Met and p.Ala306Thr. Both substitutions p.Ala306Thr and p.Ala426Thr are within the serine protease domain. These changes result in the substitution of a larger residue for a conserved amino acid, predicted to result in the destabilization of the protein. The difference in severity of the effect of the two mutations might well be explained by the location of the substituted residue. The p.Ala306Thr is located close to one of the active residues, Asp304, and therefore this substitution can be predicted to directly disturb the function of the serine protease domain. Moreover, the alanine at position 426 is predicted to be semi-buried in the protein, and the side chain of threonine is only slightly bigger than the alanine side chain. Therefore, there could be enough space for the side chain of the threonine at this position (Fig. 5a). Both substitutions p.Ala138Glu and p.Val199Met occur within the scavenger receptor cysteine-rich (SRCR) domain, which is thought to be involved in interactions with extracellular molecules. Also, these two mutations affect evolutionary conserved residues and substitute a larger amino acid for the wild-type residue, which is predicted to cause steric clashes with other residues and hence to destabilize the protein (Fig. 5b). The 3D modeling could not directly explain the difference in the effect of p.Ala138Glu and p.Val199Met mutations since the exact function and binding partners of the SRCR domain are still unknown.
FIG. 5.
Molecular modeling for TMPRSS3 missense mutations. Graphic representation of the effect of the p.Ala306Thr and p.Ala426Thr mutations in the serine protease domain (A) and of the p.Ala138Glu and p.Val199Met in the SRCR domain (B). The wild-type residues are depicted in green, while the mutant residues are shown in red. The yellow structure represents a substrate for the serine protease domain (figure made using the model for TMPRSS and YASARA).
Good speech reception after cochlear implantation
Eight family members underwent cochlear implantation in our hospital. Implantation was uneventful and the implant was fully inserted on the left side in all cases. Individuals B3, C4, and H3 were implanted with a Nucleus Freedom (Cochlear), individuals E8 and G3 with a Nucleus Contour CI24R (Cochlear), and individuals E9, E10 and E13 with a Clarion AB-5100H (Advanced Bionics). Individual 4 of family D was implanted in another hospital with a Nucleus CI24M (Cochlear), and complete insertion was reported. Phoneme and word scores were unavailable for individuals D4 and H3. Individual H3 was implanted less than a year ago. The available phoneme and word scores of sound presented at 70 dB SPL 1 year after activation of the speech processor are presented in Table 2. We compared these results with the results of our two reference groups 12 months after use. The phoneme and word scores in the seven patients were above the average phoneme and word scores of our two reference groups. The mean phoneme score of 84.1% (SD = 5.4) is significantly higher than the phoneme score of reference group 1 (p < 0.0001) and reference group 2 (p = 0.0005). Elbracht et al. (2007) also reported good results with bilateral cochlear implantation, but unfortunately, there were no phoneme or word scores available to compare with our results.
TABLE 2.
Available phoneme and word scores preoperative and 1 year after the activation of the speech processor of the cochlear implant
| Individual | Preoperative | Postoperative | |
|---|---|---|---|
| Phoneme score (%) | Phoneme score (%) | Word score (%) | |
| B3 | – | 91 | – |
| C4 | – | 80 | – |
| D4 | 5 | – | – |
| E8 | 20 | 89 | 75 |
| E9 | 5 | 76 | 60 |
| E10 | 0 | 82 | 58 |
| E13 | 0 | 83 | 62 |
| G3 | 2.5 | 88 | 68 |
| H3 | 10 | – | – |
Sound was presented at 70 dB sound pressure level
Discussion
This report presents the clinical and genetic analyses of eight Dutch DFNB8/10 families with compound heterozygous mutations in TMPRSS3 (Table 1). Our study suggests genotype–phenotype correlations for the detected mutations.
The reported age of onset and severity of hearing impairment in the present families showed a wide variation. Also within the families, variation was seen. This is frequently demonstrated in hereditary hearing impairment, however mainly in the dominantly inherited forms. At a young age, TMPRSS3-associated hearing impairment was more pronounced at the high frequencies, and sooner or later, depending on the mutation, thresholds for the low frequencies deteriorated, eventually resulting in a flatter audiogram configuration (i.e., residual hearing). Prelingual as well as postlingual hearing impairments were reported. A ski-slope audiogram configuration was indicative for postlingual hearing impairment.
Twenty-two families with a mutation in TMPRSS3 had been reported previously (Shahin et al. 2010; Bonne-Tamir et al. 1996; Veske et al. 1996; Ben-Yosef et al. 2001; Masmoudi et al. 2001; Wattenhofer et al. 2002, 2005; Ahmed et al. 2004; Hutchin et al. 2005; Walsh et al. 2006; Elbracht et al. 2007; Table 3). Since the clinical data provided for most of these TMPRSS3 families are very limited, a thorough comparison with our data is not possible. However, the available clinical data seem to be in line with those of the present families. All families with the TMPRSS3 mutations described in the literature showed either severe to profound prelingual (DFNB10) or postlingual (DFNB8) progressive bilateral sensorineural hearing impairment (Shahin et al. 2010; Bonne-Tamir et al. 1996; Veske et al. 1996; Ben-Yosef et al. 2001; Masmoudi et al. 2001; Wattenhofer et al. 2002, 2005; Ahmed et al. 2004; Hutchin et al. 2005; Elbracht et al. 2007).
TABLE 3.
Overview of the literature on mutations in the TMPRRS3 gene
The gray rows represent families with postlingual hearing impairment (DFNB8) and the blank rows the families with prelingual hearing impairment (DFNB10). Mutations in bold are the mutations with a mild effect on the phenotype, and mutations in italic are the mutations with a more severe effect on the phenotype
aThis is the notation of the IVS4-6G>A mutation with NP_076927.1 as a reference sequence
bThis is the notation of the IVS8+8insT mutation with NM_024022 as a reference sequence; the precise effect of this mutant allele is unknown
In the homozygous state, the previously described TMPRSS3 mutations lead to either postlingual progressive (DFNB8) or prelingual severe to profound (DFNB10) hearing impairment (Table 3), with one exception, namely the p.Pro404Leu mutation (Masmoudi et al. 2001; Wattenhofer et al. 2005). Although Wattenhofer et al. (2005) described an age of detection of hearing impairment of 6–7 years in a family with a homozygous p.Pro404Leu mutation, the average threshold levels (0.5–4 kH) at that age were already 85–99 dB. Since members of this family were reported to have had a normal hearing until the age of detection, it can be concluded that hearing impairment in this family exhibited a very fast progression in childhood. Wattenhofer et al. (2005) did not report on the speech development of the affected family members and hearing impairment might still be classified as prelingual, which is defined as a delayed speech development. Hearing impairment in the second family with a homozygous p.Pro404Leu mutation was reported to be congenital and severe to profound and therefore probably prelingual (Masmoudi et al. 2001). Based on these data, we conclude that p.Pro404Leu is likely to be a severe mutation. Modifying genetic factors may explain the difference in onset between the two families with this mutation.
Both in the present and previously described families, the severe classified mutations p.Ala306Thr, p.Val199Met, and p.Thr70fs result in postlingual hearing impairment when present in combination with mutations only described to be associated with postlingual hearing impairment (p.Arg216Cys, p.Ala138Glu, and p.Ala426Thr mutations). Prelingual hearing impairment is seen by us and others when the Ala306Thr, Val199Met, and Thr70fs mutations are present in the compound heterozygous state with other mutations associated with prelingual hearing impairment (p.Thr70fs and p.Ala306Thr). However, there is an inconsistency in this classification of the c.323-6G>A (p.Cys107fs) mutation. According to our classification, the c.323-6G>A mutation is (relatively) severe, which means that a homozygous c.323-6G>A mutation could be expected to result in prelingual (DFNB10) hearing impairment. However, a homozygous c.323-6G>A mutation has been described to be the underlying cause of postlingual (DFNB8) hearing impairment by Veske et al. (1996). When the Berkeley Drosophila Genome Project Splice Site Prediction Program (http://www.fruitfly.org/seq_tools/splice.html) was used, the c.323G>A mutation is predicted to introduce a novel splice acceptor site with a score of 0.96 (on a scale from 0 to 1) in addition to the normal splice acceptor site (score of 0.94). Therefore, both normal and abnormal splicing may occur in relative amounts of transcripts that vary between individuals, which may then lead to phenotypic variation. Testing this hypothesis on patient samples is not possible since the level of transcription of TMPRSS3 is low in blood cells.
We can conclude that our study, combined with previous data, suggests the classification of TMPRSS3 mutations into relatively mild and severe, which are associated with DFNB8 (postlingual hearing impairment) or DFNB10 (prelingual hearing impairment), respectively. Furthermore, our study suggests that compound heterozygosity for a mild and severe mutation leads to postlingual hearing impairment. The intrafamilial variation indicates that these data do not allow prediction of the phenotypic outcome for individual cases. Analysis of more families is necessary to confirm our conclusions and to address whether a subclassification of the mutations associated with DFNB8 is possible. Also, other (epi)genetic and nongenetic factors are likely to influence the severity of the phenotype. This has to be considered especially for families with a single affected individual. For the families of this type in our study, these factors did not seem to have a major effect since the phenotype corresponded to the previously reported phenotype of patients with the same mutations.
Conclusion
In patients with progressive hearing impairment and a possible autosomal recessive mode of inheritance, TMPRSS3 mutations should be considered. The ski-slope audiogram configuration is suggestive for mutations in this gene. The age of onset and the rate of progression are variable. Our analyses suggest that mutations in TMPRSS3 can probably be classified as mild and severe mutations according to their phenotypic effect. Furthermore, our results demonstrate that cochlear implantation is a good treatment option for patients with TMPRSS3 mutations since satisfactory speech reception was observed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 86 kb)
(PDF 69 kb)
Acknowledgment
We thank the families for their participation. This work was financially supported by the Heinsius Houbolt foundation, RNID (GR36), Fonds NutsOhra (Project SNO-T-0702-102), EUROHEAR (LSHG-CT-2004-512063), The Oticon Foundation (09–3742), and ZonMW (40-00812-98-09047). We would like to thank Mrs. W. van Drunen for support in collecting families.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Nicole J. D. Weegerink, Margit Schraders, Hannie Kremer, and Henricus P. M. Kunst contributed equally.
Contributor Information
Nicole J. D. Weegerink, Email: N.Weegerink@kno.umcn.nl
Margit Schraders, Email: M.Schraders@antrg.umcn.nl.
Jaap Oostrik, Email: J.Oostrik@antrg.umcn.nl.
Patrick L. M. Huygen, Email: P.Huygen@kno.umcn.nl
Tim M. Strom, Email: TimStrom@helmholtz-muenchen.de
Susanne Granneman, Email: S.Granneman@antrg.umcn.nl.
Ronald J. E. Pennings, Email: R.Pennings@kno.umcn.nl
Hanka Venselaar, Email: H.Venselaar@antrg.umcn.nl.
Lies H. Hoefsloot, Email: L.Hoefsloot@antrg.umcn.nl
Mariet Elting, Email: M.Elting@vumc.nl.
Cor W. R. J. Cremers, Email: C.Cremers@kno.umcn.nl
Ronald J. C. Admiraal, Email: R.Admiraal@kno.umcn.nl
Hannie Kremer, Email: H.Kremer@antrg.umcn.nl.
Henricus P. M. Kunst, Phone: +31-24-3614450, FAX: +31-24-3540251, Email: H.Kunst@kno.umcn.nl
References
- Ahmed ZM, Li XC, Powell SD, Riazuddin S, Young TL, Ramzan K, Ahmad Z, Luscombe S, Dhillon K, MacLaren L, Ploplis B, Shotland LI, Ives E, Riazuddin S, Friedman TB, Morell RJ, Wilcox ER. Characterization of a new full length TMPRSS3 isoform and identification of mutant alleles responsible for nonsyndromic recessive deafness in Newfoundland and Pakistan. BMC Med Genet. 2004;5:24. doi: 10.1186/1471-2350-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yosef T, Wattenhofer M, Riazuddin S, Ahmed ZM, Scott HS, Kudoh J, Shibuya K, Antonarakis SE, Bonne-Tamir B, Radhakrishna U, Naz S, Ahmed Z, Riazuddin S, Pandya A, Nance WE, Wilcox ER, Friedman TB, Morell RJ. Novel mutations of TMPRSS3 in four DFNB8/B10 families segregating congenital autosomal recessive deafness. J Med Genet. 2001;38:396–400. doi: 10.1136/jmg.38.6.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bom SJ, De Leenheer EM, Lemaire FX, Kemperman MH, Verhagen WI, Marres HA, Kunst HP, Ensink RJ, Bosman AJ, Van CG, Cremers FP, Huygen PL, Cremers CW. Speech recognition scores related to age and degree of hearing impairment in DFNA2/KCNQ4 and DFNA9/COCH. Arch Otolaryngol Head Neck Surg. 2001;127:1045–1048. doi: 10.1001/archotol.127.9.1045. [DOI] [PubMed] [Google Scholar]
- Bonne-Tamir B, DeStefano AL, Briggs CE, Adair R, Franklyn B, Weiss S, Korostishevsky M, Frydman M, Baldwin CT, Farrer LA. Linkage of congenital recessive deafness (gene DFNB10) to chromosome 21q22.3. Am J Hum Genet. 1996;58:1254–1259. [PMC free article] [PubMed] [Google Scholar]
- Cremers C, Van Rijn P, Ter Haar B. Autosomal recessive progressive high-frequency sensorineural deafness in childhood. Arch Otolaryngol Head Neck Surg. 1987;113:1319–1324. doi: 10.1001/archotol.1987.01860120065010. [DOI] [PubMed] [Google Scholar]
- De Leenheer EM, Huygen PL, Coucke PJ, Admiraal RJ, Van Camp G, Cremers CW. Longitudinal and cross-sectional phenotype analysis in a new, large Dutch DFNA2/KCNQ4 family. Ann Otol Rhinol Laryngol. 2002;111:267–274. doi: 10.1177/000348940211100312. [DOI] [PubMed] [Google Scholar]
- Elbracht M, Senderek J, Eggermann T, Thurmer C, Park J, Westhofen M, Zerres K. Autosomal recessive postlingual hearing loss (DFNB8): compound heterozygosity for two novel TMPRSS3 mutations in German siblings. J Med Genet. 2007;44:e81. doi: 10.1136/jmg.2007.049122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinas I, Jones KR, Tessarollo L, Vigers AJ, Huang E, Kirstein M, de CDC, Coppola V, Backus C, Reichardt LF, Fritzsch B. Spatial shaping of cochlear innervation by temporally regulated neurotrophin expression. J Neurosci. 2001;21:6170–6180. doi: 10.1523/JNEUROSCI.21-16-06170.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guipponi M, Vuagniaux G, Wattenhofer M, Shibuya K, Vazquez M, Dougherty L, Scamuffa N, Guida E, Okui M, Rossier C, Hancock M, Buchet K, Reymond A, Hummler E, Marzella PL, Kudoh J, Shimizu N, Scott HS, Antonarakis SE, Rossier BC. The transmembrane serine protease (TMPRSS3) mutated in deafness DFNB8/10 activates the epithelial sodium channel (ENaC) in vitro. Hum Mol Genet. 2002;11:2829–2836. doi: 10.1093/hmg/11.23.2829. [DOI] [PubMed] [Google Scholar]
- Guipponi M, Toh MY, Tan J, Park D, Hanson K, Ballana E, Kwong D, Cannon PZ, Wu Q, Gout A, Delorenzi M, Speed TP, Smith RJ, Dahl HH, Petersen M, Teasdale RD, Estivill X, Park WJ, Scott HS. An integrated genetic and functional analysis of the role of type II transmembrane serine proteases (TMPRSSs) in hearing loss. Hum Mutat. 2008;29:130–141. doi: 10.1002/humu.20617. [DOI] [PubMed] [Google Scholar]
- Herter S, Piper DE, Aaron W, Gabriele T, Cutler G, Cao P, Bhatt AS, Choe Y, Craik CS, Walker N, Meininger D, Hoey T, Austin RJ. Hepatocyte growth factor is a preferred in vitro substrate for human hepsin, a membrane-anchored serine protease implicated in prostate and ovarian cancers. Biochem J. 2005;390:125–136. doi: 10.1042/BJ20041955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgert N, Smith RJ, Van CG. Forty-six genes causing nonsyndromic hearing impairment: which ones should be analyzed in DNA diagnostics? Mutat Res. 2009;681:189–196. doi: 10.1016/j.mrrev.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann K, Lindner TH. easyLINKAGE-Plus—automated linkage analyses using large-scale SNP data. Bioinformatics. 2005;21:3565–3567. doi: 10.1093/bioinformatics/bti571. [DOI] [PubMed] [Google Scholar]
- Hutchin T, Coy NN, Conlon H, Telford E, Bromelow K, Blaydon D, Taylor G, Coghill E, Brown S, Trembath R, Liu XZ, Bitner-Glindzicz M, Mueller R. Assessment of the genetic causes of recessive childhood non-syndromic deafness in the UK—implications for genetic testing. Clin Genet. 2005;68:506–512. doi: 10.1111/j.1399-0004.2005.00539.x. [DOI] [PubMed] [Google Scholar]
- Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA—a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- Kunst H, Huybrechts C, Marres H, Huygen P, Van Camp G, Cremers C. The phenotype of DFNA13/COL11A2: nonsyndromic autosomal dominant mid-frequency and high-frequency sensorineural hearing impairment. Am J Otol. 2000;21:181–187. doi: 10.1016/S0196-0709(00)80006-X. [DOI] [PubMed] [Google Scholar]
- Lee YJ, Park D, Kim SY, Park WJ. Pathogenic mutations but not polymorphisms in congenital and childhood onset autosomal recessive deafness disrupt the proteolytic activity of TMPRSS3. J Med Genet. 2003;40:629–631. doi: 10.1136/jmg.40.8.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masmoudi S, Antonarakis SE, Schwede T, Ghorbel AM, Gratri M, Pappasavas MP, Drira M, Elgaied-Boulila A, Wattenhofer M, Rossier C, Scott HS, Ayadi H, Guipponi M. Novel missense mutations of TMPRSS3 in two consanguineous Tunisian families with non-syndromic autosomal recessive deafness. Hum Mutat. 2001;18:101–108. doi: 10.1002/humu.1159. [DOI] [PubMed] [Google Scholar]
- Peters TA, Levtchenko E, Cremers CW, Curfs JH, Monnens LA. No evidence of hearing loss in pseudohypoaldosteronism type 1 patients. Acta Otolaryngol. 2006;126:237–239. doi: 10.1080/00016480500388893. [DOI] [PubMed] [Google Scholar]
- Petersen MB, Willems PJ. Non-syndromic, autosomal-recessive deafness. Clin Genet. 2006;69:371–392. doi: 10.1111/j.1399-0004.2006.00613.x. [DOI] [PubMed] [Google Scholar]
- Schraders M, Lee K, Oostrik J, Huygen PL, Ali G, Hoefsloot LH, Veltman JA, Cremers FP, Basit S, Ansar M, Cremers CW, Kunst HP, Ahmad W, Admiraal RJ, Leal SM, Kremer H. Homozygosity mapping reveals mutations of GRXCR1 as a cause of autosomal-recessive nonsyndromic hearing impairment. Am J Hum Genet. 2010;86:138–147. doi: 10.1016/j.ajhg.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott HS, Kudoh J, Wattenhofer M, et al. Insertion of beta-satellite repeats identifies a transmembrane protease causing both congenital and childhood onset autosomal recessive deafness. Nat Genet. 2001;27:59–63. doi: 10.1038/83768. [DOI] [PubMed] [Google Scholar]
- Shahin H, Walsh T, Rayyan AA, Lee MK, Higgins J, Dickel D, Lewis K, Thompson J, Baker C, Nord AS, Stray S, Gurwitz D, Avraham KB, King MC, Kanaan M. Five novel loci for inherited hearing loss mapped by SNP-based homozygosity profiles in Palestinian families. Eur J Hum Genet. 2010;18:407–413. doi: 10.1038/ejhg.2009.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics. 2010;11:548. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhaegen VJ, Mylanus EA, Cremers CW, Snik AF. Audiological application criteria for implantable hearing aid devices: a clinical experience at the Nijmegen ORL clinic. Laryngoscope. 2008;118:1645–1649. doi: 10.1097/MLG.0b013e31817b013a. [DOI] [PubMed] [Google Scholar]
- Veske A, Oehlmann R, Younus F, Mohyuddin A, Muller-Myhsok B, Mehdi SQ, Gal A. Autosomal recessive non-syndromic deafness locus (DFNB8) maps on chromosome 21q22 in a large consanguineous kindred from Pakistan. Hum Mol Genet. 1996;5:165–168. doi: 10.1093/hmg/5.1.165. [DOI] [PubMed] [Google Scholar]
- Vriend G. WHAT IF: a molecular modeling and drug design program. J Mol Graph. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-V. [DOI] [PubMed] [Google Scholar]
- Walsh T, Abu RA, SeJ A, Shahin H, Shepshelovich J, Lee MK, Hirschberg K, Tekin M, Salhab W, Avraham KB, King MC, Kanaan M. Genomic analysis of a heterogeneous Mendelian phenotype: multiple novel alleles for inherited hearing loss in the Palestinian population. Hum Genom. 2006;2:203–211. doi: 10.1186/1479-7364-2-4-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattenhofer M, Di Iorio MV, Rabionet R, Dougherty L, Pampanos A, Schwede T, Montserrat-Sentis B, Arbones ML, Iliades T, Pasquadibisceglie A, D’Amelio M, Alwan S, Rossier C, Dahl HH, Petersen MB, Estivill X, Gasparini P, Scott HS, Antonarakis SE. Mutations in the TMPRSS3 gene are a rare cause of childhood nonsyndromic deafness in Caucasian patients. J Mol Med. 2002;80:124–131. doi: 10.1007/s00109-001-0310-6. [DOI] [PubMed] [Google Scholar]
- Wattenhofer M, Sahin-Calapoglu N, Andreasen D, Kalay E, Caylan R, Braillard B, Fowler-Jaeger N, Reymond A, Rossier BC, Karaguzel A, Antonarakis SE. A novel TMPRSS3 missense mutation in a DFNB8/10 family prevents proteolytic activation of the protein. Hum Genet. 2005;117:528–535. doi: 10.1007/s00439-005-1332-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 86 kb)
(PDF 69 kb)