Abstract
One potentially important mechanism for regulating class Ia phosphoinositide 3-kinase (PI 3-kinase) activity is autophosphorylation of the p85α adapter subunit on Ser608 by the intrinsic protein kinase activity of the p110 catalytic subunit, as this downregulates the lipid kinase activity in vitro. Here we investigate whether this phosphorylation can occur in vivo. We find that p110α phosphorylates p85α Ser608 in vivo with significant stoichiometry. However, p110β is far less efficient at phosphorylating p85α Ser608, identifying a potential difference in the mechanisms by which these two isoforms are regulated. The p85α Ser608 phosphorylation was increased by treatment with insulin, platelet-derived growth factor, and the phosphatase inhibitor okadaic acid. The functional effects of this phosphorylation are highlighted by mutation of Ser608, which results in reduced lipid kinase activity and reduced association of the p110α catalytic subunit with p85α. The importance of this phosphorylation was further highlighted by the finding that autophosphorylation on Ser608 was impaired, while lipid kinase activity was increased, in a p85α mutant recently discovered in human tumors. These results provide the first evidence that phosphorylation of Ser608 plays a role as a shutoff switch in growth factor signaling and contributes to the differences in functional properties of different PI 3-kinase isoforms in vivo.
Numerous studies have documented the fundamental importance of class IA phosphoinositide 3-kinases (PI 3-kinases) for a multitude of cellular functions including cell survival, growth, proliferation, intermediary metabolism, and cytoskeletal rearrangements (7, 37, 45).
PI 3-kinases catalyze the transfer of phosphate to the 3′-OH position of inositol lipids to produce phosphatidylinositol-3,4-bisphosphate and phosphatidylinositol-3,4,5-trisphosphate (PIP3), which in turn act as second messengers by recruiting proteins containing pleckstrin homology (PH) domains to the plasma membrane to assemble signaling complexes (44). In addition to the lipid kinase activity, in vitro experiments have demonstrated that class I PI 3-kinases possess an intrinsic protein serine kinase activity (9, 15, 40, 42). This protein kinase activity has attracted much interest, but its functional consequences in vivo have not been defined (24).
The typical form of class IA PI 3-kinase is a heterodimer with an 85-kDa regulatory subunit and a 110-kDa catalytic subunit (37). Two isoforms of the 85-kDa regulatory subunit have been identified: p85α and p85β, which are products of different genes. Also, several splice variants of p85α exist. Furthermore, a third gene product, termed p55γ, has been identified. Three isoforms of the 110-kDa subunit have been identified in complex with p85: p110α, p110β, and p110δ. The α and β isoforms are widely expressed, whereas the δ isoform is expressed predominantly in leukocytes.
It is well recognized that the activity of class-Ia PI 3-kinase is regulated by a range of mechanisms acting via the various modular domains of the subunits. The p85 subunit comprises one SH3 and two SH2 domains, a BH (breakpoint cluster region homology) domain, and two proline-rich domains, all of which appear to contribute to the regulation. The SH3 domain binds and stimulates the activity of the GTP-binding protein dynamin (19), and it may also interact with proline-rich motifs within the p85 protein itself (20, 26). The small GTPases Rac and Cdc42 have been shown to interact with the BH domain and activate the lipid kinase as a result (5, 48).
The SH2 domains are of fundamental importance for the enzymatic function of PI 3-kinase, since they bind to phosphotyrosine residues of activated growth factor receptors and adapter molecules such as the platelet-derived growth factor (PDGF) receptors and the insulin receptor substrate IRS-1. In this manner, they dock the enzyme on the plasma membrane. Synthetic peptides corresponding to tyrosine-phosphorylated regions of either the PDGF receptor or IRS-1 have shown that these activate the enzyme lipid kinase activity (8, 29, 36, 47). However, this activation was evident only when phosphatidylinositol was used as a substrate and not with phosphatidylinositol-4,5-bisphosphate, which is considered to be the main physiological substrate of class I PI 3-kinase. Furthermore, no effect was observed on the protein kinase activity of the enzyme (29). In addition, tyrosine phosphorylation of the regulatory subunit has been reported to activate PI 3-kinase (11, 21, 22, 27, 46).
All p110 catalytic subunits contain a domain which interacts with activated Ras (34). In the case of p110α, this interaction has been shown to increase the lipid kinase activity above that caused by binding of the regulatory subunit to phosphotyrosine.
With respect to the negative regulation of the PI 3-kinase activity, it has been reported that the p85 C-terminal SH2 domain binds to the enzymatic product PIP3. PIP3 binding and tyrosine-phosphorylated peptide binding in the SH2 C-terminal domain have been shown to be mutually exclusive (32). Thus, a redistribution of PI 3-kinase in the plasma membrane, caused by the displacement of phosphotyrosine by PIP3, was suggested as a potential feedback mechanism for the regulation of enzymatic activity.
Perhaps the most profound effect on PI 3-kinase enzymatic activity is that caused by intersubunit serine phosphorylation. Indeed, it has been shown that the intrinsic protein kinase activity of PI 3-kinase is capable of phosphorylating the Ser608 on the p85α subunit in vitro at a stoichiometry of 1 mol of phosphate per mol of p85 (9, 15). Phosphorylation of Ser608 resulted in an 80% decrease in PI 3-kinase activity, which could subsequently be reversed upon treatment with protein phosphatase 2A.
So far, the physiological relevance of this phosphorylation has not been established, and there is currently no evidence that phosphorylated Ser608 exists in cells. In the present study, we demonstrate that p85α Ser608 can be phosphorylated both in cell lines and in animal tissues and that this phosphorylation is mainly mediated by the p110α isoform. Insulin and PDGF stimulate this phosphorylation, with the stoichiometry of Ser608 phosphorylation in the total cellular pool of p85α reaching 0.12 to 0.15 after insulin stimulation in the whole cells, although the stoichiometry will be significantly higher in the p85α/p110α pool. This provides evidence for a feedback mechanism involved in growth factor regulation of one of the major isoforms of PI 3-kinase. Finally, we demonstrate that impaired autophosphorylation of a human p85α deletion mutant closely correlates with constitutive activation of PI 3-kinase, which is responsible for the potentially oncogenic properties of this mutant. Overall, these data demonstrate that the protein kinase activity of PI 3-kinase must be considered an important component of the mechanisms regulating PI 3-kinase activity in vivo.
MATERIALS AND METHODS
Cell culture, transfection, and plasmids.
HEK 293, NIH 3T3, and p85α−/− mouse embryonic fibroblasts (kindly provided by D. Fruman, University of California) were cultured in Dulbecco's modified Eagle's essential medium (DMEM). CHO-IR cells were cultured in Ham F-12 nutrient medium. Media were supplemented with 10% fetal calf serum and antibiotics. HEK 293 cells were transfected by a method involving calcium phosphate precipitation (Invitrogen). CHO-IR cells were transfected using LipofectAMINE (Invitrogen) or Fugene-6 (Roche). Construction of Myc-tagged p85α and Flag-tagged p110α and p110β expression plasmids, as well as construction of V5-tagged wild-type and Δexon13 human p85α, has been described previously (4, 31). p85α Ser608 point mutations and the p110α R916P (kinase-dead) mutation were inserted by using the QuikChange site-directed mutagenesis kit (Stratagene). The accuracy of the mutations was verified by DNA sequencing. Recombinant baculoviruses were provided by M. D. Waterfield (Ludwig Institute for Cancer Research, University College London). A polyclonal anti-p85 antibody, raised against the N-terminal SH2 domain of bovine p85α, was generated in house. An anti-IRS-1 pre-CT polyclonal antibody was purchased from Upstate. The fluorescein isothiocyanate (FITC)-conjugated swine anti-rabbit immunoglobulin G (IgG) was from DAKO. Antibody 9E10 against the Myc tag was provided by David Quinn (University of Cambridge). The Flag-M2 monoclonal antibody was purchased from Sigma. Recombinant protein kinase CK2, human recombinant PDGF-BB, okadaic acid, and LY294002 were from Calbiochem. All other chemicals were obtained from Sigma and were of the highest grade available.
Generation of a phosphospecific Ser608 antibody.
Phosphorylation-state-specific antibodies against p85α Ser608 were generated by immunizing rabbits with the synthetic peptide CTEDQYSpLVED (where Sp stands for phosphoserine) conjugated to keyhole limpet hemocyanin by using m-maleimidobenzoyl-N-hydroxysuccinimide ester. The antiserum was purified by affinity chromatography. First it was passed through a column of the corresponding dephosphopeptide coupled to SulfoLink resin (Pierce) in order to remove clones reacting with nonphosphorylated Ser608. The flowthrough was further purified by passage through a second column made of the phosphopeptide coupled to SulfoLink. The purified antibody preparation was characterized by immunoblotting against in vitro-phosphorylated recombinant p85α/p110α heterodimers.
Protein expression, isolation, and analysis.
Recombinant p85α/p110α and p85α/p110β heterodimers were expressed in baculovirally infected Sf9 insect cells and purified by using an affinity matrix prepared by coupling the phosphopeptide YpVPMLG (where Yp stands for phosphotyrosine), corresponding to the Tyr751 of the human PDGF receptor-β, to Actigel ALD (Sterogene, Carlsbad, Calif.), according to the manufacturer's instructions. Assays were performed using Actigel-bound p85α/p110 complexes.
CHO-IR cells were serum starved for 16 h prior to insulin stimulation and then treated with 100 nM insulin for 10 min at 37°C. NIH 3T3 cells were serum starved for 16 h prior to PDGF stimulation and then treated with 50 ng of PDGF-BB/ml for 15 min at 37°C. Cells were washed once in phosphate-buffered saline (PBS) and lysed in a buffer containing 50 mM Tris-HCl (pH 7.4), 5 mM EDTA, 150 mM NaCl, 50 mM NaF, and 1% Triton X-100 supplemented with 2 μg of aprotinin/ml, 1 μM pepstatin, 1 ng of leupeptin/ml, 1 mM phenylmethylsulfonyl fluoride, and 1 mM sodium orthovanadate. Immunoprecipitations were performed from the Triton-soluble fraction by using the indicated antibodies. Dilutions were 1:1,000 for the polyclonal anti-p85 antibody, 1:100 for 9E10, 1 μg of the Flag-M2 antibody per lysate, and 2 μg of anti-IRS-1 antibody per lysate. Immune complexes were collected with protein G-agarose beads, washed with lysis buffer (three times) followed by kinase assay buffer (three times), and further processed for various assays. For experiments involving mouse tissue, 5 IU of human insulin (Sigma) was injected via the portal vein into overnight-fasted, terminally anesthetized mice. Livers were removed at 3 min after injection and instantly frozen in liquid nitrogen. Protein extraction from livers was performed by homogenization in the above-described lysis buffer. Preparation, incubation, and protein extraction of rat soleus muscles were performed as previously described (39).
Immunoblotting.
Isolated protein or immunoprecipitates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon P; Millipore). Blots were probed with the indicated antibodies, followed by a horseradish peroxidase-conjugated secondary antibody. Visualization was performed with enhanced chemiluminescence (ECL). Images were analyzed with a Fuji LAS-1000 Luminescent Image Analyser and Fuji Image Gauge software.
Protein kinase and PI 3-kinase lipid kinase assays.
PI 3-kinase autophosphorylation assays were performed as previously described (15). Reactions were run for 20 min at 25°C (or 1 h for Sf9-expressed heterodimers) and terminated by the addition of 4× electrophoresis sample buffer and boiling. Protein kinase CK2 kinase assays were performed as for PI 3-kinase by using 20 U of recombinant enzyme per assay and GTP instead of ATP as a phosphoryl donor. Reaction products were analyzed by SDS-PAGE and either autoradiography or immunoblotting with the phospho-Ser608 antibody. PI 3-kinase lipid kinase assays were performed as described elsewhere (17). Images of radiolabeled protein and lipid products were analyzed using a Fuji FLA-2000 phosphorimager and Fuji Image Gauge software.
Immunofluorescence and confocal microscopy.
For immunofluorescence staining, NIH 3T3 or p85α−/− fibroblasts growing on glass coverslips were serum starved for 20 h and then stimulated with 50 ng of PDGF-BB/ml for 15 min at 37°C. Cells were rinsed in ice-cold PBS, fixed in 4% paraformaldehyde in PBS for 10 min, and permeabilized with 0.2% Triton X-100 in PBS for 5 min. After a 1-h incubation in 5% bovine serum albumin (BSA) in PBS, cell monolayers were incubated with an affinity-purified anti-phospho-Ser608 antibody (10 μg/ml) in 1% BSA in Tris-buffered saline (TBS) for 16 h at 4°C, washed three times with TBS, and then incubated with an FITC-conjugated swine anti-rabbit IgG antibody (1/50) for 1 h at room temperature. The coverslips were washed and mounted on glass slides with Vectashield immunofluorescence medium (Vector Laboratories, Burlingame, Calif.). Images of cells were obtained by using a Zeiss LSM 510 confocal laser-scanning microscope (Welwyn, Garden City, United Kingdom) with the accompanying LSM 510 software and were processed in Adobe Photoshop.
Estimation of the stoichiometry of protein phosphorylation.
Recombinant p85α/p110α heterodimers isolated on phosphopeptide beads were allowed to phosphorylate in the presence of 1 mM ATP (plus 10 μCi of [γ32P]ATP) and 2 mM MnCl2 for 16 h at 25°C. The reaction was terminated by the addition of electrophoresis sample buffer. An aliquot of the mix was analyzed on an 8% acrylamide-SDS gel along with a range of BSA standards. Following electrophoresis, the gel was stained with Coomassie brilliant blue. The amount of p85 protein was estimated by comparing the intensity of the corresponding band with the BSA standards. The band was excised from the gel, and incorporation of radioactivity was measured by Cherenkow counting. Samples of recombinant p85α/p110α heterodimers, for which the stoichiometry of phosphorylation had been calculated as described above, were used as standards in the same immunoblots with experimental samples. Equal amounts of recombinant p85α/p110α, which had not been left to autophosphorylate in vitro, were also loaded to account for the fraction of the heterodimers which had already been phosphorylated by their coexpression in insect cells. The stoichiometry of the phosphorylation of the experimental samples was estimated by comparing the intensity of the signal in immunoblots with that of the recombinant p85α standard after normalizing for total p85 expression. All quantitations were performed using Image Gauge software (Fuji).
RESULTS
Characterization of a phosphospecific antibody for p85α Ser608.
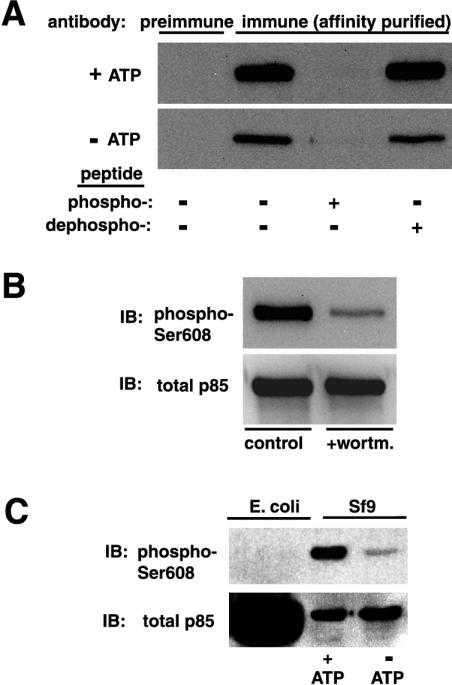
To study Ser608 phosphorylation in vivo, we generated a phosphorylation-state-dependent antibody, which was affinity purified as described in Materials and Methods. The affinity-purified antibody preparation was characterized by immunoblotting against recombinant p85α/p110α which was allowed to autophosphorylate in vitro in the presence of ATP and Mn2+. Figure 1A shows that the affinity-purified anti-phospho-Ser608 antibody strongly recognized p85α after in vitro phosphorylation in the presence of ATP. Antibody binding was fully competed by the corresponding phosphopeptide but not by the dephosphopeptide. Furthermore, p85α/p110α incubated in the presence of the PI 3-kinase inhibitor wortmannin during the kinase assays was recognized to a much lesser extent (Fig. 1B). Although the affinity-purified antibody reacted to a low extent with p85α incubated in the absence of ATP in all of the above experiments, it seems that this was due to in vivo prephosphorylation resulting from coexpression with the catalytic subunit in insect cells, since no binding was evident when an excessive amount of Escherichia coli-expressed p85α was used instead (Fig. 1C).
FIG. 1.
Characterization of the phospho-Ser608-specific antibody. Recombinant p85α/p110α was subjected to an in vitro kinase assay in the presence or the absence of ATP. Reaction products were analyzed by SDS-PAGE and transferred to a PVDF membrane. (A) Blots were incubated either with preimmune serum or with the affinity-purified anti-phospho-Ser608 antibody diluted 1:1,000. In some experiments, the immune serum was incubated with the immunizing phosphopeptide or the corresponding dephosphopeptide at 1 mM for 1 h at 25°C before further processing. (B) In some experiments, wortmannin (wortm.) at 100 nM was included during the kinase assay. IB, immunoblot. (C) Recombinant p85α/p110α which had been subjected to an in vitro autokinase assay in the presence or absence of ATP (Sf9) or bacterially expressed p85α (E. coli) was analyzed by immunoblotting using the affinity-purified anti-phospho-Ser608 antibody.
PI 3-kinase catalytic subunits phosphorylate p85α Ser608 in vivo.
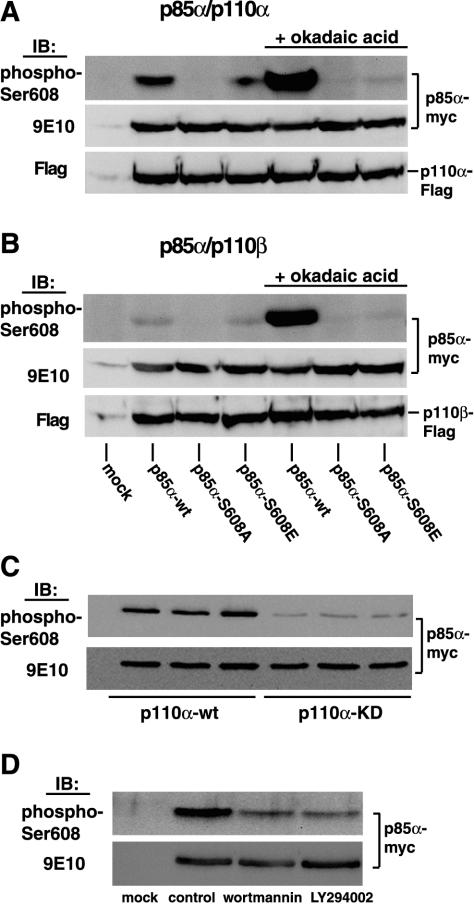
Subsequently, we used these reagents to investigate phosphorylation of p85 on Ser608 in vivo, by cotransfecting HEK 293 cells with each of the p85α Ser608 mutants and either the α or the β isoform of the p110 catalytic subunit. Lysates from the transfected cells were immunoprecipitated with antibody 9E10 followed by immunoblotting with the anti-phospho-Ser608 antibody. As shown in Fig. 2A, the Myc-tagged wild-type p85α cotransfected with the Flag-tagged p110α catalytic subunit was strongly recognized by the phosphospecific antibody. No signal was obtained with the Ser608Ala mutant, whereas some binding was evident with the phosphomimetic mutant Ser608Glu. Furthermore, treatment of cells with the phosphatase inhibitor okadaic acid resulted in strong antibody binding to wild-type p85α, consistent with an accumulation of phosphate. However, there was no increase in reactivity with the Ser608 mutants. When the same p85α mutants were coexpressed with the p110β catalytic subunit, wild-type p85 was barely recognized by the phosphospecific antibody, although okadaic acid treatment elevated the phosphorylation to nearly the same level as that in p110α (Fig. 2B). This result implies that p110β phosphorylates p85α with much less efficiency than p110α. This is consistent with the previous finding that p110β is a less efficient protein kinase than p110α (4). It is also consistent with the finding that in an in vitro autokinase assay, recombinant p110β preferentially autophosphorylates instead of phosphorylating the p85 regulatory subunit (17). The site of p110β autophosphorylation has recently been mapped as Ser1070 (12). Also, when we coexpressed the p85α subunit with a catalytically inactive p110α, recognition by the phosphospecific antibody was barely observed (Fig. 2C). Moreover, treatment of cells expressing p85α/p110α with the PI 3-kinase inhibitor wortmannin or LY294002 substantially reduced Ser608 phosphorylation (Fig. 2D). These results demonstrate that at least in the basal state, phosphorylation of Ser608 is mainly due to the autokinase activity.
FIG. 2.
Phosphorylation of Ser608 after coexpression with either p110α or p110β in HEK 293 cells. (A) Myc-tagged p85α (wild type or Ser608 mutants) was coexpressed with Flag-tagged p110α in HEK 293 cells. Cells were lysed, and lysates were immunoprecipitated with antibody 9E10. Immunoprecipitates were analyzed by SDS-PAGE and immunoblotting with the anti-phospho-Ser608 antibody. In some experiments, cells were treated with 500 nM okadaic acid for 45 min at 37°C prior to lysis. Uniform expression of Myc-tagged p85α and Flag-tagged p110 was confirmed after stripping of the blot and reprobing with antibody 9E10 and the Flag-M2 antibody, respectively (lower panels). (B) The same experiments as those described for panel A, except that p85α was cotransfected with p110β. (C) Cells were transfected with Myc-tagged p85α and either wild-type Flag-tagged p110α (p110α-wt) or catalytically inactive Flag-tagged p110α (p110α-KD [kinase dead]) and were further processed as described above. IB, immunoblot. (D) Cells were cotransfected with Myc-tagged p85α and Flag-tagged p110α. Transfected cells were treated with either 100 nM wortmannin or 30 μM LY294002 for 45 min at 37°C and were further processed as described above.
Insulin and PDGF stimulate Ser608 phosphorylation in cultured cells.
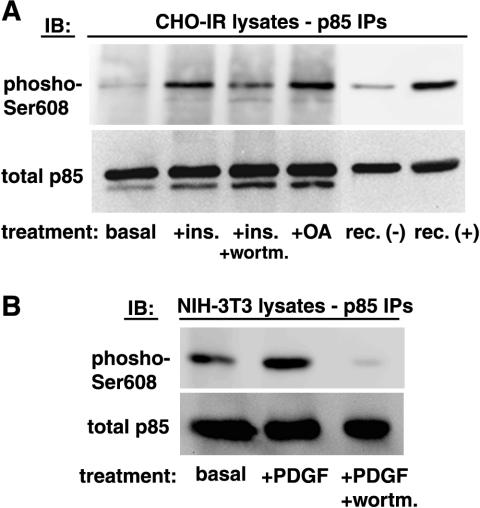
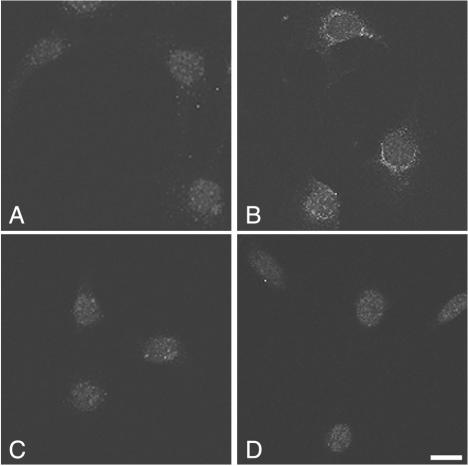
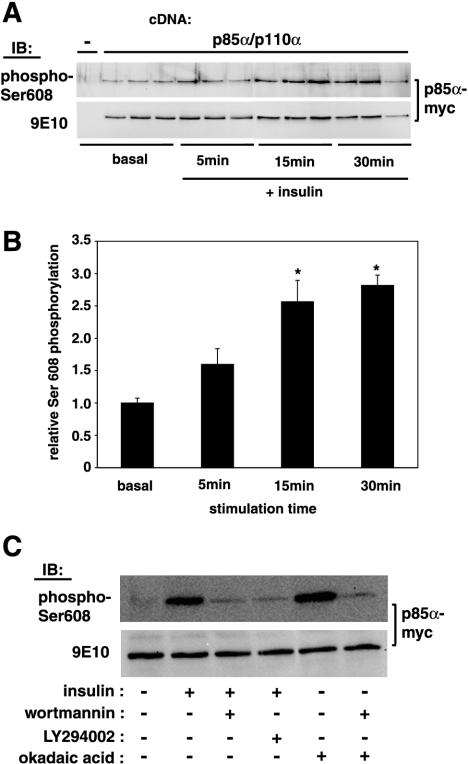
We sought to determine a possible effect of insulin and PDGF, two well-known activators of PI 3-kinase, on p85α Ser608 phosphorylation. CHO-IR cells were treated with 100 nM insulin for 30 min in the presence or absence of wortmannin. Cells were lysed, the lysates were immunoprecipitated with anti-p85 antibodies, and the immunoprecipitates were analyzed by immunoblotting with the anti-phospho-Ser608 antibody. As shown in Fig. 3A, the antibody detected Ser608 phosphorylation on endogenous p85 at the basal level. Insulin treatment caused a sixfold increase in Ser608 phosphorylation. Treatment of cells with wortmannin prior to insulin stimulation reduced Ser608 phosphorylation by half, whereas treatment with okadaic acid elevated Ser608 phosphorylation to the same level as insulin stimulation. Moreover, we were able to detect endogenous p85 Ser608 phosphorylation in 3T3 fibroblasts both by immunoblotting (Fig. 3B) and by indirect immunofluorescence (Fig. 4). In these cells, PDGF stimulation caused an approximately threefold increase in Ser608 phosphorylation, which was almost completely abolished by treatment with wortmannin (Fig. 3B). The PDGF-stimulated increase in Ser608 phosphorylation was evident also by immunofluorescence (Fig. 4). Ser608-phosphorylated p85 appeared to accumulate in a perinuclear site (Fig. 4B) and colocalized well with staining for total p85α (data not shown). The PDGF-induced increase in immunoreactivity was sensitive to treatment with LY294002 (Fig. 4C) and wortmannin (data not shown). The specificity of the anti-phospho-Ser608 antibody in immunofluorescence detection was further tested by using p85α-deficient fibroblasts (Fig. 4D). In the latter case, only a background nuclear staining, which likely represents nonspecific binding to a nuclear component, was detected. In addition to its occurrence on endogenous p85, Ser608 phosphorylation occurred in CHO-IR cells transfected with p85 α/p110 α heterodimers. As shown in Fig. 5A, insulin treatment elevated the level of Ser608 phosphorylation in a time-dependent manner, with a ca. 2.7-fold increase being evident after a 30-min stimulation (Fig. 5B). The insulin-induced Ser608 phosphorylation in these cells was much diminished by treatment of the cells with either wortmannin or LY294002 prior to stimulation (Fig. 5C). The elevation of Ser608 phosphorylation levels caused by treatment of cells with okadaic acid was also wortmannin sensitive. These data indicate that PI 3-kinase activity is necessary for both insulin- and okadaic acid-stimulated Ser608 phosphorylation in CHO-IR cells.
FIG. 3.
Inducible p85 Ser608 phosphorylation in cultured cells. (A) CHO-IR cells were serum starved for 16 h and then stimulated with 100 nM insulin (ins.) for 30 min at 37°C. In some experiments, cells were treated with 100 nM wortmannin (wortm.) for 30 min prior to insulin stimulation or with 500 nM okadaic acid (OA) for 30 min. Following stimulation, cells were lysed, and each lysate (2 mg of total protein) was immunoprecipitated with polyclonal anti-p85 antibodies. Immunoprecipitates (IPs) were analyzed by SDS-PAGE and immunoblotting with the anti-phospho-Ser608 antibody, followed by stripping of the blot and reprobing with an antibody against total p85. IB, immunoblot; rec (+) and rec (−), in vitro-phosphorylated and nonphosphorylated recombinant p85α/p110α, respectively. (B) NIH 3T3 fibroblasts were serum starved for 16 h and then stimulated with 50 ng of PDGF-BB/ml for 15 min at 37°C. In some experiments, cells were treated with 100 nM wortmannin for 30 min prior to PDGF stimulation. Cells were lysed, and the lysates (2 mg of total protein) were further processed as described above.
FIG. 4.
Immunofluorescence detection of Ser608-phosphorylated p85α. NIH 3T3 cells (A, B, and C) or p85α−/− fibroblasts (D) were serum starved for 20 h. Cells were either left untreated (A), stimulated with 50 ng of PDGF-BB/ml for 15 min at 37°C (B and D), or treated with 10 μM LY294002 for 30 min and then stimulated with PDGF (C). Following stimulation, cells were fixed, permeabilized, and incubated with affinity-purified rabbit anti-phospho-Ser608 antibodies. Antibody binding was revealed by using FITC-conjugated anti-rabbit IgG. Images were acquired using a confocal microscope. Bar, 10 μm.
FIG. 5.
Insulin stimulates phosphorylation of Ser608 in a time-dependent and PI 3-kinase inhibitor-sensitive manner. CHO-IR cells were cotransfected with Myc-tagged p85α and Flag-tagged p110α. Cells were serum starved for 16 h, followed by stimulation with 100 nM insulin at 37°C for the indicated lengths of time. Cell were lysed and immunoprecipitated with antibody 9E10, and the immunoprecipitates were analyzed by SDS-PAGE and immunoblotting with the anti-phospho-Ser608 antibody, followed by stripping of the blot and reprobing with an antibody against total p85. (A) Results of a representative experiment in triplicate are shown. (B) Phosphorylation of Ser608 was quantitated and plotted in a graph. Values are means ± standard errors of the means; *, P < 0.01 for insulin-stimulated versus basal phosphorylation. (C) Transfected CHO-IR cells were treated with either 100 nM wortmannin, 30 μM LY294002, or 500 nM okadaic acid as indicated for 30 min at 37°C, followed by a 10-min stimulation with 100 nM insulin, lysis, and immunoprecipitation with antibody 9E10. Immunoprecipitates were processed as described for panel A.
Insulin induces p85α Ser608 phosphorylation in animal tissues in vivo.
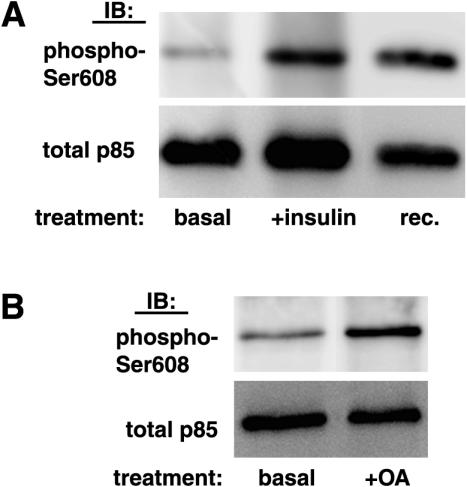
To assess whether p85α Ser608 is a physiological target for phosphorylation, protein extracts from livers of mice injected with a bolus of insulin or saline via the portal vein were analyzed for p85α Ser608 phosphorylation by immunoprecipitation with an anti-p85 antibody and immunoblotting with the anti-phospho-Ser608 antibody. As shown in Fig. 6A, substantial p85α Ser608 phosphorylation could be detected at the basal level in mouse liver protein extracts. Importantly, insulin stimulated Ser608 phosphorylation approximately 2.3-fold. Furthermore, p85 α Ser608 phosphorylation was detectable in the basal state in rat soleus muscle lysates and was elevated after treatment with okadaic acid (Fig. 6B).
FIG. 6.
Detection of inducible p85α Ser608 phosphorylation in animal tissues. (A) Protein extracts from livers of mice injected with a bolus of insulin or saline, as described in Materials and Methods, were immunoprecipitated with anti-p85 antibodies. The immunoprecipitates were analyzed by SDS-PAGE and immunoblotting with the anti-phospho-Ser608 antibody, followed by stripping of the blot and reprobing with an antibody against total p85. IB, immunoblot; rec., in vitro phosphorylated recombinant p85α/p110α. (B) Rat soleus muscle strips were isolated and incubated in the presence or absence of 500 nM okadaic acid (OA) for 45 min. Following incubations, muscle strips were snap-frozen and homogenized, and the homogenates were analyzed as described for panel A.
p85α Ser608 becomes phosphorylated at a substantial stoichiometry in vivo.
Previous studies have shown that in an in vitro autokinase assay with recombinant p85α/p110α, phosphorylation occurs exclusively on Ser608 (15). Therefore, in order to obtain an approximate estimate of the stoichiometry of Ser608 phosphorylation in vivo, we calculated the stoichiometry of phosphorylation of recombinant p85α/p110α heterodimers in vitro as described in Materials and Methods, assuming that the vast majority of incorporated 32P counts would represent phosphorylation at Ser608. Under these conditions, we estimated a stoichiometry of 0.21 mol of phosphate per mol of p85α. This is very different from the 1:1 stoichiometry reported previously (15). Most likely, the reason for this discrepancy is the fact that in insect cells, p85α is expressed at much higher levels than p110α, so that isolation of the heterodimers by p85α pulldown results in an excess of p85α over p110, whereas in the previous study, the measurements were performed on p110 immunoprecipitates, which ensured a 1:1 ratio of p85α to p110α. Subsequently, we used recombinant heterodimers with a known stoichiometry of p85α phosphorylation as standards on the same immunoblots with our experimental samples (see Materials and Methods). In this manner, we estimated that in CHO-IR cells (Fig. 3A), the stoichiometry of p85α rises from approximately 0.02 mol of phosphate per mol of p85α at the basal state to approximately 0.12 after insulin stimulation. In mouse livers (Fig. 6A), the stoichiometry of p85α phosphorylation increased from 0.065 mol of phosphate per mol of p85α at the basal state to 0.15 after insulin stimulation. The stoichiometry in the p110α-associated pool of p85 will be significantly higher, since the p110β-associated pool is unlikely to be highly phosphorylated. Further, a significant proportion of p85 in the cell may be monomeric and free of p110 (30, 41), and this pool is also likely to have low levels of Ser608 phosphorylation.
Ser608-phosphorylated p85α associates with signaling complexes.
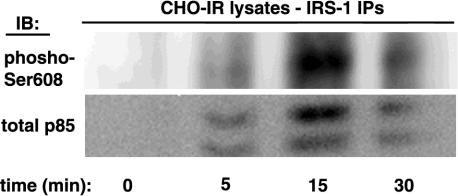
Given the ability of insulin and PDGF to stimulate p85α Ser608 phosphorylation, we then examined whether Ser608-phosphorylated p85α associates with signaling complexes. CHO-IR cells were stimulated with 100 nM insulin for various lengths of time and then lysed; the lysates were immunoprecipitated with anti-IRS-1 antibodies and further analyzed by electrophoresis and immunoblotting with the anti-phospho-Ser608 antibody. As shown in Fig. 7, association of p85 with IRS-1 was maximal 15 min after stimulation. Importantly, Ser608 phosphorylation could also be detected in IRS-1 immunoprecipitates and followed the same pattern as p85 association.
FIG. 7.
Association of Ser608-phosphorylated p85α with signaling complexes. CHO-IR cells were serum starved for 16 h and then stimulated with 100 nM insulin at 37°C for the indicated times. Following stimulation, cells were lysed, and each lysate (4 mg of total protein) was immunoprecipitated with polyclonal anti-IRS-1 antibodies. Immunoprecipitates (IPs) were analyzed by SDS-PAGE and immunoblotting using the anti-phospho-Ser608 antibody, followed by stripping of the blot and reprobing with an antibody against total p85. IB, immunoblot.
Protein kinase CK2 phosphorylates Ser608 on free but not on p110α-associated p85α.
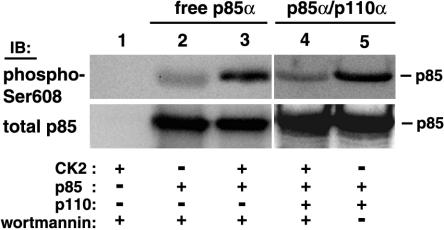
We also addressed the possibility that cellular kinases other than PI 3-kinase may be able to phosphorylate Ser608 and thus regulate PI 3-kinase activity. The Ser608 site is surrounded by acidic residues and fits very well within the consensus target motif for protein kinase CK2 (1). Indeed, we have found that protein kinase CK2 phosphorylates a peptide encompassing residues 598 to 616 of p85α (data not shown). Protein kinase CK2 activity can be distinguished from PI 3-kinase by the fact that the former is able to use GTP as a phosphoryl donor and is insensitive to wortmannin (13) (also, we have found that wortmannin concentrations up to 1 μM do not affect protein kinase CK2 activity). Under these conditions, we demonstrated that protein kinase CK2 phosphorylates free recombinant p85α on Ser608 but does not phosphorylate p85α when it is dimerized with p110α (Fig. 8).
FIG. 8.
Protein kinase CK2 phosphorylates p85α on Ser608 when p85α is free but not when it is complexed with p110α. Recombinant free p85α (lanes 2 and 3) or p85α/p110α heterodimers (lanes 4 and 5) isolated on Tyr751-phosphopeptide beads and GTP were used as substrates in recombinant protein kinase CK2 in vitro kinase assays in the presence of 100 nM wortmannin (lanes 1 to 4). Reaction products were analyzed by SDS-PAGE and immunoblotting using the anti-phospho-Ser608 antibody, followed by stripping of the blot and reprobing with an antibody against total p85. An autophosphorylation assay with ATP as a substrate in the absence of wortmannin was also performed as a positive control (lane 5).
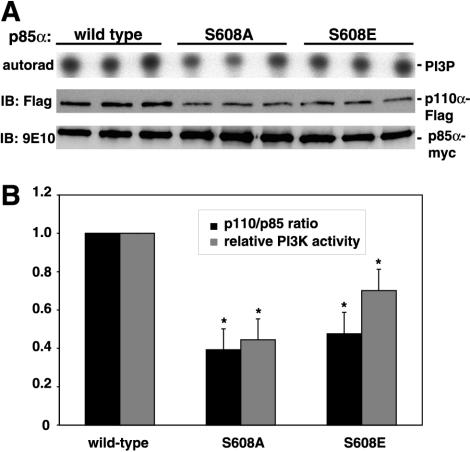
Mutation of Ser608 affects enzymatic activity and heterodimerization.
As p85α Ser608 phosphorylation has been shown to downregulate the lipid kinase activity of the enzyme in vitro (15), we attempted to assess whether phosphorylation at this site in vivo would have similar effects. For this purpose, wild-type Myc-tagged p85α and variants in which Ser608 was mutated to alanine or glutamic acid were coexpressed with the p110α catalytic subunit in HEK 293 cells. Recombinant heterodimers were isolated by immunoprecipitation with antibody 9E10 and subjected to in vitro kinase assays using [γ-32P]ATP and phosphatidylinositol as the substrates. As expected, the phosphomimetic mutant Ser608Glu was found to be less active than the wild type (Fig. 9). However, rather surprisingly, the same effect was observed with the Ser608Ala mutant, which would be expected to mimic dephosphorylated p85. Most interestingly, when the same immunoprecipitates were analyzed by immunoblotting to normalize for p85 and p110 expression, it became evident that for both the Ser608Ala mutants and the Ser608Glu mutants, the amount of the associated p110 subunit was markedly less than that for the wild-type p85α. The apparent mechanism of the reduced lipid kinase activity demonstrated by this experiment is further discussed below.
FIG. 9.
Mutation of Ser608 interferes with lipid kinase activity and heterodimer formation. Each of the Ser608 mutants of Myc-tagged p85α was coexpressed together with Flag-tagged p110α in HEK 293 cells, and the heterodimers were isolated by immunoprecipitation with antibody 9E10. Immunoprecipitates were split in two. One half of each immunoprecipitate was assayed for lipid kinase activity by using phosphatidylinositol and [γ-32P]ATP as the substrates. Reaction products were analyzed by thin-layer chromatography and autoradiography (autorad). The other half was assayed by immunoblotting with antibody 9E10, followed by stripping of the blot and reprobing with an anti-Flag antibody. (A) A representative experiment. IB, immunoblot. (B) Data from three experiments were plotted in a graph. Values are means ± standard errors of the means; *, P < 0.05 for the mutant versus the wild type.
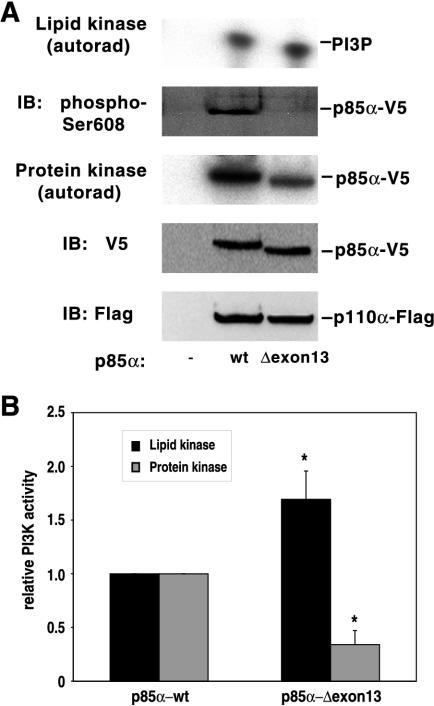
Impaired Ser608 phosphorylation in a human oncogenic p85α mutant.
Finally, we sought to decipher the basis of the constitutive activation of a PI 3-kinase comprising a p85α deletion mutant present in human primary colon and ovarian tumors and cancer cell lines (31). In this mutant, which results from the disruption of a splice site on one allele of the p85α gene, leading to the skipping of exon 13 during transcription, amino acids Met582 to Asp605 are replaced with a single Ile residue. The proximity of the deleted region to Ser608 suggested a possible interference with regulatory Ser608 autophosphorylation (31). To test this hypothesis, we coexpressed the V5-tagged cDNA of mutant p85 (p85Δexon13) together with Flag-tagged p110α in HEK 293 cells. Lysates were immunoprecipitated with an anti-V5 antibody, and the immunoprecipitates were assayed for Ser608 phosphorylation and lipid kinase activity (Fig. 10A). Immunoblot analysis of the immunoprecipitates with the phosphospecific antibody failed to detect any Ser608 phosphorylation of the p85Δexon13 mutant. However, the anti-phospho-Ser608 antibody has been developed against a peptide with the sequence CTEDQYSpLVED, and the exon 13 deletion changes this sequence to YLIQYSpLVED. Since replacement of the acidic motif TED with the hydrophobic motif YLI at the N terminus of this region could affect binding of some of the immunoglobulins present in the polyclonal antisera, we employed a [γ-32P]ATP autokinase assay to assess autophosphorylation. Indeed, we found that the autokinase activity of the p85Δexon13 mutant was severely impaired. A 70% decrease in the autophosphorylation of the p85Δexon13 mutant relative to that of wild-type p85α was accompanied by a corresponding increase in the lipid kinase activity (Fig. 10B). Thus, in this case, it seems very plausible that constitutive activation of PI 3-kinase is the consequence of defective Ser608 autophosphorylation.
FIG. 10.
An oncogenic human p85α deletion mutant displays impaired autophosphorylation and elevated lipid kinase activity. (A) Either V5-tagged wild-type p85α (wt) or the deletion mutant (Δexon13) was coexpressed with Flag-tagged p110α in HEK 293 cells. Cells were lysed, and lysates were immunoprecipitated with the anti-V5 antibody. Immunoprecipitates were split into four aliquots. One was assayed for lipid kinase activity by using phosphatidylinositol and [γ-32P]ATP as the substrates. Reaction products were analyzed by thin-layer chromatography and autoradiography (autorad). Another aliquot was subjected to an in vitro autokinase assay in the presence of [γ-32P]ATP and Mn2+. Reaction products (protein kinase) were analyzed by SDS-PAGE and autoradiography. Another aliquot was assayed for Ser608 phosphorylation by immunoblotting (IB) with the phosphospecific antibody (phospho-Ser608). The last aliquot was assayed by immunoblotting with the anti-V5 antibody, followed by stripping of the blot and reprobing with the anti-Flag antibody. (A) A representative experiment. (B) Data from three experiments were plotted in a graph. Values are means ± standard errors of the means; *, P < 0.05 for the mutant versus the wild type.
DISCUSSION
Despite a large body of data concerning the regulation of the heterodimeric p85/p110 PI 3-kinase, the precise mechanisms by which this regulation is achieved in a cellular context have still not been fully defined. Potentially important, although overlooked, features of the p85/p110α PI 3-kinase are its ability to operate as a dual-specificity kinase, phosphorylating both lipids and its regulatory subunit, and the negative impact that phosphorylation of the latter has on its lipid kinase activity. In the present study, we have investigated serine autophosphorylation of p85α in cells and its implications for the regulation of the lipid kinase activity of the enzyme. To date, the only evidence for such a phosphorylation has been obtained from in vitro experiments, and the finding that protein kinase activity required Mn2+ in vitro had been cited as evidence that in vivo phosphorylation was unlikely. However, our findings prove that the phosphorylation does occur in a cellular context, both in cell lines and in animal tissue in vivo, at a substantial stoichiometry and in a hormone- and growth factor-inducible manner. However, higher stoichiometries are almost certainly achieved in the p85α/p110α pool, as Ser608 phosphorylation is likely to be very low in the free p85α pool or in p85α associated with p110β. The finding that phosphorylation is to a large extent blocked by wortmannin and by coexpression of a catalytically inactive p110α indicates that the intrinsic kinase activity of p85/p110 heterodimers contributes a large part of this in vivo phosphorylation. However, some wortmannin-resistant phosphorylation is always observed, so it seems possible that other cellular kinases such as protein kinase CK2 can phosphorylate this site in vivo. Our data would indicate that this could happen only when heterodimers are dissociated and p85 is free, but it now appears that significant amounts of free p85 are indeed present in cells (30, 41).
The finding that insulin and PDGF, two well-known activators of PI 3-kinase, stimulate the phosphorylation of Ser608 provides evidence for an autoregulatory mechanism which is likely to represent a feedback mechanism to shut off the hormone or growth factor signal specifically to the p110α pool of class IA PI 3-kinase. This would be the first mechanism described for differential regulation of class Ia PI 3-kinase isoforms, but it is likely to have physiological significance given the differences in kinetic (4) and functional (2, 33, 43) properties described for the different PI 3-kinase isoforms.
The mechanism for the insulin- or PDGF-induced increase in phosphorylation at Ser608 is unlikely to involve stimulation of the protein kinase activity of p110, as it has previously been found that engagement of the p85 SH2 domains does not lead to an increase in the protein kinase activity of p110 (29). Our evidence suggests that this increase in Ser608 phosphorylation is also unlikely to involve exogenous kinases such as protein kinase CK2, which would recognize the serine in such an acidic motif. Although there is some evidence that protein kinase CK2 is activated by insulin (16, 38), we did not observe such an activation in CHO-IR cells (data not shown), and we find that CK2 only poorly phosphorylates p85 in heterodimers. Further, the effects of both insulin and okadaic acid are blocked by PI 3-kinase inhibitors, indicating that exogenous kinases are probably not involved. The fact that the kinetics of the increase in Ser608 phosphorylation are slow suggests that inhibition of phosphatases is a more likely mechanism. It is known that insulin acutely inhibits the activity of protein phosphatase 2A (6), with the maximal effect observed after 20 min of insulin stimulation. This correlates very well with our data, where phosphorylation of Ser608 reached a peak after 30 min of insulin stimulation. Phosphatase inhibition would also explain why both p110α and p110β ultimately reach the same level of phosphorylation, despite the fact that p110β is a less efficient kinase. Further, we find that the protein phosphatase 2A inhibitor okadaic acid greatly increases Ser608 phosphorylation, which fits with reports that PI 3-kinase activity is inhibited by okadaic acid (35).
Three mechanisms by which Ser608 phosphorylation might regulate PI 3-kinase activity can be envisaged: (i) regulation of the catalytic activity of p110 within intact heterodimers, (ii) modulation of p85 interactions with p110, and (iii) modulation of the interaction of p85 with tyrosine-phosphorylated peptide motifs. Our finding that Ser608-phosphorylated p85 is recruited to IRS-1 following insulin stimulation indicates that only the two former mechanisms are likely to operate in vivo.
Ser608 resides in the inter-SH2 domain of p85. This is predicted to be an independently folded module of a coiled-coil of two long antiparallel α-helices, and a motif mapped within this region binds the p110 subunit (14). A recent electron paramagnetic resonance spectroscopic study has confirmed the above prediction (18). The data of this study support a model in which the inter-SH2 domain serves as a rigid tether for p110 rather than a conformational switch. It is reasonable to hypothesize that events such as reversible phosphorylation in this domain could interfere with intersubunit interactions. To this end, we tested the effect of mutation of Ser608 on the lipid kinase activity. Mutation of Ser608 to the phosphomimetic glutamic acid resulted in the expected reduction in lipid kinase activity. Surprisingly, mutation of Ser608 to alanine had the same effect. At present, precise information regarding structural features of this region of p85 is lacking. Thus, we can only speculate that the Ser608 residue might be of special importance for the conformation of the p85 protein and that replacement by another residue, with different bonding properties, might have a profound impact on the conformation. In addition, a marked reduction in the association of the p110 catalytic subunit with the p85 mutants was also evident, indicating that Ser608 is crucial for tight intersubunit interaction. This observation provides a plausible explanation for the reduced lipid kinase activity associated with mutation of this site: the reduction is effected by promoting the dissociation of the heterodimer. Further, our data indicate that when p85 is free, Ser608 becomes the target for other cellular kinases, and these may affect re-formation of the heterodimers. Since free p110 is unstable in mammalian cells (23, 28), this would eventually result in a buildup of free p85, a situation that is indeed observed in several cell types (30, 41).
The crucial role of the adapter subunit in regulating PI 3-kinase function is demonstrated by the effects of various mutations of p85α that have been identified in humans. For example, a mutation which causes impairment of PI 3-kinase function and is associated with insulin resistance has been identified previously (3). Conversely, several mutations which activate PI 3-kinase and have oncogenic properties have been identified in p85α. The first oncogenic p85 mutant to be identified was the product of a C-terminal deletion in which the region following the first 571 amino acids of p85α had been replaced by a 24-amino acid tail, related to the eph receptor tyrosine kinase family (25). This mutant (p65-PI3K) induced constitutive activation of PI 3-kinase and was capable of transforming NIH 3T3 fibroblasts in culture. More recently, further studies have defined the importance of this deletion, showing that it contains a modular inhibitory domain which confers on wild-type p85 the ability to block the stimulatory effect of Ras (10). Therefore, our results suggest that any alterations in the region of Ser608, either by mutation or by phosphorylation, can enhance the inhibitory properties of this domain.
Another oncogenic p85 mutant was identified in primary colon and ovarian tumors (31). In this case, the region comprising amino acids 582 to 605 had been deleted and replaced by a single isoleucine residue. Again, the mutated PI 3-kinase was shown to exhibit elevated activity, thus causing constitutive activation of PKB/Akt. We tested the hypothesis that the proximity of the deletion to Ser608 interferes with its phosphorylation. Indeed, we found that phosphorylation of Ser608 was reduced and that this reduction was accompanied by an increase in the lipid kinase activity of the mutant. This finding provides additional evidence for the importance of Ser608 phosphorylation as a regulatory mechanism of PI 3-kinase in vivo. It also demonstrates the detrimental effects that deregulation caused by impaired autophosphorylation can have on critical cellular functions.
The preceding data provide the first evidence of the importance of the serine autophosphorylation of the PI 3-kinase regulatory subunit in the regulation of this enzyme in vivo. Factors that affect the phosphorylation of Ser608 are therefore likely to have a major impact on PI 3-kinase activity in vivo, given crucial role that PI 3-kinase plays in many cellular processes.
Acknowledgments
We thank M. D. Waterfield for supplying reagents. We thank I. Krascenicova, A. Munkert, and A. Philp for assistance in some experimental procedures and L. Li and P. Wang for providing animal tissues. We also thank Kelly Nikolaidou for expert assistance with confocal microscopy and Bart Vanhaesebroeck for critical reading of the manuscript.
This work was funded by Diabetes UK. L.C.F. was funded by the Hellenic State Scholarships Foundation.
REFERENCES
- 1.Allende, J. E., and C. C. Allende. 1995. Protein kinase CK2: an enzyme with multiple substrates and a puzzling regulation. FASEB J. 9:313-323. [DOI] [PubMed] [Google Scholar]
- 2.Asano, T., A. Kanda, H. Katagiri, M. Nawano, T. Ogihara, K. Inukai, M. Anai, Y. Fukushima, Y. Yazaki, M. Kikuchi, R. Hooshmand-Rad, C.-H. Heldin, Y. Oka, and M. Funaki. 2000. p110β is up-regulated during differentiation of 3T3-L1 cells and contributes to the highly insulin-responsive glucose transport activity. J. Biol. Chem. 275:17671-17676. [DOI] [PubMed] [Google Scholar]
- 3.Baynes, K. C., C. A. Beeton, G. Panayotou, R. Stein, M. Soos, T. Hansen, H. Simpson, S. O'Rahilly, P. R. Shepherd, and J. P. Whitehead. 2000. Natural variants of human p85α phosphoinositide 3-kinase in severe insulin resistance: a novel variant with impaired insulin-stimulated lipid kinase activity. Diabetologia 43:321-331. [DOI] [PubMed] [Google Scholar]
- 4.Beeton, C. A., E. M. Chance, L. C. Foukas, and P. R. Shepherd. 2000. Comparison of the kinetic properties of the lipid- and protein-kinase activities of the p110α and p110β catalytic subunits of class-Ia phosphoinositide 3-kinases. Biochem. J. 350:353-359. [PMC free article] [PubMed] [Google Scholar]
- 5.Beeton, C. A., P. Das, M. D. Waterfield, and P. R. Shepherd. 1999. The SH3 and BH domains of the p85α adapter subunit play a critical role in regulating class Ia phosphoinositide 3-kinase function. Mol. Cell. Biol. Res. Commun. 1:153-157. [DOI] [PubMed] [Google Scholar]
- 6.Begum, N., and L. Ragolia. 1996. cAMP counter-regulates insulin-mediated protein phosphatase-2A inactivation in rat skeletal muscle cells. J. Biol. Chem. 271:31166-31171. [DOI] [PubMed] [Google Scholar]
- 7.Cantley, L. C. 2002. The phosphoinositide 3-kinase pathway. Science 296:1655-1657. [DOI] [PubMed] [Google Scholar]
- 8.Carpenter, C. L., K. R. Auger, M. Chanudhuri, M. Yoakim, B. Schaffhausen, S. Shoelson, and L. C. Cantley. 1993. Phosphoinositide 3-kinase is activated by phosphopeptides that bind to the SH2 domains of the 85-kDa subunit. J. Biol. Chem. 268:9478-9483. [PubMed] [Google Scholar]
- 9.Carpenter, C. L., K. R. Auger, B. C. Duckworth, W. Hou, B. Schaffhausen, and L. C. Cantley. 1993. A tightly associated serine/threonine protein kinase regulates phosphoinositide 3-kinase activity. Mol. Cell. Biol. 13:1657-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan, T. O., U. Rodeck, A. M. Chan, A. C. Kimmelman, S. E. Rittenhouse, G. Panayotou, and P. N. Tsichlis. 2002. Small GTPases and tyrosine kinases coregulate a molecular switch in the phosphoinositide 3-kinase regulatory subunit. Cancer Cell 1:181-191. [DOI] [PubMed] [Google Scholar]
- 11.Cuevas, B. D., Y. Lu, M. Mao, J. Zhang, R. LaPushin, K. Siminovitch, and G. B. Mills. 2001. Tyrosine phosphorylation of p85 relieves its inhibitory activity on phosphatidylinositol 3-kinase. J. Biol. Chem. 276:27455-27461. [DOI] [PubMed] [Google Scholar]
- 12.Czupalla, C., M. Culo, E. C. Muller, C. Brock, H. P. Reusch, K. Spicher, E. Krause, and B. Nurnberg. 2003. Identification and characterization of the autophosphorylation sites of phosphoinositide 3-kinase isoforms beta and gamma. J. Biol. Chem. 278:11536-11545. [DOI] [PubMed] [Google Scholar]
- 13.Davies, S. P., H. Reddy, M. Caivano, and P. Cohen. 2000. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dhand, R., K. Hara, I. Hiles, B. Bax, I. Gout, G. Panayotou, M. J. Fry, K. Yonezawa, M. Kasuga, and M. D. Waterfield. 1994. PI 3-kinase: structural and functional analysis of intersubunit interactions. EMBO J. 13:511-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dhand, R., I. Hiles, G. Panayotou, S. Roche, M. J. Fry, I. Gout, N. F. Totty, O. Truong, P. Vicendo, K. Yonezawa, M. Kasuga, S. A. Courtneidge, and M. D. Waterfield. 1994. PI 3-kinase is a dual specificity enzyme: autoregulation by an intrinsic protein-serine kinase activity. EMBO J. 13:522-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diggle, T. A., C. Schmitz-Peiffer, A. C. Borthwick, G. I. Welsh, and R. M. Denton. 1991. Evidence that insulin activates casein kinase 2 in rat epididymal fat-cells and that this may result in the increased phosphorylation of an acid-soluble 22 kDa protein. Biochem. J. 279:545-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foukas, L. C., N. Daniele, C. Ktori, K. E. Anderson, J. Jensen, and P. R. Shepherd. 2002. Direct effects of caffeine and theophylline on p110δ and other phosphoinositide 3-kinases; differential effects on lipid kinase and protein kinase activities. J. Biol. Chem. 277:37124-37130. [DOI] [PubMed] [Google Scholar]
- 18.Fu, Z., E. Aronoff-Spencer, J. M. Backer, and G. J. Gerfen. 2003. The structure of the inter-SH2 domain of class IA phosphoinositide 3-kinase determined by site-directed spin labeling EPR and homology modeling. Proc. Natl. Acad. Sci. USA 100:3275-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gout, I., R. Dhand, I. D. Hiles, M. J. Fry, G. Panayotou, P. Das, O. Truong, N. F. Totty, J. Hsuan, and G. W. Booker. 1993. The GTPase dynamin binds to and is activated by a subset of SH3 domains. Cell 75:25-36. [PubMed] [Google Scholar]
- 20.Harpur, A. G., M. J. Layton, P. Das, M. J. Bottomley, G. Panayotou, P. C. Driscoll, and M. D. Waterfield. 1999. Intermolecular interactions of the p85α regulatory subunit of phosphatidylinositol 3-kinase. J. Biol. Chem. 274:12323-12332. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi, H., S. Kamahara, Y. Nishioka, F. Kanai, N. Miyake, Y. Fukui, F. Shibasaki, T. Takenawa, and Y. Ebina. 1992. Insulin treatment stimulates tyrosine phosphorylation of the α-type 85 kDa subunit of phosphatidylinositol 3-kinase in vivo. J. Biol. Chem. 267:22575-22580. [PubMed] [Google Scholar]
- 22.Hayashi, H., Y. Nishioka, S. Kamohara, F. Kanai, K. Ishii, Y. Fukui, F. Shibasaki, T. Takenawa, H. Kido, N. Katsunuma, and Y. Ebina. 1993. The α type 85 kDa subunit of phosphatidylinositol 3-kinase is phosphorylated at tyrosines 368, 580 and 607 by the insulin receptor. J. Biol. Chem. 268:7107-7117. [PubMed] [Google Scholar]
- 23.Hiles, I. D., M. Otsu, S. Volinia, M. J. Fry, I. Gout, R. Dhand, G. Panayotou, F. Ruiz-Larrea, A. Thompson, N. F. Totty, J. J. Hsuan, S. A. Courtneidge, P. J. Parker, and M. D. Waterfield. 1992. Phosphatidylinositol 3-kinase: structure and expression of the 110 kd catalytic subunit. Cell 70:419-429. [DOI] [PubMed] [Google Scholar]
- 24.Hunter, T. 1995. When is a lipid kinase not a lipid kinase? When it is a protein kinase. Cell 83:1-4. [DOI] [PubMed] [Google Scholar]
- 25.Jimenez, C., D. R. Jones, P. Rodriguez-Viciana, A. Gonzalez-Garcia, E. Leonardo, S. Wennstrom, C. von Kobbe, J. L. Toran, L. R.-Borlado, V. Calvo, S. G. Copin, J. P. Albar, M. L. Gaspar, E. Diez, M. A. R. Marcos, J. Downward, C. Martinez-A., I. Merida, and A. C. Carrera. 1998. Identification and characterization of a new oncogene derived from the regulatory subunit of phosphoinositide 3-kinase. EMBO J. 17:743-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapeller, R., K. V. Prasad, O. Janssen, W. Hou, B. S. Schaffhausen, C. E. Rudd, and L. C. Cantley. 1994. Identification of two SH3-binding motifs in the regulatory subunit of phosphatidylinositol 3-kinase. J. Biol. Chem. 269:1927-1933. [PubMed] [Google Scholar]
- 27.Kavanaugh, W. M., C. W. Turck, A. Klippel, and L. T. Williams. 1994. Tyrosine 508 of the 85-kilodalton subunit of phosphatidylinositol 3-kinase by the platelet-derived growth factor receptor. Biochemistry 33:11046-11050. [DOI] [PubMed] [Google Scholar]
- 28.Klippel, A., J. A. Escobedo, M. Hirano, and L. T. Williams. 1994. The interaction of small domains between the subunits of phosphatidylinositol 3-kinase determines enzyme activity. Mol. Cell. Biol. 14:2675-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Layton, M. J., A. G. Harpur, G. Panayotou, P. I. H. Bastiaens, and M. D. Waterfield. 1998. Binding of a diphosphotyrosine-containing peptide that mimics activated platelet-derived growth factor receptor β induces oligomerization of phosphatidylinositol 3-kinase. J. Biol. Chem. 273:33379-33385. [DOI] [PubMed] [Google Scholar]
- 30.Mauvais-Jarvis, F., K. Ueki, D. A. Fruman, M. F. Hirshman, K. Sakamoto, L. J. Goodyear, M. Iannacone, D. Accili, L. C. Cantley, and C. R. Kahn. 2002. Reduced expression of the murine p85α subunit of phosphoinositide 3-kinase improves insulin signaling and ameliorates diabetes. J. Clin. Investig. 109:141-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Philp, A. J., I. G. Campbell, C. Leet, E. Vincan, S. P. Rockman, R. H. Whitehead, R. J. S. Thomas, and W. A. Phillips. 2001. The phosphatidylinositol 3′-kinase p85α gene is an oncogene in human ovarian and colon tumors. Cancer Res. 61:7426-7429. [PubMed] [Google Scholar]
- 32.Rameh, L. E., C.-S. Chen, and L. C. Cantley. 1995. Phosphatidylinositol (3,4,5)P3 interacts with SH2 domains and modulates PI 3-kinase association with tyrosine-phosphorylated proteins. Cell 83:821-830. [DOI] [PubMed] [Google Scholar]
- 33.Roche, S., J. Downward, P. Raynal, and S. A. Courtneidge. 1998. A function for phosphatidylinositol 3-kinase beta (p85α-p110β) in fibroblasts during mitogenesis: requirement for insulin- and lysophosphatidic acid-mediated signal transduction. Mol. Cell. Biol. 18:7119-7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez-Viciana, P., P. H. Warne, R. Dhand, B. Vanhaesebroeck, I. Gout, M. J. Fry, M. D. Waterfield, and J. Downward. 1994. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature 370:527-532. [DOI] [PubMed] [Google Scholar]
- 35.Rondinone, C. M., and U. Smith. 1996. Okadaic acid exerts a full insulin-like effect on glucose transport and glucose transporter 4 translocation in human adipocytes. J. Biol. Chem. 271:18148-18153. [DOI] [PubMed] [Google Scholar]
- 36.Rordorf-Nikolic, T., D. J. V. Horn, D. Chen, M. F. White, and J. M. Backer. 1995. Regulation of phosphatidylinositol 3′-kinase by tyrosyl phosphoproteins. Full activation requires occupancy of both SH2 domains in the 85-kDa regulatory subunit. J. Biol. Chem. 270:3662-3666. [DOI] [PubMed] [Google Scholar]
- 37.Shepherd, P. R., D. J. Withers, and K. Siddle. 1998. Phosphoinositide 3-kinase: the key switch mechanism in insulin signalling. Biochem. J. 333:471-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sommercorn, J., J. A. Mulligan, F. J. Lozeman, and E. G. Krebs. 1987. Activation of casein kinase II in response to insulin and to epidermal growth factor. Proc. Natl. Acad. Sci. USA 84:8834-8838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soos, M. A., J. Jensen, R. A. Brown, S. O'Rahilly, P. R. Shepherd, and J. P. Whitehead. 2001. Class II phosphoinositide 3-kinase is activated by insulin but not by contraction in skeletal muscle. Arch. Biochem. Biophys. 396:244-248. [DOI] [PubMed] [Google Scholar]
- 40.Stoyanova, S., G. Bulgarelli-Leva, C. Kirsch, T. Hanck, R. Klinger, R. Wetzker, and M. P. Wymann. 1997. Lipid kinase and protein kinase activities of G-protein coupled phosphoinositide 3-kinase γ: structure-activity analysis and interactions with wortmannin. Biochem. J. 324:489-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ueki, K., D. A. Fruman, S. M. Brachman, Y.-H. Tseng, L. C. Cantley, and C. R. Kahn. 2002. Molecular balance between the regulatory and catalytic subunits of phosphoinositide 3-kinase regulates cell signalling and survival. Mol. Cell. Biol. 22:965-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vanhaesebroeck, B., K. Higashi, C. Raven, M. Welham, S. Anderson, P. Brennan, S. G. Ward, and M. D. Waterfield. 1999. Autophosphorylation of p110δ phosphoinositide 3-kinase: a new paradigm for the regulation of lipid kinases in vitro and in vivo. EMBO J. 18:1292-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vanhaesebroeck, B., G. E. Jones, W. E. Allen, D. Zicha, R. Hooshmand-Rad, C. Sawyer, C. Wells, M. D. Waterfield, and A. J. Ridley. 1999. Distinct PI(3)Ks mediate mitogenic signalling and cell migration in macrophages. Nat. Cell Biol. 1:69-71. [DOI] [PubMed] [Google Scholar]
- 44.Vanhaesebroeck, B., S. J. Leevers, K. Ahmadi, J. Timms, R. Katso, P. C. Driscoll, R. Woscholski, P. J. Parker, and M. D. Waterfield. 2001. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 70:535-602. [DOI] [PubMed] [Google Scholar]
- 45.Vanhaesebroeck, B., and M. D. Waterfield. 1999. Signaling by distinct classes of phosphoinositide 3-kinases. Exp. Cell Res. 253:239-254. [DOI] [PubMed] [Google Scholar]
- 46.von Willebrand, M., S. Williams, M. Saxena, J. Gilman, P. Tailor, T. Jascur, G. P. Amarente-Mendes, D. R. Green, and T. Mustelin. 1998. Modification of phosphatidylinositol 3-kinase SH2 domain binding properties by Abl- or Lck-mediated tyrosine phosphorylation at Tyr-688. J. Biol. Chem. 273:3994-4000. [DOI] [PubMed] [Google Scholar]
- 47.Yu, J., C. Wjasow, and J. M. Backer. 1998. Regulation of the p85/p110α phosphatidylinositol 3′-kinase. Distinct roles for the N-terminal and C-terminal SH2 domains. J. Biol. Chem. 273:30199-30203. [DOI] [PubMed] [Google Scholar]
- 48.Zheng, Y., S. Bagrodia, and R. A. Cerione. 1994. Activation of phosphoinositide 3-kinase activity by Cdc42Hs binding to p85. J. Biol. Chem. 269:18727-18730. [PubMed] [Google Scholar]