Abstract
Iron regulatory protein 2 (IRP2), a central posttranscriptional regulator of cellular and systemic iron metabolism, undergoes proteasomal degradation in iron-replete cells. The prevailing model postulates that the mechanism involves site-specific oxidation of 3 cysteine residues (C168, C174, and C178) within a 73-amino-acid (73-aa) degradation domain. By expressing wild-type and mutated versions of IRP2 in H1299 cells, we find that a C168S C174S C178S triple mutant, or a deletion mutant lacking the entire “73-aa domain,” is sensitive to iron-mediated degradation, like wild-type IRP2. The antioxidants N-acetylcysteine, ascorbate, and α-tocopherol not only fail to stabilize IRP2 but, furthermore, promote its proteasomal degradation. The pathway for IRP2 degradation is saturable, which may explain earlier data supporting the “cysteine oxidation model,” and shows remarkable similarities with the degradation of the hypoxia-inducible factor 1α (HIF-1α): dimethyl-oxalylglycine, a specific inhibitor of 2-oxoglutarate-dependent oxygenases, stabilizes IRP2 following the administration of iron to iron-deficient cells. Our results challenge the current model for IRP2 regulation and provide direct pharmacological evidence for the involvement of 2-oxoglutarate-dependent oxygenases in a pathway for IRP2 degradation.
Iron is an essential nutrient but, when present in excess, may turn into a biohazard by catalyzing the generation of aggressive radicals. Bacteria and eukaryotes have developed elegant homeostatic mechanisms to satisfy their metabolic needs for iron and to prevent the toxicity of iron overload. In mammals, the expression of several proteins of iron metabolism is coordinately regulated at the posttranscriptional level by a mechanism which involves binding of iron-regulatory proteins IRP1 and IRP2 to mRNA “iron-responsive elements” (IREs) (reviewed in references 3, 13, and 29). In iron-starved cells, IRE-IRP interactions stabilize transferrin receptor (TfR) mRNA and inhibit translation of ferritin (H and L) mRNAs, thereby promoting cellular iron uptake and preventing iron sequestration.
By contrast, IRE-binding activity decreases in iron-replete cells, resulting in opposite homeostatic responses. Under these conditions, the IRE-binding activity of IRP1 is inactivated upon assembly of a cubane iron-sulfur cluster (9), while IRP2 undergoes degradation by the proteasome (8, 16). The regulatory function of IRP1 and IRP2 in mammalian iron metabolism has been examined by gene-targeting experiments. IRP1−/− mice have been reported to lack any obvious phenotype (30), while IRP2−/− mice display aberrant iron homeostasis and accumulate iron in the intestinal mucosa and the central nervous system (23). Moreover, these mice develop a progressive neurodegenerative disorder, suggesting a prominent role of IRP2 in controlling neuronal iron metabolism.
IRP1 and IRP2 share extensive sequence homology and belong to the family of iron-sulfur cluster isomerases (6). By analogy to mitochondrial aconitase, a well-characterized enzyme of the citric acid cycle and a member of this family, both IRP1 and IRP2 are projected to contain three compact domains, linked to a fourth domain by a flexible hinge region. However, IRP2 contains an insertion of 73 amino acids (aa), which is encoded by a unique exon, within its domain 1. This cysteine- and proline-rich sequence has been implicated in the mechanism for iron sensing by IRP2 and its iron-mediated degradation. In particular, removal of the 73 aa from IRP2 has been reported to yield a stable molecule (16). Conversely, insertion of this sequence into an equivalent position within domain 1 apparently rendered IRP1 sensitive to iron-dependent degradation (16).
Further mapping of the 73-aa domain showed that mutation of the cysteines at positions 168, 174, and 178 to serines abolished the iron-dependent instability of the IRP1-IRP2 hybrid molecule (16). These data paved the way for the “cysteine oxidation model” to explain the iron-dependent degradation of IRP2 (15). According to this model, iron mediates the site-specific oxidation of C168, C174, and C178, which tags the protein for ubiquitination and degradation by the proteasome (15). In support of this model, recent experiments showed that iron can directly bind to the 73-aa domain in vitro and that this interaction yields oxidized cysteine species (19). It should also be noted that heme has earlier (7) and more recently (38) been proposed to serve as a signal for IRP2 degradation.
To better understand the function of IRP2 as an iron sensor in vivo, we generated stably transfected cell lines expressing epitope-tagged wild-type or mutated IRP2. We provide evidence that neither the three cysteines C168, C174, and C178 nor the entire 73 aa domain is critical for the iron-dependent degradation of IRP2. Instead, we demonstrate that the machinery for IRP2 degradation can be saturated when the protein is expressed at high levels, which may help to reconcile previous data. We show that, unexpectedly, antioxidants can also drive IRP2 toward degradation, and we reveal that the iron-mediated degradation of IRP2 may involve the activity of 2-oxoglutarate-dependent oxygenase(s).
MATERIALS AND METHODS
Materials.
Hemin, ferric ammonium citrate (FAC), ascorbate, α-tocopherol, MG132, and N-acetylcysteine (NAC) were purchased from Sigma (St. Louis, Mo.). Desferrioxamine (DFO) was obtained from Novartis (Dorval, Canada). Dimethyl-oxalylglycine (DMOG) was synthesized and kindly provided by Andreas Schleifenbaum and Karsten Schulz (European Molecular Biology Laboratory [EMBL], Heidelberg, Germany).
Construction of plasmids. (i) Wild-type IRP2.
To introduce a hemagglutinin (HA) epitope at the C terminus of IRP2, a 557-bp fragment at the 3′ end of human IRP2 cDNA was PCR amplified with primers 5′-CTCGTGAATT CAACTCTTAC G-3′ (forward) and 5′-CGTAGGATCC TTACTAAGCG TAATCTGGAA CATCGTATGG GTATGAGAAT TTTCGTGCCAC-3′ (reverse) and then cloned into the pGEM-T vector (Promega). This manipulation retains the SacI site at the 5′ end of the amplified fragment and introduces a BamHI site downstream of the stop codon. The 557-bp SacI/BamHI fragment was cloned into the pUHD10-3 vector (37) together with a 2.4-kb BamHI/SacI fragment, which was excised from pBluescript-IRP2 and contains the remaining sequences of human IRP2, to yield pUHD-IRP2wt. The pcDNA3-IRP2wt plasmid was generated by subcloning the 2.9-kb BamHI/BamHI fragment from pUHD-IRP2wt into pcDNA3 (Invitrogen). Cloning was confirmed by automated DNA sequencing.
(ii) IRP2 mutants.
For expression of IRP2 harboring C168S, C174S, and C178S substitutions (IRP23CS), the 557-bp C-terminal SacI/BamHI fragment was cloned into the pUHD10-3 vector together with a 2.4-kb BamHI/SacI fragment, excised from pcDNA3-IRP23CS (a kind gift from Prem Ponka, Montreal, Canada), to yield pUHD-IRP23CS. Mutants with deletions in the 73-aa domain of IRP2 were generated with the ExSite PCR-based protocol (Stratagene) by using pUHD-IRP2wt as the template. The reverse primer 5′-TTTACTGAAG TCAATTTGTA AA-3′ was used in combination with forward primer 5′-CAGCCTAAGA AGCTTCCCTG CA-3′, 5′-TCAGGAACAT TTTCTTCGCA GA-3′, or 5′-GAACCTGAAA CAGTGTTAAA AA-3′ to yield pUHD-IRP2Δ25, pUHD-IRP2Δ50, or pUHD-IRP2Δ73, respectively. Mutagenesis was confirmed by automated DNA sequencing. The pUHD-IRP23CS plasmid was also sequenced manually with the T7 sequencing kit (U.S. Biochemicals) by using primer 5′-AAATCCTGGA GGTGGTGAC-3′.
(iii) GFP indicators.
The green fluorescent protein (GFP) fusion constructs containing the 73-aa domain of IRP2 were inserted into a plasmid derived from the simian virus 40 promoter-driven expression vector pSG5. This recipient vector encodes a bicistronic RNA in which the encephalomyocarditis virus internal ribosome entry site drives the translation of the luciferase firefly reporter fused to a FLAG tag at its N terminus. A polylinker containing an AUG start codon followed by NheI, PstI, and SpeI sites and a FLAG tag was added upstream of the encephalomyocarditis virus sequence for the insertion of the upstream cistron. A GFP open reading frame was PCR amplified with primers 5′-AAAGAATTCG CTAGCGTGAG CAAGGGCGAG GAGC-3′ (forward) and 5′-AAAGAATTCA CTAGTCTTGT ACAGCTCGTC CATGC-3′ (reverse) and inserted into the plasmid described above after digestion with NheI/SpeI to yield the pSG5-GFP construct. In the resulting plasmid, the GFP open reading frame is separated from the in-frame start codon by the NheI site and from the C-terminal FLAG tag by the SpeI site. The 73-aa domain of mouse IRP2 was PCR amplified with primers 5′-AAAGCTAGCT GTGCAATACA GAATGCAC-3′ (forward) and 5′-AAAACTAGTA GGCACTGGTT GCAAATG-3′ (reverse) so that it was flanked with an NheI site at the 5′ end and an SpeI site at the 3′ end. The PCR product digested with NheI/SpeI was then inserted at the NheI site or at the SpeI site of pSG5-GFP to yield pSG5-73aa-GFP and pSG5-GFP-73aa, respectively. Cloning was confirmed by automated sequencing.
Cell culture and transfections.
H1299 (human lung cancer) and B6 (murine fibroblast) cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 U of penicillin/ml, and 0.1 ng of streptomycin/ml. For tetracycline-inducible expression (TET-off system), each of the wild-type and mutant IRP2 constructs cloned into the pUHD vector was cotransfected with the puromycin resistance vector pBabe into H1299 cells by the calcium phosphate method. Stable transfectants were selected and maintained as described by Wang and Pantopoulos (37). Similarly, pSG5-GFP, pSG5-73aa-GFP, and pSG5-GFP-73aa were each cotransfected with pBabe into H1299 cells, and stable clones were selected in media containing 2 μg of puromycin/ml. Transient transfections with the pcDNA3-IRP2wt and pcDNA3-IRP23CS plasmids were performed with the Lipofectamine Plus reagent (Gibco BRL).
Electrophoretic mobility shift assay.
Cells were lysed in cytoplasmic lysis buffer (1% Triton X-100, 25 mM Tris-Cl [pH 7.4], 40 mM KCl) containing 1 mM dithiothreitol, and IRE-binding activity was analyzed by an electrophoretic mobility shift assay with a 32P-labeled IRE probe (26). Supershifts were performed by addition of an anti-HA antibody (Santa Cruz) to the binding reaction.
Western blotting.
Cells were washed twice in phosphate-buffered saline (PBS) and lysed in cytoplasmic lysis buffer. Cell debris was cleared by centrifugation, and protein concentrations were determined with the Bradford reagent (Bio-Rad). Cell lysates (30 μg) were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 7.5% (unless otherwise indicated) gels, and proteins were transferred to nitrocellulose filters. The blots were saturated with 10% nonfat milk in PBS and probed with antibodies against HA (Santa Cruz), TfR (Zymed), β-actin (Sigma), hypoxia-inducible factor 1α (HIF-1α) (Santa Cruz), M2-FLAG (Sigma), or IRP2 (31), diluted in PBS containing 0.5% Tween 20. The anti-IRP2 antibody was a generous gift from Tracey Rouault (Bethesda, Md.). Dilutions were 1:1,000 for antibodies against HA, TfR, β-actin, M2-FLAG, and IRP2 and 1:500 for the anti-HIF-1α antibody. Following a wash with PBS containing 0.5% Tween 20, the blots with monoclonal anti-FLAG or anti-TfR antibodies were incubated with peroxidase-coupled rabbit anti-mouse immunoglobulin G (1:4,000 dilution). The blots with all other (polyclonal) antibodies were incubated with peroxidase-coupled goat anti-rabbit immunoglobulin G (1:5,000 dilution). Peroxidase-coupled antibodies were detected by the enhanced chemiluminescence (ECL) method (Amersham). The immunoreactive bands were quantified by densitometric scanning.
Pulse-chase and immunoprecipitation.
Cells were metabolically labeled for 2 h with Tran35S-label (50 μCi/ml), a 70:30 mixture of [35S]methionine-cysteine (ICN). The radioactive medium was then removed, and the cells were chased in a cold medium. The chase was terminated by a wash in PBS. Cells were lysed in a buffer containing 1% Triton X-100, 50 mM Tris-Cl (pH 7.4), and 300 mM NaCl. Cell debris was cleared by centrifugation, and cell lysates were subjected to quantitative immunoprecipitation with the anti-HA antibody (Santa Cruz). Immunoprecipitated proteins were analyzed by SDS-PAGE and visualized by autoradiography. Radioactive bands were quantified by phosphorimaging.
RESULTS
Mutation of C168, C174, and C178 does not stabilize IRP2 against iron-mediated degradation.
H1299 cells were recently utilized to express IRP1C437S, and it was found that cell density and serum factors modulate the capacity of this constitutive IRP1 mutant to inhibit ferritin synthesis (37). These findings prompted us to examine whether this phenotype can be recapitulated with a constitutive IRP2 mutant. To this end, we generated stable clones of H1299 cells for tetracycline-inducible expression of IRP23CS, a mutant previously reported to be resistant to iron-mediated degradation (16), in which the crucial cysteines at positions 168, 174, and 178 have been replaced with serines (Fig. 1A). The triple mutation was verified by sequencing. In addition, we also generated stable clones of H1299 cells for tetracycline-inducible expression of wild-type IRP2 as a control.
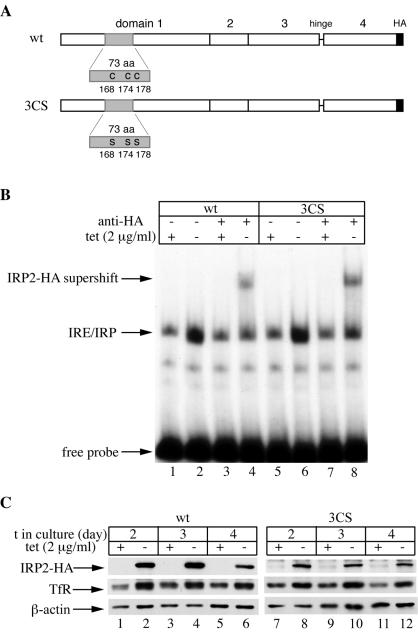
FIG. 1.
Tetracycline-inducible expression of wild-type IRP2 (IRP2wt) or IRP23CS in H1299 cells. (A) Schematic representation of IRP2wt (top) and IRP23CS (bottom) showing domains 1 to 4, the hinge which links domains 3 and 4, and the C-terminal HA tag. The 73-aa sequence within domain 1 is shaded. The cysteines at positions 168, 174, and 178 of IRP2wt have been mutated to serines in IRP23CS. (B) H1299 cells engineered to express IRP2wt (lanes 1 to 4) or IRP23CS (lanes 5 to 8) were grown for 48 h with (+) or without (−) tetracycline at 2 μg/ml, and cytoplasmic extracts were analyzed for IRE-binding activity with a 32P-labeled IRE probe in the absence or presence of 0.2 μg of a purified polyclonal anti-HA antibody. The positions of the IRE-IRP2 complex, the HA supershift, and excess free IRE probe are indicated by arrows. (C) IRP2wt (lanes 1 to 6) or IRP23CS (lanes 7 to 12) transfectants were grown for 2, 3, or 4 days with (+) or without (−) tetracycline at 2 μg/ml, and lysates were subjected to Western blotting with antibodies against HA (top), TfR (center), and β-actin (bottom). t, time.
The IRE-binding activities of chimeric wild-type IRP2 and IRP23CS, each of which was tagged with an HA epitope at its C terminus, were analyzed by an electrophoretic mobility shift assay (Fig. 1B). Removal of tetracycline for 2 days, to allow chimeric protein expression, was associated with a strong increase in the IRE-binding activities (Fig. 1B, lanes 1, 2, 5, and 6). The RNA-protein complexes could be supershifted with an anti-HA antibody (Fig. 1B, lanes 4 and 8), showing that both HA-tagged wild-type IRP2 and HA-tagged IRP23CS are active IRE-binding proteins. Their expression levels were directly assessed by Western blotting with the anti-HA antibody. Growth of cells in the absence of tetracycline for 2 to 4 days resulted in a profound induction of chimeric IRP2 expression (Fig. 1C, top). This induction was associated with an increase in the steady-state levels of TfR (Fig. 1C, center), while levels of β-actin, used as a control, remained unaffected (bottom), further demonstrating that the chimeric wild-type and mutant proteins are functional.
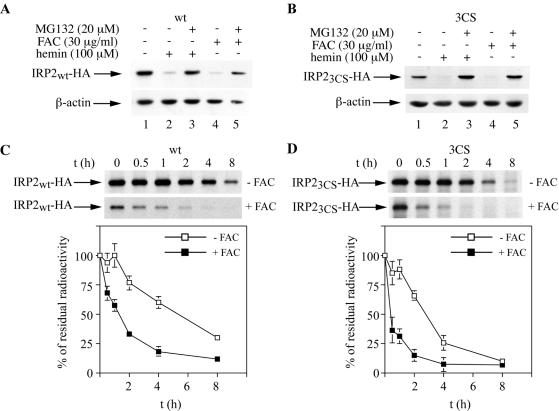
We then examined the responses of wild-type IRP2 and IRP23CS to iron. As expected, overnight treatment of cells with 100 μM hemin or 30 μg of FAC/ml resulted in a marked decrease in the levels of wild-type IRP2, and this effect could be completely blocked by the proteasome inhibitor MG132 (Fig. 2A). On the basis of the previous data (15, 16), we anticipated that iron would not affect the expression of IRP23CS. However, in all four independent clones of IRP23CS-expressing cells that were analyzed, we observed that treatment with either hemin or FAC caused a clear reduction in the steady-state levels of this mutant, which is also antagonized by MG132. Western blot analysis of a representative clone is depicted in Fig. 2B. To directly examine whether iron modulates the half-life of IRP23CS, control or iron-loaded cells (pretreated overnight with 30 μg of FAC/ml) were metabolically labeled for 2 h with [35S]methionine-cysteine. Subsequently, the cells were chased with cold medium for different time intervals in the presence or absence of 30 μg of FAC/ml. Wild-type IRP2 or IRP23CS was then analyzed by quantitative immunoprecipitation with the anti-HA antibody. As expected, iron treatment decreased the half-life of wild-type IRP2 from ∼5.5 to ∼1.5 h (Fig. 2C). Surprisingly, iron treatment also decreased the half-life of IRP23CS from ∼3 to ∼0.5 h (Fig. 2D). This result demonstrates that IRP23CS is sensitive to iron-dependent degradation and casts doubt on the cysteine oxidation model.
FIG. 2.
Iron-dependent turnover of wild-type IRP2 (IRP2wt) and IRP23CS. (A and B) Cells expressing IRP2wt (A) or IRP23CS (B) were treated overnight with 100 μM hemin (lanes 2 and 3) or 30 μg of FAC/ml (lanes 4 and 5) in the absence or presence of 20 μM MG132, and lysates were subjected to Western blotting with an anti-HA (top) or anti-β-actin (bottom) antibody. (C and D) Pulse-chase of IRP2wt (C) and IRP23CS (D) in the absence (top) or presence (bottom) of FAC. Control (untreated) or iron-loaded (pretreated overnight with 30 μg of FAC/ml) cells were metabolically labeled for 2 h with [35S]methionine-cysteine. Subsequently, the control or iron-loaded cells were chased for the indicated time intervals in cold medium either in the absence or in the presence of 30 μg of FAC/ml, respectively. Cytoplasmic lysates (500 μg) were subjected to quantitative immunoprecipitation with 1 μg of a purified polyclonal anti-HA antibody (Santa Cruz). Immunoprecipitated proteins were analyzed by SDS-PAGE on a 7.5% gel and visualized by autoradiography (arrows). The radioactive bands were quantified by phosphorimaging. In the graphs below the gels, the percentage of residual radioactivity from three independent experiments (mean ± standard deviation) is plotted against time.
Regulation of IRP2 expression by antioxidants.
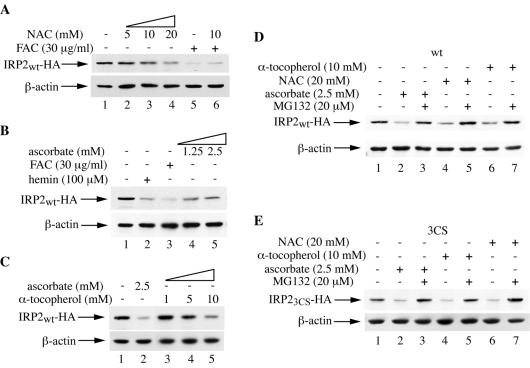
If the signal tagging IRP2 for degradation were a site-specific oxidation of cysteines, it is reasonable to hypothesize that antioxidants should protect IRP2 against iron-mediated degradation. To address this issue, cells were exposed for 10 h to 30 μg of FAC/ml in the presence of 10 mM NAC, a well-established antioxidant, and the expression of wild-type IRP2 was analyzed by Western blotting (Fig. 3A). This antioxidant not only failed to protect IRP2 against degradation by iron (Fig. 3A, lanes 5 and 6) but even promoted a dose-dependent decrease in IRP2 expression on its own (lanes 1 to 4). We then examined the effects of ascorbate (vitamin C) and α-tocopherol (vitamin E), two vitamins with antioxidant properties, on IRP2 expression. Both ascorbate (Fig. 3B) and α-tocopherol (Fig. 3C) eventually decreased IRP2 expression. The proteasomal inhibitor MG132 efficiently antagonized the negative effects of NAC, ascorbate, and α-tocopherol on the expression of both wild-type IRP2 (Fig. 3D) and IRP23CS (Fig. 3E), suggesting that these antioxidants promote the degradation of wild-type and mutant IRP2 by the proteasome. The antioxidant efficiency of the drugs under the experimental conditions described above was confirmed by employing the redox-sensitive dye 2′,7′-dichlorodihydrofluorescein diacetate (26) as a control (data not shown).
FIG. 3.
The antioxidants NAC, ascorbate, and α-tocopherol promote the proteasomal degradation of wild-type IRP2 (IRP2wt) and IRP23CS. (A) Cells expressing IRP2wt were treated for 10 h with the indicated concentrations of NAC (lanes 2 to 4) or with FAC in the absence (lane 5) or presence (lane 6) of 10 mM NAC. Lysates were analyzed by Western blotting with the anti-HA (top) or anti-β-actin (bottom) antibody. (B) Cells were treated overnight with the indicated concentrations of hemin (lane 2), FAC (lane 3), or ascorbate (lanes 4 and 5), and lysates were analyzed as for panel A. (C) Cells were treated overnight with the indicated concentrations of ascorbate (lane 2) or α-tocopherol (lanes 3 to 5), and lysates were analyzed as for panel A. (D and E) Cells expressing IRP2wt (D) or IRP23CS (E) were treated for 8 h with 2.5 mM ascorbate, 20 mM NAC, or 10 mM α-tocopherol in the absence or presence of 20 μM MG132, and lysates were analyzed as for panel A.
The pathway for IRP2 degradation by iron is saturable.
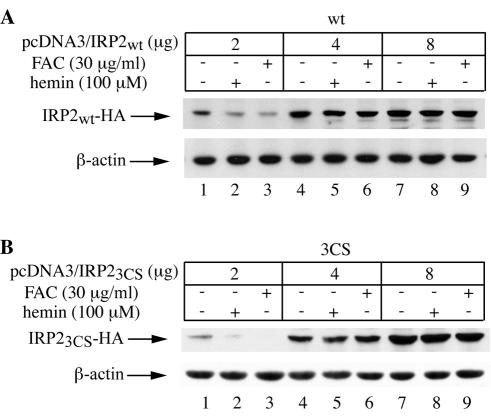
How can one explain the earlier data (15, 16), which have reported a function for C168, C174, and C178 within the 73-aa “degradation domain” of IRP2 in iron sensing? We hypothesized that the expression levels of chimeric IRP2 may interfere with the capacity of iron to promote its degradation. To explore this hypothesis, cells were transiently transfected with different doses of wild-type IRP2 or IRP23CS, and their responses to iron were assessed by immunoblotting. This experiment clearly showed that the degradation of both wild-type IRP2 (Fig. 4A) and IRP23CS (Fig. 4B) by iron is impaired when the protein is expressed at high levels. Thus, the machinery for IRP2 degradation by iron can be saturated at high doses of the substrate.
FIG. 4.
Dose-dependent saturation of the IRP2 degradation machinery. A total of 3 × 106 H1299 cells were transiently transfected with the indicated amounts of plasmid pcDNA3-IRP2wt (A) or pcDNA3-IRP23CS (B) in 100-mm-diameter dishes and were incubated for 36 h to allow expression of the chimeric proteins. Subsequently, the cells were either left untreated (lanes 1, 4, and 7) or treated overnight with either 100 μM hemin (lanes 2, 5, and 8) or 30 μg of FAC/ml (lanes 3, 6, and 9), and lysates were analyzed by Western blotting with an anti-HA (top) or anti-β-actin (bottom) antibody.
Interestingly, a similar effect was observed earlier for the degradation of HIF-1α in normoxic cells (33). We normalized the levels of transfected IRP2 to those of endogenous β-actin in order to estimate the threshold substrate concentration that saturates the degradation machinery. Immunoreactive IRP2 and β-actin bands were quantified by densitometric scanning, and the ratios were corrected for the transfection efficiency (∼80%) and for the duration of the chemiluminescence reaction required to yield the signal (visualization of HA-tagged IRP2 requires a ∼60-times-longer exposure than visualization of β-actin). For transfections with 2, 4, and 8 μg of plasmid, this semiquantitative calculation yields IRP2/β-actin ratios of 0.004, 0.045, and 0.11 for wild type IRP2 and 0.002, 0.06, and 0.23 for IRP23CS, respectively. Thus, it appears that saturation is achieved at IRP2/β-actin ratios of >0.04. The IRP2/β-actin ratios in the stable clones analyzed in Fig. 1 to 3 were 0.021 and 0.019 for wild-type IRP2 and IRP23CS, respectively. (In the calculations with stable clones, the densitometry values were corrected only for the different exposure times of chemiluminescence reactions for IRP2 and β-actin.)
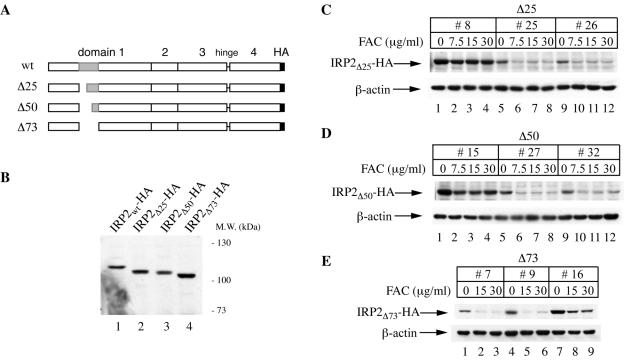
The 73-aa domain of IRP2 is neither necessary nor sufficient to promote iron-dependent degradation.
The experiments for which results are shown in Fig. 2 clearly indicate that C168, C174, and C178 are not required for the degradation of IRP2 by iron. To map functional residues within the 73-aa domain, we generated the Δ25, Δ50, and Δ73 deletion mutants (Fig. 5A). In the first two, the 73-aa domain is truncated at the N terminus by 25 or 50 aa, respectively. The Δ73 mutant gives rise to IRP2 lacking the entire domain. These constructs were stably transfected into H1299 cells for tetracycline-inducible expression, and different clones corresponding to each construct were selected. The expression of the deletion mutants, along with that of wild-type IRP2, was analyzed by Western blotting with the anti-HA antibody (Fig. 5B). Similarly, the responses of the deletion mutants to iron were analyzed following overnight treatment of cells with different doses of FAC. This treatment decreased the expression of IRP2Δ25, IRP2Δ50, and, importantly, IRP2Δ73 (Fig. 5C, D, and E, respectively) in different clones of each construct. It should be noted that the effect of iron was more pronounced in clones expressing lower levels of the chimeric proteins (IRP2/β-actin ratios are provided in the legend to Fig. 5). Conversely, the iron-mediated decrease in the IRP2 signal was weaker in clones expressing higher protein levels (Fig. 5C and D; compare lanes 1 to 4 with lanes 5 to 8 and 9 to 12), in agreement with the conclusion that the degradation machinery for IRP2 is saturable.
FIG. 5.
Iron-dependent decrease in the expression of IRP2 mutants harboring deletions within the 73-aa domain. (A) Schematic representation of IRP2 deletion mutants. (B) Lysates from cell clones expressing wild-type IRP2 (IRP2wt) and its Δ25, Δ50, and Δ73 mutants were analyzed by Western blotting with the anti-HA antibody. The positions of molecular size standards are indicated on the right. (C, D, and E) Iron-dependent decrease in the expression of IRP2Δ25, IRP2Δ50, and IRP2Δ73, respectively. For each of the mutants depicted schematically in panel A, three representative clones expressing that mutant were treated overnight with the indicated concentrations of FAC. Lysates were analyzed by Western blotting with an anti-HA (top) or anti-β-actin (bottom) antibody. For IRP2Δ25 clones 8, 25, and 26, the ratios of chimeric IRP2 to endogenous β-actin, calculated as described in Results, were 0.022, 0.003, and 0.003, respectively. For IRP2Δ50 clones 15, 27, and 32, the respective values were 0.027, 0.003, and 0.002, and for IRP2Δ73 clones 7, 9, and 16, the respective values were 0.002, 0.004, and 0.020.
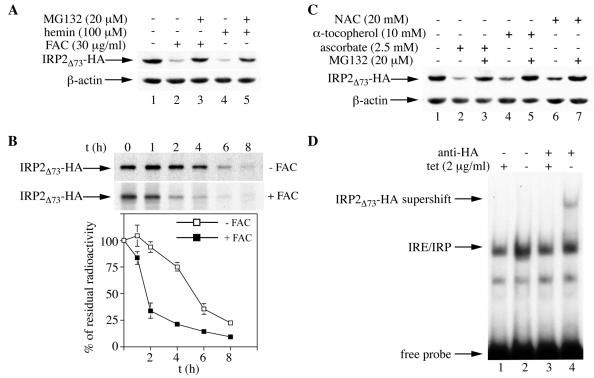
The proteasomal inhibitor MG132 efficiently blocked the decrease in the expression of IRP2Δ73 (clone 16) following treatment with hemin or FAC (Fig. 6A), in agreement with the view that this mutant is sensitive to iron-dependent degradation. The effects of iron on the stability of IRP2Δ73 were directly assessed by a pulse-chase experiment. The result depicted in Fig. 6B demonstrates that treatment of cells with FAC decreased the half-life of IRP2Δ73 from ∼5.5 to ∼1.5 h. Furthermore, as with wild-type IRP2 and IRP23CS (Fig. 3), NAC, ascorbate, and α-tocopherol each negatively affected IRP2Δ73 expression by a mechanism that was sensitive to MG132 (Fig. 6C). An electrophoretic mobility shift assay revealed that IRP2Δ73 is an active IRE-binding protein (Fig. 6D). In conclusion, an IRP2 mutant lacking the entire 73-aa domain retains its activity and its capacity to undergo proteasomal degradation in response to iron or antioxidants.
FIG. 6.
The 73-aa domain is not necessary for the iron- or antioxidant-dependent proteasomal degradation of IRP2, or for IRE-binding activity. (A) Cells expressing IRP2Δ73 were treated overnight with 30 μg of FAC/ml (lanes 2 and 3) or 100 μM hemin (lanes 4 and 5) in the absence or presence of 20 μM MG132, and lysates were subjected to Western blotting with an anti-HA (top) or anti-β-actin (bottom) antibody. (B) Pulse-chase of IRP2Δ73 in the absence (top) or in the presence (bottom) of FAC at 30 μg/ml. Experimental conditions were the same as those in Fig. 2C and D. The radioactive bands corresponding to IRP2Δ73 (arrows) were quantified by phosphorimaging. In the graph below the gel, the percentage of residual radioactivity from two independent experiments (mean ± standard deviation) is plotted against time. (C) Cells expressing IRP2Δ73 were treated for 8 h with 2.5 mM ascorbate (lanes 2 and 3), 10 mM α-tocopherol (lanes 4 and 5), or 20 mM NAC (lanes 6 and 7) in the absence or presence of 20 μM MG132, and lysates were analyzed as described for panel A. (D) Cells were grown for 48 h with (+) or without (−) tetracycline at 2 μg/ml to allow expression of IRP2Δ73, and cytoplasmic extracts were analyzed for IRE-binding activity with a 32P-labeled IRE probe in the absence (lanes 1 and 2) or presence (lanes 3 and 4) of 0.2 μg of a purified polyclonal anti-HA antibody. The positions of the IRE-IRP2Δ73 complex, the HA-supershift, and excess free IRE probe are indicated by arrows.
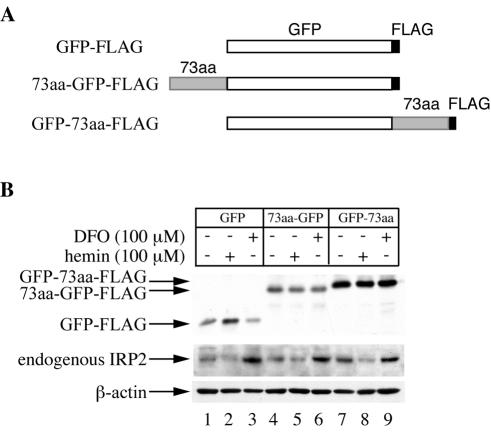
We then examined whether the 73-aa domain is sufficient to function as an iron sensor and promote iron-dependent degradation. For this purpose, we generated indicator constructs of GFP, fused at either the N or the C terminus with the 73-aa domain of IRP2. These FLAG epitope-tagged constructs, including a GFP control (Fig. 7A), were transfected into H1299 cells. Stable clones were selected and analyzed for the response of GFP to iron perturbations. The cells were exposed overnight either to hemin or to the iron chelator DFO, and the expression of GFP was analyzed by Western blotting with an antibody against the FLAG epitope (Fig. 7B, top). Clearly, the hemin treatment did not result in decreased expression of the control GFP (Fig. 7B, lanes 1 to 3) or of the GFP fusion constructs (lanes 4 to 9). Similar results were also obtained when FAC was used as the iron donor and when additional clones of these constructs were analyzed (data not shown). Importantly, hemin and DFO modulate the expression of endogenous IRP2 (Fig. 7B, center), suggesting that the degradation machinery is not saturated under these experimental conditions. The calculated GFP/β-actin ratios (0.001 for control GFP, 0.003 for 73aa-GFP, and 0.021 for GFP-73aa) were also well below the estimated saturation threshold. Collectively, the data presented in Fig. 5 to 7 demonstrate that the 73-aa domain of IRP2 is neither necessary nor sufficient for iron-dependent degradation.
FIG. 7.
The 73-aa domain is not sufficient for iron-dependent degradation of a GFP indicator. (A) Schematic representation of GFP control and 73-aa fusion constructs. (B) Representative clones expressing each of the fusion proteins depicted schematically in panel A were treated overnight with 100 μM hemin or DFO. Lysates (30 μg) were resolved by SDS-PAGE on an 11% gel and analyzed by Western blotting with an antibody against FLAG (top) or β-actin (bottom). To visualize endogenous IRP2, additional aliquots containing 30 μg of lysates were resolved by SDS-PAGE on a 7.5% gel and analyzed by Western blotting with the IRP2 antisera (center).
Iron-mediated degradation of IRP2 entails a 2-oxoglutarate-dependent step.
Recent work has revealed that a crucial step in the recognition of HIF-1α by the degradation machinery is the hydroxylation of P402 and P564 (24) by prolyl 4-hydroxylases (4, 14, 17). These enzymes, which are members of the broader family of 2-oxoglutarate-dependent oxygenases (2, 32), utilize oxygen, iron, and ascorbate as cofactors. Their enzymatic activity is inhibited by hypoxia, iron chelators, or cobalt (which mimics hypoxia, probably by competing for iron binding) (4, 25). Previous data showing the stabilization of IRP2 by hypoxia, iron chelators, or cobalt (10), the stimulatory effects of iron and ascorbate on IRP2 degradation (Fig. 2 and 3), and the results shown in Fig. 4 highlight intriguing analogies between the pathways for IRP2 and HIF-1α degradation.
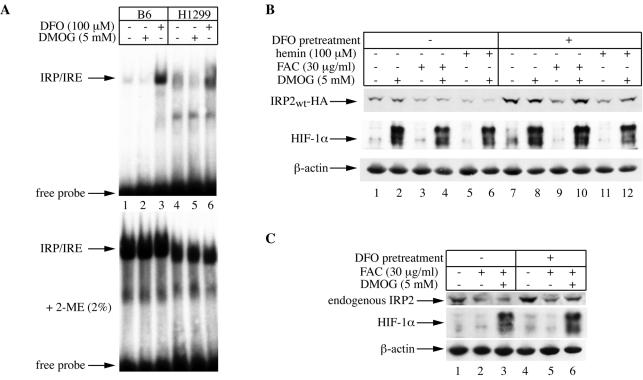
We therefore designed experiments to address whether the signal for IRP2 degradation by iron could involve the activity of 2-oxoglutarate-dependent oxygenases. To this end, we utilized DMOG, a cell-permeant inhibitor of these enzymes which efficiently blocks the degradation of HIF-1α (4, 17). Considering that some known prolyl 4-hydroxylase inhibitors, such as ethyl-3,4-dihydroxybenzoate (EDHB), also have iron-chelating properties (36), we first evaluated the capacity of DMOG to modulate the IRE-IRP system in two different cell lines. Exposure of mouse B6 fibroblasts or parent H1299 cells to 5 mM DMOG for 6 h did not perturb IRE-binding activity, while the iron chelator DFO activated it, as expected (Fig. 8A, top). Analysis of the extracts in the presence of 2-mercaptoethanol (Fig. 8A, bottom), which is known to activate latent IRP1 (13), confirmed equal loading. These results suggest that DMOG does not function as an iron chelator in cultured cells, at least under our experimental conditions.
FIG. 8.
Conditional inhibition of iron-mediated decrease in IRP2 expression by DMOG. (A) DMOG has no apparent iron-chelating properties. B6 and parent H1299 cells were either left untreated (lanes 1 and 4) or treated for 6 h with 5 mM DMOG (lanes 2 and 5) or 100 μM DFO (lanes 3 and 6). Cytoplasmic extracts from B6 (10 μg) or H1299 (30 μg) cells were analyzed for IRE-binding activity with a 32P-labeled IRE probe in the absence (top) or presence (bottom) of 2% 2-mercaptoethanol (2-ME). The positions of the IRE-IRP complexes and excess free IRE probe are indicated by arrows. (B) Effects of DMOG on steady-state levels of chimeric wild-type IRP2. H1299 cells were incubated in the absence of tetracycline for 2 days to express chimeric wild-type IRP2. Half of the cells were left untreated, and the other half received 100 μM DFO overnight to further stimulate IRP2 expression. Tetracycline was then added back to the medium to shut off IRP2 expression, and the cells were either left untreated or pretreated with 5 mM DMOG for 2 h. Subsequently, the cells were either left untreated (lanes 1, 2, 7, and 8) or treated in the presence or absence of 5 mM DMOG for 4 h with 30 μg of FAC/ml (lanes 3, 4, 9, and 10) or 100 μM hemin (lanes 5, 6, 11, and 12). Lysates were analyzed by Western blotting with an antibody against HA (top), HIF-1α (center), or β-actin (bottom). (C) Effects of DMOG on steady-state levels of endogenous IRP2. Parent H1299 cells were either left untreated (lanes 1 to 3) or pretreated overnight with 100 μM DFO (lanes 4 to 6). The cells were then either left untreated (lanes 1 and 4) or treated for 4 h with 30 μg of FAC/ml in the presence (lanes 3 and 6) or absence (lanes 2 and 5) of 5 mM DMOG. The inhibitor was administered 2 h prior to addition of FAC. Lysates were analyzed by Western blotting with an antibody against IRP2 (top), HIF-1α (center), or β-actin (bottom).
We then analyzed the effects of DMOG on IRP2 degradation. Figure 8B shows that DMOG failed to prevent a decrease in IRP2 steady-state levels following treatment of H1299 cells with either hemin or FAC (lanes 1 to 6, top). However, DMOG efficiently antagonized the negative effects of hemin or FAC on the expression of IRP2 in iron-depleted cells which had been pretreated with DFO (Fig. 8B, lanes 7 to 12, top). As expected, DMOG profoundly stimulated the expression of HIF-1α (Fig. 8B, center) under all experimental conditions (note the residual activation of HIF-1α expression as a result of DFO pretreatment in lane 7). Similar results were obtained with endogenous IRP2 in parent H1299 cells (Fig. 8C). Thus, DMOG is capable of modulating the effects of iron on IRP2 expression, but this ability appears to be restricted in iron-deficient cells.
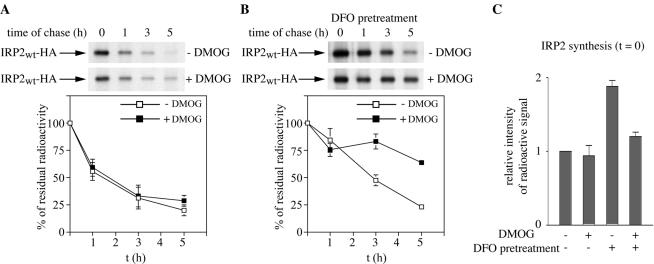
On the basis of this result, and considering that DFO also stimulates de novo synthesis of IRP2 (12, 27), it is conceivable that DMOG may differentially affect the rate of IRP2 synthesis and protein stability. By metabolic labeling of cells with [35S]methionine-cysteine in the presence or absence of DMOG, and quantification of 35S-labeled polypeptides, we found that this drug does not affect global protein synthesis (data not shown). To directly evaluate the effects of DMOG on IRP2 synthesis and stability, a pulse-chase experiment was performed (Fig. 9). Control cells (Fig. 9A) or cells pretreated overnight with 100 μM DFO (Fig. 9B) were metabolically labeled for 2 h with [35S]methionine-cysteine. Subsequently, the cells were chased for different time intervals in a “cold” medium containing 30 μg of FAC/ml in the presence or absence of 5 mM DMOG, and levels of radiolabeled IRP2 were assessed by quantitative immunoprecipitation. Quantification of IRP2 at time zero of the chase (Fig. 9C) confirmed that pretreatment with DFO stimulated a ∼2-fold increase in de novo IRP2 synthesis; however, DMOG impaired this effect by ∼35%. In addition, this result showed that DMOG does not affect IRP2 synthesis in the absence of DFO pretreatment. Analysis of the half-life of IRP2 confirmed directly that DMOG stabilizes IRP2 against iron-mediated degradation in cells pretreated with DFO (Fig. 9B) but the drug is far less efficient in control cells (Fig. 9A). This finding, which is in agreement with the results shown in Fig. 8B and C, demonstrates that the pathway for IRP2 degradation triggered by administration of iron to iron-starved cells can be efficiently inhibited by DMOG. These data provide pharmacological evidence for the involvement of a 2-oxoglutarate-dependent oxygenase activity in IRP2 degradation mediated by iron.
FIG. 9.
DMOG stabilizes IRP2 following exposure of iron-depleted cells to iron. (A and B) Pulse-chase experiment to analyze the effects of DMOG on IRP2 degradation by iron. H1299 cells expressing chimeric wild-type IRP2 (IRP2wt) were either pretreated overnight with 100 μM DFO (B) or left untreated (A). Cells were then metabolically labeled for 2 h with [35S]methionine-cysteine in the presence or absence of 5 mM DMOG. Subsequently, cells were chased for the indicated time intervals in a cold medium in the presence of 30 μg of FAC/ml. Cytoplasmic lysates were analyzed as for Fig. 2C. The radioactive bands corresponding to IRP2 (arrows) were quantified by phosphorimaging. In the graphs below the gels, the percentage of residual radioactivity from two independent experiments (mean ± standard deviation) is plotted against time. (C) Quantification of IRP2 synthesis at time zero from two independent pulse-chase experiments. Error bars, standard deviations.
DISCUSSION
A growing body of evidence indicates that IRP2 plays a central role in the control of iron homeostasis. This was not obvious when IRP2 was initially cloned, mainly due to its relatively lower abundance (relative to IRP1) and its apparent tissue-specific pattern of expression (31). IRP1 and IRP2 repress the translation of IRE-containing mRNAs with similar efficiencies (20), suggesting that their functions may, at least in part, be redundant. However, the abundance of IRP2 in tissues and cells may have been underestimated (5) due to technical difficulties (30). It is also pertinent that in murine RAW 264.7 macrophages, the exclusive modulation of IRP2 was sufficient to regulate the expression of TfR mRNA (21) and ferritin synthesis (22), regardless of IRP1 activity. Moreover, the pathological abnormalities of the intestine and the central nervous system in IRP2−/− mice (23) establish a major regulatory function of IRP2 in systemic iron metabolism.
The cysteine oxidation model and the role of the 73-aa domain in IRP2 regulation.
Thus, the understanding of the molecular basis for IRP2 regulation by iron is becoming increasingly important. It is well established (8, 16), and confirmed by our findings here, that IRP2 is regulated at the level of protein stability and undergoes degradation by the proteasome in iron-replete cells. It has been proposed that the recognition of IRP2 by the degradation machinery involves site-specific oxidation of C168, C174, and C178, possibly as a result of direct binding of iron (15, 16, 19). We present data which challenge this cysteine oxidation model. First, we demonstrate that a C168S C174S C178S triple mutant, like wild-type IRP2, is sensitive to iron-dependent proteasomal degradation (Fig. 2). Second, the antioxidant NAC, which effectively inhibits the oxidative stress-mediated activation of IRP1 (28), fails to stabilize wild type IRP2 in the presence of iron (Fig. 3A). Finally, the antioxidants NAC, ascorbate, and α-tocopherol rather promote the degradation of wild-type IRP2 and IRP23CS by the proteasome (Fig. 3D and E). We are currently investigating whether iron and antioxidants modulate IRP2 by similar or distinct pathways. Note that the vitamin ascorbate has earlier been shown to enhance ferritin synthesis (35). This effect of ascorbate, which was attributed only to a switch of IRP1 into the cytosolic aconitase form (34), is consistent with increased degradation of IRP2.
The data presented in Fig. 2 cast doubt on the interpretation of previous findings that claimed a critical role of C168, C174, and C178 in IRP2 degradation (15, 16). Although recent results have shown that C168, C174, and C178 can be converted to oxidized species by iron in vitro (19), we propose that this modification is not critical for the regulation of IRP2 in vivo. We suggest that the previously observed stabilization of IRP1-IRP2 hybrid molecules, bearing C168S, C174S, and C178S substitutions, in iron-replete cells (16) may be due to high expression levels of the protein that exceed the capacity of a limiting factor(s) of the degradation machinery. This view is directly supported by the experiment for which results are shown in Fig. 4, which reveals a dose-dependent saturable pattern for IRP2 (both wild-type IRP2 and IRP23CS) degradation. In fact, while this work was under review, Bourdon et al. reported that the previous in vivo data with the 3CS mutant (16) were obtained from a single transfected clone and could not be reproduced (1). We found that deletion of the entire 73-aa domain yields a protein that is still regulated by iron (Fig. 5 and 6). These data directly support the very recent results by Hanson et al. (11) and are in contrast with the previous findings (15, 16). We conclude that the recognition elements that are necessary for the degradation of IRP2 by iron reside outside the 73-aa domain of the protein.
The 73-aa domain also fails to drive a GFP fusion construct to iron-dependent degradation, regardless of whether it is fused to the N or the C terminus of the indicator (Fig. 7). This result suggests that the 73-aa domain is not sufficient to operate as an iron sensor. It is possible, however, that the stability of GFP could mask a potential function of this element. Nevertheless, we would expect at least a minimal effect of a fused iron-sensing domain on the regulation of even a stable indicator. It is currently unclear why the 73-aa domain was apparently capable of destabilizing a 73aa-IRP1 fusion construct in an iron-dependent manner (16). Assuming that this result is reproducible, one has to note that in this hybrid molecule, the 73-aa peptide was embedded within domain 1 of IRP1 (16), in a position that is quite equivalent to its natural context in IRP2. We speculate that the previously described function of this peptide as an iron sensor could be complemented by structural elements within IRP1, which is highly homologous to IRP2.
Iwai and coworkers recently proposed that the 73-aa domain interacts with the RING finger protein HOIL-1, which functions as an E3 ligase (38). They also proposed that heme binds directly to IRP2 and serves as a signal for its degradation, without excluding the involvement of additional E3 ligase activities (38). The data presented here show that IRP2 responds to multiple stimuli (see also below). We hypothesize that specificity could be provided by the existence of multiple recognition signals, which may not necessarily be restricted to a single degradation domain.
Iron-mediated degradation of IRP2 involves 2-oxoglutarate-dependent oxygenases.
The pathways for IRP2 and HIF-1α degradation share striking similarities, including the dependence on iron and oxygen, as well as the stimulatory effects of ascorbate. In addition, the degradation of HIF-1α exhibits a dose-dependent saturable pattern (33), similar to what is shown for IRP2 in Fig. 4. Thus, it is conceivable that these pathways may share additional elements, possibly including a common recognition signal for the degradation machinery. The mechanism of HIF-1α degradation has been a subject of intense studies, which led to the identification of prolyl hydroxylation as a crucial signal for HIF-1α recognition by the proteasome. The hydroxylation of P402 and P564 (24), catalyzed by members of the broader family of 2-oxoglutarate-dependent oxygenases (2, 32), is necessary for the interaction of HIF-1α with the von Hippel-Lindau (VHL) complex. This results in HIF-1α ubiquitination and degradation by the proteasome (14, 17).
On the basis of this information and the parallels between HIF-1α and IRP2 degradation, we designed experiments to address whether the recognition signal for IRP2 degradation by the proteasome also involves posttranslational modification by a 2-oxoglutarate-dependent oxygenase(s). We employed the substrate analog DMOG, a known inhibitor of HIF-1α prolyl 4-hydroxylases (4, 17), and evaluated its potential to protect IRP2 against iron-mediated degradation. An important control experiment showed that DMOG lacks any apparent iron-chelating properties (Fig. 8A).
The experiments with DMOG for which results are shown in Fig. 8 and 9 provide unequivocal pharmacological evidence for iron-mediated degradation of IRP2 by a pathway involving a 2-oxoglutarate-dependent oxygenase(s). While this work was under review, Hanson et al. reported that DMOG antagonizes the iron-dependent degradation of chimeric IRP2 (both the wild type and a mutant lacking the 73-aa domain) in previously iron-depleted HEK293 cells (11). Our data are in perfect agreement with these findings; they suggest, furthermore, that the pathway involving the 2-oxoglutarate-dependent oxygenase(s) operates only when iron-deficient cells are exposed to iron. The inhibitor is far less efficient in protecting IRP2 under conditions when iron is administered to previously non-iron-perturbed “control” cells (Fig. 9A). Thus, our data indicate that there is more than one signal targeting IRP2 for degradation in response to iron. We speculate that the nature of the signals possibly correlates with the iron status of the cell, which is reflected in the so-called “labile iron pool” (LIP), a fraction of intracellular iron presumably bound to low-molecular-weight chelates (18). The nature of the signals may also correlate with the oxygen status of the cell.
We propose that iron administration to cells with a depleted LIP results in IRP2 degradation by a pathway involving the activity of a 2-oxoglutarate-dependent oxygenase(s). Increase in the LIP could activate alternative pathways. Such a model can explain the partial (∼25 to 30%) degradation of IRP2 observed in the presence of FAC and DMOG (Fig. 9B). Our model also accommodates the possibility that the expression of the putative IRP2-modifying 2-oxoglutarate-dependent oxygenase(s) could be iron dependent. Our preliminary data suggest that the expression of human prolyl 4-hydroxylase alpha subunit mRNA (NCBI accession number M24487) is profoundly stimulated in iron-deficient cells (data not shown). Interestingly, the iron-dependent degradation of IRP2 in iron-deficient cells is sensitive to the protein synthesis inhibitors cycloheximide and anisomycin (12). This observation led to the proposal that IRP2 degradation requires the synthesis of another protein. Since DMOG does not affect global protein synthesis, the cycloheximide- and anisomycin-sensitive factor could be a 2-oxoglutarate-dependent oxygenase. The targets of this oxygenase within IRP2 remain to be identified.
This work also raises the possibility that the VHL complex could be involved in IRP2 degradation. It was recently found that chimeric IRP2 and pVHL interact in cotransfection and coimmunoprecipitation experiments (J. Wang and K. Pantopoulos, submitted for publication). Furthermore, overexpression of pVHL could relieve the saturation in the IRP2 degradation machinery. However, endogenous IRP2 is efficiently degraded by iron in VHL−/− cells, even after pretreatment with DFO, suggesting that pVHL itself is not necessary for IRP2 degradation. Future work will have to clarify whether the IRP2-pVHL interaction is physiologically important.
Acknowledgments
We thank Tracey Rouault for generously providing antibodies against IRP2, Prem Ponka for the pcDNA3-IRP23CS plasmid and for helpful suggestions, and Andreas Schleifenbaum and Karsten Schulz for the synthesis of DMOG. We also thank Matthias Pawlak for helpful experimental contributions. J.W. holds a fellowship from the Canadian Institutes of Health Research (CIHR). M.W.H. acknowledges funds from the Gottfried Wilhelm Leibniz Prize. K.P. is a scholar of CIHR and a researcher of the Canada Foundation for Innovation (CFI).
This work was supported by grants from the Canadian Institutes for Health Research (CIHR) and the Fonds de la Recherche en Santé du Quebéc (FRSQ).
REFERENCES
- 1.Bourdon, E., D. K. Kang, M. C. Ghosh, S. K. Drake, J. Wey, R. L. Levine, and T. A. Rouault. 2003. The role of endogenous heme synthesis and degradation domain cysteines in cellular iron-dependent degradation of IRP2. Blood Cells Mol. Dis. 31:247-255. [DOI] [PubMed] [Google Scholar]
- 2.Bruick, R. K., and S. L. McKnight. 2001. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294:1337-1340. [DOI] [PubMed] [Google Scholar]
- 3.Eisenstein, R. S. 2000. Iron regulatory proteins and the molecular control of mammalian iron metabolism. Annu. Rev. Nutr. 20:627-662. [DOI] [PubMed] [Google Scholar]
- 4.Epstein, A. C., J. M. Gleadle, L. A. McNeill, K. S. Hewitson, J. O'Rourke, D. R. Mole, M. Mukherji, E. Metzen, M. I. Wilson, A. Dhanda, Y. M. Tian, N. Masson, D. L. Hamilton, P. Jaakkola, R. Barstead, J. Hodgkin, P. H. Maxwell, C. W. Pugh, C. J. Schofield, and P. J. Ratcliffe. 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107:43-54. [DOI] [PubMed] [Google Scholar]
- 5.Erlitzki, R., J. C. Long, and E. C. Theil. 2002. Multiple, conserved iron-responsive elements in the 3′-untranslated region of transferrin receptor mRNA enhance binding of iron regulatory protein 2. J. Biol. Chem. 277:42579-42587. [DOI] [PubMed] [Google Scholar]
- 6.Frishman, D., and M. W. Hentze. 1996. Conservation of aconitase residues revealed by multiple sequence analysis. Implications for structure/function relationships. Eur. J. Biochem. 239:197-200. [DOI] [PubMed] [Google Scholar]
- 7.Goessling, L. S., D. P. Mascotti, and R. E. Thach. 1998. Involvement of heme in the degradation of iron-regulatory protein 2. J. Biol. Chem. 273:12555-12557. [DOI] [PubMed] [Google Scholar]
- 8.Guo, B., J. D. Phillips, Y. Yu, and E. A. Leibold. 1995. Iron regulates the intracellular degradation of iron regulatory protein 2 by the proteasome. J. Biol. Chem. 270:21645-21651. [DOI] [PubMed] [Google Scholar]
- 9.Haile, D. J., T. A. Rouault, C. K. Tang, J. Chin, J. B. Harford, and R. D. Klausner. 1992. Reciprocal control of RNA-binding and aconitase activity in the regulation of the iron-responsive element binding protein: role of the iron-sulfur cluster. Proc. Natl. Acad. Sci. USA 89:7536-7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanson, E. S., L. M. Foot, and E. A. Leibold. 1999. Hypoxia post-translationally activates iron-regulatory protein 2. J. Biol. Chem. 274:5047-5052. [DOI] [PubMed] [Google Scholar]
- 11.Hanson, E. S., M. L. Rawlins, and E. A. Leibold. 2003. Oxygen and iron regulation of iron regulatory protein 2. J. Biol. Chem. 278:40337-40342. [DOI] [PubMed] [Google Scholar]
- 12.Henderson, B. R., and L. C. Kühn. 1995. Differential modulation of the RNA-binding proteins IRP1 and IRP2 in response to iron. IRP2 inactivation requires translation of another protein. J. Biol. Chem. 270:20509-20515. [DOI] [PubMed] [Google Scholar]
- 13.Hentze, M. W., and L. C. Kühn. 1996. Molecular control of vertebrate iron metabolism: mRNA-based regulatory circuits operated by iron, nitric oxide and oxidative stress. Proc. Natl. Acad. Sci. USA 93:8175-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 15.Iwai, K., S. K. Drake, N. B. Wehr, A. M. Weissman, T. LaVaute, N. Minato, R. D. Klausner, R. L. Levine, and T. A. Rouault. 1998. Iron-dependent oxidation, ubiquitination, and degradation of iron regulatory protein 2: implications for degradation of oxidized proteins. Proc. Natl. Acad. Sci. USA 95:4924-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwai, K., R. D. Klausner, and T. A. Rouault. 1995. Requirements for iron-regulated degradation of the RNA binding protein, iron regulatory protein 2. EMBO J. 14:5350-5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 18.Kakhlon, O., and Z. I. Cabantchik. 2002. The labile iron pool: characterization, measurement, and participation in cellular processes. Free Radic. Biol. Med. 33:1037-1046. [DOI] [PubMed] [Google Scholar]
- 19.Kang, D. K., J. Jeong, S. K. Drake, N. Wehr, T. A. Rouault, and R. L. Levine. 2003. Iron-regulatory protein 2 as iron sensor: iron-dependent oxidative modification of cysteine. J. Biol. Chem. 278:14857-14864. [DOI] [PubMed] [Google Scholar]
- 20.Kim, H.-Y., R. D. Klausner, and T. A. Rouault. 1995. Translational repressor activity is equivalent and is quantitatively predicted by in vitro RNA binding for two iron-responsive element-binding proteins, IRP1 and IRP2. J. Biol. Chem. 270:4983-4986. [DOI] [PubMed] [Google Scholar]
- 21.Kim, S., and P. Ponka. 1999. Control of transferrin receptor expression via nitric oxide-mediated modulation of iron-regulatory protein 2. J. Biol. Chem. 274:33035-33042. [DOI] [PubMed] [Google Scholar]
- 22.Kim, S., and P. Ponka. 2002. Nitrogen monoxide-mediated control of ferritin synthesis: implications for macrophage iron homeostasis. Proc. Natl. Acad. Sci. USA 99:12214-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaVaute, T., S. Smith, S. Cooperman, K. Iwai, W. Land, E. Meyron-Holtz, S. K. Drake, G. Miller, M. Abu-Asab, M. Tsokos, R. Switzer III, A. Grinberg, P. Love, N. Tresser, and T. A. Rouault. 2001. Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat. Genet. 27:209-214. [DOI] [PubMed] [Google Scholar]
- 24.Masson, N., C. Willam, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBO J. 20:5197-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 26.Mueller, S., and K. Pantopoulos. 2002. Activation of iron regulatory protein-1 (IRP1) by oxidative stress. Methods Enzymol. 348:324-337. [DOI] [PubMed] [Google Scholar]
- 27.Pantopoulos, K., N. Gray, and M. W. Hentze. 1995. Differential regulation of two related RNA-binding proteins, iron regulatory protein (IRP) and IRPB. RNA 1:155-163. [PMC free article] [PubMed] [Google Scholar]
- 28.Pantopoulos, K., and M. W. Hentze. 1995. Rapid responses to oxidative stress mediated by iron regulatory protein. EMBO J. 14:2917-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouault, T., and J. B. Harford. 2000. Translational control of ferritin synthesis, p. 655-670. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Rouault, T. A. 2002. Post-transcriptional regulation of human iron metabolism by iron regulatory proteins. Blood Cells Mol. Dis. 29:309-314. [DOI] [PubMed] [Google Scholar]
- 31.Samaniego, F., J. Chin, K. Iwai, T. A. Rouault, and R. D. Klausner. 1994. Molecular characterization of a second iron-responsive element binding protein, iron regulatory protein 2. J. Biol. Chem. 269:30904-30910. [PubMed] [Google Scholar]
- 32.Schofield, C. J., and Z. Zhang. 1999. Structural and mechanistic studies on 2-oxoglutarate-dependent oxygenases and related enzymes. Curr. Opin. Struct. Biol. 9:722-731. [DOI] [PubMed] [Google Scholar]
- 33.Tanimoto, K., Y. Makino, T. Pereira, and L. Poellinger. 2000. Mechanism of regulation of the hypoxia-inducible factor-1 alpha by the von Hippel-Lindau tumor suppressor protein. EMBO J. 19:4298-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toth, I., and K. R. Bridges. 1995. Ascorbic acid enhances ferritin mRNA translation by an IRP/aconitase switch. J. Biol. Chem. 270:19540-19544. [DOI] [PubMed] [Google Scholar]
- 35.Toth, I., J. T. Rogers, J. A. McPhee, S. M. Elliott, S. L. Abramson, and K. R. Bridges. 1995. Ascorbic acid enhances iron-induced ferritin translation in human leukemia and hepatoma cells. J. Biol. Chem. 270:2846-2852. [DOI] [PubMed] [Google Scholar]
- 36.Wang, J., J. L. Buss, G. Chen, P. Ponka, and K. Pantopoulos. 2002. The prolyl 4-hydroxylase inhibitor ethyl-3,4-dihydroxybenzoate generates effective iron deficiency in cultured cells. FEBS Lett. 529:309-312. [DOI] [PubMed] [Google Scholar]
- 37.Wang, J., and K. Pantopoulos. 2002. Conditional derepression of ferritin synthesis in cells expressing a constitutive IRP1 mutant. Mol. Cell. Biol. 22:4638-4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamanaka, K., H. Ishikawa, Y. Megumi, F. Tokunaga, M. Kanie, T. A. Rouault, I. Morishima, N. Minato, K. Ishimori, and K. Iwai. 2003. Identification of the ubiquitin-protein ligase that recognizes oxidized IRP2. Nat. Cell Biol. 5:336-340. [DOI] [PubMed] [Google Scholar]