Abstract
The rapid induction of alpha interferon (IFN-α) and IFN-β expression plays a critical role in the innate immune response against viral infection. We studied the effects of transforming growth factor β (TGF-β) and its intracellular effectors, the Smads, on the function of IRF-7, an essential transcription factor for IFN-α and -β induction. IRF-7 interacted with Smads, and IRF-7, but not IRF-3, cooperated with Smad3 to activate IFN-β transcription. This transcriptional cooperation occurred at the IRF-binding sequences in the IFN-β promoter, and dominant-negative interference with TGF-β receptor signaling and Smad3 function decreased IRF-7-mediated transcription. Furthermore, elimination of Smad3 expression in Smad3−/− fibroblasts delayed and decreased double-stranded RNA-induced expression of endogenous IFN-β, whereas restoration of Smad3 expression enhanced IFN-β induction. The IRF-7-Smad3 cooperativity resulted from the regulation of the transactivation activity of IRF-7 by Smad3, and dominant-negative interference with Smad3 function decreased IRF-7 activity. Consistent with the regulation by Smad3, the transcriptional activity of IRF-7 depended on and was regulated by TGF-β signaling. Our studies underscore a role of TGF-β/Smad3 signaling in IRF-7-mediated induction of IFN-β expression.
Upon infection by viruses, cells activate the expression of alpha and beta interferons (IFN-α and -β) as key components of the innate immune response (25, 42, 77, 78). Following transcriptional activation of the corresponding genes, infected cells secrete these IFNs, which then bind to their cell surface receptors. Consequent activation of the Jak-STAT signaling pathway leads to induction of IFN target genes responsible for a broad array of biological effects, including the antiviral response (40, 75, 77). The Jak-STAT signaling mechanisms and transcriptional activation by STATs are well characterized. Much less is known about the signaling pathways that lead from viral infection to transcriptional activation of the IFN-α and -β genes. A double-stranded RNA intermediate generated in the course of infection by many DNA and RNA viruses is often considered a key component in the ability of viruses to activate type I IFN expression; accordingly, synthetic double-stranded poly(I-C) is commonly used as a potent inducer of IFN-α and -β (10).
In contrast to the multiple IFN-α genes, only one IFN-β gene exists in mice and humans. Extensive research during the last few years has aimed at elucidating the mechanisms that lead to the rapid transcriptional activation of the IFN-α and -β genes in response to viral infection and has focused on the IFN-β promoter. Key players in their transcriptional activation are IRF-3 and IRF-7, two structurally related members of the IRF (IFN regulatory factors) family of transcription factors (66, 76, 77, 90, 95). IRF-3 and IRF-7 are both required for efficient induction of IFN-β, and they cooperate with each other as DNA-binding transcription factors at the promoter (25, 41, 42, 77, 78). Both IRF-3 and IRF-7 are activated by phosphorylation within their C-terminal segment and consequent homo- or heterodimerization following virus infection (3, 21, 49, 66, 68, 90). However, whereas IRF-3 is fully latent in the absence of viral infection, IRF-7 has a basal activity that is enhanced upon infection (21, 45, 66, 68, 90). Other extracellular stimuli, including lipopolysaccharide, DNA damaging agents, and UV light, also activate IRF-3 and IRF-7, most likely through different mechanisms (67). While IRF-3 is constitutively expressed in most cells, IRF-7 expression is induced by exposure of cells to IFN (42, 77, 78). Most cells are thought to express low levels of IFN-α and -β in the absence of virus infection, thus providing a readiness to mediate a full IFN-α/β response upon virus infection through a positive feedback mechanism (77).
The IFN-β promoter contains several positive and negative regulatory cis elements. Among the four positive regulatory domains (PRDs), designated PRDI through PRDIV, PRDIII and PRDI are binding sequences for IRF-7 and IRF-3 as well as other IRFs. The PRDII sequence is recognized by the transcriptional activator NF-κB, and the PRDIV sequence binds a heterodimer of the basic leucine zipper proteins ATF-2 and c-Jun (47, 56). Viral infection activates these transcription factors and leads to recruitment of the transcription coactivator p300/CBP, the architectural factor HMGI (Y), and the assembly of a multiprotein enhanceosome complex on the promoter (25, 47, 56). While the ability of poly(I-C) and viral infection to induce IFN-α and -β transcription is well established, no signaling pathways are known to cross talk with the pathway that leads to IFN-β expression or to regulate the transcriptional activation of IRF-3 or -7.
One signaling pathway, which engages in cross talk with other signaling pathways and is involved in immune response modulation, is initiated by the cytokine transforming growth factor β (TGF-β). TGF-β is the prototype member of a large family of structurally related growth and differentiation factors that includes activins and bone morphogenetic proteins (11). TGF-β1 is prominently expressed in hematopoietic and immune cells, and TGF-β1 expression and activation are upregulated at sites of tissue injury and in tumorigenesis (12, 14, 39, 82). At the cellular level, TGF-β regulates proliferation, differentiation, apoptosis, adhesion, motility, and extracellular matrix deposition, and the effects are often cell type and context dependent. Through its broad array of cellular activities, TGF-β regulates cell-fate determination, tissue homeostasis, and wound healing and modulates many aspects of immune functions (12, 39). TGF-β also plays a critical role in the immune response to pathogens, including viruses, bacteria, yeast, and protozoa (61), and TGF-β1 is transcriptionally induced following viral infections, e.g., by cytomegalovirus (CMV) (27, 53, 91), human immunodeficiency virus type 1 (24, 30), and hepatitis B (92). Many studies suggest that TGF-β has a negative influence on the host response and a beneficial effect on the survival and growth of intracellular pathogens (61). However, other studies correlate TGF-β with enhanced resistance to microbes (51, 52, 74). Consistent with its context-dependent effect on many cellular functions, the complex role of TGF-β in the host defense against pathogens remains to be fully characterized.
The Smads act as intracellular effectors of gene expression responses to TGF-β, and a general model for how TGF-β signaling activates transcription of target genes has been established (13, 33, 50, 55, 71). At the cell surface, TGF-β binds to a complex of type I and type II transmembrane receptor serine/threonine kinases, resulting in transphosphorylation and consequent activation of the type I receptor by the type II receptor kinase. The activated type I TGF-β receptors phosphorylate Smad2 and Smad3 at C-terminal serines, and these receptor-activated Smads then form a complex with Smad4. The TGF-β-activated, heteromeric Smad complexes then translocate into the nucleus, where they induce or repress transcription of defined genes. While most Smads have the capacity to bind DNA at a favorable sequence context, the receptor-activated Smads naturally activate transcription through physical interaction and functional cooperation with transcription factors with defined DNA sequence binding (33, 50, 71, 79, 98). For example, the TGF-β-activated Smad3 can cooperate with the basic leucine zipper protein c-Jun (43, 60, 84, 97), the basic helix-loop-helix protein TFE3 (31, 32), the Zn finger protein Sp1 (8, 20, 58), and AML/RUNX transcription factors (28, 57, 99), depending on the target gene. The receptor-activated Smads also interact through their C termini with CBP or p300, two closely related coactivators (19, 34, 69, 80). This interaction is stabilized by Smad4, providing a mechanism by which Smad4 acts as a Smad coactivator (19).
We now provide evidence that Smad3 physically and functionally interacts with IRF-7 and that TGF-β/Smad3 signaling regulates the transcriptional activity of IRF-7 at the IFN-β promoter. Our results strongly suggest that the transcriptional activation of the IFN-β gene by IRF-7 depends on and is regulated by the endogenous level of TGF-β-activated Smad3.
MATERIALS AND METHODS
Yeast two-hybrid screen and identification of IRF-7.
The LexA-based Saccharomyces cerevisiae two-hybrid system (26, 93) was used to screen for proteins that specifically interact with Smad3. Full-length, Flag-tagged human Smad3 in the bait plasmid pEG202 (85) and a cDNA library derived from HeLa cell mRNA in the prey plasmid pJG4-5 were transformed into yeast EGY48 by using the Alkali Cation kit (Bio 101, Inc.). Protein-protein interactions were determined by scoring β-galactosidase activity on 5-bromo-4-chloro-3-indolyl-β-d-galactoside plates containing galactose or glucose.
Expression plasmids.
Smad2, -3, and -4 and the Smad3 deletion mutants Smad3C and Smad3ΔC were expressed as Flag-tagged proteins from the CMV promoter in pRK5 expression plasmids (96, 97). The constitutively active TβRI and TβRII chimera, R(II-I)C, and the dominant-negative TβRII, RII-DN, have been described previously (6, 18). Full-length IRF-7 was expressed as an N-terminally Flag-tagged or C-terminally Myc-tagged version. The DNA fragments encoding wild-type human IRF-7 were generated by PCR using pcDNA-IRF-7A (94) as a template and inserted into the BamHI-SalI sites of the mammalian expression vectors pXF1F and pRK5M (17), thus generating pXF1F-IRF-7 and pRK5M-IRF-7, respectively. pRK5M-IRF-7 N417 contains the N-terminal amino acids 1 to 417 of IRF-7 and was expressed as a C-terminally Myc-tagged protein. The DNA fragments encoding the N417 segment were excised from pRK5M-IRF-7 by EcoRI and ligated into the EcoRI sites of pRK5M. pXF1M-IRF-7 C415, containing amino acids 415 to 503, was generated by digesting pXF1F-IRF-7 with EcoRI-SalI, and the corresponding 0.3-kb fragment was ligated into EcoRI-SalI sites of expression vector pXF1M. Full-length human IRF-3 was expressed as an N-terminally Myc-tagged protein. The DNA fragments encoding wild-type IRF-3 were generated by PCR using pHA-IRF-3 (35), provided by P. M. Pitha (The Johns Hopkins University School of Medicine, Baltimore, Md.), as a template and inserted into the EcoRI-SalI sites of the mammalian expression vector pXF1M (17). The murine IRF-7 coding sequence with EcoRI and XhoI recognition sites was generated by PCR using pBabe/His-tag-IRF-7 (64) as a template and inserted into the EcoRI-SalI sites of pXF1M. All expression plasmids were verified by DNA sequencing.
In vitro GST protein binding assay.
Plasmids that direct expression of glutathione S-transferase (GST)-fused Smad2, -3, and -4 and Smad3NL in Escherichia coli have been described elsewhere (97). The Smad proteins fused to GST were expressed in E. coli and semipurified by glutathione-Sepharose 4B adsorption according to the manufacturer's recommendations (Amersham-Pharmacia Biotech). 35S-labeled IRF-7 was generated by in vitro transcription from the pRK5-based expression vector described above and translation in the presence of [35S]methionine. In vitro protein binding assays to test the ability of the radiolabeled IRF-7 to interact with GST-fused Smads were performed as described previously (60). Specifically associated proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by autoradiography. Parallel Coomassie blue staining revealed the integrity and loading of the GST fusion proteins.
Luciferase reporter plasmids.
The p-125Luc plasmid contains the −125 to +19 segment of the human IFN-β gene promoter, which drives the expression of a luciferase reporter (88). The p-125AALuc reporter is identical, except that it contains mutations in the NF-κB-binding sequence, PRDII (89). Both plasmids were obtained from T. Fujita (The Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan). The reporter plasmid p31x2-Luc, containing two copies of PRDIII-PRDI sequences of human IFN-β, was made by ligation of oligonucleotides corresponding to the PRDIII-PRDI sequences into the HindIII site of the minimal TATA box promoter, pTA-Luc (99). The oligonucleotides used were the following: sense, 5′ AGC TTA GGA AAA CTG AAA GGG AGA AGT GAA AGT GA 3′; antisense, AGC TTC ACT TTC ACT TCT CCC TTT CAG TTT TCC TA 3′. The corresponding mutant reporter, pm31x2-Luc, was similarly created by ligating the mutant oligonucleotides (lowercase letters denote mutations) with the following sequences: sense, 5′ AGC TTA GGA gcA CTG AAA GGG AGA AGT GAg cGT GA 3′; antisense, AGC TTC ACg cTC ACT TCT CCC TTT CAG Tgc TCC TA 3′. All other promoter mutants were made by two-round PCR-based mutagenesis. First, a 5′ vector primer together with a primer containing specific mutations were used to generate the fragment containing the desired mutations, and then this fragment was used as a primer together with a 3′-vector-specific primer to produce the second fragment. The final PCR product was digested with HindIII/SalI and ligated into the HindIII/SalI sites of p-125Luc. These mutant reporters were generated using the following mutation-containing primers: (i) mPRDIV-Luc reverse, 5′ GTT TTC CTA TGT CcT TTA CAT TTT AGT AG 3′ (−98G primer; following reference 16); (ii) mPRDIII-Luc reverse, 5′ CAC TTC TCC CTT TCA GTg cTC CTA TGT CAT TTA C 3′ (−88G/−87C primer); (iii) mPRDIII-mPRDI primer reverse, 5′ GGA ATT TCC CAC gcT CAC TTC TCC CTT TCA GTg cTC CTA AGT CAT TTA C 3′ (−68G/−67C/−88G/−87C primer). All promoter mutants were verified by DNA sequencing.
Cells and cell culture.
Mouse embryonic fibroblasts (MEFs) from Smad3 knockout mice and wild-type littermates (9), obtained from X.-F. Wang (Duke University Medical School, Durham, N.C.), were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 15% fetal bovine serum (FBS), 100 IU of ampicillin/ml, and 100 μg of streptomycin/ml (1× P/S). HeLa cells, HepG2 cells, COS1 cells, and the ecotropic retroviral packaging cell line Phoenix E (obtained from G. Nolan, Stanford University) were maintained in DMEM supplemented with 10% FBS and 1× P/S. All cells were routinely maintained at 37°C in the presence of 5% CO2.
Transient transfections and luciferase reporter assays.
Transient transfections of HeLa and HepG2 cells were performed in six-well tissue culture plates using Fugene 6 according to the manufacturer's instructions (Roche). For each transfection, 0.5 μg of the luciferase reporter plasmid and, when indicated, 0.1 μg of the Smad expression plasmid and 8 to 10 ng of the IRF-7 expression plasmid were used. The total amount of transfected DNA was kept constant by adding empty pRK5 vector, as needed. Cotransfection of 20 ng of the β-galactosidase expression plasmid (pRK5-β-Gal) allowed all transfections to be normalized to the β-galactosidase activity. TGF-β treatment and luciferase assays were carried out as described elsewhere (17). All experiments were carried out in duplicate and repeated at least three times.
To evaluate the effect of poly(I-C) on IRF-7 activity, we transfected HeLa cells with 0.25 μg of the reporter plasmid p31x2-Luc, 10 ng of pRK5-β-Gal, and different amounts of IRF-7 expression plasmid using Fugene 6 (Roche). At 12 h after transfection, cells were incubated with 50 μg of poly(I-C)/ml and DEAE dextran (400 μg/ml) in phosphate-buffered saline (PBS) for 2 h, then washed with PBS and incubated with DMEM with 2% FBS. At 24 h later, luciferase assays were performed as described previously (17).
Immunoprecipitation and Western blot analyses.
COS1 cells were transfected with expression plasmids for Flag-tagged Smads and Myc-tagged IRF-7 or derivatives, using Lipofectamine (GIBCO-BRL) according to the manufacturer's instructions. At 48 h posttransfection, cells were lysed in MLB buffer consisting of 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% Triton X-100, and Complete protease inhibitors (Roche). Cell lysates were precleared with anti-mouse immunoglobulin G (IgG; Jackson Laboratories) and protein A-Sepharose 4B (Amersham-Pharmacia), and immunoprecipitations were performed using the M2 anti-Flag monoclonal antibody (Sigma) or anti-Myc 9E10 antibody (Covance). Immunoprecipitated proteins were subjected to SDS-PAGE, transferred to polyvinylidene difluoride membrane (Perkin-Elmer), and blotted with anti-Myc 9E10 (Covance) or anti-Flag M2 (Sigma) antibodies. Proteins bound with antibodies were visualized using enhanced chemiluminescence (ECL; Amersham-Pharmacia).
For IRF-7/Smad3 endogenous interaction, HeLa cells were grown to ∼80% confluence in 150-mm tissue culture dishes. Cells were washed once with PBS and then placed in PBS containing 0.4 mg of DEAE-dextran/ml, 50 μg of poly(I-C) (Amersham-Pharmacia)/ml with or without 5 ng of TGF-β/ml. After 2 h at 37°C, cells were harvested and processed as described previously (72) except that the lysis buffer consisted of 1% NP-40, 50 mM Tris (pH 8.0), 137 mM NaCl, and protease inhibitor cocktail (Roche). Lysates clarified by 20 min of centrifugation in a microcentrifuge at 20,800 × g were then immunoprecipitated either with rabbit anti-mouse IgG (Jackson ImmunoResearch) or rabbit anti-Smad3 (Zymed) and protein A-Sepharose. Immunoprecipitated proteins were separated on SDS-PAGE. Western blotting was performed using anti-IRF-7 antibody (Santa Cruz). Bands were visualized using ECL+ reagents (Amersham-Pharmacia).
In vivo [32P]phosphate labeling.
MEFs were transfected with XF1M-mIRF-7 to express Myc-tagged murine IRF-7. At 12 h after transfection, cells were incubated in phosphate-free DMEM with 0.2% FBS for 3 h and then incubated with 1 mCi of [32P]orthophosphate/ml. Five micrograms of poly(I-C) and Lipofectamine mix or Lipofectamine alone was added to cultures 1 h later, and cells were incubated for 4 h. Cells were lysed, and whole-cell lysates were subjected to immunoprecipitation. The immunoprecipitated proteins were separated by SDS-6% PAGE. Results were visualized by autoradiography.
Biotinylated DNA affinity precipitation.
Cell lysates were incubated with 15 nmol of biotinylated double-stranded oligonucleotides bound on streptavidin-coated MagneSphere paramagnetic particles and 5 μg of poly(dI-dC) at 4°C for 1 h. The paramagnetic particle/DNA/protein complexes were washed four times, eluted in sample buffer, and separated on SDS-PAGE. Proteins that specifically interacted with the DNA probe were detected by Western blotting as described above.
Northern blotting and RT-PCR analyses.
MEFs near 70% confluence were treated with 50 μg of poly(I-C)/ml and DEAE dextran (400 μg/ml) in PBS for 2 h and then washed with PBS and incubated with DMEM with 2% FBS. Total RNA was isolated from the cells at the indicated times after addition of poly(I-C) using the TRIzol kit (Life Technologies). A 15-μg aliquot of total RNA was denatured and electrophoresed in 1% formaldehyde-agarose gels and blotted onto Biotrans nylon membranes (ICN). The cDNA for mouse IFN-β was obtained from P. M. Pitha and labeled with [α-32P]dCTP using random primed labeling. Hybridization with 32P-labeled DNA probes was performed at 65°C as described previously (4). Results were visualized by autoradiography and quantified using a PhosphorImager (Molecular Dynamics). Reverse transcription-PCR (RT-PCR) of IRF-7 mRNA was performed using total RNA and the following primers: sense, 5′-CAGCGAGTGCTGTTTGGAGAC-3′; antisense, 5′-AAGTTCGTACACCTTATGCGG-3′. Twenty-nanogram aliquots of RT products were used as templates for PCRs.
Generation of stably infected cell lines.
Stable cell lines infected with LPCX-based retroviruses were established essentially as described previously (7). The LPCX and LPCX-Smad3 retroviral constructs were previously described (7). Briefly, Smad3−/− MEFs were plated at 4 × 104 cells/well in six-well plates the day before infection. To generate retroviruses, Phoenix E cells were transfected with LPCX or LPCX-Smad3 plasmid DNA using the calcium phosphate method. At 48 h after transfection, the conditioned medium containing retroviruses was collected and filtered through 0.45-μm-pore-size filters, supplemented with 8 μg of Polybrene (Sigma)/ml, and applied to Smad3−/− MEFs. Cells infected with the retroviruses were centrifuged at 1,800 rpm (GPR centrifuge; Beckman) for 45 min at room temperature. The viral supernatant was aspirated, fresh viruses plus Polybrene were added to the cells, and the centrifugation procedure was repeated. After infection, the cells were maintained in fresh growth medium at 37°C. At 48 h after infection, 2 μg of puromycin (Calbiochem)/ml was used for selection of stably infected cells.
Gal4 transactivation assays.
To create the plasmid for IRF-7 fused to the Gal4 DNA-binding domain, the DNA fragment encoding IRF-7 was excised from pXF1F-IRF-7 (described above) and ligated into the BamHI-SalI sites of the mammalian expression vector pXF1Gal4, a pRK5 derivative with the Gal4 DNA binding domain (amino acids 1 to 147) inserted between the ClaI/EcoRI sites of pRK5 (19). The abilities of IRF-7 to transactivate the heterologous Gal4 promoter reporter, pFR-Luc (Stratagene), were assayed as described elsewhere (19).
RESULTS
Interaction of Smad3 with IRF-7.
To better understand the functions of TGF-β in a broad array of cellular processes, we sought to identify proteins that interact with Smad3, a key effector of TGF-β signaling, using a yeast two-hybrid screen (26, 93) with full-length Smad3 as bait (85). One isolated cDNA encoded the C-terminal amino acids 362 to 503 of the human IRF-7 transcription factor (data not shown). This identification led us to characterize the physical and functional interactions of the TGF-β-activated Smads with full-length IRF-7, which was initially identified as IRF-7A (94).
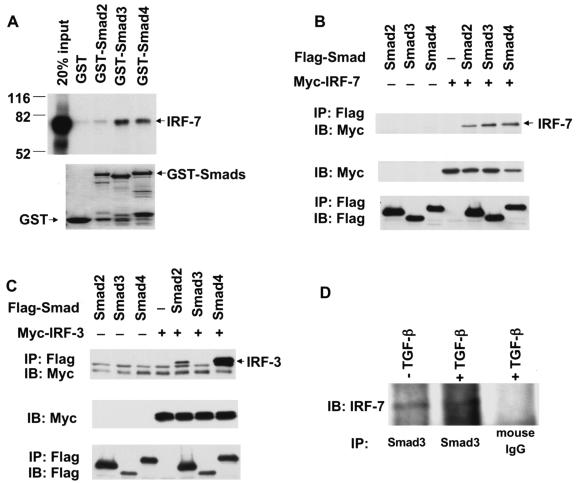
To assess whether IRF-7 interacts directly with Smad3, we evaluated the ability of in vitro-translated, 35S-labeled IRF-7 to interact with Smad proteins fused to GST. These GST adsorption experiments revealed that Smad3 and Smad4 were able to directly interact with IRF-7, whereas Smad2, which is also activated by TGF-β, displayed a much weaker interaction and the GST segment alone did not interact (Fig. 1A). Thus, among the TGF-β-activated Smads, Smad3 interacted efficiently with IRF-7, while Smad2 had only a low affinity.
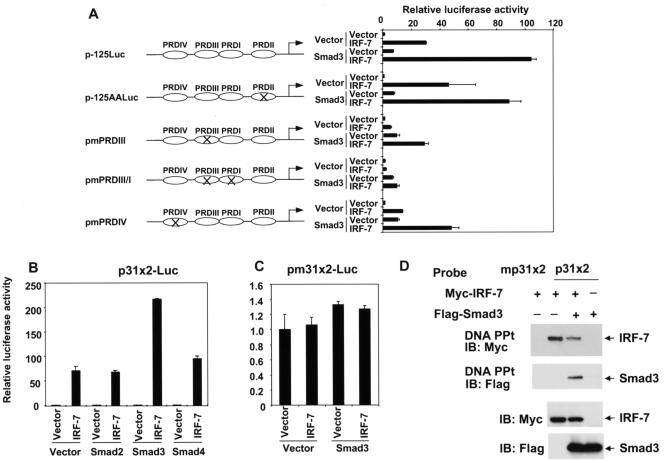
FIG. 1.
IRF-7 interacts with Smad3 in vitro and in vivo. (A) Direct interaction of 35S-labeled, in vitro-translated IRF-7 with GST-Smad fusion proteins, but not GST. Interacting IRF-7 was visualized following electrophoresis and autoradiography. IRF-7 interacted with Smad3 and 4, and to a lesser extent with Smad2. The lower panel represents Coomassie blue-stained gels showing equal loading of the fusion proteins. (B) Interaction of Smad3 with IRF-7 in vivo. COS1 cells were transfected with Flag-tagged Smads and Myc-tagged IRF-7. Cell lysates were immunoprecipitated (IP) with anti-Flag antibody, followed by immunoblotting (IB) with anti-Myc antibody to detect Smad-bound IRF-7. IRF-7 interacted efficiently with Smad3 and -4 and less efficiently with Smad2. (C) Interaction of Smads with IRF-3 in vivo. COS1 cells were transfected with Flag-tagged Smads and Myc-tagged IRF-3, and cells were processed as described for panel B. IRF-3 did not interact with Smad3, and it interacted efficiently with Smad4 and less efficiently with Smad2. Nonspecific IgG bands are visible in all lanes. The lower portions of panels B and C show the expression levels of the transfected proteins in the lysates, as assessed by immunoblotting. (D) Endogenous IRF-7 and Smad3 interact. HeLa cells were subjected to anti-Smad3 immunoprecipitations followed by Western blotting with anti-IRF-7. In a control experiment, the anti-Smad3 antibody was replaced with anti-mouse IgG.
To evaluate the in vivo interactions between Smads and IRF-7, we coexpressed Myc-tagged IRF-7 with Flag-tagged Smads in transfected COS cells, which allow high levels of transfection and expression yet have poor TGF-β responsiveness. Cell lysates were subjected to immunoprecipitation using anti-Flag antibodies followed by Western blotting with anti-Myc antibodies to detect the association. As shown in Fig. 1B, IRF-7 coprecipitated with Smad3 and Smad4 in anti-Flag immunoprecipitation experiments. The less-efficient interaction of Smad2 with IRF-7 was consistent with the weaker interaction in the GST adsorption assays. In contrast to IRF-7, IRF-3 did not detectably interact with Smad3 but was able to interact with Smad2 (Fig. 1C). IRF-7 and IRF-3 both interacted with Smad4 (Fig. 1B and C). Finally, as shown in Fig. 1D, Smad3 also interacted with IRF-7 at endogenous levels in HeLa cells, which express low levels of IRF-7 (data not shown). TGF-β treatment did not enhance the Smad3-IRF-7 interaction, which was likely due to similar distributions of IRF-7 and Smad3 in both nuclear and cytoplasmic fractions in the absence or presence of TGF-β and the poor responsiveness of HeLa cells to exogenous TGF-β (data not shown).
Defining the domains of Smad3 and IRF-7 that mediate the physical association.
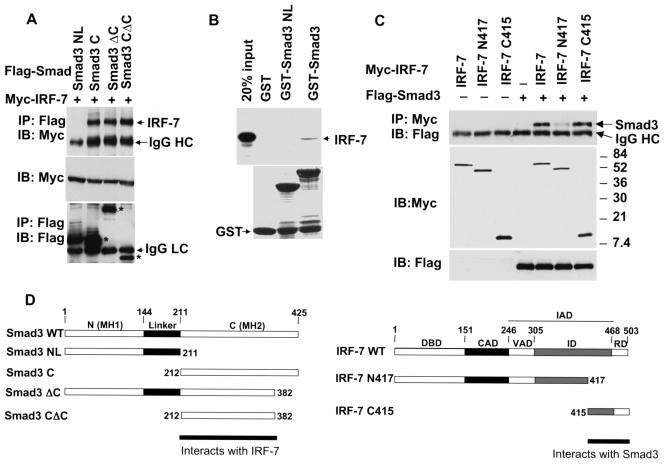
To define which segment of Smad3 interacts with IRF-7, we tested the interactions of several Smad3 deletion mutants with IRF-7 in coimmunoprecipitation assays (Fig. 2A). Deletion of the MH1 domain and the linker region of Smad3 still allowed the MH2 domain (Smad3C) to efficiently interact with IRF-7. Deletion of the C-terminal 43 amino acids, which are required for Smad trimerization and transcriptional activity (70, 85, 96), in the Smad3CΔC segment did not affect the interaction with IRF-7. Smad3NL, which comprises the MH1 domain and linker segment, did not interact with IRF-7. Accordingly, Smad3NL did not interact with IRF-7, while full-size Smad3 did, in GST adsorption assays (Fig. 2B). We conclude that Smad3 interacts through its MH2 domain with IRF-7.
FIG. 2.
The interaction of IRF-7 and Smad3 is mediated by the C-terminal domains of both proteins. (A) Smad3C, but not Smad3NL, interacts with IRF-7. COS1 cells were transfected with Flag-tagged Smad3 deletion mutants and Myc-tagged IRF-7. Cell lysates were subjected to immunoprecipitation with anti-Flag antibody as described in the legend for Fig. 1, followed by Western blotting (IB) using the antibodies indicated. The relative positions of the different Smad3 proteins are marked with an asterisk in the bottom panel. These results demonstrate an interaction of the MH2 domain of Smad3 with IRF-7. (B) Direct interaction of 35S-labeled, in vitro-translated IRF-7 with GST-Smad fusion proteins. Interacting IRF-7 was visualized following electrophoresis and autoradiography. The lower panel represents Coomassie blue-stained gels showing equal loading of the fusion proteins. IRF-7 interacted with Smad3, but not with Smad3NL. (C) Smad3 interacted with IRF-7 C415, i.e., amino acids 415 to 503 of IRF-7. COS1 cells were transfected with Myc-tagged IRF-7 deletion mutants and Flag-tagged Smad3, and cell lysates were subjected to immunoprecipitation with anti-Myc antibody, similar to the experiment shown in panel A. (D) Schematic diagram of Smad3 and IRF-7 and of the deletion mutants used in panels A and C. The interacting protein segments in Smad3 and IRF-7 are shown.
The IRF-7 segment that interacted with Smad3 in the yeast two-hybrid screen corresponded to the C-terminal amino acids 362 to 503. We generated a further truncation of that segment and, as shown in Fig. 2C, the C-terminal segment of IRF-7, containing amino acids 415 to 503, in the IRF-7 C415 mutant retained the ability to interact with Smad3. Truncation of amino acids 418 to 503 in the IRF-7 N417 mutant almost completely abolished the interaction of IRF-7 with Smad3. These data indicate that amino acids 415 to 503 of IRF-7 are sufficient and required for efficient interaction with Smad3 (Fig. 2D). This segment of IRF-7 corresponds to the signal-regulated domain of IRF-7, preceded by part of its inhibitory domain, and represents most of the sequence that is involved in IRF-7 homodimerization or heterodimerization with IRF-3 (3, 45, 66). Since this segment of IRF-7 is also part of its transactivation domain, we conclude that Smad3 and IRF-7 interact with each other through sequences in their transactivation domains.
Smad3, but not Smad2, cooperates with IRF-7 to activate transcription from the IFN-β promoter.
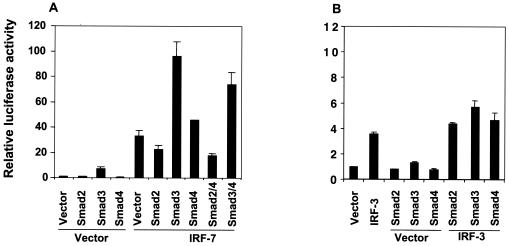
IRF-7 functions as both a transcriptional activator and repressor, depending on the promoter context (76, 94, 95). To date, the best-characterized function of IRF-7 is its role in transcriptional induction of the IFN-α and -β genes following viral infection. Similar to IRF-7, IRF-3 also plays a critical role in transcriptional activation of the IFN-β gene (25, 66, 76-78). We therefore studied the effects of ectopically expressed Smads on the transcriptional regulation of the IFN-β promoter by IRF-7 using a luciferase reporter, p-125Luc (88). This plasmid contains the luciferase reporter gene under the control of the −125 to +19 segment of the human IFN-β promoter and is commonly used to study the transcriptional regulation of the IFN-β gene (88, 89). Consistent with previous observations (45), expression of IRF-7 activated transcription from the IFN-β promoter in transfected HeLa cells (Fig. 3A). In the absence of coexpressed IRF-7, Smad2 did not activate transcription from the p-125Luc promoter, whereas Smad3 expression provided a moderate transcriptional activation. Coexpression of Smad3, but not Smad2, strongly enhanced the transcriptional activation of the IFN-β promoter by IRF-7 (Fig. 3A). Smad2 decreased the IRF-7-mediated transcription, reminiscent of the antagonism between Smad2 and Smad3 at the goosecoid promoter (37). A similar but weaker cooperation of Smad3 with IRF-7 was also observed in HepG2 cells (data not shown; see Fig. 5C).
FIG. 3.
IRF-7, but not IRF-3, cooperates with Smad3 to activate the human IFN-β promoter in HeLa cells. HeLa cells were transfected with the p-125Luc reporter plasmid and expression plasmids for Smads, IRF-7 (A) or IRF-3 (B), as indicated. Forty hours after transfection, cells were harvested and reporter activities were measured. Values, normalized for transfection efficiency, are shown as fold induction relative to basal promoter activity as described in Materials and Methods. Note the much lower luciferase expression scale and values in panel B compared with those in panel A.
FIG. 5.
Dominant-negative interference with Smad3 function inhibits transactivation by IRF-7. (A and B) The two dominant-negative mutants, Smad3ΔC and Smad3C, inhibited IRF-7-mediated transcription from p-125Luc (A) or p31x2-Luc (B) by IRF-7 in HeLa cells. HeLa cells were transfected with p-125Luc or p31x2-Luc, along with expression plasmids for IRF-7, Smad3, or mutants, as indicated. Luciferase activities were scored as described in the legend for Fig. 3. (C and D) Smad3ΔC and Smad3C inhibited IRF-7-mediated transcription from p-125Luc (C) or p31x2-Luc (D) in HepG2 cells. HepG2 cells were transfected with p-125Luc or p31x2-Luc, along with expression plasmids for IRF-7, Smad3, or relevant mutants, as indicated. Twenty hours after transfection, cells were treated with (solid bars) or without (open bars) TGF-β, and luciferase activities were measured. Normalized luciferase activities are represented relative to those of control vector-transfected cells in the absence of TGF-β.
Since receptor-activated Smads often form heteromeric complexes with Smad4, we also examined the effect of Smad4 on the transcriptional activation by IRF-7. Smad4 alone did not enhance transcription, but coexpression of Smad4 with IRF-7 consistently displayed a slightly higher transcriptional activity than that observed with IRF-7 alone (Fig. 3A). Coexpression of Smad4 did not further enhance the Smad3/IRF-7 cooperativity and did not significantly affect the inhibitory effect of Smad2 (Fig. 3A).
In contrast to IRF-7, IRF-3 exerted only a low level of transcriptional activation from the p-125Luc promoter, which is likely explained by its stringent dependence on virus activation (42, 90). This low activity was only minimally, if at all, enhanced by Smad3 or -4 (Fig. 3B). Together, these results indicate that Smad3, but not Smad2, selectively cooperates with IRF-7 to activate transcription from the human IFN-β promoter.
The IRF-7/IRF-3 binding sequences, PRDIII-PRDI, are necessary and sufficient to mediate the cooperation between Smad3 and IRF-7.
The −125 to +19 segment of the human IFN-β promoter contains four PRDs for transcriptional activation, named PRDI through PRDIV. Among these, PRDIII and PRDI are recognized by IRF-7 and IRF-3 (47, 76). PRDII is a binding site for the transcriptional activator NF-κB, composed of p65 and p50, while PRDIV interacts with a heterodimer of the basic leucine zipper proteins ATF-2 and c-Jun (47). Since Smad3 has been reported to functionally cooperate with c-Jun (43, 60, 84, 97), ATF-2 (63), and NF-κB (36, 46) in other promoter contexts, we examined whether the transcriptional cooperation of Smad3 and IRF-7 depended on these transcription factors.
As a first approach, we selectively inactivated these different binding sites in the −125 to +19 promoter segment by introducing mutations which abolished binding of the corresponding transcription factors without affecting the other binding sites. These mutant versions of p-125Luc were transfected into HeLa cells, and the transcriptional cooperation of IRF-7 and Smad3 was evaluated. As shown in Fig. 4A, inactivating mutations in the PRDII (16) or PRDIV (88) sites decreased the transcriptional activation by IRF-7, yet did not affect the cooperation of IRF-7 and Smad3. Two base substitutions in the highly conserved GAAA motif in the PRDIII sequence, one of the IRF-3/IRF-7 binding sites (44, 65), strongly decreased the transactivation by IRF-7 but did not abolish the transcriptional cooperation of IRF-7 and Smad3. However, inactivating mutations in both the PRDIII and PRDI sequences for IRF-3/IRF-7 binding (44, 65, 83) abolished the transcriptional activation by IRF-7 as well as the cooperation of Smad3 and IRF-7. These results indicate that the transcriptional cooperativity of Smad3 and IRF-7 depends on the ability of IRF-7 to interact with the PRDIII-PRDI sequence and that NF-κB and ATF-2/c-Jun binding to the promoter are not required for this cooperation.
FIG. 4.
The PRDIII-PRDI sequences in the IFN-β promoter are required and sufficient for Smad3/IRF-7 transcriptional cooperativity. (A) Mutational analysis of the human IFN-β promoter. PRDs were mutated individually, or in combinations, as depicted. The p-125Luc reporter plasmid or mutant reporter plasmids were transfected into HeLa cells, along with expression plasmids for Smad3 and/or IRF-7, as indicated. The transcriptional cooperation of Smad3 and IRF-7 was abolished following mutational inactivation of the PRDIII and PRDI sequences. (B) PRDIII-PRDI sequences are sufficient to mediate IRF-7/Smad3 transcriptional cooperation. A reporter plasmid containing two copies of the PRDIII-PRDI sequences of the IFN-β promoter, p31x2-Luc, was transfected into HeLa cells with various expression plasmids, as indicated. Smad3, but not the other Smads, strongly enhanced the transcription in the presence of IRF-7. (C) A reporter plasmid containing two copies of mutant PRDIII-PRDI sequences, pm31x2-Luc, was unable to be activated by IRF-7 or IRF-7/Smad3. Luciferase activities were scored as for Fig. 3. Note the much lower scale of luciferase activity compared to that in panel B. (D) DNA precipitation demonstrates the binding of IRF-7 to the p31x2 DNA sequence, but not to a mutated p31x2 sequence. Smad3 alone did not bind to the p31x2 sequence, but it did do so in the presence of IRF-7. The lower panels show the expression levels of IRF-7 and Smad3 in the lysates.
To further investigate whether the IRF-7-binding sequence per se is sufficient to mediate Smad3-IRF-7 cooperation, we generated an artificial luciferase reporter, p31x2-Luc, which contains two tandem repeats of the PRDIII-PRDI sequence of the human IFN-β promoter upstream from a minimal TATA box promoter. As seen with p-125Luc (Fig. 3A), IRF-7 activated transcription from the PRDIII-PRDI sequence in HeLa cells, while Smad2, -3, or -4 did not affect the basal level of transcription (Fig. 4B). However, Smad3 and IRF-7 synergized to induce transcriptional activation, which was much higher than with IRF-7 alone. This transcriptional cooperativity was not seen in the presence of Smad2, whereas coexpression of Smad4 with IRF-7 resulted in a consistent minimal enhancement above the IRF-7-mediated transcription (Fig. 4B). This slight enhancement of IRF-7-mediated transcription by Smad4 was similar to what was seen with the p-125Luc promoter-reporter plasmid (Fig. 3A). Inactivating mutations of the IRF-7 binding sites in the pm31x2-Luc plasmid abolished both the transcriptional activation by IRF-7 and cooperativity of Smad3 with IRF-7 (Fig. 4C). Together, these results indicate that the IRF-7 binding sites alone are sufficient and necessary to mediate the cooperation of IRF-7 and Smad3.
The transcriptional cooperation of Smad3 and IRF-7 resulted from the interaction of both transcription factors at the PRDIII-PRDI sequence. Using DNA interaction assays with a biotinylated oligonucleotide, we showed that IRF-7 interacted with the PRDIII-PRDI sequence, as expected, and that this interaction allowed recruitment of Smad3 to the DNA. Smad3 was unable to bind to this sequence in the absence of IRF-7 (Fig. 4D).
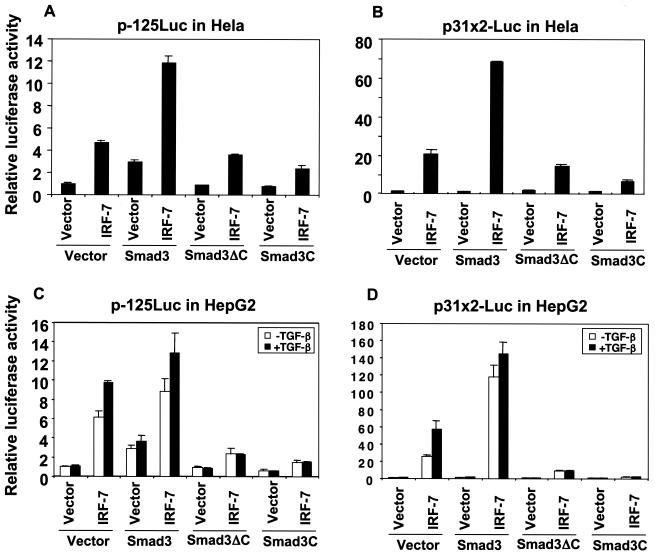
Dominant-negative interference with Smad3 function decreases IRF-7-mediated transcription.
To begin to characterize the role of endogenous Smad3 in IRF-7-mediated transcriptional activation, we assessed the effects of two Smad3 mutants, Smad3ΔC and Smad3C, on the transcriptional activation of both the p-125Luc and p31x2-Luc reporters. These assays were carried out in HeLa cells and HepG2 cells. HeLa cells have only a low-level responsiveness to exogenous TGF-β in transient-transfection reporter assays (reference 20 and data not shown), while HepG2 cells are highly responsive to TGF-β (19, 20, 32, 63, 99). In addition, the endogenous expression of TGF-β1 by HepG2 cells (and HeLa cells) makes it likely that they are subject to autocrine TGF-β stimulation in the absence of exogenous TGF-β, as has been reported for other cell lines (2, 7, 23, 54, 86).
The Smad3ΔC mutant lacks the C-terminal 43 amino acids of Smad3, which comprise the activating phosphorylation sites (96). This C-terminal sequence is required for Smad trimerization (5, 70, 85) and interaction with the CBP/p300 coactivators (19). Overexpression of Smad3ΔC has been shown to interfere in a dominant-negative manner with Smad3-mediated transcriptional activation (96). While Smad3 cooperated with IRF-7 in HeLa cells at the p-125Luc and p31x2-Luc promoters, Smad3ΔC did not, and it slightly decreased the transcription induced by IRF-7 (Fig. 5A and B). In HepG2 cells (Fig. 5C and D), expression of Smad3ΔC strongly repressed the transcriptional activation of both promoters by IRF-7. These results demonstrate that Smad3ΔC does not cooperate and interferes with IRF-7-mediated transcription in these assays. Its dominant-negative interference strongly suggests a role for endogenous Smad3 in IRF-7-mediated transcription from both the p-125Luc and p31x2-Luc promoters.
We also evaluated the effect of Smad3C on IRF-7-mediated transcription from both promoters. Smad3C corresponds to the MH2 domain of Smad3, without its DNA-binding MH1 domain and linker segment, and is able to interact with IRF-7 (Fig. 2A). Overexpression of Smad3C has been shown to exert strong transcriptional inhibition on TGF-β-responsive promoters, presumably resulting from its sequestration of coactivator CBP/p300 and/or titration of endogenous Smad3 and Smad4 (69). In HeLa cells, Smad3C was unable to cooperate with IRF-7 to induce transcription from either promoter and repressed the transcriptional activity of IRF-7 (Fig. 5A and B). The interference effect of Smad3C was stronger than that of Smad3ΔC, although this may have been due to a higher expression level of Smad3C versus Smad3ΔC (data not shown). In the TGF-β-responsive HepG2 cells, Smad3C strongly repressed both the basal level and TGF-β-induced level of IRF-7-mediated transcription at either promoter (Fig. 5C and D). Together, these data illustrate that endogenous Smad3 regulates IRF-7-mediated transcriptional activation from the PRDIII-PRDI sequence and strongly suggest that the level of transcription activation induced by IRF-7 depends on the levels of functional Smad3. Thus, interference with Smad3 function by Smad3ΔC or Smad3C inhibits the transcriptional activity of IRF-7.
Smad3 levels regulate the induction of endogenous IFN-β expression by poly(I-C).
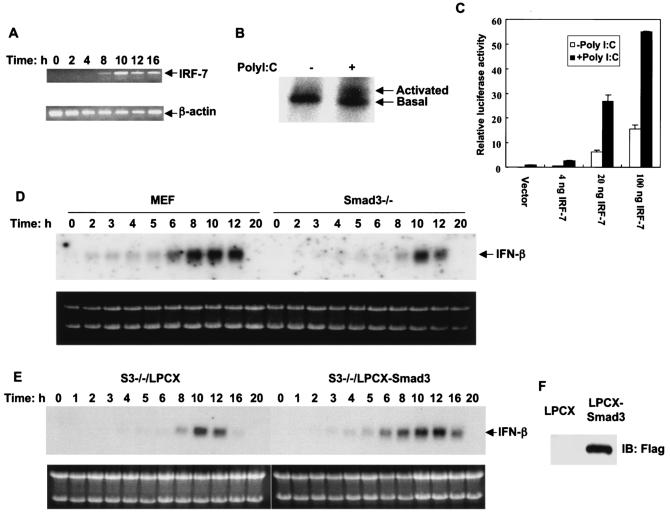
The transcriptional cooperation of Smad3 and IRF-7, and the dominant-negative inhibition of IRF-7 activity by Smad3C and Smad3ΔC as outlined above, suggested a possible role of Smad3 in the transcriptional regulation of endogenous IFN-β expression. We therefore examined whether inhibition of Smad3 expression would affect the induction of endogenous IFN-β mRNA by poly(I-C), which mimics the double-stranded RNA generated during viral infection and strongly induces IFN-β expression. In addition to the constitutive expression of IRF-3, poly(I-C) also induced IRF-7 expression in mouse embryonic fibroblasts (Fig. 6A). Consistent with its ability to activate endogenous IRF-3 (25, 66, 67), poly(I-C) is thought to activate IRF-7 through phosphorylation, presumably through the activity of the PKR kinase (15). We therefore evaluated the effect of poly(I-C) on IRF-7 phosphorylation in vivo and on IRF-7 transcriptional activity. We showed that poly(I-C) rapidly induced a mobility shift of a fraction of IRF-7 (Fig. 6B) similarly to the mobility shift of a fraction of IRF-7 in response to viral infection (73), thus reflecting increased phosphorylation of IRF-7. Poly(I-C) also enhanced the transcriptional activity of IRF-7 at the IRF-3/IRF-7-specific p31x2 promoter, proportionally with the different expression levels of IRF-7 (Fig. 6C). The slight enhancement by poly(I-C) of the basal transcription in the absence of coexpressed IRF-7 most likely resulted from the endogenous expression of IRF-3 and IRF-7 (Fig. 6A). Together with the published observations (15), these results strongly suggest that the induction of IFN-β expression by poly(I-C) is mediated by IRF-3/IRF-7 activation.
FIG. 6.
IFN-β mRNA induction by poly(I-C) depends on Smad3 expression. Induction of IFN-β mRNA was monitored by Northern hybridization. (A) Poly(I-C) treatment of MEFs induced expression of IRF-7 mRNA, as assessed by RT-PCR at the indicated times following initiation of treatment. (B) Effect of poly(I-C) on IRF-7 labeled with [32P]orthophosphate in vivo. Poly(I-C) induced a mobility shift of 32P-labeled IRF-7, as assessed after 4 h of treatment, similar to what is observed in response to viral infection. Similar results were also observed after 2 h of treatment with poly(I-C) (data not shown). (C) Effect of poly(I-C) on the transcriptional activity of IRF-7 at the p31x2-Luc promoter/reporter. Poly(I-C) enhanced the transcriptional activity of IRF-7 at different expression levels in transfected MEFs. The poly(I-C)-induced increase of transcription in vector control MEFs was likely a reflection of endogenous IRF-3 and IRF-7 expression (A). (D) Smad3−/− and matched normal MEFs were treated with poly(I-C), and RNA was isolated at the indicated times following addition of poly(I-C). The absence of Smad3 delayed and decreased early-phase IFN-β induction. (E) Expression of Smad3 in Smad3−/− cells, infected with the LPCX-Smad3 vector, conferred earlier induction of endogenous IFN-β expression in response to poly(I-C) treatment. LPCX-infected control cells and Smad3-expressing cells were treated with poly(I-C), and total RNA was isolated at the indicated times after treatment. The top portions of panels D and E show IFN-β mRNA under the same exposure condition on the same X-ray film, while the lower portions of the panels show the ethidium bromide-stained gels. (F) Expression of Smad3 in Smad3−/− cells infected with the retroviral LPCX-Smad3 vector, as shown by Western blotting to detect Flag-tagged Smad3.
Having established the ability of poly(I-C) to activate IRF-7, we examined whether inhibition of Smad3 expression would affect the induction of endogenous IFN-β mRNA by poly(I-C). MEFs derived from Smad3−/− mouse embryos (9) and wild-type littermates were transfected with poly(I-C) (15), and total RNA was isolated at different time points. The induction of IFN-β mRNA expression was determined by Northern blot analysis (Fig. 6D). In normal MEFs, IFN-β mRNA was readily detectable after 2 h and, following a lag phase of several hours, enhanced to reach peak induction at 8 to 10 h posttreatment. In contrast, IFN-β mRNA showed a much lower induction level in the Smad3−/− MEFs and perhaps a slight delay in induction. It started being detectable at 4 h, was enhanced at 8 h, and reached peak induction at 10 h posttreatment (Fig. 6D). At 12 h, however, its expression was already significantly declined in Smad3−/− cells, whereas it still persisted at a high level in wild-type MEFs. These data indicated that loss of Smad3 expression overall decreased IFN-β mRNA induction by poly(I-C).
To confirm that this lower induction of IFN-β expression by poly(I-C) is caused by loss of Smad3 expression, we established a derivative cell line of Smad3−/− MEFs, i.e., Smad3−/−/LPCX-Smad3 cells, which constitutively expresses Smad3 under the control of a CMV promoter in the LPCX retroviral vector (7). A control cell line which was infected with the empty LPCX vector was also generated. Immunoblotting analyses detected the expression of Smad3 in cells stably infected with the Smad3 expression vector, but not in the empty vector control cells (Fig. 6F). These stably transduced cells were used to assess the role of Smad3 in the induction of IFN-β by poly(I-C). Consistent with our results with normal MEFs, expression of Smad3 in Smad3−/− MEFs conferred a more rapid and a higher induction of IFN-β mRNA by poly(I-C), in comparison with control, LPCX vector-infected cells (Fig. 6E).
Smad3/TGF-β signaling enhances the transactivation function of IRF-7.
We next evaluated whether the ability of Smad3 to cooperate with IRF-7 was due to an inherent ability of Smad3 to enhance the transcriptional activity of IRF-7. For this purpose, we fused IRF-7 to the Gal4 DNA-binding domain to create a transcription factor that activates the expression of a luciferase reporter gene dependent on its binding to multiple copies of a Gal4 DNA-binding sequence. This Gal4 transactivation assay enabled us to assess the effects of Smads on the transcription activity of IRF-7, independent of its capacity to bind DNA.
As shown in Fig. 7A, expression of Smad3 enhanced the transcription activity of IRF-7. In contrast, coexpression of Smad2 or Smad4 did not affect the activity of Gal4-IRF-7. Furthermore, expression of Smad3ΔC and Smad3C decreased the activity of Gal4-IRF-7. These data illustrate that the transcription activity of IRF-7 depends on the Smad3 activity levels. The inability of Smad2 and Smad4 to affect the transcriptional activity of Gal4-IRF-7 is consistent with the lack of cooperation of Smad2 and the very low level of cooperation of Smad4 with IRF-7 at the p-125Luc promoter (Fig. 3A) and p31x2-Luc promoter (Fig. 4B). These data also explain why Smad3ΔC and Smad3C dominantly inhibit the IRF-7 activity at these two promoters (Fig. 5).
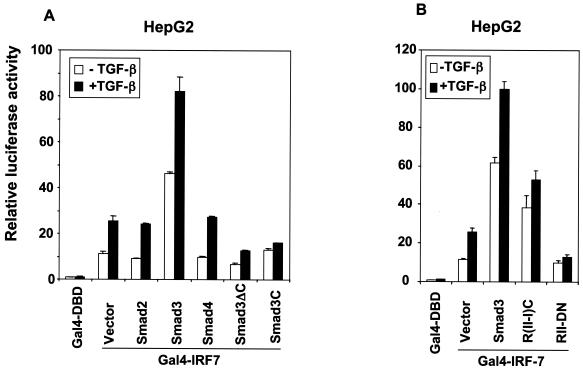
FIG. 7.
Smad3 and TGF-β signaling regulate the transactivation function of IRF-7. (A) Smad3 and TGF-β stimulated the transcriptional activity of Gal4-IRF-7, and Smad3ΔC and Smad3C inhibited it. TGF-β enhanced the activity of Gal4-IRF-7. (B) Smad3 signaling and TGF-β signaling through the cytoplasmic TβRI/RII chimera, R(II-I)C, stimulated the transcriptional activity of IRF-7, and a dominant-negative TβRII, RII-DN, inhibited it. HepG2 cells were cotransfected with Gal4-IRF-7 and the luciferase reporter pFR-Luc and expression plasmids for Smads and TGF-β receptors. Luciferase activities were scored as described for Fig. 5C and D.
We further assessed the effect of TGF-β on the transcription activity of Gal4-fused IRF-7. As shown in Fig. 7, TGF-β enhanced the transcription activity of Gal4-IRF-7, and this enhancement was further increased in the presence of Smad3. In contrast, TGF-β-mediated enhancement was inhibited by overexpressing the dominant-negative Smad3C or Smad3ΔC mutants (Fig. 7A). Furthermore, expression of the constitutively active chimeric TGF-β receptor R(II-I)C, in which the TβRII and TβRI cytoplasmic domains are covalently linked with an intervening linker sequence (18), further enhanced the transcriptional activity of IRF-7 (Fig. 7B).
Conversely, we examined the effect of perturbation of TGF-β receptor signaling on the transcriptional activity of Gal4-IRF-7. As shown in Fig. 7B, dominant-negative interference with TGF-β signaling through expression of RII-DN, which contains only the extracellular domain and the transmembrane domain of the type II TGF-β receptor without its cytoplasmic domain (6), decreased the transcriptional activity of Gal4-IRF-7 and blocked TGF-β-induced transcriptional activation of IRF-7.
TGF-β signaling enhances transcription from the IFN-β promoter.
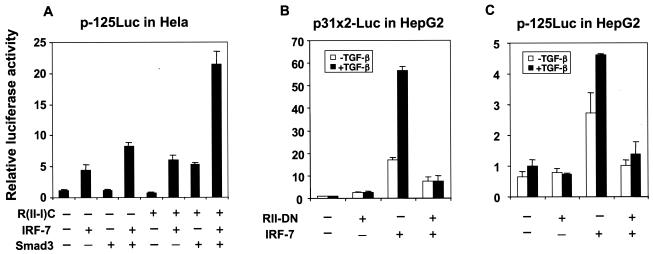
The effect of TGF-β/Smad3 signaling on the transactivation function of Gal4-IRF-7 suggested that TGF-β signaling regulates the transcription from the IFN-β promoter through activation of Smad3. We therefore tested the effect of TGF-β on transcriptional activation of p-125Luc in HeLa and HepG2 cells. HeLa cells have only a limited responsiveness to exogenous TGF-β, even though they display autocrine TGF-β responsiveness, while treatment of HepG2 cells with TGF-β activates transcription of TGF-β-responsive promoters. The lack of a convincing effect of TGF-β on the transcription from the IFN-β promoter in HeLa cells (data not shown) may have been due to the low number of receptors and autocrine TGF-β signaling and, therefore, the limited ability of TGF-β signaling to activate endogenous Smad3, compared to the high level of transfected reporter plasmid. We therefore assessed the regulation of the transfected p-125Luc reporter by TGF-β signaling using the cotransfected chimeric TGF-β receptor protein R(II-I)C. As shown in Fig. 8A, the R(II-I)C chimera cooperated with Smad3 and IRF-7 to activate transcription from the p-125Luc promoter. Additionally, the cooperation of Smad3 with IRF-7 was further enhanced by coexpression of R(II-I)C. These results suggest that activation of Smad3, as a result of TGF-β signaling, enhances the cooperativity of both endogenous and cotransfected Smad3 with IRF-7.
FIG. 8.
TGF-β signaling enhances the transcriptional cooperation of IRF-7 and Smad3 at the IFN-β promoter. (A) TGF-β receptor signaling increased Smad3-IRF-7 cooperativity in HeLa cells. HeLa cells were transfected with p-125Luc and expression plasmids for IRF-7, Smad3, and the cytoplasmic TβRI/RII chimera, R(II-I)C. (B) TGF-β enhanced and the dominant-negative TβRII receptor inhibited transcription from p31x2-Luc by IRF-7 in transfected HepG2 cells. (C) TGF-β enhanced and the dominant-negative TβRII receptor inhibited transcription from p-125Luc by IRF-7 in transfected HepG2 cells. Luciferase activities were scored as described in the legend for Fig. 3.
We further assessed the role of TGF-β signaling in the regulation of IFN-β expression by IRF-7 in HepG2 cells both using the dominant-negative type II TGF-β receptor, RII-DN, and in response to exogenous TGF-β. As shown in Fig. 8B, IRF-7 activated transcription from the p31x2-Luc promoter, and this activity was enhanced in response to added TGF-β. The basal IRF-7 activity in the absence of added TGF-β was decreased by blocking the type II receptor signaling, i.e., in the presence of RII-DN, which is consistent with autocrine TGF-β signaling in these cells. RII-DN also blocked the response to exogenous TGF-β. Similar results were observed using the p-125Luc promoter in HepG2 cells (Fig. 8C). These results further indicate that TGF-β signaling, both exogenously and through autocrine signaling, is a major determinant of the IRF-7 activity at the IFN-β promoter.
DISCUSSION
The rapid induction of IFN-α and -β plays a key role in the cell's response against viral infection. Extensive studies have focused on the mechanisms of transcriptional induction of these IFN genes following viral infection, and they have revealed that IRF-3 and IRF-7 are essential for transcriptional activation of the IFN-β gene (25, 41, 42, 77, 78). However, it is not known whether IFN-β expression, or the activity of IRF-7 or IRF-3, are regulated through cross talk with other signaling pathways. We now provide evidence that TGF-β signaling regulates the transcription activity of IRF-7 through direct interaction and functional cooperation of Smad3 with IRF-7. The regulation of the IRF-7 function by TGF-β/Smad3 at the IFN-β promoter suggests a possible role for TGF-β/Smad signaling in the induction of IFN-β expression and possibly in the innate immune response.
TGF-β/Smad3 regulates IRF-7 function.
The conclusion that Smad3 regulates IRF-7-mediated transcription is based on mutational analysis of the IFN-β promoter and on physical and functional interaction studies. The 125-bp IFN-β promoter segment used in our assays contains binding sequences for c-Jun/ATF-2 (16), NF-κB (22, 38, 81), and IRF-3/7 (44, 83). Since TGF-β/Smad3 signaling can cooperate with c-Jun (45, 60, 84, 97), ATF-2 (63), and NF-κB (36, 46) at other promoters, we addressed the contributions of these binding sequences in transcriptional activation. Inactivation of the PRDII sequence, which is able to bind NF-κB, did not affect the transcriptional activation by IRF-7 and cooperation with Smad3. Inactivation of the PRDIV sequence, which binds c-Jun/ATF-2, decreased the overall level of transcriptional activation but did not affect the cooperation of IRF-7 and Smad3 either. However, mutational inactivation of the PRDIII-PRDI sequence, i.e., the IRF-3/IRF-7 binding site, abolished both the induction by IRF-7 and the cooperation of Smad3 and IRF-7 (Fig. 4A). Furthermore, two tandem copies of the PRDIII-PRDI sequence in the minimal reporter plasmid p31x2-Luc fully supported the cooperation of IRF-7 and Smad3, and mutation of the IRF-7 binding site in this reporter abolished the cooperation (Fig. 4B and C). Together, these data demonstrate that the PRDIII-PRDI sequence is required and sufficient for IRF-7/Smad3 cooperation at the IFN-β promoter, and they raise the possibility that IRF-7/Smad3 cooperation regulates transcription from other promoters.
In addition to the cooperation of Smad3 with IRF-7 and the enhancement of the transcription activity of IRF-7 by Smad3 (Fig. 3, 4, 5, and 7), two dominant-negative mutants of Smad3, i.e., Smad3C and Smad3ΔC, repressed IRF-7-mediated promoter activation (Fig. 5) and the transactivation of Gal4-IRF-7 (Fig. 7A). While Smad3C may sequester CBP/p300 and endogenous Smads (69), its mechanism of dominant-negative interference on IRF-7 function may also resemble that of an IRF-7 C-terminal segment. The C terminus of IRF-7 mediates the interactions with IRF-7 or IRF-3 and, when expressed alone, interferes with IRF-7 activity through the formation of a nonfunctional IRF-7 complex (3). Since the MH2 domain of Smad3 interacts with the C-terminal segment of IRF-7 (Fig. 2), both the MH2 domain of Smad3 and the C-terminal domain of IRF-7 may interfere with the IRF-7/Smad3 complex formation. In this regard, it is worth noting that that the C-terminal domains of IRF-3, which is closely related to IRF-7, and Smads share considerable similarity in their three-dimensional structures (59).
In our study, IRF-3 activated at a low level the transcription from the PRDIII-PRDI sequence in p-125Luc (Fig. 3B) or p31x2-Luc (data not shown), and Smad3 did not enhance IRF-3-mediated transcription. The latter observation is consistent with the lack of physical interaction of Smad3 and IRF-3 in coimmunoprecipitation assays (Fig. 1C). However, our experiments were performed in the absence of virus, and IRF-3 activation is dependent on virus infection; in contrast, IRF-7 has a basal activity in the absence of virus (45, 66, 67, 90). We therefore do not exclude the possibility that virus-activated IRF-3 may be regulated by Smad3. We are currently investigating the role of TGF-β/Smad3 signaling on virus-activated IRF-3/IRF-7 function.
Consistent with the functional interaction of Smad3 with IRF-7, TGF-β signaling regulates IRF-7 function. This regulation is illustrated by the ability of a dominant-negative TGF-β receptor to strongly decrease the transactivation function of IRF-7 fused to a Gal4 DNA-binding domain (Fig. 7B) and to decrease the transcription activity of IRF-7 at the IFN-β and p31x2 promoters (Fig. 8B and C) and by the ability of added TGF-β to enhance IRF-7-mediated transcription from the IFN-β and p31x2 promoters (Fig. 8B and C). In addition, enhanced TGF-β signaling through a constitutively active receptor chimera strongly stimulated IRF-7 function and the IRF-7/Smad3 cooperation at the IFN-β promoter (Fig. 7B and 8A). Thus, the function of IRF-7 is regulated by both the autocrine responsiveness to endogenous TGF-β, observed in most cells in culture, and the response to exogenous TGF-β.
Our present findings that Smad3 physically associates and functionally cooperates with IRF-7 present a new angle in our understanding of the mechanism of action of IRFs. They indicate that the transcription function of IRF-7 is regulated by TGF-β/Smad3 signaling and that IRF-7 can activate transcription through interaction with another class of transcription factors, i.e., the Smads.
Mechanism of IRF-7 regulation by Smad3.
Smads, especially Smad3, interact and synergize with several types of DNA binding transcription factors (33, 50, 79, 98). The direct interaction and functional cooperation of Smad3 with IRF-7 are consistent with this model for Smad-mediated transcriptional activation, yet extend it to another class of transcription factors. Since Smad3 enhances the DNA binding of certain transcription factors, e.g., Sp1 (20, 58), CBFA1 (1), and c-Jun (J. Qing and R. Derynck, unpublished results), we evaluated the effect of Smad3 on the DNA binding of IRF-7. However, IRF-7 binding to the PRDIII-PRDI sequence was not affected by the presence of Smad3, as assessed using an electrophoretic mobility shift assay and DNA precipitation, and Smad3 by itself did not detectably bind to this DNA sequence (Fig. 4D and data not shown). On the other hand, TGF-β/Smad3 enhanced the transcriptional activity of IRF-7, as determined in Gal4-IRF-7 transactivation assays, and dominant-negative mutants of the TGF-β receptor and Smad3 decreased the transactivation by Gal4-IRF-7 (Fig. 7). The Smad3-mediated increase of the transcriptional activity of Gal4-IRF-7 bears similarity to the effect of Smad3 on the transcriptional activity of Gal4-fused Sp1 (20, 58). Since Smad3 and other receptor-activated Smads interact with CBP/p300 (19, 34, 69, 80) and Sp1 does not (62), Smad3 is likely to enhance the activity of Sp1 through its ability to recruit CBP/p300 in response to TGF-β (19). Considering the low-affinity interaction of IRF-7 for p300 (87), it is conceivable that the interaction of TGF-β-activated Smad3 with CBP/p300 helps stabilize the recruitment of CBP/p300 in the multiprotein transcription complex, thus enhancing transcription in response to TGF-β.
Role of Smad3/TGF-β signaling in IFN-β induction.
The regulation of the transcription activity of IRF-7 by Smad3 suggests a role for TGF-β/Smad3 signaling in the induction of IFN-β expression. In the absence of viral infection, cells express IFN-β at very low levels, most likely resulting from constitutive expression of IRF-3 and a very low level of IRF-7 that results from a spontaneous, low level of IFN-α/β expression and signaling (29, 77). Following viral infection, IFN-β expression is rapidly induced and IRF-7, which has a short half-life, plays a key role in this induction. A two-phase model of IFN-β induction suggests that IRF-3 is mainly responsible for the initial inefficient induction of IFN-β which, following receptor binding, activates Jak-Stat signaling, thus leading to IRF-7 expression. IRF-7 then cooperates with IRF-3 to launch a robust induction of IFN-β expression (25, 41, 48, 64, 77). Based on this model, the expression level and activity of IRF-7 play a critical role in the maximal induction of IFN-β through cooperation with constitutively expressed IRF-3. Therefore, the ability of TGF-β/Smad3 signaling to target the activity of IRF-7 may impact IFN-β expression in response to viral infection.
To evaluate the relevance of the regulation of IRF-7 by TGF-β/Smad3 signaling in endogenous IFN-β induction, we examined its induction in response to poly(I-C) in Smad3−/− and wild-type fibroblasts. Poly(I-C) is thought to activate IRF-3 (25, 66, 67) and induces IRF-7 expression (Fig. 6A). We also observed that poly(I-C) rapidly induced a mobility shift of in vivo 32P-labeled IRF-7 (Fig. 6B), similarly to what has been reported for virus-activated IRF-7 (73), and activated the transcription function of IRF-7 (Fig. 6C). The peak of IFN-β mRNA expression coincided with poly(I-C)-induced IRF-7 expression and was consistent with the poly(I-C)-induced activation of IRF-7. In these experiments, inhibition of Smad3 expression resulted in lower induction of IFN-β expression following poly(I-C) transfection (Fig. 6D), whereas reintroduction of Smad3 conferred an earlier onset and higher level of induction (Fig. 6E). Although TGF-β/Smad3 signaling regulates IRF-7 function and transcription from the IFN-β promoter, we were unable to see a significant effect of TGF-β on the poly(I-C)-mediated induction of endogenous IFN-β mRNA expression in MEFs (data not shown). This lack of detectable effect was likely primarily due to the low levels of available TGF-β receptors in these cells (data not shown), thus allowing only a minimal effect of exogenous TGF-β on activation of endogenous Smad3 in these cells, and our use of a very robust protocol for induction of IFN-β, i.e., poly(I-C) treatment. Nevertheless, the endogenous Smad3 levels influenced the induction of IFN-β expression, presumably reflecting activation of Smad3 from autocrine TGF-β receptor signaling. While the exact mechanism of regulation of poly(I-C)-induced IFN-β expression by Smad3 requires further characterization, our results nevertheless demonstrate an important role of Smad3 in defining the quantitative regulation of IFN-β expression in response to poly(I-C). Efforts to study the role of TGF-β/Smad3 signaling on virus-activated IRF-3/IRF-7 function and IFN-β expression are under way.
In summary, considering the essential role of IRF-7 in the induction of IFN-β expression, our finding that TGF-β/Smad3 signaling regulates the activity of IRF-7 suggests a role for TGF-β/Smad3 signaling in the induction of IFN-β expression. Since TGF-β levels increase rapidly after viral infection, IFN-β induction is likely to depend on the endogenous Smad3 level, the level of autocrine TGF-β/Smad3 signaling, and the responsiveness of the cells to elevated TGF-β. Future experiments will address the dependence of the transcription activity of IRF-7 and expression of IFN α and β upon viral infection on TGF-β/Smad3 signaling.
Acknowledgments
We thank Takashi Fujita, Tadatsugu Taniguchi, Xiao-Fan Wang, and Paula Pitha for reagents used in this study. We give special thanks to Pierre Lee and Dong Liu in the Derynck lab for their insights and discussion of the projects in the course of this research.
This research is supported by grant CA63101 from the National Institutes of Health to R.D. J. Qing was supported by a postdoctoral fellowship from the American Heart Association, L.C. was supported by KO1 grant DK02877, and R.-Y.W was supported by a Leukemia and Lymphoma Society of America postdoctoral fellowship.
REFERENCES
- 1.Alliston, T., L. Choy, P. Ducy, G. Karsenty, and R. Derynck. 2001. TGF-β-induced repression of CBFA1 by Smad3 decreases cbfa1 and osteocalcin expression and inhibits osteoblast differentiation. EMBO J. 20:2254-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arteaga, C. L., R. J. Coffey, Jr., T. C. Dugger, C. M. McCutchen, H. L. Moses, and R. M. Lyons. 1990. Growth stimulation of human breast cancer cells with anti-transforming growth factor β antibodies: evidence for negative autocrine regulation by transforming growth factor β. Cell Growth Differ. 1:367-374. [PubMed] [Google Scholar]
- 3.Au, W. C., W. S. Yeow, and P. M. Pitha. 2001. Analysis of functional domains of IFN regulatory factor 7 and its association with IRF-3. Virology 280:273-282. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. E. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 5.Chacko, B. M., B. Qin, J. J. Correia, S. S. Lam, M. P. de Caestecker, and K. Lin. 2001. The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization. Nat. Struct. Biol. 8:248-253. [DOI] [PubMed] [Google Scholar]
- 6.Chen, R. H., R. Ebner, and R. Derynck. 1993. Inactivation of the type II receptor reveals two receptor pathways for the diverse TGF-β activities. Science 260:1335-1338. [DOI] [PubMed] [Google Scholar]
- 7.Choy, L., J. Skillington, and R. Derynck. 2000. Roles of autocrine TGF-β receptor and Smad signaling in adipocyte differentiation. J. Cell Biol. 149:667-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datta, P. K., M. C. Blake, and H. L. Moses. 2000. Regulation of plasminogen activator inhibitor-1 expression by transforming growth factor-β-induced physical and functional interactions between Smads and Sp1. J. Biol. Chem. 275:40014-40019. [DOI] [PubMed] [Google Scholar]
- 9.Datto, M. B., J. P. Frederick, L. Pan, A. J. Borton, Y. Zhuang, and X.-F. Wang. 1999. Targeted disruption of Smad3 reveals an essential role in transforming growth factor β-mediated signal transduction. Mol. Cell. Biol. 19:2495-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeMaeyer, E. and J. DeMaeyer-Guignard. 1988. Interferons and other regulatory cytokines. John Wiley and Sons, New York, N.Y.
- 11.Derynck, R., and X.-H. Feng. 1997. TGF-β receptor signaling. Biochim. Biophys. Acta 1333:F105-F150. [DOI] [PubMed] [Google Scholar]
- 12.Derynck, R., and L. Choy. 1998. Transforming growth factor-β and its receptors, p. 593-636. In A. Thompson (ed.), The cytokine handbook, 3rd ed. Academic Press, San Diego, Calif.
- 13.Derynck, R., Y. Zhang, and X.-H. Feng. 1998. Smads: transcriptional activators of TGF-β responses. Cell 95:737-740. [DOI] [PubMed] [Google Scholar]
- 14.Derynck, R., R. J. Akhurst, and A. Balmain. 2001. TGF-β signaling in tumor suppression and cancer progression. Nat. Genet. 29:117-129. [DOI] [PubMed] [Google Scholar]
- 15.Diebold, S. S., M. Montoya, H. Unger, L. Alexopoulou, P. Roy, L. E. Haswell, A. Al-Shamkhani, R. Flavell, P. Borrow, and C. Reis e Sousa. 2003. Viral infection switches non-plasmacytoid dendritic cells into high IFN producers. Nature 424:324-328. [DOI] [PubMed] [Google Scholar]
- 16.Du, W., and T. Maniatis. 1992. An ATF/CREB binding site is required for virus induction of the human IFN β gene. Proc. Natl. Acad. Sci. USA 89:2150-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng, X.-H., E. H. Filvaroff, and R. Derynck. 1995. Transforming growth factor-β (TGF-β)-induced down-regulation of cyclin A expression requires a functional TGF-β receptor complex. Characterization of chimeric and truncated type I and type II receptors. J. Biol. Chem. 270:24237-24245. [DOI] [PubMed] [Google Scholar]
- 18.Feng, X.-H., and R. Derynck. 1996. Ligand-independent activation of transforming growth factor (TGF) β signaling pathways by heteromeric cytoplasmic domains of TGF-β receptors. J. Biol. Chem. 271:13123-13129. [DOI] [PubMed] [Google Scholar]
- 19.Feng, X.-H., Y. Zhang, R. Y. Wu, and R. Derynck. 1998. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for Smad3 in TGF-β-induced transcriptional activation. Genes Dev. 12:2153-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng, X.-H., X. Lin, and R. Derynck. 2000. Smad2, Smad3 and Smad4 cooperate with Sp1 to induce p15Ink4B transcription in response to TGF-β. EMBO J. 19:5178-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKɛ and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 22.Fujita, T., M. Miyamoto, Y. Kimura, J. Hammer, and T. Taniguchi. 1989. Involvement of a cis-element that binds an H2TF-1/NFκB like factor(s) in the virus-induced IFN-β gene expression. Nucleic Acids Res. 17:3335-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gandrillon, O., U. Schmidt, H. Beug, and J. Samarut. 1999. TGF-β cooperates with TGF-α to induce the self-renewal of normal erythrocytic progenitors: evidence for an autocrine mechanism. EMBO J. 18:2764-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibellini, D., G. Zauli, M. C. Re, D. Milani, G. Furlini, E. Caramelli, S. Capitani, and M. La Placa. 1994. Recombinant human immunodeficiency virus type-1 (HIV-1) Tat protein sequentially up-regulates IL-6 and TGF-β1 mRNA expression and protein synthesis in peripheral blood monocytes. Br. J. Haematol. 88:261-267. [DOI] [PubMed] [Google Scholar]
- 25.Grandvaux, N., B. R. tenOever, M. J. Servant, and J. Hiscott. 2002. The IFN antiviral response: from viral invasion to evasion. Curr. Opin. Infect. Dis. 15:259-267. [DOI] [PubMed] [Google Scholar]
- 26.Gyuris, J., E. Golemis, H. Chertkov, and R. Brent. 1993. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75:791-803. [DOI] [PubMed] [Google Scholar]
- 27.Haagmans, B. L., K. J. Teerds, A. J. van den Eijnden-van Raaij, M. C. Horzinek, and V. E. Schijns. 1997. Transforming growth factor β production during rat cytomegalovirus infection. J. Gen. Virol. 78:205-213. [DOI] [PubMed] [Google Scholar]
- 28.Hanai, J., L. F. Chen, T. Kanno, N. Ohtani-Fujita, W. Y. Kim, W. H. Guo, T. Imamura, Y. Ishidou, M. Fukuchi, M. J. Shi, et al. 1999. Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Cα promoter. J. Biol. Chem. 274:31577-31582. [DOI] [PubMed] [Google Scholar]
- 29.Hata, N., M. Sato, A. Takaoka, M. Asagiri, N. Tanaka, and T. Taniguchi. 2001. Constitutive IFN-α/β signal for efficient IFN-α/β gene induction by virus. Biochem. Biophys. Res. Commun. 285:518-525. [DOI] [PubMed] [Google Scholar]
- 30.Hu, R., N. Oyaizu, S. Than, V. S. Kalyanaraman, X. P. Wang, and S. Pahwa. 1996. HIV-1 gp160 induces transforming growth factor-β production in human PBMC. Clin. Immunol. Immunopathol. 80:283-289. [DOI] [PubMed] [Google Scholar]
- 31.Hua, X., X. Liu, D. O. Ansari, and H. F. Lodish. 1998. Synergistic cooperation of TFE3 and Smad proteins in TGF-β-induced transcription of the plasminogen activator inhibitor-1 gene. Genes Dev. 12:3084-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hua, X., Z. A. Miller, H. Benchabane, J. L. Wrana, and H. F. Lodish. 2000. Synergism between transcription factors TFE3 and Smad3 in transforming growth factor-β-induced transcription of the Smad7 gene. J. Biol. Chem. 275:33205-33208. [DOI] [PubMed] [Google Scholar]
- 33.Itoh, S., F. Itoh, M. J. Goumans, and P. Ten Dijke. 2000. Signaling of transforming growth factor-β family members through Smad proteins. Eur. J. Biochem. 267:6954-6967. [DOI] [PubMed] [Google Scholar]
- 34.Janknecht, R., N. J. Wells, and T. Hunter. 1998. TGF-β-stimulated cooperation of Smad proteins with the coactivators CBP/p300. Genes Dev. 12:2114-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Juang, Y., W. Lowther, M. Kellum, W. C. Au, R. Lin, J. Hiscott, and P. M. Pitha. 1998. Primary activation of IFN α and IFN β gene transcription by IFN regulatory factor 3. Proc. Natl. Acad. Sci. USA 95:9837-9842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kon, A., L. Vindevoghel, D. J. Kouba, Y. Fujimura, J. Uitto, and A. Mauviel. 1999. Cooperation between SMAD and NF-κB in growth factor regulated type VII collagen gene expression. Oncogene 18:1837-1844. [DOI] [PubMed] [Google Scholar]
- 37.Labbé, E., C. Silvestri, P. A. Hoodless, J. L. Wrana, and L. Attisano. 1998. Smad2 and Smad3 positively and negatively regulate TGF-β-dependent transcription through the forkhead DNA-binding protein FAST1/2. Mol. Cell 2:109-120. [DOI] [PubMed] [Google Scholar]
- 38.Lenardo, M. J., C. M. Fan, T. Maniatis, and D. Baltimore. 1989. The involvement of NF-κB in β-IFN gene regulation reveals its role as widely inducible mediator of signal transduction. Cell 57:287-294. [DOI] [PubMed] [Google Scholar]
- 39.Letterio, J. J., and A. B. Roberts. 1998. Regulation of immune responses by TGF-β. Annu. Rev. Immunol. 16:137-161. [DOI] [PubMed] [Google Scholar]
- 40.Levy, D. E., and J. E. Darnell, Jr. 2002. Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell. Biol. 3:651-662. [DOI] [PubMed] [Google Scholar]
- 41.Levy, D. E., I. Marié, E. Smith, and A. Prakash. 2002. Enhancement and diversification of IFN induction by IRF-7-mediated positive feedback. J. Interferon Cytokine Res. 22:87-93. [DOI] [PubMed] [Google Scholar]
- 42.Levy, D. E., I. Marié, and A. Prakash. 2003. Ringing the IFN alarm: differential regulation of gene expression at the interface between innate and adaptive immunity. Curr. Opin. Immunol. 15:52-58. [DOI] [PubMed] [Google Scholar]
- 43.Liberati, N. T., M. B. Datto, J. P. Frederick, X. Shen, C. Wong, E. M. Rougier-Chapman, and X.-F. Wang. 1999. Smads bind directly to the Jun family of AP-1 transcription factors. Proc. Natl. Acad. Sci. USA 96:4844-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin, R., P. Genin, Y. Mamane, and J. Hiscott. 2000. Selective DNA binding and association with the CREB binding protein coactivator contribute to differential activation of α/β IFN genes by IFN regulatory factors 3 and 7. Mol. Cell. Biol. 20:6342-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin, R., Y. Mamane, and J. Hiscott. 2000. Multiple regulatory domains control IRF-7 activity in response to virus infection. J. Biol. Chem. 275:34320-34327. [DOI] [PubMed] [Google Scholar]
- 46.Lopez-Rovira, T., E. Chalaux, J. L. Rosa, R. Bartrons, and F. Ventura. 2000. Interaction and functional cooperation of NF-κB with Smads. Transcriptional regulation of the JunB promoter. J. Biol. Chem. 275:28937-28946. [DOI] [PubMed] [Google Scholar]
- 47.Maniatis, T., J. V. Falvo, T. H. Kim, T. K. Kim, C. H. Lin, B. S. Parekh, and M. G. Wathelet. 1998. Structure and function of the IFN-β enhanceosome. Cold Spring Harbor Symp. Quant. Biol. 63:609-620. [DOI] [PubMed] [Google Scholar]
- 48.Marié, I., J. E. Durbin, and D. E. Levy. 1998. Differential viral induction of distinct IFN-α genes by positive feedback through IFN regulatory factor-7. EMBO J. 17:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marié, I., E. Smith, A. Prakash, and D. E. Levy. 2000. Phosphorylation-induced dimerization of IFN regulatory factor 7 unmasks DNA binding and a bipartite transactivation domain. Mol. Cell. Biol. 20:8803-8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Massagué, J. 2000. How cells read TGF-β signals. Nat. Rev. Mol. Cell. Biol. 1:169-178. [DOI] [PubMed] [Google Scholar]
- 51.McCartney-Francis, N. L., M. Frazier-Jessen, and S. M. Wahl. 1998. TGF-β: a balancing act. Int. Rev. Immunol. 16:553-580. [DOI] [PubMed] [Google Scholar]
- 52.McKiel, V., Z. Gu, M. A. Wainberg, and J. Hiscott. 1995. Inhibition of human immunodeficiency virus type 1 multiplication by transforming growth factor β1 and AZT in HIV-1-infected myeloid cells. J. Interferon Cytokine Res. 15:849-855. [DOI] [PubMed] [Google Scholar]
- 53.Michelson, S., J. Alcami, S. J. Kim, D. Danielpour, F. Bachelerie, L. Picard, C. Bessia, C. Paya, and J. L. Virelizier. 1994. Human cytomegalovirus infection induces transcription and secretion of transforming growth factor β1. J. Virol. 68:5730-5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mossalayi, M. D., F. Mentz, F. Ouaaz, A. H. Dalloul, C. Blanc, P. Debre, and F. W. Ruscetti. 1995. Early human thymocyte proliferation is regulated by an externally controlled autocrine transforming growth factor-β1 mechanism. Blood 85:3594-3601. [PubMed] [Google Scholar]
- 55.Moustakas, A., S. Souchelnytskyi, and C.-H. Heldin. 2001. Smad regulation in TGF-β signal transduction. J. Cell Sci. 114:4359-4369. [DOI] [PubMed] [Google Scholar]
- 56.Munshi, N., Y. Yie, M. Merika, K. Senger, S. Lomvardas, T. Agalioti, and D. Thanos. 1999. The IFN-β enhancer: a paradigm for understanding activation and repression of inducible gene expression. Cold Spring Harbor Symp. Quant. Biol. 64:149-159. [DOI] [PubMed] [Google Scholar]
- 57.Pardali, E., X. Q. Xie, P. Tsapogas, S. Itoh, K. Arvanitidis, C. H. Heldin, P. ten Dijke, T. Grundström, and P. Sideras. 2000. Smad and AML proteins synergistically confer transforming growth factor β1 responsiveness to human germ-line IgA genes. J. Biol. Chem. 275:3552-3560. [DOI] [PubMed] [Google Scholar]
- 58.Pardali, K., A. Kurisaki, A. Morén, P. ten Dijke, D. Kardassis, and A. Moustakas. 2000. Role of Smad proteins and transcription factor Sp1 in p21Waf1/Cip1 regulation by transforming growth factor-β. J. Biol. Chem. 275:29244-29256. [DOI] [PubMed] [Google Scholar]
- 59.Qin, B. Y., C. Liu, S. S. Lam, H. Srimath, R. Delston, J. J. Correia, R. Derynck, and K. Lin. 2003. Crystal structure of IRF-3 transactivation domain reveals mechanism of autoinhibition and virus-induced phospho-activation. Nat. Struct. Biol. 10:913-921. [DOI] [PubMed] [Google Scholar]
- 60.Qing, J., Y. Zhang, and R. Derynck. 2000. Structural and functional characterization of the transforming growth factor-β-induced Smad3/c-Jun transcriptional cooperativity. J. Biol. Chem. 275:38802-38812. [DOI] [PubMed] [Google Scholar]
- 61.Reed, S. G. 1999. TGF-β in infections and infectious diseases. Microbes Infect. 1:1313-1325. [DOI] [PubMed] [Google Scholar]
- 62.Ryu, S., S. Zhou, A. G. Ladurner, and R. Tjian. 1999. The transcriptional cofactor complex CRSP is required for activity of the enhancer-binding protein Sp1. Nature 397:446-450. [DOI] [PubMed] [Google Scholar]
- 63.Sano, Y., J. Harada, S. Tashiro, R. Gotoh-Mandeville, T. Maekawa, and S. Ishii. 1999. ATF-2 is a common nuclear target of Smad and TAK1 pathways in transforming growth factor-β signaling. J. Biol. Chem. 274:8949-8957. [DOI] [PubMed] [Google Scholar]
- 64.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-α/β gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 65.Schafer, S. L., R. Lin, P. A. Moore, J. Hiscott, and P. M. Pitha. 1998. Regulation of type I IFN gene expression by IFN regulatory factor-3. J. Biol. Chem. 273:2714-2720. [DOI] [PubMed] [Google Scholar]
- 66.Servant, M. J., B. tenOever, and R. Lin. 2002. Overlapping and distinct mechanisms regulating IRF-3 and IRF-7 function. J. Interferon Cytokine Res. 22:49-58. [DOI] [PubMed] [Google Scholar]
- 67.Servant, M. J., N. Grandvaux, and J. Hiscott. 2002. Multiple signaling pathways leading to the activation of IFN regulatory factor 3. Biochem. Pharmacol. 64:985-992. [DOI] [PubMed] [Google Scholar]
- 68.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin,and J. P. Hiscott. 2003. Triggering the IFN antiviral response through a IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 69.Shen, X., P. P. Hu, N. T. Liberati, M. B. Datto, J. P. Frederick, and X.-F. Wang. 1998. TGF-β-induced phosphorylation of Smad3 regulates its interaction with coactivator p300/CREB-binding protein. Mol. Biol. Cell 9:3309-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi, Y., A. Hata, R. S. Lo, J. Massagué, and N. P. Pavletich. 1997. A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature 388:87-93. [DOI] [PubMed] [Google Scholar]
- 71.Shi, Y., and J. Massagué. 2003. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113:685-700. [DOI] [PubMed] [Google Scholar]
- 72.Shum, L., S. A. Reeves, A. Kuo, E. S. Fromer, and R. Derynck. 1994. Association of the transmembrane transforming growth factor-α precursor with a protein kinase complex. J. Cell Biol. 125:903-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith, E. J., I. Marié, A. Prakash, A. Carcia-Sastre, and D. E. Levy. 2003. IRF-3 and IRF-7 phosphorylation in virus-infected cells does not require double stranded RNA-dependent protein kinase R or IκB kinase, but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 23:8951-8957. [DOI] [PubMed] [Google Scholar]
- 74.Spaccapelo, R., L. Romani, L. Tonnetti, E. Cenci, A. Mencacci, G. Del Sero, R. Tognellini, S. G. Reed, P. Puccetti, and F. Bistoni. 1995. TGF-β is important in determining the in vivo patterns of susceptibility or resistance in mice infected with Candida albicans. J. Immunol. 155:1349-1360. [PubMed] [Google Scholar]
- 75.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to IFNs. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 76.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623-655. [DOI] [PubMed] [Google Scholar]
- 77.Taniguchi, T., and A. Takaoka. 2001. A weak signal for strong responses: IFN-α/β revisited. Nat. Rev. Mol. Cell. Biol. 2:378-386. [DOI] [PubMed] [Google Scholar]
- 78.Taniguchi, T., and A. Takaoka. 2002. The IFN-α/β system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr. Opin. Immunol. 14:111-116. [DOI] [PubMed] [Google Scholar]
- 79.ten Dijke, P., K. Miyazono, and C.-H. Heldin. 2000. Signaling inputs converge on nuclear effectors in TGF-β signaling. Trends Biochem. Sci. 25:64-70. [DOI] [PubMed] [Google Scholar]
- 80.Topper, J. N., M. R. DiChiara, J. D. Brown, A. J. Williams, D. Falb, T. Collins, and M. A. Gimbrone, Jr. 1998. CREB binding protein is a required coactivator for Smad-dependent, transforming growth factor β transcriptional responses in endothelial cells. Proc. Natl. Acad. Sci. USA 95:9506-9511. (Erratum, 95:12735.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Visvanathan, K. V., and S. Goodbourn. 1989. Double-stranded RNA activates binding of NF-κB to an inducible element in the human β-IFN promoter. EMBO J. 8:1129-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wakefield, L. M., and A. B. Roberts. 2002. TGF-β signaling: positive and negative effects on tumorigenesis. Curr. Opin. Genet. Dev. 12:22-29. [DOI] [PubMed] [Google Scholar]
- 83.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 84.Wong, C., E. M. Rougier-Chapman, J. P. Frederick, M. B. Datto, N. T. Liberati, J. M. Li, and X.-F. Wang. 1999. Smad3-Smad4 and AP-1 complexes synergize in transcriptional activation of the c-Jun promoter by transforming growth factor β. Mol. Cell. Biol. 19:1821-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu, R. Y., Y. Zhang, X.-H. Feng, and R. Derynck. 1997. Heteromeric and homomeric interactions correlate with signaling activity and functional cooperativity of Smad3 and Smad4/DPC4. Mol. Cell. Biol. 17:2521-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu, S. P., D. Theodorescu, R. S. Kerbel, J. K. Willson, K. M. Mulder, L. E. Humphrey, and M. G. Brattain. 1992. TGF-β1 is an autocrine-negative growth regulator of human colon carcinoma FET cells in vivo as revealed by transfection of an antisense expression vector. J. Cell Biol. 116:187-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang, H., C. H. Lin, G. Ma, M. O. Baffi, and M. G. Wathelet. 2003. Interferon regulatory factor (IRF)-7 synergizes with other transcription factors through multiple interactions with p300/CBP coactivators. J. Biol. Chem. 278:15495-15504. [DOI] [PubMed] [Google Scholar]
- 88.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Sato, K. Ozato, and T. Fujita. 1996. Autocrine amplification of type I IFN gene expression mediated by IFN stimulated gene factor 3 (ISGF3). J. Biochem. (Tokyo) 120:160-169. [DOI] [PubMed] [Google Scholar]
- 89.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I IFN system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yoneyama, M., W. Suhara, and T. Fujita. 2002. Control of IRF-3 activation by phosphorylation. J. Interferon Cytokine Res. 22:73-76. [DOI] [PubMed] [Google Scholar]
- 91.Yoo, Y. D., C. J. Chiou, K. S. Choi, Y. Yi, S. Michelson, S. Kim, G. S. Hayward, and S. J. Kim. 1996. The IE2 regulatory protein of human cytomegalovirus induces expression of the human transforming growth factor β1 gene through an Egr-1 binding site. J. Virol. 70:7062-7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yoo, Y. D., H. Ueda, K. Park, K. C. Flanders, Y. I. Lee, G. Jay, and S. J. Kim. 1996. Regulation of transforming growth factor-β1 expression by the hepatitis B virus (HBV) X transactivator. Role in HBV pathogenesis. J. Clin. Investig. 97:388-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zervos, A. S., J. Gyuris, and R. Brent. 1993. Mxi1, a protein that specifically interacts with Max to bind Myc-Max recognition sites. Cell 72:223-232. [DOI] [PubMed] [Google Scholar]
- 94.Zhang, L., and J. S. Pagano. 1997. IRF-7, a new IFN regulatory factor associated with Epstein-Barr virus latency. Mol. Cell. Biol. 17:5748-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang, L., and J. S. Pagano. 2002. Structure and function of IRF-7. J. Interferon Cytokine Res. 22:95-101. [DOI] [PubMed] [Google Scholar]
- 96.Zhang, Y., X. Feng, R. Wu, and R. Derynck. 1996. Receptor-associated Mad homologues synergize as effectors of the TGF- β response. Nature 383:168-172. [DOI] [PubMed] [Google Scholar]
- 97.Zhang, Y., X. H. Feng, and R. Derynck. 1998. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature 394:909-913. (Erratum, 396:491, 1998.) [DOI] [PubMed] [Google Scholar]
- 98.Zhang, Y., and R. Derynck. 1999. Regulation of Smad signalling by protein associations and signalling crosstalk. Trends Cell Biol. 9:274-279. [DOI] [PubMed] [Google Scholar]
- 99.Zhang, Y., and R. Derynck. 2000. Transcriptional regulation of the transforming growth factor-β-inducible mouse germ line Igα constant region gene by functional cooperation of Smad, CREB, and AML family members. J. Biol. Chem. 275:16979-16985. [DOI] [PubMed] [Google Scholar]