Abstract
BACKGROUND:
The prevalence of Helicobacter pylori infection, which may increase the risk of gastritis, peptic ulcers, and cancer, has increased worldwide. This number is estimated to be around 70–90% in developing countries and 25–50% in developed countries. It is possible that the bacterium can be transmitted via food and water as well as zoonotically and iatrogenically. Because of high prevalence of this infection in Iran, the aim of this study is to examine whether H. pylori infection might be transmitted from cow's milk and faeces.
METHODS:
The existence of the H. pylori antibody and antigen was investigated in samples of serum, milk, and faeces from 92 lactating Holstein cows in Shahrekord, Iran. The H. pylori antigen and antibody were detected using ELISA and were confirmed by PCR.
RESULTS:
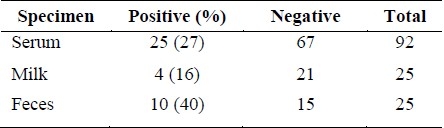
It was found that out of 92 serum specimens, 25 (27%) of the cows were positive for the H. pylori antibody and 67 specimens were negative. From these 25 seropositive cows, 10 (40%) faeces samples and four (16%) milk samples were antigen positive for H. pylori. Four of the antigen-positive milk specimens were also antigen positive for faeces. The existence of the UreC gene was also confirmed in positive samples of milk and faeces.
CONCLUSIONS:
There is a possibility that cow's milk is a transmission mode in H. pylori infection and faecal contamination and inappropriate management processes could transfer H. pylori to humans. The awareness of the H. pylori epidemiology and its method of distribution are necessary for public health measures and controlling the spread of this bacterium. Further investigation with a greater sample number is necessary to verify the ability of H. pylori transmission via milk consumption.
KEYWORDS: Helicobacter pylori, Milk, Faeces, Zoonotic
Helicobacter pylori can cause chronic gastritis, peptic ulcers, and even can-cer in humans. Its prevalence has in-creased worldwide to the point that more than seven million infections are now reported every year. The prevalence of H. pylori is 70–90% in developing countries and 25–50% in developed countries.1 Studies have shown that the prevalence of this infection in Isfahan prov-ince is 70-80%.1–2 Knowledge of the epidemiology and the ways of transmission of H. pylori is important for preventing its distribution and this can be useful for identifying high-risk populations, especially in areas that have high rates of gastritis, peptic ulcers, and gastric cancer. The source and transmission of this bacterium has not been clearly explained yet. However, Goodman and Correa stated that humans are the only natural host of H. pylori and that the bacterium can be transmitted via food and water as well as zoonotically and iatrogenically, especially among those sharing the same residence. This could also be consistent with a common source of transmission.3–5
Cow's milk is usually consumed as human food, especially by children. Therefore, one of the suggested theories is the transmission of H. pylori through milk from animals to human beings. Some studies have reported the presence and survival of H. pylori in dairy and milk products.6–10 Therefore, the high prevalence of infection in central Iran might partly be because of animal and milk contamination. As such, the goal of this study is to examine whether H. pylori infection might be transmitted from cow's milk and faeces.
Methods
In this study, which was conducted from May 2008 to September 2009 in Shahrekord (Iran), the existence of H. pylori was investigated in the serum, fresh raw milk and faeces of 92 lactating Holstein cows from 12 flocks (size range 30–40 cows). Cows were selected according to lactation and age (at least two years old). There were no differences in the environmental situa-tions or the feeding and sanitary management of the flocks. Five millilitres of blood, 10 millili-tres of milk and 10 grams of faeces from each cow was collected into sterile common bottles. The specimens were transported in ice-cooled containers to the laboratory within two hours after collection. The serum samples were fro-zen at -80°C until processing. The stool sam-ples were also stored at -20°C before the test. A small portion of the specimen was mixed with sample diluents. Milk samples were filtered and kept in -20° before the test. We used ELISA to investigate IgG Antibody titre against H. pylori in the serum samples7 according to the manufacturer's instructions (Radim Co., Iran). Milk and faeces samples of seropositive cows were tested for H. pylori antigens using the ELISA kit (11) (GA Co. Germany) . The Helicobacter pylori IgG antibody and antigen test is based on an enzyme immunoassay (ELISA) utilising a horseradish peroxidase conjugated detection antibody or antigen and the results were read by spectrophotometer (450 nm). The cut-off values were: less than 0.140 was considered negative; between 0.140 and 0.159 was considered equivocal and greater than or equal to 0.160 was considered positive. Equivocal results, according to the manufacturer's instruction, should be repeated. The results were then confirmed by a PCR test.12 DNA was extracted by the DNA extraction kit (Roche Co. Germany) and its density was assessed by optic densitometry. Extracted genomic DNA was amplified for the UreC gene and detected with the specific primers
HP-F: 5’-GAATAAGCTTTTAGGGGTGTTAGGGG-3’, HP-R: 5’GCTTACTTTCTAACACTAACGCGC-3’.
The gene product was 294 bp. PCR reactions were performed in a final volume of 50 μL containing 5 μL 10× buffer + Mg, 2 mM dNTP, 2 unit Taq DNA polymerase, 100 ng genomic DNA as a template, and 25 picomole of each primer. PCR was performed using a thermal cycler (Eppendorf Co. Germany) under the following conditions: an initial denaturation for 10 minutes at 94°C; 35 cycles for 1 minute at 94°C, 1 minute at 55°C, 1 minute at 72°C, and a final extension at 72°C for 10 minutes.12 PCR yields were electrophoresed in 1.5% agarose gels (Roche Co. Germany) containing ethidium bromide. A DNA ladder (Fermentas Co. Germany) used to detect the molecular weight of observed bands under a UV lamp. All tests were performed in triplicate. Samples inoculated with H. pylori were used as positive controls.
Results
A total of 200 subjects were studied. Based on serological tests (ELISA), it was found that out of 92 serum specimens, 25 (27%) of the cows were positive for the H. pylori IgG antibody and 67 specimens were negative. From these 25 seropositive cows 10 (40%) faeces samples and four (16%) milk samples were antigen positive for H. pylori (Table 1). Four of the antigen-positive milk specimens were also antigen positive for faeces. The UreC gene was detected in the milk and faeces samples of antigen positive cows using the PCR method. The PCR product size was 294 bp. The seropositive cows came from six different flocks and the faeces and milk antigen-positive samples came from three of these.
Table 1.
Frequency of H. pylori contamination in seropositive cows’ raw milk and feces
Discussion
The prevention and control of the prevalence of H. pylori infection and thereby the spread of gastritis, gastric ulcer, and gastric cancer in humans is very important. In central Iran, the prevalence of H. pylori is very high (78%). One of the major sources of infection in humans could be cow, sheep, and goat's milk contaminated with H. pylori.10 The consumption of milk and its products vary considerably in different regions in the world. Bovine milk and dairy products have a long tradition in human nutrition. Iranian people especially children drink cow's milk commonly. In this study, cow's serum, milk, and faeces were investigated to find the possible source of contamination. For this purpose, the antibody against H. pylori was investigated in 92 cows from 12 similar flocks. The results showed that 27% of serum samples from six flocks showed antibody against H. pylori. The existence of H. pylori antibody in cow's serum might indicate that H. pylori is a commensal in cows, which might be H. pylori's natural host as Dore et al. indicated for sheep.7 Seropositive cows were tested for H. pylori antigen in faeces and milk. Forty per-cent16 of faeces specimens and 16% (four) raw milk specimens were found to be antigen positive for H. pylori from three flocks. Four antigen-positive milk specimens were also antigen positive in the faeces samples. The cost-efficient ELISA kit used for the detection of H. Pylori IgG antibody is very sensitive (96%) and highly specific (97%) and for H. Pylori antigen detection, sensitivity of the test is 98% and its specificity is 99%. The existence of UreC gene was also confirmed in positive antigen samples of milk and faeces by PCR method.
Fujimura et al showed that the prevalence of H. pylori was 50% in cow faeces and 38% in soil samples. Contact with cow faeces and soil is the main source of milk contamination. Milk could also be contaminated during production or because of the inadequate post-processing hygienic management of the product, which could transmit the bacteria to humans.9 Azevedo et al could not prove the existence of H. pylori in milk through culturing. These findings are very important to explain the way of transmission of H. pylori to humans through milk and food.13 Fujimura et al reported that H. pylori might not be cultured in pasteurised cow's milk because it might be changed to a coccoid form through pasteurisation. They found the ureA gene of H. pylori in 13 out of 18 (72.2%) raw milk samples and in 11 out of 20 (55%) commercial pasteurised milk samples.9 Wang et al and Orozco et al showed a progressive decrease in bacterial load with an average survival of nine days in pasteurised milk and 12 days in UHT milk, with an estimated average of original inoculums of 105 and 106 cfu/ml. Other studies were unable to pick up H. pylori from the culture of pasteurized milk. Anti H. pylori effects in milk might damage H. pylori and inactivate a non culturable state that inhibits its multiplication in culture media.14,15 H. pylori changes in three different forms under environmental stress: first, viable spiral forms that are culturable, virulent, and infectious and induce inflammation in experimental animals; second, in viable coccoid forms that are nonculturable, less virulent, and less likely to colonise and induce inflammation in experimental animals; and a third form that is a nonviable degenerative forms of dying H. pylori.16 It is necessary to detect the coccid form of active bacteria by using the PCR method.17
Conclusions
This study is the first to look for the presence of bacterial antigens in cow's milk, one of the most consumed foods for children in Iran, to recognize the mode of transmission of H. pylori. According to this study and similar previous ones, faecal contamination and consuming infected cow milk might be a transmission way for H. pylori infection and unfit management could transfer H. pylori to humans. The awareness of H. pylori epidemiology and methods of its distribution is essential for public health authorities and controlling the spread of this bacterium. Further investigation with a greater number of samples is necessary to verify the ability of H. pylori to spread via milk consumption.
Authors’ Contributions
The principal idea was from HGhS who desigened and coordinated the study also participated in writing and editing the manuscript. AR provided assistance in the design of the study and conducting, assessments and coordination of the study. AZ and AR were the manager of the study and participated in designing, conducting the study and assessments. All authors have read and approved the final manuscript.
Acknowledgments
This study was supported by vice chancellor for research of Azad University of Shahrekord, Iran.
Footnotes
Conflict of Interests
Authors have no conflict of interests.
References
- 1.Ghasemian Safaei H, Tavakkoli H, Mojtahedi A, Salehei R, Soleimani B, Pishva E. Correlation of cagA positive Helicobacter pylori Infection with clinical outcomes in Alzahra hospital, Isfahan, Iran. Journal of Reaserch Medical Science. 2008;13(4):196–201. [Google Scholar]
- 2.Ghasemian Safaei H, Havaei SA, Tavakkoli H, Eshaghei M, Navabakbar F, Salehei R. Relation of bab A2 genotype of Helicobacter pylori infection with chronic active gastritis, duodenal ulcer and non-cardia active gastritis in Alzahra hospital Isfahan, Iran. Jundishapur Journal of Microbiology. 2010;3(3):93–8. [Google Scholar]
- 3.Goodman KJ, Correa P. Transmission of Helicobacter pylori among siblings. Lancet. 2000;355(9201):358–62. doi: 10.1016/S0140-6736(99)05273-3. [DOI] [PubMed] [Google Scholar]
- 4.Dunn BE, Vakil NB, Schneider BG, Miller MM, Zitzer JB, Peutz T, et al. Localization of Helicobacter pylori urease and heat shock protein in human gastric biopsies. Infect Immun. 1997;65(4):1181–8. doi: 10.1128/iai.65.4.1181-1188.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kivi M, Johansson AL, Reilly M, Tindberg Y. Helicobacter pylori status in family members as risk factors for infection in children. Epidemiol Infect. 2005;133(4):645–52. doi: 10.1017/s0950268805003900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown LM. Helicobacter pylori: epidemiology and routes of transmission. Epidemiol Rev. 2000;22(2):283–97. doi: 10.1093/oxfordjournals.epirev.a018040. [DOI] [PubMed] [Google Scholar]
- 7.Dore MP, Bilotta M, Vaira D, Manca A, Massarelli G, Leandro G, et al. High prevalence of Helicobacter pylori infection in shepherds. Dig Dis Sci. 1999;44(6):1161–4. doi: 10.1023/a:1026676223825. [DOI] [PubMed] [Google Scholar]
- 8.Dore MP, Sepulveda AR, El Zimaity H, Yamaoka Y, Osato MS, Mototsugu K, et al. Isolation of Helicobacter pylori from sheep-implications for transmission to humans. Am J Gastroenterol. 2001;96(5):1396–401. doi: 10.1111/j.1572-0241.2001.03772.x. [DOI] [PubMed] [Google Scholar]
- 9.Fujimura S, Kawamura T, Kato S, Tateno H, Watanabe A. Detection of Helicobacter pylori in cow's milk. Lett Appl Microbiol. 2002;35(6):504–7. doi: 10.1046/j.1472-765x.2002.01229.x. [DOI] [PubMed] [Google Scholar]
- 10.Vale FF, Vitor JM. Transmission pathway of Helicobacter pylori: does food play a role in rural and urban areas? Int J Food Microbiol. 2010;138(1-2):1–12. doi: 10.1016/j.ijfoodmicro.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 11.Fanti L, Mezzi G, Cavallero A, Gesu G, Bonato C, Masci E. A new simple immunoassay for detecting Helicobacter pylori infection: antigen in stool specimens. Digestion. 1999;60(5):456–60. doi: 10.1159/000007691. [DOI] [PubMed] [Google Scholar]
- 12.Lage AP, Godfroid E, Fauconnier A, Burette A, Butzler JP, Bollen A, et al. Diagnosis of Helicobacter pylori infection by PCR: comparison with other invasive techniques and detection of cagA gene in gastric biopsy specimens. J Clin Microbiol. 1995;33(10):2752–6. doi: 10.1128/jcm.33.10.2752-2756.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azevedo NF, Guimaraes N, Figueiredo C, Keevil CW, Vieira MJ. A new model for the transmission of Helicobacter pylori: role of environmental reservoirs as gene pools to increase strain diversity. Crit Rev Microbiol. 2007;33(3):157–69. doi: 10.1080/10408410701451922. [DOI] [PubMed] [Google Scholar]
- 14.Wang X, Hirmo S, Willen R, Wadstrom T. Inhibition of Helicobacter pylori infection by bovine milk glycoconjugates in a BAlb/cA mouse model. J Med Microbiol. 2001;50(5):430–5. doi: 10.1099/0022-1317-50-5-430. [DOI] [PubMed] [Google Scholar]
- 15.Orozco A, Ogura T, Hirosawa T, Garduno R, Kubo I. In hydrolyzed cow's milk Helicobacter pylori becomes nonculturable and the growth of Salmonella typhi and Escherichia coli is inhibited. J Food Sci. 2007;72(8):M306–9. doi: 10.1111/j.1750-3841.2007.00490.x. [DOI] [PubMed] [Google Scholar]
- 16.Oliver JD. The viable but nonculturable state in bacteria. J Microbiol. 2005;43(Spec No):93–100. [PubMed] [Google Scholar]
- 17.Shahamat M, Alavi M, Watts JE, Gonzalez JM, Sowers KR, Maeder DW, et al. Development of two PCR-based techniques for detecting helical and coccoid forms of Helicobacter pylori. J Clin Microbiol. 2004;42(8):3613–9. doi: 10.1128/JCM.42.8.3613-3619.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


