Abstract
Most cis-acting regulatory elements have generally been assumed to activate a single nearby gene. However, many genes are clustered together, raising the possibility that they are regulated through a common element. We show here that a single peroxisome proliferator response element (PPRE), located between the mouse PEX11α and perilipin genes, confers on both genes activation by peroxisome proliferator-activated receptor alpha (PPARα) and PPARγ. A functional PPRE 8.4 kb downstream of the promoter of PEX11α, a PPARα target gene, was identified by a gene transfection study. This PPRE was positioned 1.9 kb upstream of the perilipin gene and also functioned with the perilipin promoter. In addition, this PPRE, when combined with the natural promoters of the PEX11α and perilipin genes, conferred subtype-selective activation by PPARα and PPARγ2. The PPRE sequence specifically bound to the heterodimer of RXRα and PPARα or PPARγ2, as assessed by electrophoretic gel mobility shift assays. Furthermore, tissue-selective binding of PPARα and PPARγ to the PPRE was demonstrated in hepatocytes and adipocytes, respectively, by chromatin immunoprecipitation assay. Hence, the expression of these genes is induced through the same PPRE in the liver and adipose tissue, where the two PPAR subtypes are specifically expressed.
Eukaryotic genes are regulated by transcription factors that bind to specific DNA elements located in their vicinity. The binding site and the transcription factor for a given gene are generally supposed to act only on the gene and thus assure the specificity of its regulation. Recent genome analysis, however, revealed that an unexpectedly large number of genes of higher eukaryotes form clusters (3). They may reside in the same or opposite orientation, sometimes sharing promoters and, in some cases, even overlapping in part. Such an arrangement of genes would raise a hitherto unassumed possibility that two or more genes can be regulated through a common cis element, because the regulatory element of a given gene would inevitably be close to another gene. We report here that the mouse PEX11α and perilipin genes, which are positioned in tandem on the genome, are regulated through a common recognition site for peroxisome proliferator-activated receptors (PPARs).
The PPAR family has three subtypes, α, γ, and δ (or β), within the nuclear hormone receptor superfamily (18). PPARα is highly expressed in the liver and regulates the expression of several genes involved in lipid metabolism (14). PPARγ has two isoforms (γ1 and γ2) generated by alternative transcription start (77); PPARγ1 is expressed in many tissues, including the adipose tissue and immune cells (10), whereas PPARγ2 is exclusively expressed in adipose tissue, playing a central role in adipocyte differentiation (71). PPARβ/δ is ubiquitously expressed and is involved in the physiology of certain tissues as well as lipid homeostasis (7, 51, 74). PPAR binds to peroxisome proliferator response elements (PPREs) located in the vicinity of target genes by heterodimerizing with another nuclear hormone receptor, retinoid X receptor (RXR) (32). A PPRE is comprised of two AGGTCA or related half sites separated by a single nucleotide, termed direct repeat 1. For optimal binding, PPAR requires an extended half site constituted by AGGTCA and four extra residues on the 5′ side (28, 31, 46, 49).
PPARα was first identified as a transcription factor that mediates the action of certain drugs that cause peroxisomes to proliferate in rodent liver (29). Peroxisomes are ubiquitous organelles bounded by a single membrane, and mammalian peroxisomes have many important metabolic functions, such as β-oxidation of very-long-chain fatty acids and synthesis of cholesterol and ether lipids (35). Peroxisomes in rodent liver markedly proliferate on the administration of various hypolipidemic agents collectively termed peroxisome proliferators (8, 55). Concomitantly, peroxisomal and mitochondrial β-oxidation enzymes as well as microsomal ω-oxidation enzymes are induced. Disruption of the PPARα gene of mice results in refractivity to peroxisome proliferation in the liver upon administration of peroxisome proliferators (36). PPARα was shown to activate the expression in the liver of several genes for enzymes involved in peroxisomal and mitochondrial fatty acid β-oxidation, such as acyl coenzyme A (CoA) oxidase (AOx) (47, 72), peroxisomal bifunctional enzyme (76), and 3-ketoacyl-CoA thiolase (32, 45). On the other hand, some adipogenesis-related genes, such as those for adipocyte P2 (70) and phosphoenolpyruvate carboxykinase (69), are known as PPARγ targets. However, no gene involved in peroxisome biogenesis has been reported to date as a target of PPARα, despite the dramatic increase in the number of hepatic peroxisomes in rodents caused by the PPARα ligands.
PEX11 is one of the PEX genes (15) encoding the factors required for the biogenesis of peroxisomes. At least 26 PEX genes have been identified, based on genetic complementation using peroxisome-deficient mutants of yeast and mammalian cells (43, 53, 63, 64, 66). PEX11 was shown to promote peroxisome division in yeast and mammalian cells (1, 2, 19, 42, 50, 58, 61). Disruption of the PEX11 gene in Saccharomyces cerevisiae and Candida boidinii resulted in decreased numbers of peroxisomes that were larger than normal (19, 42, 58). Although it was suggested that peroxisomal fatty acid metabolism affects peroxisome abundance (12), a recent study showed that overexpression of the PEX11 gene is sufficient to promote peroxisome proliferation without affecting peroxisomal metabolism (39). In mammals, three subtypes of the PEX11 gene (α, β, and γ) have been identified and mapped on different chromosomes. The expression of PEX11α and PEX11γ is tissue specific, being most prominent in the liver, whereas PEX11β is ubiquitously expressed (37, 61). Interestingly, PEX11α expression is induced by peroxisome proliferators, though the expression of PEX11β and PEX11γ is not affected by these agents (1, 2, 37, 61, 65). Thus, the proliferation of peroxisomes caused by these agents is possibly attributable to the induction of PEX11α expression, and hence we inferred that PEX11α is a target gene of PPARα.
In the present study, we searched for a functional PPRE in the vicinity of the mouse PEX11α gene and identified one in the downstream region. Moreover, upon inspection of the mouse genome database, we found that the gene for perilipin is located downstream of PEX11α, close to the PPRE. Perilipin is expressed mainly in adipocytes and steroidogenic cells and coats the surfaces of intracellular lipid droplets (22, 62). Perilipin was suggested to regulate the lipolysis of triacylglycerol by blocking the access of hormone-sensitive lipase to stored lipids (13). Perilipin is produced during adipocyte differentiation (22), where PPARγ2 plays a central regulatory role. Here, we show that the PEX11α and perilipin genes are regulated by PPARα and PPARγ, respectively, through a common PPRE.
MATERIALS AND METHODS
RNA analysis.
Reverse transcription (RT)-PCR was carried out using hepatic RNA derived from wild-type and PPARα-null mice fed a diet with or without Wy14,643 for 2 weeks (36). The procedure for RT-PCR was as described previously (68). Concentrations of RNA samples were normalized based on the intensity of the signal for a ribosomal protein, 36B4, a control unaffected by peroxisome proliferators. Adequate PCR cycles were set for individual genes so that the band intensities could be compared in the exponential amplification phase.
Isolation of mouse PEX11α gene.
Screening was carried out using a mouse genomic λ phage library with a cDNA fragment of rat PEX11α as a probe. Rat PEX11α cDNA was cloned from a rat cDNA library by PCR. We obtained a positive genomic clone (clone D) containing a 2.2-kb upstream region of the PEX11α gene. To obtain the genomic DNA fragments encompassing more upstream regions than clone D, we isolated two restriction fragments (kb −12.5 to −7.5 and −9.5 to −1.5) from a mouse BAC clone, RP23-171D12, by walking from the 5′ end of clone D.
Plasmids.
Genomic DNA fragments of the PEX11α gene were subcloned into the luciferase reporter plasmid pGVP, containing a SV40 promoter or pGVB basic vector (Toyo Ink). When the genomic fragments were placed on the downstream side, they were inserted in a restriction site downstream of the poly(A) addition site of the luciferase gene. Site-directed mutagenesis was carried out using a QuickChange (Stratagene) mutagenesis kit according to the manufacturer's protocol. The mutant clones were tested for the presence of the desired mutation and the absence of any unexpected mutations by nucleotide sequencing.
pAOXPPREluc, a mouse PPARα expression vector, pNCMVPPARα, and an empty plasmid, pCMVNot, were constructed as described previously (46). Expression vectors for mouse PPARγ2 (70) and PPARδ (5) were provided by B. M. Spiegelman and P. A. Grimaldi, respectively, and the inserts were subcloned into a cloning vector, pCMX, provided by K. Umesono with the permission of R. M. Evans. PPARγ1 cDNA was obtained by RT-PCR and subcloned into pCMX. The β-galactosidase expression vector, pCMVβ, was a gift from G. MacGregor (41).
Cell culture and DNA transfection.
HeLa cells were cultured in 96-well culture plates with F-12 medium containing 10% fetal bovine serum (FBS) at 37°C under 5% CO2. Transfection was carried out by the calcium phosphate method (59). To each well, 0.1 μg of a reporter plasmid, 0.1 μg of a PPAR expression vector, and 0.175 μg of an empty vector (pCMVNot or pCMX) were cotransfected. After 4 h, the calcium phosphate precipitates were removed, and the cells were cultured for 24 h in the same medium supplemented with the ligands (100 μM Wy14,643; 1 μM BRL49,653; and 10 μM carbaprostacyclin, for PPARα, PPARγ, and PPARβ/δ, respectively) or vehicle (dimethyl sulfoxide [DMSO]).
3T3-L1 and H4IIEC3 cells were cultured in 35-mm dishes with Dulbecco's modified Eagle's medium (DMEM) containing 10% FBS. For differentiation experiments, 3T3-L1 cells were treated for 2 days with a hormone cocktail containing 1 μM dexamethasone, 0.5 mM 3-isobutyl-1-methylxanthine, and 5 μg of insulin/ml (57). The cocktail was then removed, and the cells were cultured for 3 days in the same medium supplemented with 5 μg of insulin/ml. Transfection was performed using LipofectAMINE (Invitrogen) 5 days after initiation of the cocktail treatment. Cells were maintained for 8 h with the DNA-liposome mixture in a serum-free medium, Opti-MEM (Invitrogen), and then cultured for 20 h in DMEM containing 10% FBS and 5 μg of insulin/ml supplemented with 1 μM BRL49,653 or DMSO. H4IIEC3 cells were stably transfected with the plasmids by a modified calcium phosphate method (47). For the transfection of linearized plasmids, reporter plasmids were cut at a unique Aor51HI site just behind the downstream fragment of PEX11α. The linearized plasmids were purified using NucleoSpin Extract (MACHEREY-NAGEL). HeLa cells were cultured in 35-mm dishes with F-12 medium containing 10% FBS and transfected as described above with 4 μg of a linearized reporter plasmid, 1 μg of the PPARα expression vector, and 1 μg of pCMVβ. After 6 h of transfection, the cells were subjected to a glycerol shock with 0.5 ml of 15% glycerol in HEPES-buffered saline for 30 s. The cells were washed with F-12 medium and then cultured in the same medium containing 10% FBS supplemented with 100 μM Wy14,643 or DMSO for 24 h.
Luciferase assays.
In the 96-well plates, the cells were solubilized with 20 μl of cell lysis buffer (5 mM Tris-phosphate [pH 7.8]; 2 mM dithiothreitol; 1 mM ethylenediamine-N,N,N′,N′-tetraacetic acid; 10% glycerol; 1% Triton X-100). Luciferase activity was measured using a PicaGene (Toyo Ink) reagent kit in a Lucy2 (Anthos) microplate luminometer. After transfection was performed in 35-mm dishes, cells were extracted with 200 μl of the cell lysis buffer, and the luciferase assay was carried out using 10, 20, and 40 μl of cell extract for the experiments involving stable transformants, linearized plasmids, and 3T3-L1 cells, respectively, with the same reagent kit in a Lumat LB9501 (Berthold) luminometer. The luciferase activities in the experiments using 35-mm dishes were normalized for the efficiency of transfection on the basis of β-galactosidase activity and are presented as relative values. All transfection experiments were carried out in triplicate, and the averages are shown together with the standard deviations.
Electrophoretic mobility shift assay (EMSA).
A double-stranded oligonucleotide composed of 5′-TCGACTTCCCTTGTCACCTTTCACCCACATCCTAGAATCC-3′ and 5′-TCGAGGATTCTAGGATGTGGGTGAAAGGTGACAAGGGAAG-3′ and encompassing the downstream PPRE of PEX11α was used as a probe (Underlining indicates the sequences corresponding to PPRE throughout). The mutant sequence of PEX11α PPRE was as described below. We also used a double-stranded oligonucleotide consisting of 5′-CGAACGTGACCTTTGTCCTGGTCCCCTTTTGCTCC-3′ and 5′-TCGGGAGCAAAAGGGGACCAGGACAAAGGTCACGT-3′ and containing the known PPRE of rat AOx as a positive control (46). A mutant AOx PPRE composed of 5′-CGAACGTGACCTTCTCGAGGGTCCCCTTTTGCTCC-3′ and its complement was also used. The probes were 3′ end labeled with Klenow DNA polymerase and [α-32P]dCTP. The assay was carried out with fusion proteins, maltose-binding protein-PPARα, and glutathione S-transferase (GST)-RXRα, as described previously (46), or with PPARγ2 and RXRα synthesized in a rabbit reticulocyte lysate system (Amersham). Other experimental conditions were as previously described (46).
ChIP assay.
H4IIEC3 and 3T3-L1 cells were cultured in 10-cm dishes with DMEM containing 10% FBS. H4IIEC3 was grown to 80% confluency, followed by treatment with 100 μM Wy14,643 or DMSO for 2 days. 3T3-L1 was differentiated as described above and cultured for 5 days after initiation of the cocktail treatment. Approximately 1 × 107 cells were then processed for chromatin immunoprecipitation (ChIP) assay using a reagent kit (Upstate Biotechnology) as recommended by the manufacturer. Immunoprecipitation was performed with polyclonal antibodies against PPARα, PPARγ, pan-RXR, and CBP (all from Santa Cruz Biotechnology) and a preimmune rabbit immunoglobulin G (IgG). For PCR, primer pair 5′-GAATGGCCAAGAGCCCTGCTC-3′ (positions + 2282 to + 2302, relative to the first residue of the putative polyadenylation signal of the PEX11α gene) and 5′-GCTCTGCTGACAAAGCTGGTC-3′ (+2462 to + 2482) was used for the rat PEX11α/perilipin-PPRE, primer pair 5′-GAGTGGTCAAGACCTCTGCTC-3′ (+2094 to + 2114) and 5′-GCTCTGCTGACAAAGCCGGTC-3′ (+2265 to + 2285) was used for the mouse PEX11α/perilipin-PPRE, primer pair 5′-ACCTATGCATGGATGACCACTA-3′ (−1737 to −1716) and 5′-CACAGCAATTAAACAGTGAC-3′ (−1543 to −1524) was used for a region distal to the rat PEX11α/perilipin-PPRE, and primer pair 5′-CTGTGCATGAGTGACCACTCG-3′ (−1694 to −1674) and 5′-CTAAACAGTGACTAAGGAGTCATTA-3′ (−1500 to −1476) was used for a region distal to the mouse PEX11α/perilipin-PPRE. A 5-μl aliquot out of the 50-μl solution of DNA recovered from each immunoprecipitate was used for PCR, and the products were analyzed on an agarose gel after 35 cycles of amplification.
RESULTS
PEX11α is a target gene of PPARα.
To confirm that the PEX11α gene is a target of PPARα, we first investigated the induction of PEX11α mRNA expression by Wy14,643, one of the peroxisome proliferators, using hepatic RNA from wild-type and PPARα-null mice (36) (Fig. 1). AOx, a known target gene of PPARα (47), was used as a reference. 36B4, a gene for ribosomal protein, was examined as a control that was not affected by peroxisome proliferators. In the wild-type mice, PEX11α expression was induced by the peroxisome proliferator, consistent with other reports (2, 61), but no induction was seen in the PPARα-null mice. These results closely paralleled those for AOx. Thus, PEX11α is a bona fide target gene of PPARα.
FIG. 1.
Induction of mouse PEX11α gene expression by a peroxisome proliferator. RT-PCR was carried out using hepatic RNA of wild-type and PPARα-null mice fed with or without Wy14,643, a peroxisome proliferator. AOx, a known target gene of PPARα, was used as a positive control. 36B4 was used as a control for a gene that is not induced by Wy14,643.
A PPRE is located downstream of PEX11α.
To seek the PPRE of the PEX11α gene, we isolated a DNA fragment (clone D) containing the region of mouse PEX11α from a mouse genomic λ phage library, using rat PEX11α cDNA as a probe. Clone D contained a 2-kb upstream region, a 5-kb region of the PEX11α structural gene consisting of three exons and two introns, and a 12-kb downstream region (Fig. 2A). In addition, two DNA fragments containing regions further upstream (−12.5 to −7.5 kb and −9.5 to −1.5 kb from the transcriptional initiation site, respectively) were isolated from a mouse genomic BAC clone (RP23-171D12).
FIG. 2.
Gene loci of PEX11α and perilipin. (A) Maps of the PEX11α and perilipin genes. Exons are shown by shaded boxes, together with the numbers. A DNA fragment derived from a mouse genomic library (clone D) is shown by an open box, together with the relative distance from the transcriptional initiation site of the PEX11α gene. The asterisk indicates the location of PPRE identified in the present study. The human genome map was obtained from the NCBI database (http://www.ncbi.nlm.nih.gov). (B) Sequence comparison of the downstream regions of mouse and human PEX11α genes. Matching nucleotides are shown by asterisks. The PPRE is boxed, together with arrows indicating the half sites, and a broken line following the second arrow shows the additional nucleotides required for optimal binding of PPAR. The numbers are relative to the first residue of the polyadenylation signal of the PEX11α gene. The alignment was made with the GENETYX-Mac (SDC, Tokyo, Japan) program, using the human genomic sequence of the PEX11α region (GenBank accession no. AC079075).
To search for a PPRE in the PEX11α promoter region, we constructed a luciferase reporter plasmid containing a 2.2-kb upstream region (positions −2.2 to +43), as well as several constructs containing partial fragments of the region deleted from the 5′ side stepwise. These were introduced into the HeLa human cervical cancer cell line together with a PPARα expression vector (Fig. 3A). Only basal levels of luciferase activity were observed with all the constructs tested in the presence of PPARα and/or Wy14,643 compared with that for a positive control containing the PPRE of rat AOx, which exhibited marked activation by PPARα under these assay conditions. The promoter region of the PEX11α gene appeared to be devoid of a functional PPRE, since we observed only a slight increase in luciferase activity with the PEX11α constructs by the ligand in the presence of PPARα, but a similar increase was also observed with the vector without a promoter sequence (Fig. 3A, inset). The slight activation by PPARα and the ligand was probably due to a cryptic PPRE-like sequence in the vector. In fact, the luciferase vector containing only a basal promoter of SV40 (pGVP) (Fig. 3B) or the herpes simplex virus thymidine kinase gene also exhibited a similar activation by PPARα and the ligand (data not shown).
FIG. 3.
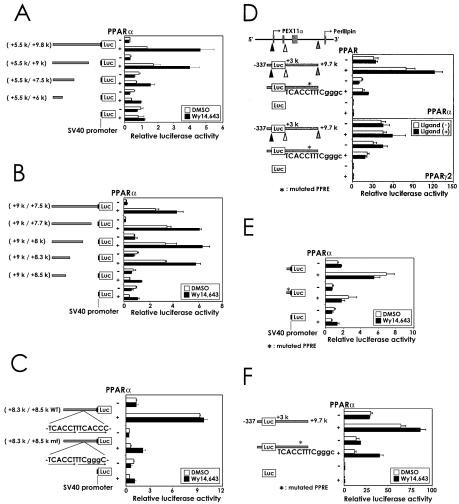
Transfection analysis of the genomic region of mouse PEX11α. The luciferase (Luc) reporter gene constructs are depicted on the left, and the results of the reporter assay are depicted on the right. The numbers in the maps of the constructs are relative to the transcriptional initiation sites of the PEX11α gene. (A) Analysis of the region proximal to the PEX11α gene promoter. pAOXPPREluc, containing the functional PPRE and promoter region of the rat AOx gene (46), was used as a positive control. Luciferase activity is shown as a relative value, with the activity of pAOXPPREluc in the absence of both ligand and PPARα expression vector as 1. The inset shows luciferase activity of a negative control vector without a promoter sequence (pGVB). (B) Analysis of various genomic regions of the PEX11α gene. Triangles indicate the positions of boundaries of regions included in the reporter vectors. Luciferase activity is given as a value relative to the activity of pGVP in the absence of both ligand and PPARα ex-pression vector. (C) Effect of the PEX11α downstream region combined with the native promoter. Luciferase activity is shown as a value relative to the activity of pGVB in the absence of both ligand and PPARα expression vector.
To locate the functional PPRE of the PEX11α gene, luciferase vectors containing various genomic regions around PEX11α were constructed and introduced into HeLa cells together with a PPARα expression vector. The SV40 basal promoter was present in these reporter vectors (Fig. 3B). Only with the reporter plasmid containing the downstream region, kb +5.5 to +9.8, did we observe a significant degree of ligand-dependent transactivation by PPARα. This construct was also activated by PPARγ1 but not PPARβ/δ (data not shown). On the other hand, there was no functional PPRE in the upstream region up to kb −12.5.
We next examined the effect of the downstream region, using a luciferase reporter plasmid containing the PEX11α native promoter (Fig. 3C). In this plasmid, a downstream fragment (kb +3 to +9.7) was placed behind the luciferase gene, keeping the distance between the transcriptional initiation site and the downstream region as close as possible to the natural distance. We observed a ligand-dependent transactivation by PPARα with the construct carrying the downstream region but not with that containing only the promoter. These results indicate that the functional PPRE of the PEX11α gene is located in the downstream region.
To determine the precise location of the PPRE, we examined a series of deletion constructs of the downstream region starting from the kb +9.8 position (Fig. 4A). The downstream region could be deleted to kb +9 without a significant decrease in activation by PPARα. Further deletion of the sequence between kb +7.5 and +9 resulted in a marked decrease in transactivation, indicating that the PPRE of the PEX11α gene is located in this region. When the sequence from kb +7.5 to +9 was deleted stepwise from the upstream side (Fig. 4B), the transcriptional activity dropped to basal level, between kb +8.3 and +8.5. Hence, the PPRE of PEX11α seemed to be located within this 200-bp region. We next tested the kb +8.3 to +8.5 fragment for activation by PPARα (Fig. 4C). Activation by PPARα was indeed observed, indicating that this region contained a functional PPRE. Wy14,643, however, did not further promote activation by PPARα, and hence this region by itself seemed to be insufficient for responsiveness to the peroxisome proliferator. A limited region between kb +8.5 and +9 containing several candidate motifs for transcription-factor binding was found to confer responsiveness when combined with the kb +8.3 to +8.5 fragment (M. Shimizu and T. Osumi, unpublished data).
FIG. 4.
Identification of the PPRE. The luciferase (Luc) reporter gene constructs are depicted on the left, and the results of the reporter assay are depicted on the right. The numbers in the maps of the constructs are relative to the transcriptional initiation sites of the PEX11α gene. The lowercase letters in the sequences are mutated residues. (A) Deletion analysis of the PEX11α downstream region from the 3′ side. (B) Analysis of the kb +9.5 to +7.5 region by sequential deletion from the 5′ side. The region was inserted in an orientation opposite to the natural one for convenience in construction. (C) Sequence-specific transactivation by PPARα conferred by the kb +8.3 to +8.5 region of PEX11α. WT and mt indicate reporter plasmids containing the wild-type and mutated forms of putative PPRE, respectively. (D) PPARα-selective transactivation of the PEX11α gene. For PPARα and PPARγ2, 100 μM Wy14,643 and 1 μM BRL49,653, respectively, were used as ligands. (E) The PPRE is sufficient for transactivation by PPARα. Double-stranded DNA fragments (WT, 5′-TCGACTTCCCTTGTCACCTTTCACCCACATCCTAGAATCC-3′; mt, 5′-TCGACTTCCCTTGTCACCTTTCGGGCACATCCTAGAATCC-3′) were inserted in pGVP, upstream of the SV40 promoter. The underlined letters in the sequences correspond to the PPRE. (F) Transfection study of linearized plasmids. Plasmids were cut with a restriction enzyme, Aor51HI, at a unique site just behind the kb +9.7 position.
A PPRE-like sequence, TCACCTTTCACCC, was found in the kb +8.4 downstream region. Within this motif, we mutated three bases that closely matched the PPRE consensus and abolished activation by PPARα (Fig. 4C). Transactivation was also suppressed by this mutation in a construct composed of the native promoter and a longer downstream region of the PEX11α gene (Fig. 4D, upper panel). Moreover, a 40-nucleotide fragment around the PPRE-like motif was sufficient for activation by PPARα (Fig. 4E). Thus, this motif is a genuine PPRE and the primary target of PPAR-mediated transactivation of the PEX11α gene. Interestingly, PPARγ2 did not support transactivation from the PPRE in this context (Fig. 4D, lower panel), indicating that the function of this element was selective for PPAR subtypes. The constructs shown in Fig. 4D were next stably transfected into a rat hepatoma cell line, H4IIEC3, which is susceptible to induction by peroxisome proliferators in culture (48). In these transformants, a significant induction by Wy14,643 was observed with the wild type but not the mutant construct (data not shown).
We further investigated whether the PPRE indeed worked with the PEX11α promoter when located downstream, because the PPRE was separated from the transcriptional initiation site of the PEX11α gene by more than 8 kb. In a supercoiled circular plasmid, the PPRE may act from the upstream side or a sterically close position, even when it is placed downstream of the reporter gene. Therefore, we linearized the reporter plasmids shown in Fig. 4D by cutting at a point just behind the downstream fragment of PEX11α and transfected them into HeLa cells together with a PPARα expression vector (Fig. 4F). We again observed significant transactivation with the wild type but not the mutant construct, thereby confirming that the PPRE worked from the downstream side.
Perilipin gene is transactivated by PPARγ2 through the PPRE.
The transcriptional initiation site of the perilipin gene is located 1.9 kb downstream from the PEX11α-PPRE in the mouse genome (Fig. 2A). Production of perilipin is induced during adipocyte differentiation (22), and the level of perilipin mRNA is increased by BRL49,653, a PPARγ ligand, in the differentiated adipocytes (56). Hence, we inferred that the perilipin gene is regulated by PPARγ2 through the same PPRE. To examine this model, the region from positions −2877 to +56 of the perilipin gene containing the PPRE and native perilipin promoter was inserted into a reporter vector and introduced into HeLa cells together with a PPARγ2 expression vector (Fig. 5A, lower panel). A ligand-dependent transactivation by PPARγ2 was observed, and mutation of PPRE completely abolished this activation. This construct was activated by PPARγ1 and marginally by PPARα (Fig. 5A, upper panel) but not at all by PPARβ/δ (data not shown).
FIG. 5.
Transactivation of the perilipin gene through the common PPRE. The numbers in the maps of the constructs are relative to the transcriptional initiation site of the perilipin gene. Luciferase (Luc) activity is shown as a relative value, with the activity of pGVB in the absence of both a ligand and a PPAR expression vector as 1. The lowercase letters in the sequences are mutated residues. (A) PPARγ2-selective transactivation of the perilipin gene. For PPARα and PPARγ2, 100 μM Wy14,643 and 1 μM BRL49,653, respectively, were used as ligands. (B) Ligand-dependent function of the PPRE in adipocytes. Reporter plasmids were introduced into 3T3-L1 preadipocytes and adipocytes. Adipocytes were the cells treated with the hormone cocktail as described in Materials and Methods. Preadipocytes were the cells cultured for the same period as adipocytes without the hormone cocktail treatment.
To investigate whether the PPRE works in adipocytes, we introduced these plasmids into 3T3-L1 preadipocytes and adipocytes (Fig. 5B). A transactivation by BRL49,653 was observed in the differentiated adipocytes where PPARγ2 is highly expressed, but the reporter expression was not significantly activated in the preadipocytes. We also observed a significant luciferase expression without BRL49,653 in the adipocytes, possibly because of endogenous ligands of PPARγ, such as fatty acids. The reporter expression was completely abolished in the mutant construct, thereby indicating that the perilipin gene is transactivated by PPARγ2 through the PPRE.
PPAR/RXR heterodimer binds to the PPRE.
We next investigated the binding of the PPARα/RXRα heterodimer to the PPRE by EMSA, using probes encompassing the PPREs of the PEX11α and AOx genes (Fig. 6A). An assay was carried out using the fusion proteins, maltose-binding protein-PPARα, and GST-RXRα (46). Shifted bands were observed with both probes only when both PPARα and RXRα were added to the reaction (Fig. 6A, lanes 4 and 10). This binding was abolished by the wild-type but not the mutant competitors (Fig. 6A, lanes 5 to 8 and 11 to 14). These results indicate that the heterodimer PPARα/RXRα directly binds to the PPRE. We also examined the binding of PPARγ2/RXRα to the PPRE by EMSA (Fig. 6B) using the proteins synthesized in a rabbit reticulocyte lysate system. PPARγ2 and RXRα, when added together, exhibited binding to the PPRE, but PPARγ2 or RXRα alone did not (Fig. 6B, lanes 2 to 4). This binding was abolished only by the wild-type competitors (Fig. 6B, lanes 5 to 8). Thus, the PPARγ2/RXRα heterodimer specifically bound to the PPRE.
FIG. 6.
Binding of the PPAR/RXR heterodimer to the PPRE in vitro. For competition experiments, unlabeled oligonucleotides in 100-fold excess were used. Sequences of probes and competitors were used as described in Materials and Methods. Competitors a, b, c, and d were wild-type and mutant PEX11α and wild-type and mutant AOx, respectively. (A) Specific PPARα/RXRα binding to the PEX11α/perilipin-PPRE. EMSA was performed using MAL-PPARα and GST-RXRα fusion proteins. (B) Specific binding of PPARγ2/RXRα to the PPRE. PPARγ2 and RXRα were synthesized in a rabbit reticulocyte lysate system.
PPARα and PPARγ bind to the PPRE tissue selectively in vivo.
To confirm that the PPARα/RXR or PPARγ/RXR heterodimer binds to the PPRE in vivo in a tissue-selective manner, we performed a ChIP assay. H4IIEC3 and 3T3-L1 cells were used as representatives of hepatocytes and adipocytes, respectively. H4IIEC3 derived from a rat hepatoma and the rat sequence of the region at a position corresponding to the PEX11α/perilipin-PPRE is different from that of mouse (Fig. 7A). Hence, we first confirmed that the rat genomic sequence encompassing the putative PPRE also conferred the PPAR-dependent transactivation in the gene reporter assay (data not shown). In addition to the binding of PPARα, PPARγ, and RXR, recruitment of CREB-binding protein (CBP), a representative coactivator of nuclear hormone receptor, was also examined as an indicator of transactivation.
FIG. 7.
Tissue-selective binding of PPARα and PPARγ to the PPRE in vivo. (A) Schematic diagram of the region around the PEX11α/perilipin-PPRE in the genomes of mouse and rat. Arrows indicate the regions amplified by PCR in the ChIP assay. (B) Selective binding of PPARα to the PPRE in the liver. H4IIEC3 cells were treated with 100 μM Wy14,643 (Wy) or DMSO (DM) for 2 days and processed for the ChIP assay. Immunoprecipitation was performed with the antibodies indicated or preimmune rabbit IgG. (C) PPARγ selectively binds to the PPRE in the adipose tissue. The ChIP assay wasperformed on the 3T3-L1 preadipocytes (Pre) and adipocytes (Ad).
In H4IIEC3 cells, we observed significant bands for PPARα, RXR, and CBP but not for PPARγ with the PPRE primer pair (Fig. 7B). These bands were negligible either with the distal region primers or for a control IgG. No dependence on the PPARα ligand was observed even for CBP, possibly reflecting the apparently ligand-independent transactivation by PPARα often observed in the culture system. In 3T3-L1 cells, strong bands were observed for PPARγ, RXR, and CBP but not for PPARα with the PPRE, and only in the differentiated adipocytes were strong bands not observed with the distal primers (Fig. 7C). These results support the view that PPARα and PPARγ selectively bind to the PPRE in the liver and the adipose tissue and activate the transcription of PEX11α and perilipin genes, respectively.
The PEX11α/perilipin-PPRE is conserved in the human genome.
We compared the nucleotide sequences around the PPRE in the mouse and human genomes. In the human genome database, we found a PPRE-like sequence in the downstream region of the human PEX11α gene (Fig. 2B). This sequence matched the PPRE of the mouse PEX11α gene except for an optional base between the two half sites. Furthermore, not only the PPRE but also the sequences around it were highly conserved between the two species. Hence, the human PEX11α gene was also suspected to be transactivated by PPARα. We examined whether expression of the human PEX11α was induced by peroxisome proliferators, using human hepatoma, HepG2. Cells were cultured in the presence or absence of ciprofibrate, a peroxisome proliferator, and the expression of endogenous PEX11α was monitored by RT-PCR. The expression was not increased by the drug (data not shown), though under the same conditions, carnitine palmitoyl transferase 1 and mitochondrial HMG-CoA synthetase were induced, in accordance with reports by other investigators (27, 34). Peroxisome numbers are not significantly increased by peroxisome proliferators in human liver (40). The response of the PEX11α gene to peroxisome proliferators is probably restricted by a unique mechanism in human liver.
DISCUSSION
We found a functional PPRE located downstream of the mouse PEX11α gene, 8.4 kb from the promoter. On the other hand, we failed to observe PPAR-dependent transactivation with the upstream region up to kb −12.5 as well as the region of the PEX11α structural gene that includes the introns. Using a linearized reporter plasmid, we observed transactivation by PPARα via the downstream PPRE, thereby indicating that the PPRE indeed functioned from the downstream location at this distance. The PPRE identified is located in the vicinity of the perilipin gene and supported the activation of transcription from the perilipin promoter. We also found that the PEX11α gene was selectively transactivated by PPARα and that the perilipin gene was transactivated by PPARγ through this common PPRE. Consistently, the expression of PEX11α is induced in the liver and the expression of perilipin is induced in the adipose tissue, where PPARα and PPARγ2 are, respectively, selectively expressed. In the ChIP assay, endogenous PPARα and PPARγ indeed bound selectively to the PPRE in hepatocytes and adipocytes, respectively. Based on these observations, we propose a mechanism of tissue-selective regulation of PEX11α and perilipin (Fig. 8). The PPRE functions as a cis element for PEX11α gene activation by PPARα in the liver and for perilipin gene expression by PPARγ in the adipose tissue. We observed transactivation selective for PPAR subtypes only with the natural PEX11α and perilipin promoters. When the SV40 promoter was used, the PPRE worked with both PPARα and PPARγ (data not shown). Thus, the combination with specific promoters seems to determine the selectivity for PPAR subtypes. Transcription machineries acting on different promoters are not always the same, and tissue-specific promoter-binding factors have been described (25). The selectivity of PEX11α-perilipin PPRE to PPAR subtypes might be determined by the combinatorial interactions between PPAR subtypes and promoter-binding factors, probably also involving coactivators.
FIG. 8.
Models of the tissue-selective regulation of PEX11α and perilipin genes. These genes are regulated through the common PPRE in a tissue-selective manner. In the liver, PPARα is highly expressed; hence, it initiates the activation of the PEX11α gene. On the other hand, PPARγ2 is limited to adipose tissue, working for perilipin gene induction.
Most PPREs reported to date (14) are located proximally to the transcriptional initiation site. The location of the PEX11α PPRE, however, is not necessarily exceptional: a few PPREs have been found in distal positions, e.g., 4 to 5 kb upstream in the adipocyte P2 (70) and CYP4A1 genes (4), in an intron in the acyl-CoA-binding protein gene (23), and in the downstream region in the apolipoprotein E gene (20). Enhancers often function irrespective of position relative to the promoter and distance from the transcriptional initiation site (6). In fact, the wing margin enhancer of the Drosophila cut locus (30) and the T-cell receptor Cα gene enhancer (75) act from positions more than 50 kb upstream, and the β-globin enhancer is functional in the downstream region (16, 17). The distance- and position-independent functions of enhancers have been attributed to the physical communication between the promoters and enhancers, bridged by transcription factor complexes (9). Evidence for such physical interactions has been emerging recently (11, 67).
The bidirectional function of a single PPRE is exceptional. Although tissue-selective activation by PPARα and PPARγ was reported for the lipoprotein lipase gene, this activation involves only a single gene (60). On the other hand, there are several precedents for bidirectional enhancers. In most of these cases, such as chicken β-globin-ɛ-globin (44) and albumin-α-fetoprotein (21) gene pairs, the target genes are related to each other, structurally and functionally, reflecting their origination by gene duplication. The functions of PEX11α and perilipin, however, are different, and their amino acid sequences are not related. The function of PEX11 remains obscure, made more confusing by a report that its primary function is in fatty acid oxidation, not peroxisomal division (73). In any case, PEX11α is expressed mainly in the liver, where lipid metabolism is one important function, and is located on the membrane of peroxisomes, organelles involved in fatty acid catabolism. On the other hand, the expression of perilipin is limited to the adipose tissue where lipid is stored. Based on these observations, the functions of PEX11α and perilipin would be the same in the area of lipid mobilization, and hence it would be reasonable that these genes are regulated by partially overlapping mechanisms.
It has generally been postulated that an enhancer serves a single gene. Recent genome analysis, however, revealed that many genes are located very close together, sometimes even sharing promoters (3). Enhancers as well as individual enhancer elements can activate target promoters located several kilobases apart. Hence, the functional distal enhancer element of a gene may be even closer to another gene. Based on these considerations, we expect that even more gene pairs will be revealed in the future to be regulated by common bidirectional enhancer elements. In such cases, combinations with promoters and other proximal elements may confer regulatory specificity to each gene, as found in the present study of the PEX11α-perilipin gene pair.
The proliferative effect of hypolipidemic agents on peroxisomes in rodent liver was first observed in the 1960s (24). However, the protein that promotes peroxisomal division in response to these agents is as yet unknown. One of the likely candidates seems to be PEX11α, though peroxisomal metabolic activities were also shown to affect the number of this organelle (12, 52, 54, 73). In this study, we showed that the PEX11α gene is a bona fide target of PPARα and identified its functional PPRE. The expression of no other PEX gene has so far been reported to be induced by peroxisome proliferators. Thus, it is tempting to speculate that the proliferation evoked by these agents is directly caused by PEX11α. It was reported, however, that peroxisome proliferation is observed in PEX11α-null mice (37). In addition, a dynamin-related protein has recently been reported to be involved in peroxisomal division (26, 33, 38). Future studies will be required in order to determine which gene is the key factor in peroxisome proliferation.
Acknowledgments
We thank B. M. Spiegelman, G. MacGregor, P. A. Grimaldi, the late K. Umesono, and R. M. Evans for the plasmids. We also thank Y. Emi for a mouse genomic library.
This work was supported in part by the 21st Century COE Program and a Grand-in-Aid for Scientific Research from the Japan Society for the Promotion of Science.
REFERENCES
- 1.Abe, I., and Y. Fujiki. 1998. cDNA cloning and characterization of a constitutively expressed isoform of the human peroxin Pex11p. Biochem. Biophys. Res. Commun. 252:529-533. [DOI] [PubMed] [Google Scholar]
- 2.Abe, I., K. Okumoto, S. Tamura, and Y. Fujiki. 1998. Clofibrate-inducible, 28-kDa peroxisomal integral membrane protein is encoded by PEX11. FEBS Lett. 431:468-472. [DOI] [PubMed] [Google Scholar]
- 3.Adachi, N., and M. R. Lieber. 2002. Bidirectional gene organization: a common architectural feature of the human genome. Cell 109:807-809. [DOI] [PubMed] [Google Scholar]
- 4.Aldridge, T. C., J. D. Tugwood, and S. Green. 1995. Identification and characterization of DNA elements implicated in the regulation of CYP4A1 transcription. Biochem. J. 306:473-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amri, E. Z., F. Bonino, G. Ailhaud, N. A. Abumrad, and P. A. Grimaldi. 1995. Cloning of a protein that mediates transcriptional effects of fatty acids in preadipocytes. Homology to peroxisome proliferator-activated receptors. J. Biol. Chem. 270:2367-2371. [DOI] [PubMed] [Google Scholar]
- 6.Banerji, J., S. Rusconi, and W. Schaffner. 1981. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27:299-308. [DOI] [PubMed] [Google Scholar]
- 7.Barak, Y., D. Liao, W. He, E. S. Ong, M. C. Nelson, J. M. Olefsky, R. Boland, and R. M. Evans. 2002. Effects of peroxisome proliferator-activated receptor delta on placentation, adiposity, and colorectal cancer. Proc. Natl. Acad. Sci. USA 99:303-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger, J., and D. E. Moller. 2002. The mechanisms of action of PPARs. Annu. Rev. Med. 53:409-435. [DOI] [PubMed] [Google Scholar]
- 9.Blackwood, E. M., and J. T. Kadonaga. 1998. Going the distance: a current view of enhancer action. Science 281:61-63. [DOI] [PubMed] [Google Scholar]
- 10.Braissant, O., F. Foufelle, C. Scotto, M. Dauca, and W. Wahli. 1996. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology 137:354-366. [DOI] [PubMed] [Google Scholar]
- 11.Carter, D., L. Chakalova, C. S. Osborne, Y. F. Dai, and P. Fraser. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32:623-626. [DOI] [PubMed] [Google Scholar]
- 12.Chang, C. C., S. South, D. Warren, J. Jones, A. B. Moser, H. W. Moser, and S. J. Gould. 1999. Metabolic control of peroxisome abundance. J. Cell Sci. 112:1579-1590. [DOI] [PubMed] [Google Scholar]
- 13.Clifford, G. M., C. Londos, F. B. Kraemer, R. G. Vernon, and S. J. Yeaman. 2000. Translocation of hormone-sensitive lipase and perilipin upon lipolytic stimulation of rat adipocytes. J. Biol. Chem. 275:5011-5015. [DOI] [PubMed] [Google Scholar]
- 14.Desvergne, B., and W. Wahli. 1999. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr. Rev. 20:649-688. [DOI] [PubMed] [Google Scholar]
- 15.Distel, B., R. Erdmann, S. J. Gould, G. Blobel, D. I. Crane, J. M. Cregg, G. Dodt, Y. Fujiki, J. M. Goodman, W. W. Just, J. A. Kiel, W. H. Kunau, P. B. Lazarow, G. P. Mannaerts, H. W. Moser, T. Osumi, R. A. Rachubinski, A. Roscher, S. Subramani, H. F. Tabak, T. Tsukamoto, D. Valle, I. van der Klei, P. P. van Veldhoven, and M. Veenhuis. 1996. A unified nomenclature for peroxisome biogenesis factors. J. Cell Biol. 135:1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dodgson, J. B., J. Strommer, and J. D. Engel. 1979. Isolation of the chicken beta-globin gene and a linked embryonic beta-like globin gene from a chicken DNA recombinant library. Cell 17:879-887. [DOI] [PubMed] [Google Scholar]
- 17.Dolan, M., B. J. Sugarman, J. B. Dodgson, and J. D. Engel. 1981. Chromosomal arrangement of the chicken beta-type globin genes. Cell 24:669-677. [DOI] [PubMed] [Google Scholar]
- 18.Dreyer, C., G. Krey, H. Keller, F. Givel, G. Helftenbein, and W. Wahli. 1992. Control of the peroxisomal beta-oxidation pathway by a novel family of nuclear hormone receptors. Cell 68:879-887. [DOI] [PubMed] [Google Scholar]
- 19.Erdmann, R., and G. Blobel. 1995. Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J. Cell Biol. 128:509-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galetto, R., M. Albajar, J. I. Polanco, M. M. Zakin, and J. C. Rodriguez-Rey. 2001. Identification of a peroxisome-proliferator-activated-receptor response element in the apolipoprotein E gene control region. Biochem. J. 357:521-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godbout, R., R. Ingram, and S. M. Tilghman. 1986. Multiple regulatory elements in the intergenic region between the α-fetoprotein and albumin genes. Mol. Cell. Biol. 6:477-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenberg, A. S., J. J. Egan, S. A. Wek, N. B. Garty, E. J. Blanchette-Mackie, and C. Londos. 1991. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J. Biol. Chem. 266:11341-11346. [PubMed] [Google Scholar]
- 23.Helledie, T., L. Grontved, S. S. Jensen, P. Kiilerich, L. Rietveld, T. Albrektsen, M. S. Boysen, J. Nohr, L. K. Larsen, J. Fleckner, H. G. Stunnenberg, K. Kristiansen, and S. Mandrup. 2002. The gene encoding the Acyl-CoA-binding protein is activated by peroxisome proliferator-activated receptor gamma through an intronic response element functionally conserved between humans and rodents. J. Biol. Chem. 277:26821-26830. [DOI] [PubMed] [Google Scholar]
- 24.Hess, R., W. Staubli, and W. Riess. 1965. Nature of the hepatomegalic effect produced by ethyl-chlorophenoxy-isobutyrate in the rat. Nature 208:856-858. [DOI] [PubMed] [Google Scholar]
- 25.Hochheimer, A., and R. Tjian. 2003. Diversified transcription initiation complexes expand promoter selectivity and tissue-specific gene expression. Genes Dev. 17:1309-1320. [DOI] [PubMed] [Google Scholar]
- 26.Hoepfner, D., M. van den Berg, P. Philippsen, H. F. Tabak, and E. H. Hettema. 2001. A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155:979-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu, M. H., U. Savas, K. J. Griffin, and E. F. Johnson. 2001. Identification of peroxisome proliferator-responsive human genes by elevated expression of the peroxisome proliferator-activated receptor alpha in HepG2 cells. J. Biol. Chem. 276:27950-27958. [DOI] [PubMed] [Google Scholar]
- 28.Ijpenberg, A., E. Jeannin, W. Wahli, and B. Desvergne. 1997. Polarity and specific sequence requirements of peroxisome proliferator-activated receptor (PPAR)/retinoid X receptor heterodimer binding to DNA. A functional analysis of the malic enzyme gene PPAR response element. J. Biol. Chem. 272:20108-20117. [DOI] [PubMed] [Google Scholar]
- 29.Issemann, I., and S. Green. 1990. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature 347:645-650. [DOI] [PubMed] [Google Scholar]
- 30.Jack, J., D. Dorsett, Y. Delotto, and S. Liu. 1991. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development 113:735-747. [DOI] [PubMed] [Google Scholar]
- 31.Juge-Aubry, C., A. Pernin, T. Favez, A. G. Burger, W. Wahli, C. A. Meier, and B. Desvergne. 1997. DNA binding properties of peroxisome proliferator-activated receptor subtypes on various natural peroxisome proliferator response elements. Importance of the 5′-flanking region. J. Biol. Chem. 272:25252-25259. [DOI] [PubMed] [Google Scholar]
- 32.Kliewer, S. A., K. Umesono, D. J. Noonan, R. A. Heyman, and R. M. Evans. 1992. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature 358:771-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koch, A., M. Thiemann, M. Grabenbauer, Y. Yoon, M. A. McNiven, and M. Schrader. 2003. Dynamin-like protein 1 is involved in peroxisomal fission. J. Biol. Chem. 278:8597-8605. [DOI] [PubMed] [Google Scholar]
- 34.Lawrence, J. W., Y. Li, S. Chen, J. G. DeLuca, J. P. Berger, D. R. Umbenhauer, D. E. Moller, and G. Zhou. 2001. Differential gene regulation in human versus rodent hepatocytes by peroxisome proliferator-activated receptor (PPAR) alpha. PPAR alpha fails to induce peroxisome proliferation-associated genes in human cells independently of the level of receptor expresson. J. Biol. Chem. 276:31521-31527. [DOI] [PubMed] [Google Scholar]
- 35.Lazarow, P. B., and Y. Fujiki. 1985. Biogenesis of peroxisomes. Annu. Rev. Cell Biol. 1:489-530. [DOI] [PubMed] [Google Scholar]
- 36.Lee, S. S., T. Pineau, J. Drago, E. J. Lee, J. W. Owens, D. L. Kroetz, P. M. Fernandez-Salguero, H. Westphal, and F. J. Gonzalez. 1995. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol. 15:3012-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, X., E. Baumgart, G. X. Dong, J. C. Morrell, G. Jimenez-Sanchez, D. Valle, K. D. Smith, and S. J. Gould. 2002. PEX11α is required for peroxisome proliferation in response to 4-phenylbutyrate but is dispensable for peroxisome proliferator-activated receptor alpha-mediated peroxisome proliferation. Mol. Cell. Biol. 22:8226-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, X., and S. J. Gould. 2003. The dynamin-like GTPase DLP1 is essential for peroxisome division and is recruited to peroxisomes in part by PEX11. J. Biol. Chem. 278:17012-17020. [DOI] [PubMed] [Google Scholar]
- 39.Li, X., and S. J. Gould. 2002. PEX11 promotes peroxisome division independently of peroxisome metabolism. J. Cell Biol. 156:643-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lock, E. A., A. M. Mitchell, and C. R. Elcombe. 1989. Biochemical mechanisms of induction of hepatic peroxisome proliferation. Annu. Rev. Pharmacol. Toxicol. 29:145-163. [DOI] [PubMed] [Google Scholar]
- 41.MacGregor, G. R., and C. T. Caskey. 1989. Construction of plasmids that express E. coli beta-galactosidase in mammalian cells. Nucleic Acids Res. 17:2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marshall, P. A., Y. I. Krimkevich, R. H. Lark, J. M. Dyer, M. Veenhuis, and J. M. Goodman. 1995. Pmp27 promotes peroxisomal proliferation. J. Cell Biol. 129:345-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsumoto, N., S. Tamura, and Y. Fujiki. 2003. The pathogenic peroxin Pex26p recruits the Pex1p-Pex6p AAA ATPase complexes to peroxisomes. Nat. Cell Biol. 5:454-460. [DOI] [PubMed] [Google Scholar]
- 44.Nickol, J. M., and G. Felsenfeld. 1988. Bidirectional control of the chicken beta- and epsilon-globin genes by a shared enhancer. Proc. Natl. Acad. Sci. USA 85:2548-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nicolas-Frances, V., V. K. Dasari, E. Abruzzi, T. Osumi, and N. Latruffe. 2000. The peroxisome proliferator response element (PPRE) present at positions −681/-669 in the rat liver 3-ketoacyl-CoA thiolase B gene functionally interacts differently with PPARalpha and HNF-4. Biochem. Biophys. Res. Commun. 269:347-351. [DOI] [PubMed] [Google Scholar]
- 46.Osada, S., T. Tsukamoto, M. Takiguchi, M. Mori, and T. Osumi. 1997. Identification of an extended half-site motif required for the function of peroxisome proliferator-activated receptor alpha. Genes Cells 2:315-327. [DOI] [PubMed] [Google Scholar]
- 47.Osumi, T., J. K. Wen, and T. Hashimoto. 1991. Two cis-acting regulatory sequences in the peroxisome proliferator-responsive enhancer region of rat acyl-CoA oxidase gene. Biochem. Biophys. Res. Commun. 175:866-871. [DOI] [PubMed] [Google Scholar]
- 48.Osumi, T., S. Yokota, and T. Hashimoto. 1990. Proliferation of peroxisomes and induction of peroxisomal beta-oxidation enzymes in rat hepatoma H4IIEC3 by ciprofibrate. J. Biochem. (Tokyo) 108:614-621. [DOI] [PubMed] [Google Scholar]
- 49.Palmer, C. N., M. H. Hsu, H. J. Griffin, and E. F. Johnson. 1995. Novel sequence determinants in peroxisome proliferator signaling. J. Biol. Chem. 270:16114-16121. [DOI] [PubMed] [Google Scholar]
- 50.Passreiter, M., M. Anton, D. Lay, R. Frank, C. Harter, F. T. Wieland, K. Gorgas, and W. W. Just. 1998. Peroxisome biogenesis: involvement of ARF and coatomer. J. Cell Biol. 141:373-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peters, J. M., S. S. Lee, W. Li, J. M. Ward, O. Gavrilova, C. Everett, M. L. Reitman, L. D. Hudson, and F. J. Gonzalez. 2000. Growth, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor β(δ). Mol. Cell. Biol. 20:5119-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poll-The, B. T., F. Roels, H. Ogier, J. Scotto, J. Vamecq, R. B. Schutgens, R. J. Wanders, C. W. van Roermund, M. J. van Wijland, A. W. Schram, et al. 1988. A new peroxisomal disorder with enlarged peroxisomes and a specific deficiency of acyl-CoA oxidase (pseudo-neonatal adrenoleukodystrophy). Am. J. Hum. Genet. 42:422-434. [PMC free article] [PubMed] [Google Scholar]
- 53.Purdue, P. E., and P. B. Lazarow. 2001. Peroxisome biogenesis. Annu. Rev. Cell Dev. Biol. 17:701-752. [DOI] [PubMed] [Google Scholar]
- 54.Qi, C., Y. Zhu, J. Pan, N. Usuda, N. Maeda, A. V. Yeldandi, M. S. Rao, T. Hashimoto, and J. K. Reddy. 1999. Absence of spontaneous peroxisome proliferation in enoyl-CoA hydratase/l-3-hydroxyacyl-CoA dehydrogenase-deficient mouse liver. Further support for the role of fatty acyl CoA oxidase in PPARα ligand metabolism. J. Biol. Chem. 274:15775-15780. [DOI] [PubMed] [Google Scholar]
- 55.Reddy, J. K., and T. Hashimoto. 2001. Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu. Rev. Nutr. 21:193-230. [DOI] [PubMed] [Google Scholar]
- 56.Rosenbaum, S. E., and A. S. Greenberg. 1998. The short- and long-term effects of tumor necrosis factor-alpha and BRL 49653 on peroxisome proliferator-activated receptor (PPAR)gamma2 gene expression and other adipocyte genes. Mol. Endocrinol. 12:1150-1160. [DOI] [PubMed] [Google Scholar]
- 57.Rubin, C. S., A. Hirsch, C. Fung, and O. M. Rosen. 1978. Development of hormone receptors and hormonal responsiveness in vitro. Insulin receptors and insulin sensitivity in the preadipocyte and adipocyte forms of 3T3-L1 cells. J. Biol. Chem. 253:7570-7578. [PubMed] [Google Scholar]
- 58.Sakai, Y., P. A. Marshall, A. Saiganji, K. Takabe, H. Saiki, N. Kato, and J. M. Goodman. 1995. The Candida boidinii peroxisomal membrane protein Pmp30 has a role in peroxisomal proliferation and is functionally homologous to Pmp27 from Saccharomyces cerevisiae. J. Bacteriol. 177:6773-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, p. 16.32-16.36. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 60.Schoonjans, K., J. Peinado-Onsurbe, A. M. Lefebvre, R. A. Heyman, M. Briggs, S. Deeb, B. Staels, and J. Auwerx. 1996. PPARalpha and PPARgamma activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. EMBO J. 15:5336-5348. [PMC free article] [PubMed] [Google Scholar]
- 61.Schrader, M., B. E. Reuber, J. C. Morrell, G. Jimenez-Sanchez, C. Obie, T. A. Stroh, D. Valle, T. A. Schroer, and S. J. Gould. 1998. Expression of PEX11beta mediates peroxisome proliferation in the absence of extracellular stimuli. J. Biol. Chem. 273:29607-29614. [DOI] [PubMed] [Google Scholar]
- 62.Servetnick, D. A., D. L. Brasaemle, J. Gruia-Gray, A. R. Kimmel, J. Wolff, and C. Londos. 1995. Perilipins are associated with cholesteryl ester droplets in steroidogenic adrenal cortical and Leydig cells. J. Biol. Chem. 270:16970-16973. [DOI] [PubMed] [Google Scholar]
- 63.Smith, J. J., M. Marelli, R. H. Christmas, F. J. Vizeacoumar, D. J. Dilworth, T. Ideker, T. Galitski, K. Dimitrov, R. A. Rachubinski, and J. D. Aitchison. 2002. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 158:259-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tam, Y. Y., and R. A. Rachubinski. 2002. Yarrowia lipolytica cells mutant for the PEX24 gene encoding a peroxisomal membrane peroxin mislocalize peroxisomal proteins and accumulate membrane structures containing both peroxisomal matrix and membrane proteins. Mol. Biol. Cell 13:2681-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tanaka, A., K. Okumoto, and Y. Fujiki. 2003. cDNA cloning and characterization of the third isoform of human peroxin Pex11p. Biochem. Biophys. Res. Commun. 300:819-823. [DOI] [PubMed] [Google Scholar]
- 66.Titorenko, V. I., and R. A. Rachubinski. 2001. The life cycle of the peroxisome. Nat. Rev. Mol. Cell Biol. 2:357-368. [DOI] [PubMed] [Google Scholar]
- 67.Tolhuis, B., R. J. Palstra, E. Splinter, F. Grosveld, and W. de Laat. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10:1453-1465. [DOI] [PubMed] [Google Scholar]
- 68.Tominaga, S., M. Morikawa, and T. Osumi. 2002. Growth hormone has dual stage-specific effects on the differentiation of 3T3-L1 preadipocytes. J. Biochem. (Tokyo) 132:881-889. [DOI] [PubMed] [Google Scholar]
- 69.Tontonoz, P., E. Hu, J. Devine, E. G. Beale, and B. M. Spiegelman. 1995. PPARγ2 regulates adipose expression of the phosphoenolpyruvate carboxykinase gene. Mol. Cell. Biol. 15:351-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tontonoz, P., E. Hu, R. A. Graves, A. I. Budavari, and B. M. Spiegelman. 1994. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 8:1224-1234. [DOI] [PubMed] [Google Scholar]
- 71.Tontonoz, P., E. Hu, and B. M. Spiegelman. 1994. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell 79:1147-11456. [DOI] [PubMed] [Google Scholar]
- 72.Tugwood, J. D., I. Issemann, R. G. Anderson, K. R. Bundell, W. L. McPheat, and S. Green. 1992. The mouse peroxisome proliferator activated receptor recognizes a response element in the 5′ flanking sequence of the rat acyl CoA oxidase gene. EMBO J. 11:433-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.van Roermund, C. W., H. F. Tabak, M. van Den Berg, R. J. Wanders, and E. H. Hettema. 2000. Pex11p plays a primary role in medium-chain fatty acid oxidation, a process that affects peroxisome number and size in Saccharomyces cerevisiae. J. Cell Biol. 150:489-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, Y. X., C. H. Lee, S. Tiep, R. T. Yu, J. Ham, H. Kang, and R. M. Evans. 2003. Peroxisome-proliferator-activated receptor delta activates fat metabolism to prevent obesity. Cell 113:159-170. [DOI] [PubMed] [Google Scholar]
- 75.Winoto, A., and D. Baltimore. 1989. A novel, inducible and T cell-specific enhancer located at the 3′ end of the T cell receptor alpha locus. EMBO J. 8:729-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang, B., S. L. Marcus, F. G. Sajjadi, K. Alvares, J. K. Reddy, S. Subramani, R. A. Rachubinski, and J. P. Capone. 1992. Identification of a peroxisome proliferator-responsive element upstream of the gene encoding rat peroxisomal enoyl-CoA hydratase/3-hydroxyacyl-CoA dehydrogenase. Proc. Natl. Acad. Sci. USA 89:7541-7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu, Y., C. Qi, J. R. Korenberg, X. N. Chen, D. Noya, M. S. Rao, and J. K. Reddy. 1995. Structural organization of mouse peroxisome proliferator-activated receptor gamma (mPPAR gamma) gene: alternative promoter use and different splicing yield two mPPAR gamma isoforms. Proc. Natl. Acad. Sci. USA 92:7921-7925. [DOI] [PMC free article] [PubMed] [Google Scholar]