Abstract
Type 1 diabetes mellitus formerly called juvenile diabetes, is an organ specific T-cell mediated autoimmune disease characterized by the progressive loss of function of the insulin producing beta–cells of the islets of Langerhans. Cytotoxic T lymphocyte-associated antigen 4 gene (CTLA-4) has been proposed as a candidate gene for conferring susceptibility to autoimmunity. Association of CTLA-4 gene polymorphism is well established in autoimmune endocrinopathies across world population. The present study was conducted to investigate the association of CTLA-4 exon 1 49A/G polymorphism with TIDM in Madurai, a city in Southern India. Fifty three clinically proven T1DM patients and 53 control subjects with no history of autoimmune disease were recruited for the study. Genomic DNA was extracted from peripheral blood. CTLA-4 exon 1 49 A/G polymorphism was assessed using PCR-RFLP methods. Our findings revealed a significant association of CTLA-4 exon 1 49 A/G polymorphism with T1DM in Madurai population.
Keywords: Autoimmune disease, Type 1 diabetes; Cytotoxic T lymphocyte-associated antigen 4 gene, Gene polymorphism, T-cell receptor, polymerase chain reaction, restriction enzyme, restriction fragment length polymorphism
Introduction
Cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) gene is a member of the immunoglobulin super family. It encodes a protein which transmits an inhibitory signal to T-cells. Because of its inhibitory role, CTLA-4 is a strong candidate susceptibility gene in autoimmunity. The human CTLA-4 gene consists of four exons and three introns.[1] There are at least three well-studied polymorphic markers: a C/T substitution at position-318 of the promoter region,[2] an A/G transition at position 49 of exon 1 (A49G) which causes a threonine-to-alanine substitution in codon 17 of the leader peptide (A17T);[3] and, an (AT)n dinucleotide repeat polymorphism located in the 3’ untranslated region of exon 4.[4] The A/G polymorphism at position +49 in exon 1 of the CTLA-4 gene has been extensively studied in autoimmune disorders and is found associated with Graves’ disease, Hashimoto's thyroiditis and multiple sclerosis including Type 1 diabetes mellitus (T1DM). Though the CTLA-4 gene polymorphism is studied in different ethnic groups, its association with TIDM in South Indian population was not established. The present study was conducted to investigate the association of CTLA-4 gene exon 1 49 A/G (codon 17 Thr/Ala) polymorphism, with T1DM was studied in Madurai population.
Type 1 diabetes
Type 1 diabetes mellitus is a T-cell mediated organ specific autoimmune disease characterized by the destruction of insulin secreting β cells of the islets of Langehans in the pancreas.[3] The worldwide prevalence of diabetes is continuing to increase. In the year 2000, there were 151 million people with diabetes. This number is expected to reach 300 million in the year 2025. Presently, India leads the world with the largest number of adults with diabetes.[5] Diabetes mellitus includes a group of metabolic disorders that is characterized by hyperglycemia and leads to long term macro and micro vascular complications.[6] The most striking increase has been found in Finland but in Asian countries, the incidence is comparatively low. The incidence of Type 1 diabetes in south India has been reported to be 10.5 cases/100,000 per year.[7] There are over 20 regions in the human genome that are associated with TIDM, but most of it make only small contribution to the susceptibility of type 1 diabetes.[8] T1DM has been reported to be strongly associated with HLA-DR3 and or DR4.[9] Insulin gene localized on chromosome 11p15, cytotoxic T- lymphocytic antigen 4 gene on chromosome 2q33 (CTLA-4), and lymphoid specific phosphatase (Lyp) gene on 1p13 and autoimmunity regulation gene (AIRE) on chromosome 21q22.3 are some of the gene associated with TIDM.[10] It has been postulated factors like, certain viral infections and, nutritional compounds, when superimposed on genetic factors, may lead to the activation of disease.[11]
Cytotoxic T lymphocyte-associated antigen 4 gene and autoimmunity
The CTLA-4 gene is located on the long arm of the chromosome 2q33.[12] It consists of 4 exons and 3 introns. Exon 1 encodes a leather peptide sequence, exon 2 codes for an immunoglobulin domain, exon 3 and 4 codes for the hydrophobic transmembrane domain, and the cytoplasmic domain respectively.[13] Several studies have shown the association of CTLA-4 gene with autoimmune diseases.[14] The CTLA-4 gene encodes T-cell receptor involved in the control of proliferation and mediates T-cell apoptosis.[15] The receptor protein is a specific T lymphocyte surface antigen that is detected on cells only after antigen presentation. Thus, CTLA-4 is directly involved in both immune and autoimmune responses.[16] Experiments showed that CTLA-4 deficient mice develop a severe lymphoproliferative disease with multiorgan lymphocytic infiltration and tissue destruction, including the pancreas, revealing an immunosuppressive role for the CTLA-4 protein. This may indicate the involvement of CTLA-4 in the pathogenesis of multiple T-cell mediated autoimmune diseases.[17]
The CTLA-4 together with CD28, a co-stimulatory molecule, is expressed on the surface of both naive and activated T cells. Both these molecule plays a significant role in the T-cell mediated immune response, which is initiated when the antigen-specific T- cell surface receptor encounters an antigen presented by the antigen presenting cells (APC) complexed with major histocompatibility complex (MHC) class II molecule. A co-stimulatory signal is necessary to accomplish this activation. In the absence of a positive co-stimulatory signal, T cell's antigen receptor stimulation causes a state of immune unresponsiveness called anergy. On the contrary, a positive co-stimulatory signal, provided mainly by the interaction of CD28 with its ligands, B7.1 (CD80) and B7.2 (CD86) on antigen-presenting cells, will complete T cell activation, leading to T-cell proliferation and cytokine production. CTLA-4 binds to the same B7 (CD80 and CD86) molecules with a greater affinity than CD28, but unlike CD28, it delivers inhibitory signals to T-cell activation and therefore down regulates T-cell proliferative immune response.[18]
Materials and Methods
Fifty three clinically proven Type 1 diabetes patients and control subjects with no history of autoimmune disease within the age group of 10–50 were selected for the study. Peripheral blood from both patient and the control group was collected in EDTA coated tubes and genomic Deoxyribonucleic Acid (DNA) was extracted from the same following the protocol from Sambrook and Russel.[19] Isolated DNA was confirmed electrophoretically (0.8%) and quantified by spectrophotometric absorption at 260 nm. CTLA-4 exon 1 position 49 (A/G: Codon 17 Thr/Ala) polymorphism was defined employing polymerase chain reaction. A 25μl polymerase chain reactions (PCR) recipe was prepared using 2μl of 5μM Forward primer (5’-GCTCTACTTCCTGAAGACCT-3’) and Reverse primer (5’-AGTCTCACTCACCTTTGCAG- 3’)[20] (Fermentas Life Sciences, Germany),~200 ng Genomic DNA, 1 U of Taq polymerase (Bangalore Genei, India) dNTP's 200μM (New England Biolabs, Beverly, MA), 10ΧPCR buffer with MgCl2 and the volume was made up with nuclease free water. Thermocycling conditions were as follows: Initial denaturation - 94°C for 4 minutes, annealing - 58°C for 45 seconds, extension - 72°C for 45 seconds, denaturation - 94°C for 45 seconds, final extension - 72°C for 4 minutes. The polymerase chain reaction was carried out using a thermocycler (Eppendorf, Germany). The amplicons were elctrophoretically confirmed (2% agarose). PCR product was subjected to restriction digestion with Bacillus stearothermophilus, (BseX1,BbV1) (Fermentas Life Sciences, Germany) restriction enzyme as per manufacture's protocol for SNP(single nucleotide polymorphism) detection. The restriction products were analyzed electrophoreticaly by running on a 2% agarose gel alongside a 100bp DNA ladder (New England Biolabs, Beverly, MA).
Results
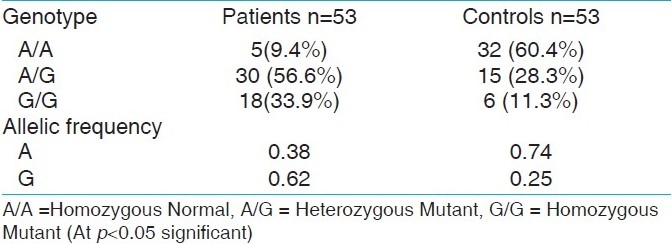
The study was conducted on a group comprising of 53 TIDM patients and 53 controls. Among the T1DM Patients 38 (71.7%) were male and 15 (28.3%) were female. The male and female ratio was maintained same in the control group too. The age group of the patients and the control population was in the range of 10-50 years. The study clearly showed that the male population was found to be more susceptible to T1DM and 30.2% of the patients showed family history of type I diabetes. In the present study, genomic DNA was isolated from the blood of both the patients and the controls and the isolated DNA was confirmed using 0.7% agarose gel. The high molecular weight and slow migrating DNA was found migrating just under the wells and indicated that our methods yielded undegraded DNA as seen in Figure 1. The extracted DNA was subjected to polymerase chain reaction. The post PCR products on gel electrophoresis showed a 162 bp amplicon corresponding to the CTLA-4 gene exon 1 as shown in Figure 2. Restriction digestion was performed on the amplicons using BseX1 (BbV1) restriction enzyme. The restriction enzyme cuts the sequence if there is a G allele at the 49th position and fragments it into 88 bp and 74 bp and in the presence of an A allele the sequence would remain uncut. Electrophoretic analysis of restriction fragment length polymorphism (RFLP) products on 2% agarose gel was carried out to determine A/G polymorphism, genotype and allelic frequency [Figure 3].
Figure 1.

Confirmation of the genomic DNA of T1D patients and control group of Madurai population
Figure 2.

Confirmation of the polymerase chain reactions product of both T1D patients and control group
Figure 3.

Confirmation of restriction fragment length polymorphism product of T1D patients and control group
In the present study, A/G genotype was observed in 30 (56.6%) patients and 15 (28.3%) from the control group. G/G genotype was observed in 18 (34%) of the patients and 6 (11.3%) subjects of the control group. A/A genotype was observed in 5 (9.4%) patients and 32 (60.4%) individuals of the control group [Table 1]. The frequency of the G allele in patients was 0.62 and control population was 0.25, whereas the frequency of the A allele was 0.38 in the patients group and 0.74 in the control group. The chi square test (χ2) value was less than 3.84 under 1 degree of freedom at 5% significance level. Hence the null hypothesis that the population is in Hardey Weinberg equilibrium is accepted.
Table 1.
The frequency of the CTLA-4 A/G genotypes and alleles in type 1 diabetic patients and normal control subjects
Discussion
The present study was carried out at Madurai, a city in southern India and the patient's represented Tamilian population . Our studies show similarity with other ethnic populations of the world in frequent base substitution of A allele with a G nucleotide in the exon 1 at position 49 of the CTLA-4 gene with a significance of P < 0.05. High frequency of G allele in type 1 diabetic patients in Chinese, Korean, Spanish,[21] Italian,[22] and Belgian population.[3] Makes it evident that CTLA-4 gene is a susceptible gene in type 1 diabetes mellitus and CTLA-4 exon 1 49 A/G polymorphism is associated with T1DM and our results indicates the same in Madurai population. However a larger sample size may better emphasize the results.
The results also prove that the population is in accord with Hardey Weinberg equilibrium.
Our results show a small percentage of the control group (28%) though not significant, carry an A/G polymorphism, but don’t suffer any autoimmune disease may be indicative of epigenetic or small RNA mediated control on T-cell regulatory function. The lack of association between T1DM and CTLA-4 gene exon 1 49 A/G polymorphism in some populations like Czech[23] also lay emphasis on an alternate pathogenesis. Polymorphisms in the CTLA-4 gene can lower the affinity of CTLA-4 for B7 molecule, cause decreased production resulting in lower surface expression on T-cells and even can alter the peptide sequence.[14] CTLA-4 monoclonal antibody based treatment may be useful to overcome the effect of mutation. However, extended molecular study is needed to establish the role of CTLA-4 gene in autoimmune disease. This may even contribute to deeper understanding of graft rejection in post- transplant patients. CTLA-4 as a potential drug target may seem promising in the treatment of autoimmune disease especially in the population showing CTLA-4 gene polymorphism. Our studies also support the need for routine genetic testing for both diagnosis and classification of autoimmune disease.
Footnotes
Source of Support: Nil
Conflict of Interest: Nil.
References
- 1.Dariavach P, Mattéi MG, Golstein P, Lefranc MP. Human Ig superfamily CTLA-4 gene: Chromosomal localization and identity of protein sequence between murine and human CTLA-4 cytoplasmic domains. Eur J Immunol. 1988;18:1901–5. doi: 10.1002/eji.1830181206. [DOI] [PubMed] [Google Scholar]
- 2.Deichmann K, Heinzmann A, Brüggenolte E, Forster J, Kuehr J. An Mse I RFLP in the human CTLA-4 promoter. Biochem Biophys Res CoMun. 1996;225:817–8. doi: 10.1006/bbrc.1996.1256. [DOI] [PubMed] [Google Scholar]
- 3.Nistico L, Buzzetti R, Pritchard LE, Van der Auwera B, Giovannini C, Bosi E, et al. The CTLA-4 gene region of chromosome 2q33 is linked to, and associated with, type 1 diabetes.Belgian Diabetes Registry. Hum Mol Genet. 1996;5:1075–80. doi: 10.1093/hmg/5.7.1075. [DOI] [PubMed] [Google Scholar]
- 4.Harper K, Balzano C, Rouvier E, Mattéi MG, Luciani MF, Golstein P. CTLA-4 and CD28 activated lymphocyte molecules are closely related in both mouse and human as to sequence, message expression, gene structure, and chromosomal location. J Immunol. 1991;147:1037–44. [PubMed] [Google Scholar]
- 5.Gossain VV. Insulin analogs and intensive insulin therapy in type 1 diabetes. Int J Diabetes Dev Ctries. 2003;23:26–36. [Google Scholar]
- 6.Arikan E, Sabuncu T, Ozu ME, Hatremia H. The characterization of latent autoimmunediabetes in adults and its relation with chronic complication in metabolically controlled Turkish patients with type 2 diabetes mellitus. J Diabetes Complications. 2004;19:254–8. doi: 10.1016/j.jdiacomp.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Ramachandra A, Snehalatha C, Krishnaswami CV. Incidence of IDDM in urban population in Southern India. Madras IDDM registry group South India. Diabetes Res Clin Pract. 1996;34:79–82. doi: 10.1016/s0168-8227(96)01338-1. [DOI] [PubMed] [Google Scholar]
- 8.Radha V, Vimaleswaran KS, Deepa R, Mohan V. The genetics of Diabetes mellitus. Indian J Med Res. 2003;117:225–35. [PubMed] [Google Scholar]
- 9.Mehra NK, Kumar N, Kaur G, Kanga U, Tandon N. Biomarkers of susceptibility to type 1 diabetes with special reference to Indian population. Indian J Med Res. 2007;125:321–44. [PubMed] [Google Scholar]
- 10.SiMonds SJ, Gough SC. Genetic insights into disease mechanism of autoiMunity. Br Med Bull. 2005;71:93–113. doi: 10.1093/bmb/ldh032. [DOI] [PubMed] [Google Scholar]
- 11.Sawka AM, Boulos P, Talib AS, Gafni A, Thabane L, Papaioannou A, et al. Low socioeconomic status and increased risk of severe hypoglycemia in type 1 diabetes: A systematic literature review. Can J Diabetes. 2007;31:233–41. [Google Scholar]
- 12.Brunet JF, Denizot F, Luciani MF. A new member of the iMunoglobulin superfamily-CTLA-4. Nature. 1987;328:267–70. doi: 10.1038/328267a0. [DOI] [PubMed] [Google Scholar]
- 13.Kucharska AM, Gorska E, Wasik M, Pyrzak B, Demkow U. Expression of CD152 (CTLA-4) in children with autoimmune thyroiditis and +49 A/G polymorphism of exon 1 of the CTLA-4 gene. J Physiol Pharmacol. 2009;60(Suppl 5):77–80. [PubMed] [Google Scholar]
- 14.Kristiansen OP, Larsen ZM, Pociot F. CTLA-4 in autoimmunediseases: A general susceptibility gene to autoiMunity? Genes IMun. 2000;1:170–84. doi: 10.1038/sj.gene.6363655. [DOI] [PubMed] [Google Scholar]
- 15.Waterhouse P, Penninger JM, TiMs E, Wakeham A, Shahinian A, Lee KP, et al. Lymphoproliferative disorders with early lethality in mice deficient in Ctla-4. Science. 1995;270:985–8. doi: 10.1126/science.270.5238.985. [DOI] [PubMed] [Google Scholar]
- 16.Chistiakov DA, Savost’anov KV, Nosikov VV. CTLA-4 gene polymorphisms are associated with, and linked to, insulin-dependent diabetes mellitus in a Russian population. BMC Genet. 2001;2:6. doi: 10.1186/1471-2156-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouqbis L, Izaabel H, Akhayat O, Pérez-Lezaun A, Calafell F, Bertranpetit J, et al. Association of the CTLA-4 promoter region (-1661G allele) with type 1 diabetes in the South Moroccan population. Genes IMun. 2003;4:132–7. doi: 10.1038/sj.gene.6363933. [DOI] [PubMed] [Google Scholar]
- 18.Vaidya B, Pearce SH. The emerging role of the CTLA-4 gene in autoimmuneendocrinopathies. Eur J Endocrinol. 2004;150:619–26. doi: 10.1530/eje.0.1500619. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Russel DW. 3rd ed. New York: Cold Spring Harbor Laboratory Press; 2001. Molecular cloning. A Laboratory Manual. [Google Scholar]
- 20.Abe T, Takino H, Yamasaki H, Ozaki M, Sera Y, Kondo H, et al. CTLA-4 gene polymorphism correlates with the mode of onset and presence of ICA512 Ab in Japanese Type 1diabetes. Diabetes Res Clin Pract. 1999;46:169–75. doi: 10.1016/s0168-8227(99)00084-4. [DOI] [PubMed] [Google Scholar]
- 21.Marron MP, Raffel LJ, Garchon HJ, Jacob CO, Serrano-Rios M, Martinez-Larrad MT, et al. Insulin-dependent diabetes mellitus (IDDM) is associated with CTLA-4 polymorphisms in multiple ethnic groups. Hum Mol Genet. 1997;6:1275–82. doi: 10.1093/hmg/6.8.1275. [DOI] [PubMed] [Google Scholar]
- 22.Donner H, Rau H, Walfish PG, Braun J, Siegmund T, Finke R, et al. CTLA-4 Alanine-17confers genetic susceptibility to Graves's disease and to type 1 diabetes mellitus. J Clin Endocrinol Metab. 1997;82:143–6. doi: 10.1210/jcem.82.1.3699. [DOI] [PubMed] [Google Scholar]
- 23.Cinek O, Sumnik DZ, Bendlovat B, Kolouskova S, Snajderova M, Vavrinec J. The CTLA 4 + 49 diamorphism is not accepted with type 1 diabetes in Czech children. Eur J IMunogenet. 2001;29:219–22. doi: 10.1046/j.1365-2370.2002.00292.x. [DOI] [PubMed] [Google Scholar]


