Abstract
The Saccharomyces cerevisiae Sir2 protein is an NAD+-dependent histone deacetylase (HDAC) that functions in transcriptional silencing and longevity. The NAD+ salvage pathway protein, Npt1, regulates Sir2-mediated processes by maintaining a sufficiently high intracellular NAD+ concentration. However, another NAD+ salvage pathway component, Pnc1, modulates silencing independently of the NAD+ concentration. Nicotinamide (NAM) is a by-product of the Sir2 deacetylase reaction and is a natural Sir2 inhibitor. Pnc1 is a nicotinamidase that converts NAM to nicotinic acid. Here we show that recombinant Pnc1 stimulates Sir2 HDAC activity in vitro by preventing the accumulation of NAM produced by Sir2. In vivo, telomeric, rDNA, and HM silencing are differentially sensitive to inhibition by NAM. Furthermore, PNC1 overexpression suppresses the inhibitory effect of exogenously added NAM on silencing, life span, and Hst1-mediated transcriptional repression. Finally, we show that stress suppresses the inhibitory effect of NAM through the induction of PNC1 expression. Pnc1, therefore, positively regulates Sir2-mediated silencing and longevity by preventing the accumulation of intracellular NAM during times of stress.
The Sir2 protein of budding yeast, Saccharomyces cerevisiae, is the founding member of a phylogenetically conserved NAD+-dependent histone-protein deacetylase family called the Sirtuins (29, 36). The number of Sirtuins varies in different organisms. S. cerevisiae contains Sir2 and four other versions called Hst1, Hst2, Hst3, and Hst4 (8, 14), whereas the human genome encodes seven Sirtuins named SIRT1 through SIRT7 (17). Each Sirtuin has a conserved catalytic core domain. Some proteins are little more than the core domain, while others contain larger N- and/or C-terminal extensions. Interestingly, histones are not the only substrates of this deacetylase family. The CobB protein of Salmonella deacetylates the active site of acetyl-coenzyme A synthetase (64), mammalian SIRT1 deacetylates the p53 tumor suppressor protein (37, 41, 70), and human SIRT2 deacetylates tubulin (48). Furthermore, some Sirtuins do not localize to the nucleus, with human SIRT3 being mitochondrial and yeast Hst2 and human SIRT2 being cytoplasmic (49, 50, 56).
Yeast Sir2 is the most widely studied NAD+-dependent histone deacetylase (HDAC) and is absolutely required for transcriptional silencing at the HMR and HML silent-mating type loci, telomeres, and the rDNA locus (for a review, see references 20 and 46). Sir2 localizes primarily to the nucleolus and perinuclear telomeric foci (23). At the HM loci and telomeres, Sir2 is associated in a complex with two other known silencing factors called Sir3 and Sir4 (47, 65). In the nucleolus, Sir2 associates with the rDNA as a component of the RENT complex, which also contains Net1, Cdc14, and Nan1 (58, 66). Each of these complexes contains NAD+-dependent HDAC activity due to the presence of Sir2 (21). RENT also contains NAD+-independent HDAC activity, which makes up the majority of the in vitro activity (21). At HMR, HDAC activity is not required for the loading of the SIR complex to the HMR-E silencer but is essential for silencing to spread from its nucleation site (27, 53). The HDAC activity is also required for continued Sir2 association with telomeres but not for association with the rDNA (3, 27). Silencing in the rDNA locus was recently shown to spread unidirectionally in an RNA polymerase I-dependent manner (10), but whether the HDAC activity of Sir2 is necessary for spreading is currently unclear. Nonetheless, loss of NAD+-dependent HDAC activity results in a phenotypic loss of silencing at all three loci (for reviews, see references 20 and 46).
NAD+ hydrolysis is an integral step of the deacetylation reaction carried out by Sirtuins (35, 69). For every deacetylated lysine residue, one molecule of NAD+ is consumed, and one molecule each of o-acetyl-ADP ribose and nicotinamide (NAM) is produced (35, 69). This NAD+ dependence potentially makes the Sirtuins susceptible to alterations in cellular NAD+ concentration or the NAD+/NADH ratio (26). Growth conditions that elevate the NAD+ concentration could stimulate HDAC activity. Consistent with this idea, mutations in the NAD+ salvage pathway gene NPT1 cause a threefold reduction in the cellular NAD+ concentration, ultimately resulting in telomeric and rDNA silencing defects (54, 62). The NAD+ salvage pathway converts NAM generated by NAD+ hydrolysis or exogenously imported nicotinic acid into nicotinic acid mononucleotide (NaMN), which is an intermediate of the de novo NAD+ synthesis pathway (Fig. 1). Deleting genes in the de novo synthesis pathway has no effect on silencing, but deletion of another NAD+ salvage pathway gene called PNC1 results in partial silencing defects yet does not cause any reduction in NAD+ concentration (54). Regulating NAD+ concentration is, therefore, not the only silencing function of the NAD+ salvage pathway.
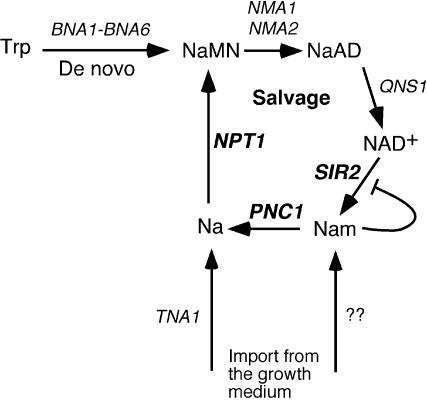
FIG. 1.
Schematic diagram of NAD+ biosynthesis and salvage in yeast cells. The de novo synthesis pathway starts with tryptophan (Trp) and is converted to NaMN by the BNA1 through BNA6 gene products. NaMN is then converted to deamido-NAD (NaAD) by the nicotinic acid/NAM mononucleotide adenylyltransferases NMA1 and NMA2. NaAD is then converted to NAD+ by NAD+ synthetase (QNS1). If NAD+ is hydrolyzed into NAM and o-acetyl-ADP ribose (not shown) by Sir2 or an NAD glycohydrolase, then the NAD+ salvage pathway converts NAM back into NaMN for reentry to the biosynthesis pathway. The PNC1 gene product is a nicotinamidase that converts NAM to nicotinic acid (Na), which is then converted to NaMN by the nicotinic acid phosphoribosyltransferase, Npt1. Nicotinic acid can also be imported to the cell by the Tna1 nicotinic acid permease. It is at present unclear how NAM is imported into the cell.
NAM is a noncompetitive inhibitor of several Sirtuins, including yeast Sir2 (5, 35). Since one of the by-products of the Sir2 HDAC reaction is NAM, large amounts of histone deacetylation during the establishment and/or maintenance of silencing could potentially generate high local concentrations of NAM, resulting in negative feedback (or product) inhibition. The PNC1 gene of the NAD+ salvage pathway codes for a nicotinamidase that is homologous to the bacterial PncA proteins (22). Lack of nicotinamidase activity could, therefore, result in elevated NAM levels in the cell that could in turn inhibit Sir2-mediated silencing. Interestingly, Sir2-mediated longevity is also modulated by NAM and the NAD+ salvage pathway (1, 5). Sir2 is required for the longevity of yeast cells, and overexpression of SIR2 or NPT1 results in extended life span (1, 32). We have now determined that PNC1 regulates silencing and longevity through the deamidation of NAM produced by Sir2 activity. We also demonstrate that the stress-induced expression of PNC1 can suppress the inhibitory effects of NAM on silencing. Importantly, we show that NAM and PNC1 regulate not only the Sir2-dependent functions but also the transcriptional repression activity of Hst1. PNC1 is therefore likely to be a general stress-induced regulator of all Sir2 family members in yeast and, potentially, in other organisms.
MATERIALS AND METHODS
Strains, plasmids, and media.
Yeast media were as previously described (54, 61). Yeast extract-peptone-dextrose (YPD) and synthetic complete (SC) media were supplemented with various concentrations of NAM (Sigma) where indicated. Counterselection against URA3 expression was carried out in SC media containing 1% 5-fluoroorotic acid (5-FOA) (Toronto Research Chemicals). All yeast strains were grown at 30°C, unless otherwise indicated. NPT1 and PNC1 were deleted and replaced with KanMX4 by means of one-step PCR-mediated gene replacement of the entire open reading frame (54). All gene deletions were confirmed by PCR. The genotypes of all strains used in this study are listed in Table 1. Plasmids pJOE31 and pJOE54 were made by PCR amplification of PNC1, including 451 bp upstream and 339 bp downstream of the open reading frame, from genomic DNA. The PCR product was digested with XhoI and ligated into plasmids pRS425 and pRS415, respectively. Plasmid pJOE48 was also made by PCR amplification of the PNC1 open reading frame from genomic DNA. The PCR product was digested with BamHI and ligated into pET-16b (Novagen). Plasmid pCG64 was made by PCR-mediated mutagenesis of plasmid pJOE48 by using a QuickChange site-directed mutagenesis kit (Stratagene). All plasmids are also listed in Table 2.
TABLE 1.
Yeast strains
| Strain | Genotype |
|---|---|
| JJSy143 | MATahis3Δ200 leu2Δ0 ura3-52 trp1Δ63 lys2Δ202 ADH4::URA3-TEL |
| JJSy137 | JJSy143 npt1Δ::kanMX4 |
| JJSy165 | JJSy143 pnc1Δ::kanMX4 |
| DSY35 | JJSy143 pRS425 |
| DSY36 | JJSy143 pSB765 |
| DSY37 | JJSy143 pJOE31 |
| DSY38 | JJsy137 pRS425 |
| DSY39 | JJSy137 pSB765 |
| DSY40 | JJSy137 pJOE31 |
| DSY41 | JJSy165 pRS425 |
| DSY42 | JJSy165 pSB765 |
| DSY43 | JJSy165 pJOE31 |
| JB740 | MATα his3Δ200 leu2Δ1 ura3-167 |
| DSY52 | JB740 pRS425, pJX43 |
| DSY53 | JB740 pRS425, pAV124 |
| DSY54 | JB740 pJOE31, pJX43 |
| DSY55 | JB740 pJOE31, pAV124 |
| JS122a | MATα his3Δ200 leu2Δ1 ura3-167 ???::Ty1-mURA3 |
| JS128a | MATα his3Δ200 leu2Δ1 ura3-167 RDN1(NTS1)::Ty1-mURA3 |
| JS163a | JS128 sir2Δ::HIS3 |
| JS587 | JS128 npt1Δ::kanMX4 |
| JS902, JS903 | JS128 pnc1Δ::kanMX4 |
| DSY98 | JS128 pRS425 |
| DSY99 | JS128 pSB765 |
| DSY100 | JS128 pJOE31 |
| DSY104 | JS902 pRS425 |
| DSY105 | JS902 pSB765 |
| DSY106 | JS902 pJOE31 |
| BY4741b | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 |
| DSY67 | BY4741 pRS415 |
| DSY68 | BY4741 pJOE54 |
| DSY50 | MATα his3Δ200 leu2Δ1 met15Δ0 trp1Δ63 ura3-167 RDN1(NTS1)::mURA3-HIS3 |
| DSY46 | DSY50 pnc1Δ::kanMX4 |
| DSY75 | DSY50 npt1Δ::kanMX4 |
| CGY93c | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hmrΔA::TRP1 pRS425 |
| CGY94c | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 hmrΔA::TRP1 pJOE31 |
| CGY95c | CGY93 sir2::HIS3 |
| CGY96c | CGY94 sir2::HIS3 |
| SY35 | MATahis1 (mating tester strain) |
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Reference |
|---|---|---|
| pRS415 | CEN/ARS LEU2 shuttle vector | 12 |
| pRS425 | 2μm LEU2 shuttle vector | 12 |
| pRS424 | 2μm TRP1 shuttle vector | 12 |
| pSB765 | SIR2 in pRS425 | 10 |
| pJOE31 | PNC1 in pRS425 | |
| pJOE54 | PNC1 in pRS415 | |
| pJSS81 | NPT1 in pRS424 | 54 |
| pDM111 | pGEX-SIR2 | 68 |
| pJOE48 | pET-16b-PNC1 | |
| pCG64 | pET-16b-pnc1 (C167A) | |
| pJX43 | MSE-lacZ reporter plasmid | 73 |
| pAV124 | LacZ reporter plasmid lacking the MSE | 73 |
Silencing assays.
Strains were patched onto SC or SC lacking leucine (SC−Leu) media and allowed to grow for approximately 15 h. Cells were resuspended in water and normalized to an optical density at 600 nm (OD600) of 1.0. All strains were spotted in fivefold serial dilutions. To assay telomeric silencing, strains were spotted onto SC or SC−Leu media to measure growth and on SC containing FOA (SC+FOA) or SC−Leu+FOA media to measure silencing of a URA3 reporter gene adjacent to the left telomere of chromosome VII. To assay rDNA silencing, strains were spotted onto SC or SC−Leu media to measure growth and on SC−Ura or SC−Leu−Ura media to measure the silencing of a mURA3 reporter gene located in NTS1 of the rDNA array. To assay HMR silencing, strains were spotted onto SC or SC−Leu media to measure growth and on SC−Trp or SC−Leu−Trp media to measure silencing of the hmrΔA::TRP1 reporter in the YLS59 strain background (67). NAM was added to the media at various concentrations where indicated. Plates were grown at 30°C for 2 to 3 days.
For the mating assay, strains were grown in SC media with corresponding amounts of NAM (0, 2.5, or 5 mM). Equal numbers of cells (∼1 × 107) from strains DSY50, DSY46, and DSY75 were aliquoted to microcentrifuge tubes along with a twofold excess of the opposite mating type tester strain (∼2 × 107) into the same microcentrifuge tube. Cells were vortexed and incubated at 30°C for 4 h. The volumes were adjusted with water to standardize the concentration of cells in each mating sample and 1:5 serial dilutions were performed. Five microliters of each dilution was then spotted onto SC and synthetic dextrose (SD) media and on SD media supplemented with various concentrations of NAM. Plates were incubated at 30°C for 2 days.
Protein purification.
Glutathione S-transferase (GST)-Sir2 was purified from BL21 (Codon Plus) cells (Stratagene) containing plasmid pDM111 as described previously (60, 68), with a few exceptions. Cells were grown to an OD600 of ∼0.7 at 30°C and then induced with 100 μM isopropylthio-β-d-galactoside. Cultures were then transferred to 25°C and grown overnight. Cell pellets were lysed in phosphate-buffered saline containing 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, 2 μg of leupeptin/ml, and 2 μg of pepstatin A/ml by sonication. Salt was then increased to 350 mM NaCl. Clarified extract was incubated with 0.5 ml 50% glutathione-Sepharose slurry (Amersham Biosciences) for 2 h at 4°C and then poured into a 20-ml Poly-Prep chromatography column (Bio-Rad). The column was washed with 20 volumes of wash buffer (50 mM Tris-Cl [pH 8.0], 350 mM NaCl) and protein was eluted with 10 volumes of elution buffer (50 mM Tris-Cl [pH 8.0], 350 mM NaCl, 20 mM reduced glutathione). Eluted protein was then dialyzed into storage buffer (50 mM Tris-Cl [pH 7.0], 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% Triton X-100) and bovine serum albumin was added to reach a concentration of 1 mg/ml for stability.
His6-Pnc1 and His6-Pnc1 (C167A) were expressed in BL21(DE3) cells (Stratagene). Cells were grown overnight and diluted 1:50 in fresh Luria broth plus ampicillin (100 μg/ml). Cells were then allowed to grow for 1 h at 37°C, induced with 1 mM isopropylthio-β-d-galactoside, and grown for an additional 4 h at 37°C. The cell pellets were washed with water and resuspended in lysis buffer A (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 10 mM imidazole) containing 20 mg of lysozyme. Samples were incubated on ice for 30 min and sonicated. Twenty micrograms of RNase A was added, and samples were incubated on ice for 15 min. Extracts were clarified and incubated with 1 ml of 50% Ni-nitrilotriacetic acid slurry (QIAGEN) for 3 h at 4°C. Samples were poured over a 20-ml Poly-Prep chromatography column and then washed with 10 volumes of buffer B (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 20 mM imidazole). Pnc1 was eluted with 5 volumes of buffer C (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 250 mM imidazole, 20% glycerol). The purity was ∼40 to 50% for both wild-type (WT) and mutant proteins.
In vitro deacetylase assays.
Deacetylase assays were performed with an HDAC Fluorescent Activity Assay-Drug Discover Kit (AK-500; Biomol). This assay involves a proprietary acetylated lysine side chain substrate that becomes sensitized when deacetylated, such that treatment with a Fluor de Lys developer produces a fluorophore. The fluorophore is excited at 360 nm and detected at 460 nm with a fluorescent plate reader. All 50-μl reaction volumes contained ∼1 μg (11.2 pmol) of GST-Sir2, 200 μM NAD+, and 250 μM Fluor de Lys substrate. Approximately 0.5 μg (17.8 pmol) of His6-Pnc1 or His6-Pnc1 (C167A) was included where indicated; the amount of NAM added is indicated for each assay. For the time course experiments, reactions were stopped by the addition of developing reagent at the indicated time points, which efficiently inactivated Sir2. All reactions were carried out at 30°C, and the final readout of activity is in arbitrary fluorescence units at 460 nM.
Northern analysis.
DSY35 and DSY37 cells were grown to an OD600 of ∼0.5 to 0.6 in SC−Leu media. Cultures either were left at 30°C, shifted to 37°C, or had methyl methanesulfonate (MMS) added to reach a concentration of 0.02% and grown for an additional 2 h. Cells were harvested and RNA was extracted by the acid-phenol method (4). Total RNA (20 μg) was separated on a 1.2% agarose-2.22% formaldehyde gel and transferred overnight in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) to Immobilon-Ny+ (Millipore). The membrane was prehybridized in QuickHyb hybridization solution (Stratagene) at 68°C for 20 min. For PNC1 detection, a denatured [α32P]dCTP (3,000 Ci/mmol; Perkin Elmer)-labeled probe corresponding to a KpnI-BsrGI fragment from plasmid pJOE30 was used. The denatured probe and 100 μl of sonicated salmon sperm DNA (10 mg/ml) was hybridized to the membrane for 1 h at 68°C. The membrane was washed two times at room temperature in 2× SSC-0.1% sodium dodecyl sulfate (SDS) and two times at 60°C in 0.1× SSC-0.1% SDS. ACT1 detection was performed as previously described (55).
Replicative life span assay.
Five-milliliter cultures of strains DSY67 and DSY68 were grown overnight at 30°C in SC−Leu and SC−Leu plus 5 mM NAM media. Cultures were diluted to an OD600 of 0.25 and continued with incubation at 30°C for ∼3 h. One hundred microliters of the resulting cultures was diluted into 900 μl of dH2O and spotted on the side of medium plates (YPD and YPD plus 5 mM NAM, respectively) Daughter cells were picked and aligned for virgin daughter isolation. Plates were incubated at 30°C for 3 h, and 40 virgin daughters were staged within the plate grid for life span determination and then continued with 1-h incubations as daughters were separated from mothers, counted, and discarded to the side of the plate. Plates were wrapped with Parafilm and placed at 4°C overnight between harvesting. Plasmid loss was determined by allowing mother cells to grow into colonies, which were then replica plated to SC−Leu plates. Growth on SC−Leu media indicated retention of the plasmid.
Liquid β-galactosidase assay.
Strains DSY52, DSY53, DSY54, and DSY55 were grown in selective media overnight at 30°C. Overnight cultures were diluted into fresh media to an OD600 of ∼0.25 and incubated at 30°C for 5 h. A total of 0.5 ml of each culture was harvested, and the cells were resuspended in 0.8 ml of Z buffer (60 mM NaHPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4) containing β-mercaptoethanol at a concentration of 50 mM. Fifty microliters of chloroform and 0.1% SDS were added to each sample. Samples were then vortexed at maximum speed for 30 seconds. Fresh o-nitrophenylgalactoside solution (a total of 0.2 ml at a concentration of 4 mg/ml dissolved in Z buffer) was added to each sample, and samples were incubated at 30°C for 20 min (until a yellow color appeared). A total of 0.4 ml of 1 M Na2CO3 was added to stop the reaction. Samples were then centrifuged for 5 min at room temperature. Supernatant absorptions were read at OD420. Units of β-galactosidase were calculated as follows: (OD420 × 1000)/(T × V × OD600), where T is the incubation time in minutes and V is the volume (in milliliters) of yeast used in the assay. OD600 and OD420 refer to absorption readings at the beginning and end of the assay, respectively.
RESULTS
The nicotinamidase activity of Pnc1 stimulates the histone deacetylase activity of Sir2 in vitro.
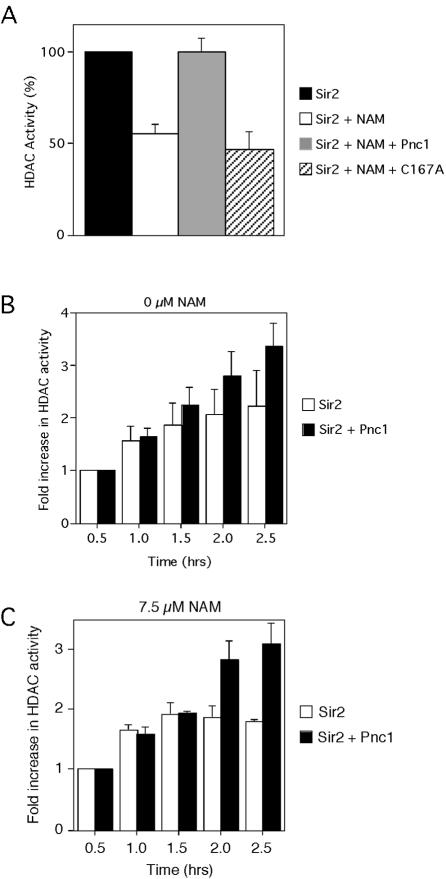
We previously determined that deletion of the PNC1 gene caused a reduction in rDNA and telomeric silencing without reducing the intracellular NAD+ concentration (54). Additionally, extra copies of PNC1 enhanced rDNA and telomeric silencing (1). These results led to the hypothesis that Pnc1 regulates silencing by deamidating the NAM inhibitor that is generated when Sir2 hydrolyzes NAD+ and not by influencing NAD+ levels (Fig. 1). We therefore predicted that the nicotinamidase activity of the recombinant Pnc1 protein would suppress the NAM-inhibition of Sir2 in vitro. To test this hypothesis, recombinant six-His-tagged Pnc1 was added to a Fluor de Lys HDAC reaction (see Materials and Methods) in which Sir2 was N-terminally tagged with GST (Fig. 2A). The recombinant Pnc1 protein was active according to an in vitro nicotinamidase assay (reference 22 and data not shown). As expected from previous studies (5), 50 μM NAM inhibited Sir2 HDAC activity by approximately 50% (Fig. 2A). However, HDAC activity was fully restored when Pnc1 was added to the reaction. Importantly, the Pnc1 protein preparation did not contain any contaminating deacetylases or Sir2 inhibitors (data not shown).
FIG. 2.
Stimulation of Sir2 HDAC activity by Pnc1 in vitro. (A) HDAC assays were carried out with a Biomol fluorescent HDAC kit (see Materials and Methods). Each reaction mixture contained 1 μg (11.2 pmol) of GST-Sir2 and 200 μM NAD+ and was incubated for 30 min at 30°C. Where indicated, 50 μM NAM and 0.5 μg (17.8 pmol) of recombinant His6-Pnc1 or C167A mutant Pnc1 was added to the reaction mixture. Activity for the Sir2-only reaction was set at 100%. (B) Sir2 activity measured over time in the absence of exogenously added NAM. The amount of activity at the 0.5-h time point without Pnc1 added was assigned an arbitrary value of 1.0. For later time points, the average fold increase in HDAC activity compared to the 0.5-h time point is plotted. (C) Sir2 activity measured over time in the presence of 7.5 μM NAM. Data are plotted in the same way as in panel B. Error bars represent the average deviation from three independent experiments.
To determine if the nicotinamidase activity of Pnc1 was important for its effect on Sir2, we mutated a conserved cysteine (C167) to alanine and tested the purified mutant protein for nicotinamidase activity and restoration of Sir2 activity (Fig. 2A). Based on a crystallographic structure of the Pyrococcus horikoshii pyrazinamidase (nicotinamidase), the equivalent conserved cysteine was predicted to directly participate in catalysis as a nucleophile (15), which is consistent with the yeast enzyme being sensitive to sulfhydril reagents (11). The purified C167A mutant Pnc1 was confirmed to be inactive in a nicotinamidase assay (data not shown) and did not rescue the inhibition of HDAC activity by NAM (Fig. 2A). These results suggested that Pnc1 stimulated Sir2 activity through its nicotinamidase activity, not through a direct physical interaction. We then confirmed by a coprecipitation assay that GST-Sir2 and His6-Pnc1 do not interact in vitro (data not shown).
Sir2 generates one molecule of NAM for every lysine residue that is deacetylated (35, 69). We therefore predicted that an in vitro Sir2 HDAC reaction would be self-limiting due to the accumulation of NAM over time. To test this hypothesis, we measured the HDAC activity of GST-Sir2 during a time course, in either the presence or absence of Pnc1 (Fig. 2B). Including Pnc1 in the reaction had no effect on Sir2 activity during early time points (0.5 and 1 h). However, Pnc1 consistently increased the HDAC activity at the later time points, suggesting that over time, NAM was beginning to accumulate and inhibit Sir2 (Fig. 2B). Including Pnc1 in the reaction always improved Sir2 activity at the later time points. The overlapping error bars are due to variations in the degree of stimulation. To confirm this result, we next determined the maximum amount of NAM that could be added to a 30-min Sir2 HDAC reaction mixture without causing any apparent inhibition, which turned out to be 7.5 μM (data not shown) (Fig. 2C, 0.5-h time point). Another time course experiment was then carried out in the presence of 7.5 μM NAM (Fig. 2C). In the absence of Pnc1, Sir2 remained active until 1.5 h, as indicated by a lack of product accumulation at the later time points. In contrast, adding Pnc1 allowed Sir2 to remain active even at the 2.5-h time point. These results indicate that Sir2 can generate enough NAM during an in vitro HDAC assay (>7.5 μM) to inhibit itself and that the nicotinamidase activity of Pnc1 can enhance Sir2 activity by clearing out the NAM.
PNC1 prevents NAM-induced inhibition of telomeric and rDNA silencing in vivo.
It was previously determined that all three forms of silencing can be inhibited in vivo by growing strains in the presence of 5 mM NAM (5). If the in vivo silencing function of Pnc1 is to limit the intracellular NAM concentration, then silencing in a pnc1Δ mutant should be hypersensitive to NAM in the growth medium. To test this hypothesis, the WT strain and npt1Δ and pnc1Δ mutants were first assayed for silencing of a telomeric URA3 reporter gene on medium containing 5-FOA and various concentrations of NAM (Fig. 3A). The pnc1Δ mutant had a minor silencing defect in comparison to silencing in the npt1Δ control when NAM was not added to the growth medium. As expected, 5 mM NAM completely inhibited silencing in each strain, as indicated by the lack of growth on 5-FOA-containing media. At a NAM concentration of only 500 μM, silencing in the WT strain was normal, but silencing in the pnc1Δ mutant was still abolished. Telomeric silencing in the pnc1Δ mutant was therefore ∼10-fold more sensitive than that in the WT strain to NAM. General growth of the WT strain and pnc1Δ mutant on SC plus 5 mM NAM medium was normal, but the npt1Δ mutant had a slow-growth phenotype that was specific to the S288C strain background used for telomere position effect (TPE) assays (data not shown). Since an hst3Δ hst4Δ double mutant has mild telomeric silencing defects (8), part of the effect of NAM on the TPE could potentially be through the inhibition of Hst3 and/or Hst4, not just Sir2.
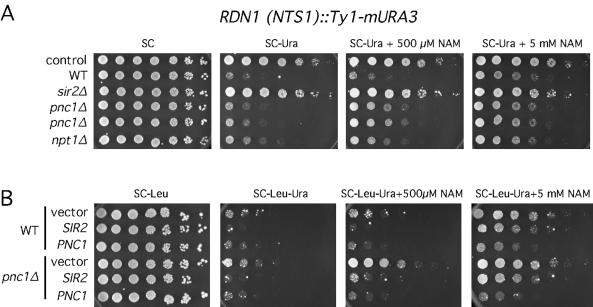
FIG. 3.
Regulation of telomeric silencing by NAM and PNC1. Silencing of a telomeric URA3 reporter gene was tested in WT (JJSy143), npt1Δ (JJSy137), and pnc1Δ (JJSy165) strains. (A) Fivefold serial dilutions of cells were spotted onto SC media or SC media containing 5-FOA. The plates were supplemented with 500 μM or 5 mM NAM where indicated. Two independent colonies for each strain type were tested. (B) WT and pnc1Δ strains were transformed with an empty vector (pRS425), a 2μm SIR2 plasmid (pSB765), or a 2μm PNC1 plasmid (pJOE31). The transformants were plated as fivefold serial dilutions onto SC-Leu or onto SC-Leu+FOA that was supplemented with 500 μM or 5 mM NAM where indicated. Photographs were taken after incubation for 2 days.
We next determined whether overexpression of PNC1 from a high-copy-number plasmid could suppress the telomeric silencing defect caused by 5 mM NAM (Fig. 3B). The PNC1 plasmid fully restored growth to the WT strain on the SC−Leu+FOA plate containing 5 mM NAM, but the empty vector had no effect (Fig. 3B). The high-copy-number PNC1 plasmid also restored silencing to the pnc1Δ mutant in the presence of 500 μM or 5 mM NAM. Suppression of the NAM inhibitory effect was specific to PNC1 overexpression because the high-copy-number SIR2 and NPT1 plasmids did not restore silencing (Fig. 3B and data not shown). Therefore, high levels of Sir2 cannot overcome elevated levels of the NAM inhibitor at telomeres. The high-copy-number PNC1 plasmid was unable to suppress the NAM-induced silencing defect in an npt1Δ mutant background (data not shown), suggesting that the conversion of NAM to nicotinic acid by Pnc1 enhances telomeric silencing only if the NAD+ salvage pathway is intact.
To determine whether Pnc1 also regulates rDNA silencing by metabolizing NAM, we first analyzed the sensitivity of rDNA silencing to various concentrations of NAM (Fig. 4A). In this reporter strain, a Ty1-mURA3 marker was integrated into the nontranscribed spacer 1 (NTS1) region of the rDNA (61), and silencing was measured by the inability to grow on SC−Ura medium. When NAM was not added, the pnc1Δ mutants had minor silencing defects and the npt1Δ mutant had a moderate silencing defect compared to silencing in the sir2Δ mutant (Fig. 4A). Silencing in the pnc1Δ mutants was significantly weakened by 500 μM NAM, whereas the WT strain and npt1Δ mutant were mostly unaffected. This NAM hypersensitivity of the pnc1Δ mutant was similar to that seen with telomeric silencing (Fig. 3A). NAM at a concentration of 5 mM weakened rDNA silencing in all strains but did not cause a complete loss of silencing compared to that of the non-rDNA control strain or the sir2Δ mutant (Fig. 4A). rDNA silencing is therefore more resistant than the TPE to the inhibitory effects of NAM. The inhibitory effect of NAM on rDNA silencing is unlikely to be through HST3 and HST4 because simultaneous deletion of these two genes does not weaken rDNA silencing (J. Smith, unpublished data; I. Celic and J. Boeke, personal communication).
FIG. 4.
Regulation of rDNA silencing by NAM and PNC1. (A) Silencing of a Ty1-mURA3 reporter integrated into the NTS1 sequence of the rDNA was tested in WT (JS128), sir2Δ (JS163), npt1Δ (JS587), and pnc1Δ (JS902 and JS903) strains. The control strain (JS122) contains Ty1-mURA3 integrated at a non-rDNA location that is not subjected to silencing. Fivefold serial dilutions of cells were spotted onto SC of SC−Ura medium. The medium was supplemented with 500 μM or 5 mM NAM where indicated. Photographs were taken after incubation for 2 days. (B) WT and pnc1Δ strains were transformed with an empty vector (pRS425), a 2μm SIR2 plasmid (pSB765), or a 2μm PNC1 plasmid (pJOE31). Serial dilutions were spotted onto SC−Leu or SC−Leu−Ura medium. The medium was supplemented with 500 μM or 5 mM NAM where indicated. Photographs were taken after incubation for 3 days.
It has previously been shown that SIR2 overexpression could suppress the rDNA silencing defect caused by an npt1Δ mutation even though the mutant had about a threefold reduction in NAD+ concentration (54). It was therefore possible that SIR2 overexpression could overcome the inhibitory effects of the pnc1Δ mutation and NAM on rDNA silencing. The high-copy-number SIR2 plasmid fully suppressed the pnc1Δ silencing defect up to a concentration of 1 mM NAM (Fig. 4B) (data not shown). However, the suppression was very weak with 5 mM NAM. As predicted, the high-copy-number PNC1 plasmid was much more efficient at suppressing the silencing defect at 5 mM NAM (Fig. 4B), although as with the TPE, the suppression by the high-copy-number PNC1 plasmid was largely dependent on NPT1 (D. Smith, unpublished data). Elevated SIR2 expression can therefore compensate for the inhibitory effects of low NAM concentrations on rDNA silencing (Fig. 4B) but not telomeric silencing (Fig. 3B).
HMR silencing does not require PNC1.
Since the silent chromatin formed at telomeres and the HM loci are believed to be similar, we predicted that NAM and PNC1 would have similar effects at these two loci. When a semiquantitative mating assay was used as a measurement of HMR silencing, the addition of 5 mM NAM fully inhibited silencing in the WT, pnc1Δ, and npt1Δ strains (Fig. 5A). Growth on the SD plates is an indicator of mating (and silencing). At a concentration of 2.5 mM NAM, silencing was partially inhibited, but silencing in the pnc1Δ mutant was not hypersensitive to the NAM (Fig. 5A). The lack of sensitivity to NAM was not due to a difference in strain background because the strains used in this mating assay were similar to those used in the rDNA silencing experiment in Fig. 4. Similar results were also observed with hmrΔA::TRP1 reporter strains that were in the strain W303 background (data not shown). Regardless of which assay was used, it took higher concentrations of NAM (2.5 mM) to begin seeing HM silencing defects compared to telomeric and rDNA silencing (1 mM).
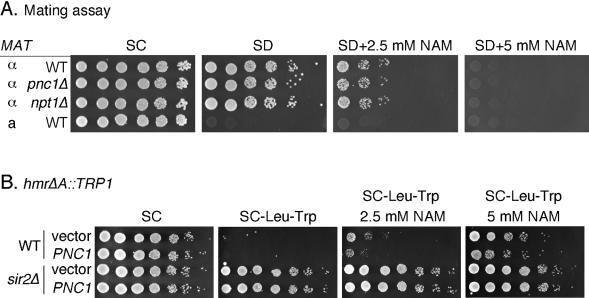
FIG. 5.
Regulation of HM silencing by NAM and PNC1. (A) Mating assay to measure the silencing of the HMR and HML loci in general. WT (DSY50), pnc1Δ (DSY46), and npt1Δ (DSY75) strains were mated to a MATa tester strain (SY35) in the presence of 0, 2.5, or 5 mM NAM for 4 h. Fivefold serial dilutions of cells were spotted. Growth of the resulting diploids on SD plates is indicative of silencing. The MATa tester strain was spotted as a negative control. (B) Silencing at HMR was measured by using WT or sir2Δ versions of a hmrΔA::TRP1 reporter strain that were transformed with either a pRS425 empty vector or the high-copy-number PNC1 vector, pJOE31.
We next tested whether PNC1 overexpression suppressed the inhibitory effect of NAM on HMR silencing by using the hmrΔA::TRP1 reporter strain. In this system, 5 mM NAM caused a loss of silencing and thus growth on plates lacking tryptophan (Fig. 5B). In this assay, 5 mM NAM did not completely eliminate silencing compared to the result for a sir2Δ mutant. Compared to the effect with an empty vector, the high-copy-number PNC1 plasmid suppressed the silencing inhibition by only about fivefold on both 2.5- and 5-mM NAM plates (Fig. 5B). NAM clearance is, therefore, not very important for HM silencing, which may be related to the greater tolerance of HMR silencing to high NAM concentrations.
Pnc1 and the regulation of life span.
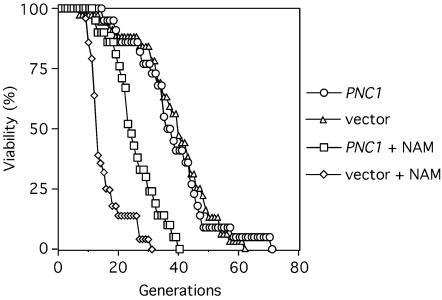
SIR2 and the NPT1 genes of the NAD+ salvage pathway are known to be important for the regulation of S. cerevisiae life span (32), specifically in response to caloric restriction-like growth conditions (39). Extra copies of SIR2 or NPT1 extend replicative life span (1, 32). Furthermore, growing yeast on medium containing 5 mM NAM significantly shortens life span, presumably by inhibiting the HDAC activity of Sir2 (5). Based on the above findings with rDNA silencing (Fig. 4), we tested whether the role of Pnc1 in life span regulation was similar to its role in rDNA silencing, which was to stimulate Sir2 activity by limiting the intracellular concentration of NAM. The replicative life span of strains harboring either an empty CEN/ARS vector or a PNC1 CEN/ARS vector was analyzed (Fig. 6). A CEN/ARS PNC1 vector (pJOE54) was chosen because it should not asymmetrically accumulate in old cells and because it suppresses the telomeric silencing defect caused by 5 mM NAM, although not as well as the 2μm plasmid (data not shown). The 2μm plasmids are not compatible with the replicative aging assays because they artificially shorten life span (16; D. Smith, Jr., and J. Smith, unpublished data). When grown on standard YPD medium, strains containing either the empty vector or PNC1 plasmids each had relatively long average life spans of ∼38 generations (Fig. 6). When the YPD medium was supplemented with 5 mM NAM, the average life span of the empty vector strain dropped to ∼14 generations (a 63% reduction). Average life span of the strain containing the PNC1 plasmid was partially elevated to ∼24 generations on NAM-containing plates (Fig. 6). There are two major reasons that only a partial restoration in life span was observed. First is the lower PNC1 expression level from a CEN plasmid compared to that for a 2μm plasmid. Second, the CEN plasmids are not completely stable. At the start of the aging assay, the PNC1 and empty CEN vectors were maintained in 78 and 76% of the virgin cells, respectively. By the 10th generation of bud removal 47% of the mothers had maintained the empty vector, and 62% of the mothers still maintained the PNC1 vector. While the effect we observed is diminished by plasmid loss, the PNC1 plasmid was still maintained well enough to extend life span through the clearance of NAM.
FIG. 6.
NAM and PNC1 regulate longevity. The replicative life span of strains containing either an empty CEN/ARS pRS415 vector (DSY67) or a CEN/ARS PNC1 pJOE54 plasmid (DSY68) was tested on rich YPD growth medium. Where indicated, 5 mM NAM was added. The data are plotted as the percentage of mother cells still viable (y axis) after each successive generation (x axis).
A role for Pnc1 in silencing regulation during stress response.
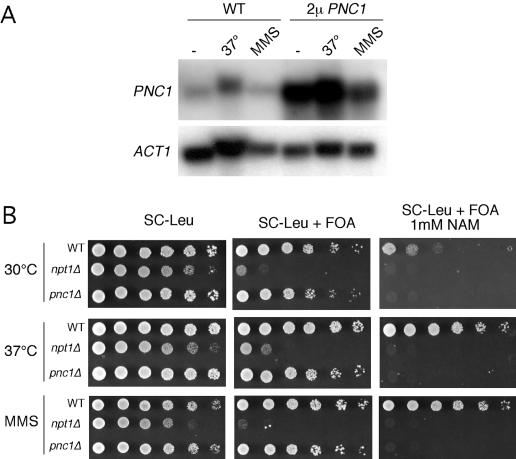
The PNC1 gene is upregulated in response to a variety of cellular stresses, including heat shock, hyperosmotic shock, and treatment with the DNA-damaging agent MMS (18, 19, 22). We were therefore interested in determining whether the amount of endogenous PNC1 expressed during heat shock or MMS treatment was enough to suppress the telomeric silencing defect caused by NAM in the growth medium. A CEN/ARS PNC1 vector suppressed the silencing defect caused by 5 mM NAM in a PNC1+ strain but not in a pnc1Δ strain (data not shown), suggesting that relatively small elevations in PNC1 expression could potentially have an effect. A Northern blot for PNC1 mRNA confirmed that PNC1 is indeed upregulated during heat shock at 37°C (Fig. 7A). However, the PNC1 expression level in a WT strain grown at 37°C was not as high as when PNC1 was present on a 2μm vector. When the ACT1 loading control is taken into account, PNC1 RNA levels were slightly elevated when WT cells were treated with 0.02% MMS (Fig. 7A).
FIG. 7.
Stress-mediated silencing regulation by PNC1. (A) Northern blot analysis of steady-state PNC1 RNA levels from cells grown normally (at 30°C), at an elevated temperature (37°C), or in the presence of 0.02% MMS. The strains contained either an empty vector (pRS425) or the 2μm PNC1 plasmid (pJOE31). The ACT1 gene was used as a loading control. (B) Telomeric silencing spot assay for silencing of a URA3 reporter. WT (JJSy143), npt1Δ (JJSy137), and pnc1Δ (JJSy165) strains were spotted onto SC−Leu medium as a growth control, and SC−Leu+FOA medium to measure silencing. To partially inhibit silencing, 1 mM NAM was added. The strains were grown either at 30 or 37°C or in the presence of 0.02% MMS.
We decided to test both conditions for suppression of the telomeric silencing defect caused by NAM in the growth medium. Neither heat shock nor MMS treatment was able to suppress 5 mM NAM (data not shown). This result was not surprising since PNC1 expression under these conditions was not as strong as with a 2μm PNC1 vector under the same conditions (Fig. 7A). When cultures were grown on 1 mM NAM at the normal 30°C temperature, silencing was completely eliminated from the npt1Δ and pnc1Δ mutants, but weak silencing was maintained in the WT strain (Fig. 7B). However, when cultures were grown at 37°C or on MMS plates, silencing was fully restored to the WT strain in the presence of 1 mM NAM (indicated by increased growth on 5-FOA-containing media). There was no increase in silencing for the pnc1Δ or npt1Δ mutant. To confirm that the stress effect was due to PNC1, we examined the effect of heat shock on a pnc1Δ mutant in the presence of 400 μM NAM. At 30°C the mutant had a partial silencing defect, but growth at 37°C did not enhance silencing (data not shown). We conclude that Sir2 activity is stimulated during times of stress due to the elevated expression of PNC1.
Hst1 is regulated by NAM and PNC1.
NAM has been shown to inhibit the deacetylase activity of several Sir2 family members, including yeast Sir2 and Hst2 (5, 35) and the human versions SIRT1, SIRT2, and SIRT3 (41, 48, 56). NAM is clearly a nonspecific inhibitor of most, if not all, class III HDACs. We were therefore interested in determining whether PNC1 would influence biological processes controlled by other Sir2 family members. Other than Sir2, the best-studied homolog in yeast is Hst1, which, along with Sum1 and Rfm1, functions in the repression of a subclass of middle-sporulation genes in which a middle-sporulation element (MSE) acts as a strong repressor site during vegetative growth (44, 73). Deletion of HST1 causes partial derepression of this class of genes due to a lack of Hst1-mediated deacetylation at the promoters (73). We examined the inhibition of Hst1 activity by using a plasmid-based reporter in which lacZ is repressed by an MSE site in the promoter (73). As expected, the MSE site on plasmid pJX43 strongly repressed lacZ expression (Table 3). Adding 5 mM NAM to the growth medium resulted in partial derepression of the lacZ gene controlled by the MSE site (Table 3, empty vector strains). Adding the high-copy-number PNC1 plasmid almost completely restored normal repression to the NAM-treated strain (Table 3). Finally, the inhibitory effect of NAM on the MSE-containing reporter was dependent on HST1 (data not shown). From these results, we conclude that NAM inhibits multiple Sirtuin-mediated processes in vivo and that Pnc1 functions to counteract these effects.
TABLE 3.
PNC1 and NAM regulate HST1 activity
| Growth conditionsa
|
Activity of promoter-lacZ reporterb
|
Fold repressionc
|
||
|---|---|---|---|---|
| Plasmid | 5 mM NAM | −MSE | +MSE | |
| Empty | − | 25.89 (2.96) | 0.29 (0.15) | 89.3 |
| PNC1 | − | 26.86 (4.82) | 0.22 (0.09) | 122.1 |
| Empty | + | 16.02 (2.03) | 2.49 (0.15) | 6.4 |
| PNC1 | + | 27.36 (5.47) | 0.43 (0.22) | 63.6 |
Strain JB740 contained either the pRS425 empty vector or the pJOE31 2μm PNC1 plasmid. Each strain was grown with (+) or without (−) 5 mM NAM.
β-galactosidase units measured from whole-cell extracts from strains containing either pAV124 (−MSE) or pJX43 (+MSE). The reported units are averaged from three independent experiments. The average deviation calculated by Microsoft Excel is provided in parentheses.
Calculated as the β-GAL activity of pAV124 strains (−MSE) divided by the β-GAL activity of the pJX43 (+MSE) strains.
DISCUSSION
Pnc1 as a central regulator of the Sir2 deacetylase family.
Sir2 is the founding member of the class III family of HDACs (8, 24). Its unusual property of coupling NAD+ hydrolysis to protein deacetylation has several important biological implications. First, in the cases of Sir2 and Hst1, the catalytic requirement for NAD+ potentially links cellular metabolism to the regulation of posttranslational histone modifications. Growth conditions that increase the intracellular NAD+ concentration or elevate the NAD+/NADH ratio could stimulate Sir2 deacetylase activity, thus enhancing silencing and extending life span (26). While there have been no clear experimental examples of yeast Sir2 being stimulated by elevated NAD+ concentration in vivo, mutations in the NAD+ salvage gene, NPT1, reduce NAD+ concentration by about threefold and cause silencing defects (54).
Another biological implication of NAD+ hydrolysis by Sir2 is that the NAM by-product is a noncompetitive inhibitor of Sir2. Excessive local accumulation of NAM has the potential to inhibit Sir2 function. Indeed, several reports have determined that Sir2 family members can be inhibited in vivo by growing cells in the presence of 5 mM NAM (41, 48, 56) or in vitro with lower concentrations (5, 35). In the present study we have determined that the conversion of NAM to nicotinic acid by the Pnc1 nicotinamidase regulates the activities of Sir2 and Hst1. Telomeric silencing is normally completely inhibited by growth in the presence of 5 mM NAM. However, telomeric silencing in a pnc1Δ mutant was fully inhibited by only 500 μM NAM (Fig. 3A). Furthermore, overexpression of PNC1 completely suppressed the inhibitory action of 5 mM NAM (Fig. 3B). While the in vivo concentration of NAM in yeast cells has not been determined, the concentration in human cerebrospinal fluid was recently measured as 54.2 μM (63), which is consistent with NAM inhibiting Sir2 with a 50% inhibitory concentration of ∼50 μM (5). Increasing the expression level of PNC1 is therefore an efficient way to stimulate Sir2 activity (see below).
PNC1 links the stress response to Sir2 activity.
The PNC1 gene is induced by a wide variety of cellular stresses, including heat shock, hyperosmotic shock, H2O2 exposure, and DNA damage (18, 19, 22). Our data indicate that one of the important functions of Pnc1 during stress is to stimulate Sir2 activity through the deamidation of NAM. During times of high Sir2 activity within the cell, there ironically is a great deal of NAM produced, having the potential to inhibit Sir2 if it overaccumulates. Therefore, inducing PNC1 expression coincident with the need for Sir2 activity helps facilitate the biological processes in which Sir2 or the other Sir2 family members are involved. For Sir2, the processes are silencing and longevity. For Hst1, the process is the repression of middle sporulation genes. Importantly, Sir2 appears to be involved in processes related to the stresses that induce PNC1 expression. For example, MMS induces double-strand DNA breaks, and the Sir proteins have been implicated in double-strand break repair (38, 42, 43, 45). Similarly, a transient heat shock or high osmolarity have been shown to extend yeast cell life span (31, 57), and Sir2 is required for longevity (32).
The link between stress and Sir2 goes beyond yeast cells. SIRT1 is the closest human homolog of yeast Sir2, and it has been shown to deacetylate the p53 tumor suppressor protein (37, 41, 70). Acetylation stimulates the transcriptional activation activity of p53 (25), which can lead to cell cycle arrest, senescence, or apoptosis. SIRT1 expression suppresses apoptosis induced by DNA-damaging agents or oxidative stress, presumably through the deacetylation of p53 by SIRT1 (37, 41, 70). These data led to the hypothesis that SIRT1 promotes cell survival during stress (41), which is consistent with the extension of life span in yeast caused by certain types of stress (31, 57). Like Sir2 and Npt1 (8, 52, 54), Pnc1 is also phylogenetically conserved, with homologs identified in Eubacteria, Archaea, Drosophila, and Caenorhabditis elegans (22). Nicotinamidase activity is also found in mammalian, including human, cells (51, 72). It is therefore possible that Pnc1 homologs in more complex organisms regulate the corresponding Sirtuins of each species.
Elevated PNC1 expression from a CEN/ARS plasmid can suppress the short life span and loss of rDNA silencing caused by 5 mM NAM (Figs. 4 and 6), indicating that PNC1 regulates longevity and silencing through the stimulation of Sir2 activity. Life span in multiple organisms can be extended by limiting calorie consumption in a process called caloric restriction (for a review, see reference 34). For yeast cells, growth on reduced glucose medium (0.1 or 0.2% glucose) extends life span (30, 39) and strengthens rDNA silencing (40). The NPT1 gene is required for this extension in life span (39). The Sinclair laboratory has independently demonstrated that PNC1 is involved in longevity regulation (2). Interestingly, researchers found that PNC1 protein levels were increased by caloric restriction growth conditions and that PNC1, like NPT1 (40), was required for an extended life span induced by caloric restriction (2). We found that NPT1 was required for the suppression of NAM-induced telomeric and rDNA silencing defects (Fig. 4 and 7) (data not shown). It is therefore apparent that an intact NAD+ salvage pathway is required for caloric restriction-induced life span extension. In other words, NAM clearance by Pnc1 will not have a large impact on silencing or longevity if NAD+ levels are critically low. Taken together, the results from the present study and from the Sinclair laboratory (2) strongly implicate PNC1 as a key link between stress, Sir2, silencing, and longevity.
One surprising aspect of the NAM results with longevity is that 5 mM NAM has a very dramatic effect on life span but only partially inhibited rDNA silencing. This finding raises the possibility that the strength of rDNA silencing does not always correlate with longevity. Longevity may be more tightly linked to the suppression of rDNA recombination than to the silencing of RNA polymerase II reporter genes. In fact, it is already known that rDNA silencing and suppression of recombination can be separated. For example, sgs1Δ mutants have elevated rDNA recombination levels and a short life span (59), but rDNA silencing is not significantly altered (9). Furthermore, fob1Δ mutants have reduced rDNA recombination levels and a long life span (13) but are defective for rDNA silencing (28). Longevity appears to fully require Sir2 deacetylase activity, but Sir2 may have other functions in terms of rDNA silencing.
PNC1 differentially regulates rDNA, telomeric, and HM silencing.
Telomeric silencing was completely inhibited by 5 mM NAM in the growth medium (Fig. 3A), but rDNA and HMR silencing were only partially inhibited compared to results for the sir2Δ mutants (Figs. 4A and 5B). These data are consistent with fluorescence microscopy images from the Sinclair laboratory showing that 5 mM NAM delocalizes Sir2 from telomeres but that a significant amount of the nucleolar Sir2 remains intact (5). Similarly, chromatin immunoprecipitation experiments have demonstrated that a catalytically inactive H364Y Sir2 mutant, among others, does not associate with telomeres but does associate with rDNA chromatin and the HMR-E silencer (3, 27, 53). Furthermore, Sir3 and Sir4 can associate with the HMR-E silencer in the absence of Sir2 activity (53) but not with telomeres (27, 53). The assembly of silent chromatin is therefore more highly dependent on Sir2 activity at telomeres than at the HM loci or rDNA.
In addition to its role as an HDAC, Sir2 could have additional structural functions at HM loci and the rDNA that are not so important at telomeres. Support for this idea at the rDNA comes from the finding that Sir2 overexpression can suppress the rDNA silencing defect of a npt1Δ mutant, even though this mutant has 2.5-fold less intracellular NAD+ (54), a condition that is presumably poor for Sir2 activity. Furthermore, SIR2 overexpression suppressed the inhibitory effect of NAM on rDNA silencing up to a concentration of at least 1 mM (Fig. 4B). It is unclear if Sir2 has any structural role at the HM loci, although the RENT complex does have the capacity to function in HMR silencing (33).
While rDNA silencing was less sensitive than telomeres to the inhibitory effect of NAM, it was still very responsive to deletion and overexpression of PNC1 (Fig. 4). HM silencing, however, was not sensitive to deletion of PNC1, and the silencing inhibition caused by 5 mM NAM was only weakly suppressed by the high-copy-number PNC1 plasmid, which was very surprising. It is possible that the relative ineffectiveness of PNC1 on HM silencing is related to the stability of HM silent chromatin. HM chromatin rarely switches from the silent to expressed state, and this stability could make the HM silent chromatin less susceptible to changes in Sir2 activity induced by fluctuations in NAM concentration. Alternatively, since it takes at least 2.5 mM NAM to observe any inhibition of HMR silencing, compared to 400 μM in telomeric silencing, the in vivo changes in NAM concentration induced by PNC1 overexpression or deletion may not disrupt Sir2 activity enough so that significant changes in HMR silencing can be observed. Clearly NAM and PNC1 regulate Sir2 activity in vivo, but they differentially regulate each type of silent chromatin.
NAM and PNC1 as global class III HDAC regulators.
It is now clear that members of the Sir2 family of protein deacetylases hydrolyze NAD+ as an integral step of the deacetylation reaction (35, 69). In addition, all Sir2-like enzymes tested to date are inhibited by the NAM by-product. It has been suggested that the o-acetyl-ADP ribose by-product is a potential signaling molecule or substrate for another type of reaction (6). Since all class III HDACs are likely to be inhibited by NAM, it is possible that they all are regulated by the activity of a nicotinamidase (Pnc1 in yeast). Pnc1 homologs have been identified in multiple species, including several metazoans (22). However, it remains to be determined if the Pnc1 homologs in other eukaryotic species are stress induced.
Multiple laboratories are starting to use NAM as an in vivo inhibitor of class III HDACs. In most cases, NAM is used to inhibit one particular homolog. However, most eukaryotes have multiple class III HDACs. For example, S. cerevisiae has five homologs and humans have seven (8, 17). Unless it is known that other Sir2-like proteins are not involved in a specific function being tested by the NAM inhibition, then it is difficult to rule out the possibility that other Sir2-like proteins are contributing. In addition, our data indicate that some Sir2-dependent activities are less sensitive to NAM than others. Another potential complication from using NAM as an in vivo inhibitor of Sir2 proteins is that poly(ADP-ribose) polymerase, which has many cellular functions, is also inhibited by NAM (71). The bottom line is that caution should be used in applying NAM as an in vivo Sirtuin inhibitor.
Two different activities of the NAD+ salvage pathway are critical Sir2 regulators. The function of Npt1 appears to be maintenance of a sufficiently high intracellular NAD+ concentration, whereas the function of Pnc1 is to minimize the concentration of intracellular NAM. Loss of either of these activities results in a cellular environment that is not optimal for Sir2 function. Therefore, modulation of Npt1 and Pnc1 expression levels in response to stress or other signals provides an excellent means for the cell to control the activity of not only Sir2 but also the other Sir2 family members.
Acknowledgments
We thank Joe Sandmeier for technical assistance, Andrew Vershon and Danesh Moazed for providing plasmids, and Marty Mayo for access to the fluorescent plate reader. We also thank Zu-Wen Sun and David Auble for critically reading the manuscript and members of the Smith laboratory for helpful discussions.
This work was supported by grants GM61692 and AG022685 to J.S.S. from the NIH. D.S. and C.G. were both supported in part by NIH training grant GM08136.
REFERENCES
- 1.Anderson, R. M., K. J. Bitterman, J. G. Wood, O. Medvedik, H. Cohen, S. S. Lin, J. K. Manchester, J. I. Gordon, and D. A. Sinclair. 2002. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem. 277:18881-18890. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R. M., K. J. Bitterman, J. G. Wood, O. Medvedik, and D. A. Sinclair. 2003. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423:181-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong, C. M., M. Kaeberlein, S. I. Imai, and L. Guarente. 2002. Mutations in Saccharomyces cerevisiae gene SIR2 can have differential effects on in vivo silencing phenotypes and in vitro histone deacetylation activity. Mol. Biol. Cell. 13:1427-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 2000. Current protocols in molecular biology, vol. 2. John Wiley & Sons, Inc., New York, N.Y.
- 5.Bitterman, K. J., R. M. Anderson, H. Y. Cohen, M. Latorre-Esteves, and D. A. Sinclair. 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277:45099-45107. [DOI] [PubMed] [Google Scholar]
- 6.Borra, M. T., F. J. O'Neill, M. D. Jackson, B. Marshall, E. Verdin, K. R. Foltz, and J. M. Denu. 2002. Conserved enzymatic production and biological effect of O-acetyl-ADP-ribose by silent information regulator 2-like NAD+-dependent deacetylases. J. Biol. Chem. 277:12632-12641. [DOI] [PubMed] [Google Scholar]
- 7.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 8.Brachmann, C. B., J. M. Sherman, S. E. Devine, E. E. Cameron, L. Pillus, and J. D. Boeke. 1995. The SIR2 gene family, conserved from bacteria to humans, functions in silencing, cell cycle progression, and chromosome stability. Genes Dev. 9:2888-2902. [DOI] [PubMed] [Google Scholar]
- 9.Bryk, M., M. Banerjee, D. Conte, Jr., and M. J. Curcio. 2001. The Sgs1 helicase of Saccharomyces cerevisiae inhibits retrotransposition of Ty1 multimeric arrays. Mol. Cell. Biol. 21:5374-5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buck, S. W., J. J. Sandmeier, and J. S. Smith. 2002. RNA polymerase I propagates unidirectional spreading of rDNA silent chromatin. Cell 111:1003-1014. [DOI] [PubMed] [Google Scholar]
- 11.Calbreath, D. F., and J. G. Joshi. 1971. Inhibition of nicotinamidase by nicotinamide adenine dinucleotide. J. Biol. Chem. 246:4334-4339. [PubMed] [Google Scholar]
- 12.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 13.Defossez, P. A., R. Prusty, M. Kaeberlein, S. J. Lin, P. Ferrigno, P. A. Silver, R. L. Keil, and L. Guarente. 1999. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell 3:447-455. [DOI] [PubMed] [Google Scholar]
- 14.Derbyshire, M. K., K. G. Weinstock, and J. N. Strathern. 1996. HST1, a new member of the SIR2 family of genes. Yeast 12:631-640. [DOI] [PubMed] [Google Scholar]
- 15.Du, X., W. Wang, R. Kim, H. Yakota, H. Nguyen, and S. H. Kim. 2001. Crystal structure and mechanism of catalysis of a pyrazinamidase from Pyrococcus horikoshii. Biochemistry 40:14166-14172. [DOI] [PubMed] [Google Scholar]
- 16.Falcon, A. A., and J. P. Aris. 2003. Plasmid accumulation reduces life span in Saccharomyces cerevisiae. J. Biol. Chem. 278:41607-41617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frye, R. A. 2000. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 273:793-798. [DOI] [PubMed] [Google Scholar]
- 18.Gasch, A. P., M. Huang, S. Metzner, D. Botstein, S. J. Elledge, and P. O. Brown. 2001. Genomic expression responses to DNA-damaging agents and the regulatory role of the yeast ATR homolog Mec1p. Mol. Biol. Cell. 12:2987-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasser, S. M., and M. M. Cockell. 2001. The molecular biology of the SIR proteins. Gene 279:1-16. [DOI] [PubMed] [Google Scholar]
- 21.Ghidelli, S., D. Donze, N. Dhillon, and R. T. Kamakaka. 2001. Sir2p exists in two nucleosome-binding complexes with distinct deacetylase activities. EMBO J. 20:4522-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghislain, M., E. Talla, and J. M. Francois. 2002. Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast 19:215-224. [DOI] [PubMed] [Google Scholar]
- 23.Gotta, M., S. Strahl-Bolsinger, H. Renauld, T. Laroche, B. K. Kennedy, M. Grunstein, and S. M. Gasser. 1997. Localization of Sir2p: the nucleolus as a compartment for silent information regulators. EMBO J. 16:3243-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray, S. G., and T. J. Ekstrom. 2001. The human histone deacetylase family. Exp. Cell Res. 262:75-83. [DOI] [PubMed] [Google Scholar]
- 25.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 26.Guarente, L. 2000. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 14:1021-1026. [PubMed] [Google Scholar]
- 27.Hoppe, G. J., J. C. Tanny, A. D. Rudner, S. A. Gerber, S. Danaie, S. P. Gygi, and D. Moazed. 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22:4167-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, J., and D. Moazed. 2003. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 17:2162-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 30.Jiang, J. C., E. Jaruga, M. V. Repnevskaya, and S. M. Jazwinski. 2000. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J. 14:2135-2137. [DOI] [PubMed] [Google Scholar]
- 31.Kaeberlein, M., A. A. Andalis, G. R. Fink, and L. Guarente. 2002. High osmolarity extends life span in Saccharomyces cerevisiae by a mechanism related to calorie restriction. Mol. Cell. Biol. 22:8056-8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaeberlein, M., M. McVey, and L. Guarente. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13:2570-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kasulke, D., S. Seitz, and A. E. Ehrenhofer-Murray. 2002. A role for the Saccharomyces cerevisiae RENT complex protein Net1 in HMR silencing. Genetics 161:1411-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koubova, J., and L. Guarente. 2003. How does calorie restriction work? Genes Dev. 17:313-321. [DOI] [PubMed] [Google Scholar]
- 35.Landry, J., J. T. Slama, and R. Sternglanz. 2000. Role of NAD+ in the deacetylase activity of the SIR2-like proteins. Biochem. Biophys. Res. Commun. 278:685-690. [DOI] [PubMed] [Google Scholar]
- 36.Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins, L. Pillus, R. Sternglanz, S. Imai, C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Proc. Natl. Acad. Sci. USA 97:5807-5811.10811920 [Google Scholar]
- 37.Langley, E., M. Pearson, M. Faretta, U. M. Bauer, R. A. Frye, S. Minucci, P. G. Pelicci, and T. Kouzarides. 2002. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 21:2383-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, S. E., F. Paques, J. Sylvan, and J. E. Haber. 1999. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol. 9:767-770. [DOI] [PubMed] [Google Scholar]
- 39.Lin, S.-J., P.-A. Defossez, and L. Guarente. 2000. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289:2126-2128. [DOI] [PubMed] [Google Scholar]
- 40.Lin, S. J., M. Kaeberlein, A. A. Andalis, L. A. Sturtz, P. A. Defossez, V. C. Culotta, G. R. Fink, and L. Guarente. 2002. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 418:344-348. [DOI] [PubMed] [Google Scholar]
- 41.Luo, J., A. Y. Nikolaev, S. Imai, D. Chen, F. Su, A. Shiloh, L. Guarente, and W. Gu. 2001. Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107:137-148. [DOI] [PubMed] [Google Scholar]
- 42.Martin, S. G., T. Laroche, N. Suka, M. Grunstein, and S. M. Gasser. 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97:621-633. [DOI] [PubMed] [Google Scholar]
- 43.McAinsh, A. D., S. Scott-Drew, J. A. Murray, and S. P. Jackson. 1999. DNA damage triggers disruption of telomeric silencing and Mec1p-dependent relocation of Sir3p. Curr. Biol. 9:963-966. [DOI] [PubMed] [Google Scholar]
- 44.McCord, R., M. Pierce, J. Xie, S. Wonkatal, C. Mickel, and A. K. Vershon. 2003. Rfm1, a novel tethering factor required to recruit the Hst1 histone deacetylase for repression of middle sporulation genes. Mol. Cell. Biol. 23:2009-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mills, K. D., D. A. Sinclair, and L. Guarente. 1999. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell 97:609-620. [DOI] [PubMed] [Google Scholar]
- 46.Moazed, D. 2001. Common themes in mechanisms of gene silencing. Mol. Cell 8:489-498. [DOI] [PubMed] [Google Scholar]
- 47.Moazed, D., A. Kistler, A. Axelrod, J. Rine, and A. D. Johnson. 1997. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc. Natl. Acad. Sci. USA 94:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.North, B. J., B. L. Marshall, M. T. Borra, J. M. Denu, and E. Verdin. 2003. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11:437-444. [DOI] [PubMed] [Google Scholar]
- 49.Onyango, P., I. Celic, J. M. McCaffery, J. D. Boeke, and A. P. Feinberg. 2002. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc. Natl. Acad. Sci. USA 99:13653-13658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perrod, S., M. M. Cockell, T. Laroche, H. Renauld, A. L. Ducrest, C. Bonnard, and S. M. Gasser. 2001. A cytosolic NAD-dependent deacetylase, Hst2p, can modulate nucleolar and telomeric silencing in yeast. EMBO J. 20:197-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petrack, B., P. Greengard, A. Craston, and F. Sheppy. 1965. Nicotinamide deamidase from mammalian liver. J. Biol. Chem. 240:1725-1730. [PubMed] [Google Scholar]
- 52.Rajavel, M., D. Lalo, J. W. Gross, and C. Grubmeyer. 1998. Conversion of a cosubstrate to an inhibitor: phosphorylation mutants of nicotinic acid phosphoribosyltransferase. Biochemistry 37:4181-4188. [DOI] [PubMed] [Google Scholar]
- 53.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell. 13:2207-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sandmeier, J. J., I. Celic, J. D. Boeke, and J. S. Smith. 2002. Telomeric and rDNA silencing in Saccharomyces cerevisiae are dependent on a nuclear NAD+ salvage pathway. Genetics 160:877-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sandmeier, J. J., S. French, Y. Osheim, W. L. Cheung, C. M. Gallo, A. L. Beyer, and J. S. Smith. 2002. RPD3 is required for the inactivation of yeast ribosomal DNA genes in stationary phase. EMBO J. 21:4959-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwer, B., B. J. North, R. A. Frye, M. Ott, and E. Verdin. 2002. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 158:647-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shama, S., C. Y. Lai, J. M. Antoniazzi, J. C. Jiang, and S. M. Jazwinski. 1998. Heat stress-induced life span extension in yeast. Exp. Cell Res. 245:379-388. [DOI] [PubMed] [Google Scholar]
- 58.Shou, W., J. H. Seol, A. Shevchenko, C. Baskerville, D. Moazed, Z. W. Chen, J. Jang, H. Charbonneau, and R. J. Deshaies. 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell 97:233-244. [DOI] [PubMed] [Google Scholar]
- 59.Sinclair, D. A., K. Mills, and L. Guarente. 1997. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science 277:1313-1316. [DOI] [PubMed] [Google Scholar]
- 60.Smith, J. S., J. Avalos, I. Celic, S. Muhammad, C. Wolberger, and J. D. Boeke. 2002. The SIR2 family of NAD+-dependent protein deacetylases. Methods Enzymol. 353:282-300. [DOI] [PubMed] [Google Scholar]
- 61.Smith, J. S., and J. D. Boeke. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11:241-254. [DOI] [PubMed] [Google Scholar]
- 62.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, and J. D. Boeke. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smythe, G. A., O. Braga, B. J. Brew, R. S. Grant, G. J. Guillemin, S. J. Kerr, and D. W. Walker. 2002. Concurrent quantification of quinolinic, picolinic, and nicotinic acids using electron-capture negative-ion gas chromatography-mass spectrometry. Anal. Biochem. 301:21-26. [DOI] [PubMed] [Google Scholar]
- 64.Starai, V. J., I. Celic, R. N. Cole, J. D. Boeke, and J. C. Escalante-Semerena. 2002. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science 298:2390-2392. [DOI] [PubMed] [Google Scholar]
- 65.Strahl-Bolsinger, S., A. Hecht, K. Luo, and M. Grunstein. 1997. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 11:83-93. [DOI] [PubMed] [Google Scholar]
- 66.Straight, A. F., W. Shou, G. J. Dowd, C. W. Turck, R. J. Deshaies, A. D. Johnson, and D. Moazed. 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97:245-256. [DOI] [PubMed] [Google Scholar]
- 67.Sussel, L., and D. Shore. 1991. Separation of transcriptional activation and silencing functions of the RAP1-encoded repressor/activator protein 1: isolation of viable mutants affecting both silencing and telomere length. Proc. Natl. Acad. Sci. USA 88:7749-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tanny, J. C., G. J. Dowd, J. Huang, H. Hilz, and D. Moazed. 1999. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99:735-745. [DOI] [PubMed] [Google Scholar]
- 69.Tanny, J. C., and D. Moazed. 2001. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc. Natl. Acad. Sci. USA 98:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vazirii, H., S. K. Dessain, E. N. Eaton, S. I. Imai, R. A. Frye, T. K. Pandita, L. Guarente, and R. A. Weinberg. 2001. hSIR2SIRT1 functions as an NAD-dependent p53 deacetylase. Cell 107:149-159. [DOI] [PubMed] [Google Scholar]
- 71.Virag, L., and C. Szabo. 2002. The therapeutic potential of poly(ADP-ribose) polymerase inhibitors. Pharmacol. Rev. 54:375-429. [DOI] [PubMed] [Google Scholar]
- 72.Wintzerith, M., A. Dierich, and P. Mandel. 1979. Nicotinamide deamidase released by neuroblastoma cell cultures. J. Neurochem. 33:677-685. [DOI] [PubMed] [Google Scholar]
- 73.Xie, J., M. Pierce, V. Gailus-Durner, M. Wagner, E. Winter, and A. K. Vershon. 1999. Sum1 and Hst1 repress middle sporulation-specific gene expression during mitosis in Saccharomyces cerevisiae. EMBO J. 18:6448-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]