Abstract
The tumor suppressor function of p53 is linked to its ability to repress gene expression, but the mechanisms of specific gene repression are poorly understood. We report that wild-type p53 inhibits an effector of the Ras oncogene/mitogen-activated protein (MAP) kinase pathway, the transcription factor Net. Tumor-associated mutant p53s are less efficient inhibitors. p53 inhibits by preventing phosphorylation of Net by MAP kinases. Loss of p53 in vivo leads to increased Net phosphorylation in response to wound healing and UV irradiation of skin. Our results show that p53 can repress specific gene expression by inhibiting Net, a factor implicated in cell cycle entry.
The tumor suppressor p53 regulates apoptosis, cell cycle arrest, and angiogenesis in response to genotoxic and other stresses. Loss of p53 function is required for malignant progression, and mutations in p53 occur in more than half of all human cancers (42, 43). p53 activates expression of genes involved in apoptosis (e.g., Bax, NOXA, and p53AIP1) and inhibition of cell cycle progression (e.g., p21WAF1/CIP1, 14-3-3 sigma, and GADD45). p53 also represses transcription, and repression is required for both apoptosis and tumor suppression (22, 36). Despite the importance of repression by p53, little is known about the mechanisms, especially about how p53 mediates the repression of specific target genes.
Cell growth is regulated by a balance of factors that promote or repress cell cycle progression. Growth factors and oncogenes such as Ras induce mitogen-activated protein (MAP) kinase signaling cascades that transduce extracellular signals from ligand-activated cell surface receptors to the nucleus. MAP kinases phosphorylate nuclear effectors, such as the three ternary complex factors Net, Elk-1, and Sap-1, that regulate immediate-early genes and cell cycle entry (38, 41). Net (Elk-3/ERP/Sap-2) is a repressor of transcription that is converted to a positive regulator by MAP kinase phosphorylation of critical residues of the C-terminal (C) domain (11, 18, 28). The A domain, located at the N terminus, mediates DNA binding, and the B domain interacts with the serum response factor to help form ternary complexes on serum response elements of immediate-early response genes. Net mutant mice develop a vascular phenotype and have altered expression of egr-1, a factor implicated in the cell cycle and vascular biology (2). This study shows that p53 inhibits Net activity and the expression of one of its target genes, egr-1. It provides a mechanism by which p53 can inhibit expression of genes required for cell cycle progression.
MATERIALS AND METHODS
Plasmids. (i) Expression vectors.
Glutathione S-transferase (GST) fusion proteins were expressed from plasmids derived from pBC-Net (28). Plasmids pBC-N6, pBC-N6δ5, pBC-N6δ6, and pBC-N6δ7 were constructed by cloning PCR fragments that encode Net sequences from positions 327 to 409, 327 to 375, 347 to 375, and 327 to 353, respectively, in the NdeI-KpnI sites of pBC (9). All PCR inserts were verified by sequencing. For Gal4 fusion proteins, pGal4-N5 and pGal4-N6 (28) were used, as well as Elk1-C (a gift from R. Treisman [29]). pTL2-Net (28) was used to express full-length Net.
Oncogenic Ha-Ras was expressed from pHa-Ras (44), constitutively active ERK2 was expressed from pERK2-MEK1-LA (a gift from M. Cobb [35]), and constitutively active MEK1 (S218D S222D) was expressed from pECE-MAPKK(S218D S222D) (a gift from J. Pouysségur [6]). p53 and common human cancer mutants with the V143A, R178H, R248Q, R273H, or R281W mutation were expressed from pC53-SN3 and derivatives (a gift from A. Levine [20]). p53 mutants 1-100, 1-313, 1-394, 94-313, 94-394, and 290-394, with an N-terminal Flag-tag, were expressed from pSG5-Puro-Flag (Institut de Génétique et de Biologie Moléculaire et Cellulaire [IGBMC] core facility) derivatives with the corresponding PCR fragments in the BamHI site. All PCR inserts were verified by sequencing.
β-Galactosidase was expressed from CMV-lacZ (52).
(ii) Reporters.
For Net, Palx8-Luc (18, 28) was used as a reporter. For Gal4, we used upstream activation sequence (UAS)-Luc, generously provided by A. Bradford and A. Gutierrez-Hartman, which contains five Gal4-binding sites in pGL2-Luc.
Antibodies.
The following antibodies were used (with sources and references given in parentheses): the anti-Net antibody 375, the anti-Gal4 antibody 2GV3, anti-GST (28), the anti-phospho-Net antibody 2F3, specific for Net phosphorylated on serine 365 (14), anti-ERK1/2 (catalog no. 9102; Cell Signaling), anti-phospho-ERK1/2 (catalog no. 9106; Cell Signaling), anti-phospho-Elk-1 (catalog no. 9181; Cell Signaling), the anti-human p53 antibody DO-1 (39), the anti-mouse p53 monoclonal antibody 240 (17), anti-Ras (sc-520; Santa Cruz), the anti-TATA-binding protein (anti-TBP) antibody 3G3 (5), and a mouse anti-Flag monoclonal antibody (epitope DYKDDDDK; IGBMC Monoclonal Common Service).
Luciferase and β-galactosidase assays.
PALx8-Luc (1.5 μg) and CMV-LacZ (250 ng) were cotransfected by the N,N-bis(2-hydroxyethyl)-2-aminoethyl sulfonic acid-buffered saline calcium phosphate technique in CHO cells in six-well plates (Corning) with 100 ng of the expression vector for Net, ERK2-MEK1, Ha-Ras, or active MEK1, and 30 or 100 ng of the expression vector for p53. For the Gal4 fusion proteins, UAS-Luc and the Gal4 expression vectors replaced PALx8-Luc and the Net vector pTL2-Net, respectively. Cells were washed 16 h after transfection and were cultured for 10 h in a medium containing 0.05% fetal calf serum (FCS). Cells were lysed in lysis buffer (25 mM Tris-phosphate [pH 7.8], 2 mM EDTA, 1 mM dithiothreitol, 10% glycerol, and 1% Triton X-100). Luciferase activity was normalized by β-galactosidase activity, and fold inductions were calculated relative to transfections lacking activators of Net. Values shown are averages from two typical experiments.
GST pulldown assay.
SAOS2 cells in 10-cm-diameter dishes were transfected with 5 μg of pC53-SN3 and 15 μg of GST-fused Net expression vectors (pBC-Net, pBC-N6, pBC-N6δ5, pBC-N6δ6, and pBC-N6δ7) by the BBS-based calcium phosphate technique. Cells were washed 16 h after transfection, and after a further 24 h they were lysed in 1 ml of NETN-200 buffer (0.2 M NaCl, 20 mM Tris-HCl [pH 7.5], 1 mM EDTA, 0.5% NP-40, 5 mM dithiothreitol, 0.4 mM phenylmethylsulfonyl fluoride). Cell lysates were cleared by centrifugation (for 10 min at 10,000 × g) and incubated with glutathione-Sepharose 4B for 2 h with gentle rocking. Beads were washed four times with NETN-100 buffer (similar to NETN-200 except that the NaCl concentration is 0.1 M). The input and precipitated proteins were analyzed by immunoblotting with the anti-p53 antibody DO-1 or with anti-GST.
Net phosphorylation in cell culture.
Mouse embryonic fibroblasts (MEFs) and human umbilical vein endothelial cells (HUVEC) were cultured overnight with Dulbecco's modified Eagle medium (DMEM) plus 0.05% FCS in the absence or presence of 10 μM mitomycin C. For serum activation, FCS was added to a concentration of 10% for 10 min. For UV induction, the cells were irradiated with 40 J of UV-B/m2 and cultured for 1 h with DMEM plus 0.05% FCS. Cells were quickly washed with phosphate-buffered saline (PBS), lysed in sodium dodecyl sulfate (SDS) sample buffer, and analyzed by immunoblotting.
Quantitative real-time RT-PCR.
HUVEC and wild type and p53−/− MEFs were cultured overnight in DMEM plus 0.05% FCS in the absence or presence of 10 μM mitomycin C (where appropriate), activated with DMEM plus 10% FCS for 30 or 45 min, and washed with PBS, and RNA was prepared with RNAsolv (Omega Biotech). cDNA was synthesized by reverse transcription (RT) from 20 ng (for c-fos or egr-1) or 100 pg (for 28S RNA) of RNA for 30 min in the presence of 1× Master Mix (1× Sigma PCR buffer, 4 mM MgCl2, 5% glycerol, 0.15 mg of bovine serum albumin/ml, and 0.2 mM deoxynucleoside triphosphates), Superscript reverse transcriptase (Invitrogen), and the appropriate antisense primer at 0.5 μM (see below). Ten percent of the RT reaction mixture was used for PCR in a LightCycler (Roche) under the following conditions: 92°C for 1 min, followed by 50 cycles of 92°C for 5 s, 62°C for 15 s, and 72°C for 15 s in the presence of 1× Master Mix, 1× SyberGreen (Roche), 0.5 μM primers, and Taq polymerase. Standard curves for the internal control (28S RNA) and the test mRNAs were generated with four different amounts of the appropriate RT reaction mixtures. The relative levels of expression of c-fos and egr-1 were determined by use of the standard curves and corrected for variations in the 28S RNA internal control. The following primers were used: for human c-fos, ACZ139 (5′-TTTCCTGGCAATAGTGTGTTC-3′) and ACZ140 (5′-TTCAGACCACCTCAACAATG-3′); for human egr-1, ADC221 (5′-CACGGGCGAGCAGCCCTACG-3′) and ADC222 (5′-TCCACCAGCACCTTCTCGTT-3′); for mouse egr-1, ACG46 (5′-GCCGAGCGAACAACCCTA-3′) and ACG47 (5′-TCCACCATCGCCTTCTCATT-3′); for mouse c-fos, ACG44 (5′-AAGGGAACGGAATAAGATGGC-3′) and ACG45 (5′-CAACGCAGACTTCTCATCTTCAA-3′); and for 28S RNA, ACD229 (5′-GGCGGCCAAGCGTTCATAGC-3′) and ACD230 (5′-ATTTGGTGTATGTGCTTGGC-3′).
Immunoprecipitation.
HUVEC were cultured overnight in DMEM plus 0.05% FCS in the presence or absence of 10 μM mitomycin C, activated with DMEM plus 10% FCS for 10 min, washed with PBS, and lysed with NETN-100 buffer. To precipitate the Net-p53 complex, the cell extracts were incubated with the anti-Net antibody 375 for 2 h, followed by protein G-Sepharose (Pharmacia) for 1 h. Beads were washed three times with NETN-100 and analyzed by immunoblotting with the anti-p53 antibody DO-1.
Wound healing.
Protocols were modified from the work of Ortega et al. (33). After 8-mm-diameter, full-thickness dorsal skin wounds were generated (on 4- to 12-week-old mice; 11 pairs in total), wound surfaces were measured and photographed every 2 days, with or without removal of the clot. After the wounds had healed totally, the same mice were wounded elsewhere (at a distance of >1 cm from the first wound), and these new wounds were used for histological analysis 9 days after wounding. For histology and immunostaining (4, 21, 52), 8-mm-thick sections were rehydrated, unmasked in 0.01 M citrate for 60 min at 94°C, cooled for 60 min, incubated with primary antibodies (the anti-phospho-Net antibody 2F3, used at 1:1,000 in PBS-Tween) for 180 min at 25°C, followed by overnight incubation at 4°C, and stained with the VECTASTAIN Elite ABC kit (Vector Laboratories, Inc.). The same conditions were used for sections from p53−/− mice as for sections from wild-type mice.
UV irradiation.
Transgenic and wild-type mice, matched with respect to age, gender, and body site, were UV irradiated (5,000 J/m2), and skin samples were processed for immunohistochemistry (IHC) (16). Samples were incubated successively with the anti-phospho-Net antibody 2F3 (used at 1:500 overnight at 4°C), a biotin-conjugated secondary antibody, streptavidin peroxidase (1:200), a Dab liquid substrate kit (5 min; Zymed), and hematoxylin. For immunoblotting, skin samples were homogenized in radioimmunoprecipitation assay buffer with an Ultrathorax. Homogenates were analyzed by SDS-polyacrylamide gel electrophoresis and immunoblotting with anti-p53 (NCL-p53-CM5), anti-phospho-Net (2F3), and anti-TBP (3G3) overnight at 4°C, followed by peroxidase-conjugated mouse anti-rabbit immunoglobulin G and chemiluminescence (Pierce).
RESULTS
p53 inhibits phosphorylation of Net.
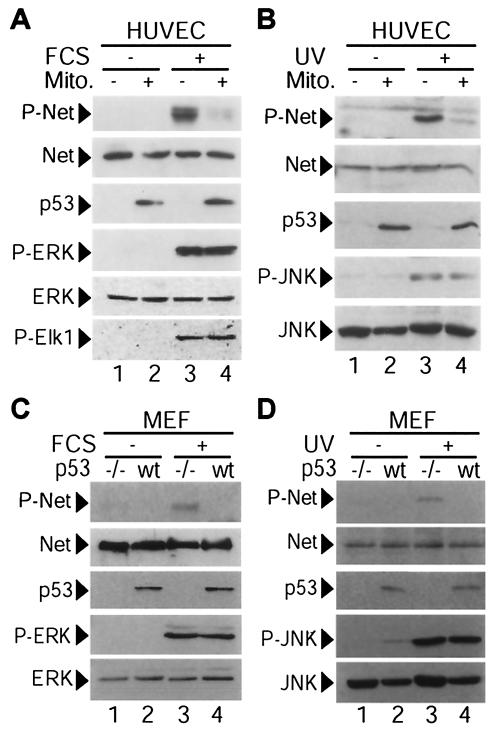
We initially studied whether p53 affects Net phosphorylation by serum and by UV irradiation. We used HUVEC because they are normal: they express relatively high levels of Net and very low levels of p53. p53 is highly induced by mitomycin C in these cells (37), and mitomycin C has been used previously to study the effects of p53 induction on transcription factor activity (37, 45). HUVEC were serum starved and treated with 10 μM mitomycin C for 12 h. There was no detectable apoptosis under these conditions, as shown by the morphology of 4′,6′-diamidino-2-phenylindole (DAPI)-stained cells, cleavage of poly(ADP-ribose) polymerase, and DNA laddering (37) (data not shown). The cells were treated with serum for 10 min to induce the extracellular signal-regulated kinase (ERK) pathway, and protein phosphorylation and expression were followed by Western blotting (Fig. 1A). Serum induced Net phosphorylation on the critical serine 365 (Fig. 1A, lane 3, P-Net), as shown with the phosphospecific antibody 2F3 (13). U0126 blocked Net phosphorylation, demonstrating that the ERK pathway is required for serum-induced Net phosphorylation (data not shown). p53 induction with mitomycin C inhibited Net phosphorylation induced by serum (Fig. 1A, lane 4, P-Net). Net phosphorylation was not detected in the absence of serum, showing that mitomycin C does not induce Net phosphorylation (Fig. 1A, lanes 1 and 2; see also Fig. 1B). Net protein levels remained constant under all conditions, whereas p53 accumulated to similar levels with mitomycin C in the presence and absence of serum. p53 accumulated in the nucleus, as shown by immunocytochemistry with antibody DO-1 (data not shown). ERK and the ERK substrate Elk-1 were phosphorylated with serum to similar levels in the presence and absence of mitomycin C, showing that p53 does not affect either the activation of ERK by upstream kinases or its activity on another substrate. These results show that p53 specifically inhibits serum-induced Net phosphorylation.
FIG. 1.
Inhibition of serum- or UV-induced Net phosphorylation by endogenous p53 in HUVEC and MEFs. (A) p53 inhibits Net phosphorylation (P-Net) by serum in HUVEC. HUVEC were incubated for 12 h with DMEM plus 0.05% FCS in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of 10 μM mitomycin C (Mito.), which induces p53. Cells were then treated with 10% serum for 10 min. (B) p53 inhibits Net phosphorylation by UV irradiation in HUVEC. Serum-starved and mitomycin C-treated (lanes 2 and 4) or untreated (lanes 1 and 3) HUVEC were either irradiated with UV-B (lanes 3 and 4) or mock irradiated (lanes 1 and 2) and then incubated for 1 h. (C) p53 inhibits Net phosphorylation by serum in MEFs. MEFs prepared from p53−/− (−/−) (lanes 1 and 3) or wild-type (wt) (lanes 2 and 4) mouse embryos were serum starved overnight and stimulated with 10% serum for 7 min (lanes 3 and 4). (D) p53 inhibits Net phosphorylation by UV irradiation in MEFs. Serum-starved p53−/− (lanes 1 and 3) or wild-type (lanes 2 and 4) MEFs were either irradiated with UV-B (lanes 3 and 4) or mock treated (lane 1 and 2) and were then incubated with DMEM plus 0.05% FCS. Protein extracts were analyzed by immunoblotting with the anti-phospho-Net antibody 2F3 (P-Net), the anti-Net antibody 375, the anti-p53 antibody DO-1, anti-phospho-ERK (P-ERK), anti-ERK, anti-phospho-Elk-1 (P-Elk1), anti-phospho-JNK (P-JNK), or anti-JNK.
We tested whether mitomycin C induction of p53 also inhibits Net phosphorylation by c-Jun NH2-terminal kinase (JNK) that has been activated by UV irradiation. HUVEC were treated with 10 μM mitomycin C for 12 h and then UV irradiated, and after 1 h, cell extracts were prepared and analyzed by immunoblotting (Fig. 1B). Mitomycin C treatment inhibited UV-induced Net phosphorylation (Fig. 1B, lanes 3 and 4, P-Net), without affecting Net protein levels (Fig. 1B, lanes 3 and 4, Net). Mitomycin C-induced p53 levels (Fig. 1B, lanes 1 and 2, p53) were not further increased by UV irradiation (lanes 3 and 4). Phosphorylation of JNK was induced by UV irradiation (Fig. 1B, lanes 1 to 4, P-JNK), and the levels were similar in the presence and absence of mitomycin C (lanes 3 and 4). Total JNK levels did not vary significantly (Fig. 1B, lanes 1 to 4, JNK).
Mitomycin C is a DNA-damaging agent that has other effects on the cell besides p53 induction. To more specifically show that p53 is required for the inhibition of Net phosphorylation, we used p53−/− MEFs. Serum (Fig. 1C) and UV irradiation (Fig. 1D) induced higher levels of phospho-Net in MEFs lacking p53 (lanes 3 and 4, P-Net), whereas levels of P-ERK and P-JNK were not affected by the loss of p53 (lanes 3 and 4). Amounts of p53 did not increase significantly 1 h after UV treatment (Fig. 1D, lanes 2 and 4). These results show that p53 inhibits Net phosphorylation by the ERK and JNK pathways without affecting the activation of ERK and JNK.
p53 inhibits Net transcriptional activity.
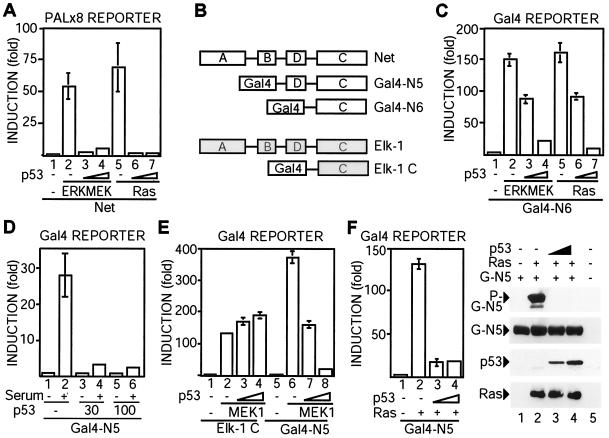
In order to analyze the effect of p53 on Net transcriptional activity, we used CHO cells, in which Net activity is highly inducible by the ERK pathway (11). CHO cells were transfected with a luciferase reporter gene containing tandem ets motifs and vectors that express Net, p53, Ha-Ras, and ERK2-MEK1-LA (a constitutively active form of ERK2 [35]). p53 expression inhibited Net activation by Ras or ERK2-MEK1-LA (Fig. 2A; compare bars 6 and 7 with bars 5 and 1, and compare bars 3 and 4 with bars 2 and 1, respectively). To delimit the sequences of Net that mediate p53 inhibition, we used fusion proteins containing the heterologous DNA binding domain of Gal4 (Fig. 2B) and a corresponding UAS-luciferase reporter. Fusion proteins that contain the C domain of Net (Gal4-N6) or the C and D domains (Gal4-N5) were efficiently stimulated by activators of the ERK pathway (ERK2-MEK1-LA, Ras, serum, and MEK1) (Fig. 2C, bars 1, 2, and 5; Fig. 2D, bars 1 and 2; Fig. 2E, bars 5 and 6; Fig. 2F, bars 1 and 2). p53 expression inhibited Net activation by all of these inducers (Fig. 2C, bars 3, 4, 6, and 7; Fig. 2D, bars 3 to 6; Fig. 2E, bars 7 and 8; Fig. 2F, bars 3 and 4) and also inhibited phosphorylation of Gal4-N5 (Fig. 2F, P-G-N5) without altering its expression level (Fig. 2F, G-N5) or that of Ras. p53 expression also inhibited Net activation in SAOS2 cells, indicating that p53 has similar effects in the two cell lines (data not shown). In contrast, p53 did not inhibit MEK1 activation of an equivalent Elk-1 fusion protein containing its C domain fused to Gal4 (Fig. 2E, bars 1 to 4). These results show that p53 specifically inhibits Net activation by the ERK pathway.
FIG. 2.
p53 inhibits Net-dependent transcription activation through the C-terminal region. (A) p53 inhibits Net-dependent transcription activation. pTL2-Net, PALx8-Luc, and CMV-lacZ were cotransfected in CHO cells with expression vectors for ERK2-MEKl-LA (bars 2, 3, and 4) or Ha-Ras (bars 5, 6, and 7) and increasing amounts of p53 (bars 3, 4, 6, and 7). The cells were washed 16 h after transfection and then cultured for 10 h in a medium containing 0.05% FCS. Cell lysates were assayed for luciferase and β-galactosidase (for standardization). (B) Schematic diagrams. Net and Elk-1 have similar DNA binding domains (rectangle A), SRF binding domains (rectangle B), transactivation domains (rectangle C), and p42/44 binding domains (rectangle D). Gal4-N5 and Gal4-N6 contain the Gal4 DNA binding domain (amino acids 1 to 147) fused to amino acids 219 to 409 or 327 to 409 of mouse Net, respectively. Elk-1 C contains the Gal4 DNA binding domain fused to amino acids 355 to 429 of human Elk-1. (C) The C-terminal domain of Net is sufficient for inhibition by p53. pGal4-N6, UAS-Luc, and CMV-lacZ were cotransfected in CHO cells, and luciferase and β-galactosidase activities were measured. (D) p53 inhibits serum induction of Net activity. By using CHO cells, pGal4-N5, UAS-Luc, and CMV-lacZ were cotransfected with increasing amounts of expression vectors for p53 (bars 3, 4, 5, and 6). Cells were washed with medium 16 h after the addition of the DNA mixture and then cultured for 10 h with a medium containing 0.05% FCS. Gal4-N5 was activated by further incubation with medium plus 10% FCS for 10 h. Extracts were assayed for luciferase and β-galactosidase activities. (E) Elk-1 activation is not inhibited by p53. By using CHO cells, pElk1-C or pGal4-N5 was cotransfected with UAS-Luc, CMV-lacZ, and expression vectors for active-MEK1 (bars 2, 3, 4, 6, 7, and 8) and p53 (bars 3, 4, 7, and 8), and luciferase and β-galactosidase activities were measured. (F) p53 inhibits Net phosphorylation. (Left) CHO cells were transfected with Ras (bars 2, 3, and 4) and p53 (bars 3 and 4) expression vectors, pGal4-N5, UAS-Luc, and CMV-lacZ, and luciferase and β-galactosidase activities were measured. (Right) Cell lysates were immunoblotted with the anti-phospho-Net antibody 2F3 (P-G-N5), the anti-Gal antibody 2GV3 (G-N5), the anti-p53 antibody DO-1, or an anti-Ras antibody.
p53 physically interacts with Net.
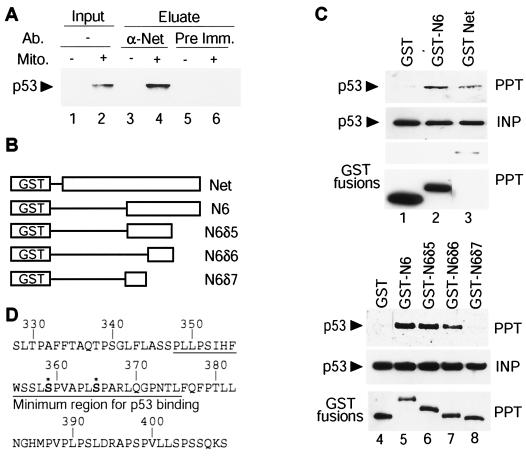
We tested by coimmunoprecipitation whether p53 interacts with Net. HUVEC were treated with mitomycin C to induce p53, and cell lysates were immunoprecipitated with the anti-Net antibody 375, followed by immunoblotting with an anti-p53 antibody (DO-1) to detect p53 in the precipitates (Fig. 3A). The Net antibody coprecipitated p53 only from mitomycin C-treated cell extracts (Fig. 3A, lanes 3 and 4), whereas the preimmune serum control did not contain p53 (lanes 5 and 6). In order to identify the domain of Net that binds to p53, we used GST-Net fusion proteins (Fig. 3B). Expression vectors for GST-Net and p53 were transfected into SAOS2 cells, which lack endogenous p53. Extracts were incubated with glutathione-Sepharose beads, and proteins on the beads were analyzed by immunoblotting with an anti-p53 antibody (DO-1). p53 was detected on the beads containing full-length Net fused to GST (Fig. 3C, lanes 1 and 3). These results show that Net interacts with p53 in SAOS2 cells as well as in HUVEC. p53 was also found to interact with GST-N6, GST-N6δ5, and GST-N6δ6 (Fig. 3C, lanes 2 and 5 to 7) but not with GST alone or with GST-N6δ7 (lanes 1, 4, and 8). These results show that the minimum sequence of Net that binds to p53, amino acids 347 to 375, contains the two major phosphorylation sites (Fig. 3D).
FIG. 3.
Net forms a complex with p53. (A) Immunoprecipitation of a Net-p53 complex. HUVEC were serum starved overnight in the presence (lanes 2, 4, and 6) or absence (lanes 1, 3, and 5) of 10 μM mitomycin C (Mito.) and were then incubated with 10% FCS for 10 min. Cell extracts were incubated with the anti-Net antibody 375 (α-Net) (lanes 3 and 4) or preimmune rabbit serum (Pre Imm.) (lanes 5 and 6) for 1 h, followed by precipitation with protein G-Sepharose. Proteins on the beads were analyzed by immunoblotting with an anti-p53 antibody (DO-1). (B) Schematic diagram of GST-fused Net deletion mutants. GST-N6, GST-N6δ5, GST-N6δ6, and GST-N6δ7 have Net sequences from 327 to 409, 327 to 375, 347 to 375, or 327 to 353, respectively. (C) GST-Net-p53 complex formation in SAOS2 cells. SAOS2 cells were transfected with expression vectors for p53 and GST, GST-Net, GST-N6, GST-N6δ5, GST-N6δ6, or GST-N6δ7. Cell lysates were incubated with glutathione-Sepharose beads, and the input (INP) and precipitated (PPT) proteins were analyzed by immunoblotting with anti-p53 (DO-1) or anti-GST. (D) Amino acid sequence of the C-terminal region of Net. Sequences that bind to p53 are underlined. Asterisks indicate the two major phosphorylation sites.
Mutant forms of p53 are less effective inhibitors of Net transcriptional activity.
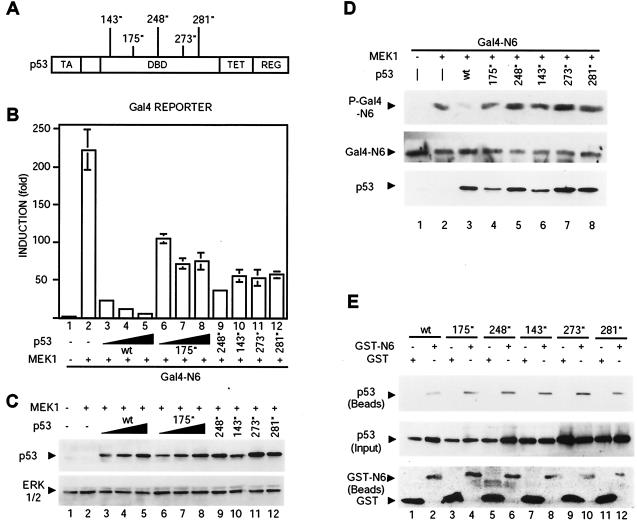
We analyzed the effects of mutant p53s commonly found in human cancers (V143A, R175H, R248Q, R273H, and R281W) (Fig. 4A) on Net transcriptional activation. We cotransfected expression vectors for p53, Gal4-Net N6, MEK1, and the Gal4 reporter gene. The mutants were all less effective than wild-type p53 at inhibiting Net activity (Fig. 4B; compare bars 6 to 12 with bars 3 to 5). They were expressed at similar levels (Fig. 4C). The mutant p53s did not inhibit Net phosphorylation, as shown by immunoblotting of cell lysates with an anti-phospho-Net antibody (Fig. 4D; compare phosphorylated Gal4-N6 with total Gal4-N6). Wild-type and mutant p53 were expressed at similar levels (Fig. 4C and D, bottom panel, lanes 3 to 6). We also studied whether the mutant p53s interact with Net. Expression vectors for GST fused to the C domain of Net (GST-N6) and the p53 mutants were transfected in SAOS2 cells, cell extracts were incubated with glutathione-Sepharose beads, and the proteins on the beads were analyzed by immunoblotting (Fig. 4E). All the p53 mutants were found to bind to Net [Fig. 4E; compare p53 (Beads) with the controls p53 (Input) and GST-N6 and GST (Beads)]. These results indicate that complex formation is not sufficient for the p53 mutants to inhibit Net phosphorylation.
FIG. 4.
Reduced inhibition of Net transactivation by mutant p53s. (A) p53 mutants. The p53 mutations (indicated by asterisks), which are commonly found in human tumors, are V143A, R175H, R248Q, R273H, and R281W. TA, transactivation domain; DBD, DNA binding domain; TET, tetramerization domain; REG, regulatory domain. (B) Effects of mutant p53s on Net transactivation. By using CHO cells, Gal4-N6 was transfected with the Gal4 reporter, CMV-lacZ, and expression vectors for active MEK1 (bars 2 to 12) and for wild-type (wt) or mutant p53 (bars 3 to 12). (C) p53 expression levels in the transfected cells for which results are shown in panel B were determined by immunoblotting with DO-1. ERK1/2 is a loading control. (D) Phosphorylation of Net in the presence of mutant p53s. Gal4-N6, active MEK1 (lanes 2 to 8), and wild-type or mutant p53s (lanes 3 to 8) were expressed in CHO cells. After a wash, cells were incubated for 10 h in MEM alpha 1900 plus 0.05% FCS. Cell lysates were analyzed by immunoblotting with the anti-phospho-Net antibody 2F3 (P-Gal4-N6), the anti-Net antibody 375 (Gal4-N6), and the anti-p53 antibody DO-1. (E) Mutant p53s bind to Net. SAOS2 cells were transfected with expression vectors for wild-type or mutant p53s and GST or GST-N6. A GST pulldown assay was performed as for Fig. 3C. p53 (Beads), p53 retained on the glutathione-Sepharose beads; GST-N6 and GST (Beads), GST proteins on the beads.
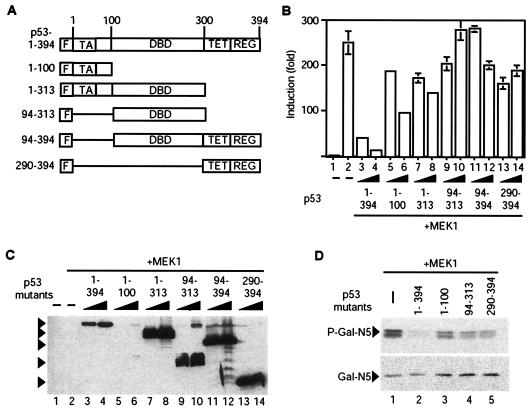
In order to investigate which sequences of p53 are required for inhibition of Net activation and phosphorylation, we prepared N-terminally Flag-tagged deletion mutants of p53 (Fig. 5A) that contain either the transactivation domain (amino acids 1 to 100), the DNA-binding domain (amino acids 94 to 313), the C-terminal domain (including the tetramerization and regulatory domains, amino acids 290 to 394), or combinations of adjacent domains (amino acids 1 to 313 and 94 to 394). The effects of the mutants were studied in CHO cells by using Gal4-N5 Net and the UAS-luciferase reporter. Compared to the striking inhibition by wild-type p53 of Gal4-N5 activation by MEK1 (Fig. 5B, bars 1 to 4), the p53 mutants were much less efficient at inhibiting Net activation (bars 5 to 14). All of the p53 mutants were efficiently expressed relative to wild-type expression (Fig. 5C; compare lanes 7 to 14 with lanes 3 and 4), except for the 1-100 mutant (lanes 5 and 6), which was detectable only upon longer exposure of the autoradiograms (data not shown). It appears unlikely that the lack of inhibition by the 1-100 mutant is due to its low level of expression, since p53 1-313, which contains this region and is efficiently expressed, is not a more efficient inhibitor (Fig. 5B, bars 5 to 8). The p53 deletion mutants had relatively little effect on the phosphorylation of Gal4-N5 (Fig. 5D, lanes 1 and 3 to 5) compared to the effect of wild-type p53 (lane 2). These results show that individual fragments of p53, like the p53 point mutants, have little effect on Net activation and phosphorylation and that wild-type p53 is required for efficient inhibition of Net.
FIG. 5.
Effects of deletion mutants of p53 on Net activation and phosphorylation. (A) Structures of the p53 deletion mutants. The full-length and mutant p53s contain an N-terminal Flag tag (F) fused to the indicated sequences of p53. TA, transactivation domain; DBD, DNA binding domain; TET, tetramerization domain; REG, regulatory domain. (B and C) Effects of p53 deletion mutants on Net activation. CHO cells in six-well plates were transfected with expression vectors for Flag-tagged full-length p53 and p53 deletion mutants (100 or 300 ng; lanes 3 to 14) and for active MEK1 (100 ng; lanes 2 to 14), 100 ng of pGal4-N5, 1.5 μg of UAS-luciferase, and 250 ng of CMV-lacZ. Cell extracts were assayed for luciferase and β-galactosidase (B), and expression levels were assayed by immunoblotting with the anti-Flag antibody (C). Arrowheads in panel C point to prominent bands migrating with the expected mobility. (D) Effects of p53 deletion mutants on Net phosphorylation. CHO cells in 9-cm-diameter plates were transfected with expression vectors for Flag-tagged full-length p53 and p53 deletion mutants (1.5 μg; lanes 2 to 5) and for active MEK1 (500 ng; all lanes), 500 ng of pGal4-N5, 7.5 μg of UAS-luciferase, and 1.25 μg of CMV-lacZ. Cell extracts were analyzed by Western blotting with the anti-phospho-Net antibody 2F3 (P-Gal4-N5) and the anti-Gal4 antibody 2GV3 (Gal4-N5).
Inhibition of egr-1 expression in the presence of p53.
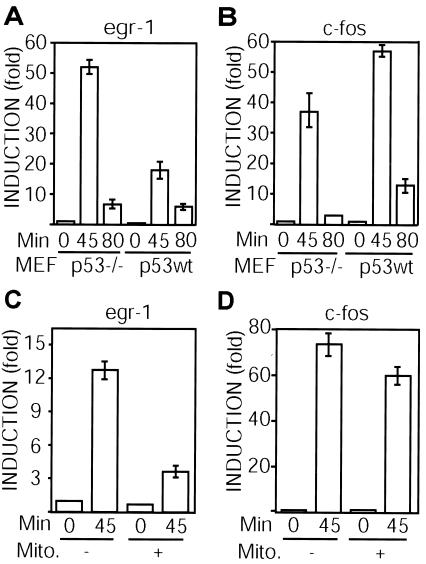
We investigated whether p53 affects the induction by serum of egr-1, a gene that is regulated by Net. Serum-starved p53−/− and wild-type MEFs were stimulated with serum for 45 and 80 min, and egr-1 mRNA expression was measured by quantitative RT-PCR. egr-1 levels after 45 min were lower in the wild-type p53 cells (Fig. 6A). In contrast, c-fos levels were higher in the wild-type p53 cells (Fig. 6B). Similarly, HUVEC were serum starved, treated with mitomycin C, and stimulated with serum for 45 min. Mitomycin C inhibited egr-1 induction and had a much smaller effect on c-fos (Fig. 6C and D). These results show that p53 inhibits egr-1 induction by serum, possibly through its ability to inhibit Net activity.
FIG. 6.
p53 inhibits egr-1 induction. (A and B) p53−/− and wild-type MEFs were serum starved and activated with DMEM plus 10% FCS for 45 or 80 min. (C and D) HUVEC were serum starved, incubated with or without mitomycin C (Mito.), and activated with DMEM plus 10% FCS for 45 min. mRNA levels were measured by quantitative real-time RT-PCR using specific primers for mouse egr-1 (A), mouse c-fos (B), human egr-1 (C), and human c-fos 1 (D).
Faster skin wound healing in p53-null mice.
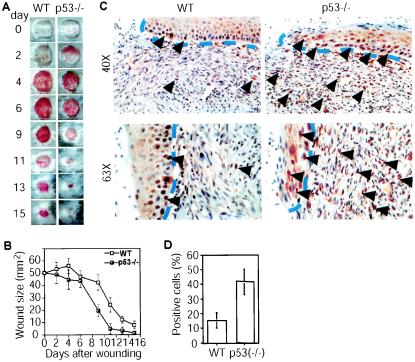
We have found that skin wound healing is delayed in Net mutant mice (52), showing that Net is required for this process. p53 is known to have a role in skin biology, raising the possibility that in p53−/− animals, relief of p53 repression of Net would accelerate wound healing. Full-thickness wounds were generated on the dorsal skin of wild-type and p53−/− animals, and the wounds were photographed (Fig. 7A) and measured (Fig. 7B) at regular intervals. The wounded skin of p53−/− mice healed faster than wild-type skin. Similar results were observed with 11 pairs of littermates. The absence of p53 could lead to increased phosphorylation of Net. Using IHC, we observed significantly more cells that expressed phospho-Net in p53−/− mouse skin 6 days after wounding (Fig. 7C and D; P < 0.001 for wild-type versus p53-null mice; n = 20). Increased phospho-Net levels were observed both in the keratinocytes of the regenerating epidermis and in mononuclear cells of the granulation tissue (Fig. 7C). These observations suggest that p53 inhibits Net activation in vivo during wound healing.
FIG. 7.
Accelerated wound healing and up-regulation of phospho-Net expression in p53−/− mice. (A) Macroscopic view of healing dorsal skin at different times after wounding of p53−/− and wild-type (WT) littermates. (B) Time course of wound healing. The sizes of the wounds were measured at different times after wounding. Eleven pairs of mice were tested. P < 0.01. (C) Phospho-Net expression (brown nuclear staining) of wounds 9 days after wounding. Dashed blue lines separate the newly formed migrating frontiers of the epidermis from the granulation tissue of the wounds. Arrowheads indicate nuclei of cells that express phospho-Net. In p53−/− mice, both keratinocytes and granulation tissue mononuclear cells express more phospho-Net than those of their wild-type littermates. (D) Statistical analysis of proportions of phospho-Net-positive cells in the wounds of wild-type and p53−/− mice. Five randomly selected fields from three sections from three pairs of mice were counted blindly by two investigators. P < 0.001.
Increased Net phosphorylation in UV-irradiated p53−/− mouse skin.
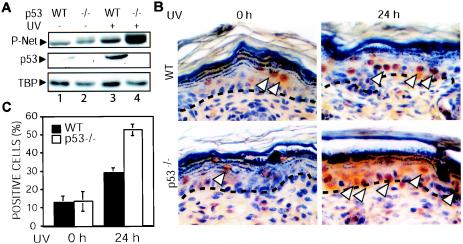
UV irradiation of skin activates p53, which may inhibit Net phosphorylation. We investigated whether loss of p53 would lead to increased levels of Net phosphorylation in response to UV irradiation. We irradiated the skin of p53−/− mice and wild-type littermates with UV-B and studied the phosphorylation of Net 24 h later by immunoblotting (Fig. 8A) and IHC (Fig. 8B and C). Immunoblots of proteins extracted from skin showed increases in levels of phospho-Net as well as p53 in wild-type mice following UV irradiation (Fig. 8A, lanes 1 and 3). The increase in phospho-Net levels was higher in p53−/− skin (Fig. 8A, lanes 2 and 4). Phospho-Net was detected in the basal and suprabasal layers of the epidermis (Fig. 8B), the layers in which p53 is induced (16). There was a statistically significant increase in the number of phospho-Net-positive cells in the p53−/− epidermis, as shown by counting of the positive cells (Fig. 8C) (P < 0.001). These results show that p53 inhibits Net phosphorylation in vivo, in UV-irradiated mouse skin.
FIG. 8.
Increased phosphorylation of Net in UV-treated skin of p53−/− mice. p53−/− and wild-type mice were irradiated, and 24 h later, skin samples were harvested. (A) Immunoblots. Total-cell extracts were prepared by using radioimmunoprecipitation assay buffer, fractionated by SDS-polyacrylamide gel electrophoresis, and immunoblotted with antibodies against p53 (NCL-p53-CM5), anti-phospho-Net (2F3), and TBP (used as a loading control) (3G3). P-Net, phospho-Net. (B) IHC of phospho-Net (with antibody 2F3). Arrowheads indicate sites of Net phosphorylation. (C) Quantification of phospho-Net-positive cells. Positive cells were counted blindly by two investigators using samples from three different mice. Error bars, standard deviations. There was a statistically significant difference in the number of 2F3-positive cells between wild-type and p53−/− animals (P < 0.001).
DISCUSSION
This study demonstrates that p53 inhibits MAP kinase-dependent Net transcriptional activation by modulating its level of phosphorylation. p53 inhibits Net activation and phosphorylation in different cell lines, indicating that the effects of p53 on Net could be a general phenomenon. Inhibition is observed under physiological conditions, in normal cells in culture, and in vivo in mice. Inhibition is specific, in that phosphorylation and activation of the highly related factor Elk-1 are not inhibited. There are several possible mechanisms by which p53 could inhibit Net activation, including a direct mechanism involving p53-Net complex formation and an indirect mechanism involving p53-induced expression of a Net inhibitor. p53 inhibits Net phosphorylation by two different kinases, ERK and JNK, without affecting their activation, indicating that the mechanism involves a step downstream from MAP kinase activation. p53 interacts with Net in vivo, and the minimum binding domain is the region of the C-terminal domain that includes the two phosphorylation sites. These observations suggest that p53 inhibits by a mechanism involving steric hindrance of MAP kinase access to the phosphorylation sites. However, mutant p53s that bind to Net do not inhibit phosphorylation, showing that binding alone is not sufficient. Wild-type p53 may recruit, into a complex with Net, other factors that are required for inhibition of phosphorylation, and mutant p53s may not be able to recruit these factors. p53 has been shown to bind to JNK (46), raising the possibility that JNK and, by analogy, ERK may be part of such a complex. The p53 point and deletion mutants that we studied cannot activate transcription, raising the possibility that gene expression is required for Net inhibition. The corresponding gene products would have to have specific effects on Net and not on the related factor Elk-1. Mutant p53s are less efficient than wild-type p53 at inhibiting Net transactivation. They do retain some activity, raising the possibility that binding to Net may inhibit transactivation to some extent, perhaps by preventing interactions with coactivators. However, additional properties of the wild-type protein are required for full inhibition. Further experiments will be required to determine the precise mechanisms by which p53 inhibits Net activation.
We found that p53 inhibits serum induction of an immediate-early gene, egr-1. Similarly, p53 has been shown to inhibit egr-1 induction by UV irradiation (50). It has previously been shown that Net regulates egr-1 expression in vivo (2), indicating that Net may mediate p53 repression of egr-1 induction. Interestingly, serum induction of c-fos is not sensitive to p53, showing that the effects of p53 are specific for certain immediate-early genes. Net may not be a major physiological regulator of c-fos, since we did not detect changes in c-fos expression in Net mutant mice (2). p53-insensitive factors such as Elk-1 may mediate c-fos induction. Alternatively, c-fos regulation may be more complex, since p53 has been shown to both activate and inhibit c-fos expression (15).
p53 inhibits more than 100 different genes (23, 48, 51), raising the question of how they are inhibited. Unlike activation of transcription, the mechanisms of gene repression by p53 are poorly defined. p53 appears to repress transcription by different mechanisms. p53 has a general suppressive activity on transcription by all three RNA polymerases (I, II, and III), perhaps due to its ability to bind to the common factor TBP (7, 8, 10, 30, 34, 49). General repression may also be mediated by recruitment of the corepressor mSin3a and histone deacetylation (31). Promoter-specific effects could result from binding to particular p53 motifs (12, 19, 24, 32) and specific interactions with certain factors, including Sp1 (3, 47), CBF (1), AP-1 (40), WT-1 (27), STAT-3 (25, 26), and the glucocorticoid receptor (37). Although the mechanisms of repression are still poorly understood, they include inhibition of DNA binding by Sp1 (3), inhibition of phosphorylation of STAT-3 by unknown mechanisms (25, 26), and cytoplasmic sequestration of the glucocorticoid receptor (37). p53 interactions with particular factors probably lead to inhibition of defined physiological functions. Repression of Net might be expected to be important for a key function of p53, inhibition of cell proliferation (42, 43).
Net is required for efficient wound healing (52). Interestingly, p53 mutant mice have a faster rate of wound healing and increased activation of Net through phosphorylation (this study), as expected from loss of inhibition by p53. Net is a regulator of immediate-early genes that control cell cycle entry (38, 41). As expected, Net mutant MEFs have a defect in proliferation, with more cells in the G1 phase (G. Buchwalter, C. Gross, A. Ayadi, S. Sengupta, and B. Wasylyk, unpublished data). The importance of our results is that they show how p53 prevents other factors from “applying the accelerator” of the cell cycle, in addition to providing better understanding of how it “applies the brakes” through induction of molecules such as CDKN1A (p21WAF1/CIP1).
Acknowledgments
We thank A. Ayadi and S. Sengupta for materials and helpful discussions, C. Wasylyk for initial observations and contributions to some experiments, and the IGBMC core facilities for help and support.
We thank the French Foreign Ministry and the Ligue Regionale Contre le Cancer for fellowships for K.N., the MRT for fellowships for G.B and C.G., and the Ligue Regionale Contre le Cancer for a fellowship for G.G. We also thank BioAvenir (Aventis, Rhone-Poulenc), the Centre National de la Recherche Scientifique, the Institut National de la Santé et de la Recherche Médicale, the Hôpital Universitaire de Strasbourg, the Association pour la Recherche sur le Cancer, the Fondation pour la Recherche Médicale, the Ligue Nationale Française contre le Cancer (Equipe labellisée), the Ligue Régionale contre le Cancer (Haut-Rhin and Bas-Rhin), and the EU (FP5 project QLK6-2000-00159) for financial assistance.
REFERENCES
- 1.Agoff, S. N., J. Hou, D. I. Linzer, and B. Wu. 1993. Regulation of the human hsp70 promoter by p53. Science 259:84-87. [DOI] [PubMed] [Google Scholar]
- 2.Ayadi, A., H. Zheng, P. Sobieszczuk, G. Buchwalter, P. Moerman, K. Alitalo, and B. Wasylyk. 2001. Net-targeted mutant mice develop a vascular phenotype and up-regulate egr-1. EMBO J. 20:5139-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bargonetti, J., A. Chicas, D. White, and C. Prives. 1997. p53 represses Sp1 DNA binding and HIV-LTR directed transcription. Cell. Mol. Biol. (Noisy-le-grand) 43:935-949. [PubMed] [Google Scholar]
- 4.Bergers, G., K. Javaherian, K. M. Lo, J. Folkman, and D. Hanahan. 1999. Effects of angiogenesis inhibitors on multistage carcinogenesis in mice. Science 284:808-812. [DOI] [PubMed] [Google Scholar]
- 5.Brou, C., S. Chaudhary, I. Davidson, Y. Lutz, J. Wu, J. M. Egly, L. Tora, and P. Chambon. 1993. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 12:489-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunet, A., G. Pages, and J. Pouyssegur. 1994. Constitutively active mutants of MAP kinase kinase (MEK1) induce growth factor-relaxation and oncogenicity when expressed in fibroblasts. Oncogene 9:3379-3387. [PubMed] [Google Scholar]
- 7.Budde, A., and I. Grummt. 1999. p53 represses ribosomal gene transcription. Oncogene 18:1119-1124. [DOI] [PubMed] [Google Scholar]
- 8.Cairns, C. A., and R. J. White. 1998. p53 is a general repressor of RNA polymerase III transcription. EMBO J. 17:3112-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatton, B., A. Bahr, J. Acker, and C. Kedinger. 1995. Eukaryotic GST fusion vector for the study of protein-protein associations in vivo: application to interaction of ATFa with Jun and Fos. BioTechniques 18:142-145. [PubMed] [Google Scholar]
- 10.Chesnokov, I., W. M. Chu, M. R. Botchan, and C. W. Schmid. 1996. p53 inhibits RNA polymerase III-directed transcription in a promoter-dependent manner. Mol. Cell. Biol. 16:7084-7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Criqui-Filipe, P., C. Ducret, S. M. Maira, and B. Wasylyk. 1999. Net, a negative Ras-switchable TCF, contains a second inhibition domain, the CID, that mediates repression through interactions with CtBP and de-acetylation. EMBO J. 18:3392-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Souza, S., H. Xin, S. Walter, and D. Choubey. 2001. The gene encoding p202, an interferon-inducible negative regulator of the p53 tumor suppressor, is a target of p53-mediated transcriptional repression. J. Biol. Chem. 276:298-305. [DOI] [PubMed] [Google Scholar]
- 13.Ducret, C., S. M. Maira, A. Dierich, and B. Wasylyk. 1999. The Net repressor is regulated by nuclear export in response to anisomycin, UV, and heat shock. Mol. Cell. Biol. 19:7076-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ducret, C., S. M. Maira, Y. Lutz, and B. Wasylyk. 2000. The ternary complex factor Net contains two distinct elements that mediate different responses to MAP kinase signalling cascades. Oncogene 19:5063-5072. [DOI] [PubMed] [Google Scholar]
- 15.Elkeles, A., T. Juven-Gershon, D. Israeli, S. Wilder, A. Zalcenstein, and M. Oren. 1999. The c-fos proto-oncogene is a target for transactivation by the p53 tumor suppressor. Mol. Cell. Biol. 19:2594-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganguli, G., J. Abecassis, and B. Wasylyk. 2000. MDM2 induces hyperplasia and premalignant lesions when expressed in the basal layer of the epidermis. EMBO J. 19:5135-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gannon, J. V., R. Greaves, R. Iggo, and D. P. Lane. 1990. Activating mutations in p53 produce a common conformational effect. A monoclonal antibody specific for the mutant form. EMBO J. 9:1595-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giovane, A., A. Pintzas, S. M. Maira, P. Sobieszczuk, and B. Wasylyk. 1994. Net, a new ets transcription factor that is activated by Ras. Genes Dev. 8:1502-1513. [DOI] [PubMed] [Google Scholar]
- 19.Han, X., A. B. Patters, and R. W. Chesney. 2002. Transcriptional repression of taurine transporter gene (TauT) by p53 in renal cells. J. Biol. Chem. 277:39266-39273. [DOI] [PubMed] [Google Scholar]
- 20.Hinds, P. W., C. A. Finlay, R. S. Quartin, S. J. Baker, E. R. Fearon, B. Vogelstein, and A. J. Levine. 1990. Mutant p53 DNA clones from human colon carcinomas cooperate with ras in transforming primary rat cells: a comparison of the “hot spot” mutant phenotypes. Cell Growth Differ. 1:571-580. [PubMed] [Google Scholar]
- 21.Holash, J., S. J. Wiegand, and G. D. Yancopoulos. 1999. New model of tumor angiogenesis: dynamic balance between vessel regression and growth mediated by angiopoietins and VEGF. Oncogene 18:5356-5362. [DOI] [PubMed] [Google Scholar]
- 22.Hsieh, J. K., F. S. Chan, D. J. O'Connor, S. Mittnacht, S. Zhong, and X. Lu. 1999. RB regulates the stability and the apoptotic function of p53 via MDM2. Mol. Cell 3:181-193. [DOI] [PubMed] [Google Scholar]
- 23.Kannan, K., N. Amariglio, G. Rechavi, J. Jakob-Hirsch, I. Kela, N. Kaminski, G. Getz, E. Domany, and D. Givol. 2001. DNA microarrays identification of primary and secondary target genes regulated by p53. Oncogene 20:2225-2234. [DOI] [PubMed] [Google Scholar]
- 24.Lee, K. C., A. J. Crowe, and M. C. Barton. 1999. p53-mediated repression of alpha-fetoprotein gene expression by specific DNA binding. Mol. Cell. Biol. 19:1279-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin, J., X. Jin, K. Rothman, H. J. Lin, H. Tang, and W. Burke. 2002. Modulation of signal transducer and activator of transcription 3 activities by p53 tumor suppressor in breast cancer cells. Cancer Res. 62:376-380. [PubMed] [Google Scholar]
- 26.Lin, J., H. Tang, X. Jin, G. Jia, and J. T. Hsieh. 2002. p53 regulates Stat3 phosphorylation and DNA binding activity in human prostate cancer cells expressing constitutively active Stat3. Oncogene 21:3082-3088. [DOI] [PubMed] [Google Scholar]
- 27.Maheswaran, S., S. Park, A. Bernard, J. F. Morris, F. J. Rauscher III, D. E. Hill, and D. A. Haber. 1993. Physical and functional interaction between WT1 and p53 proteins. Proc. Natl. Acad. Sci. USA 90:5100-5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maira, S. M., J. M. Wurtz, and B. Wasylyk. 1996. Net (ERP/SAP2), one of the Ras-inducible TCFs, has a novel inhibitory domain with resemblance to the helix-loop-helix motif. EMBO J. 15:5849-5865. [PMC free article] [PubMed] [Google Scholar]
- 29.Marais, R., J. Wynne, and R. Treisman. 1993. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell 73:381-393. [DOI] [PubMed] [Google Scholar]
- 30.Martin, D. W., R. M. Munoz, M. A. Subler, and S. Deb. 1993. p53 binds to the TATA-binding protein-TATA complex. J. Biol. Chem. 268:13062-13067. [PubMed] [Google Scholar]
- 31.Murphy, M., J. Ahn, K. K. Walker, W. H. Hoffman, R. M. Evans, A. J. Levine, and D. L. George. 1999. Transcriptional repression by wild-type p53 utilizes histone deacetylases, mediated by interaction with mSin3a. Genes Dev. 13:2490-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ori, A., A. Zauberman, G. Doitsh, N. Paran, M. Oren, and Y. Shaul. 1998. p53 binds and represses the HBV enhancer: an adjacent enhancer element can reverse the transcription effect of p53. EMBO J. 17:544-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ortega, S., M. Ittmann, S. H. Tsang, M. Ehrlich, and C. Basilico. 1998. Neuronal defects and delayed wound healing in mice lacking fibroblast growth factor 2. Proc. Natl. Acad. Sci. USA 95:5672-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ragimov, N., A. Krauskopf, N. Navot, V. Rotter, M. Oren, and Y. Aloni. 1993. Wild-type but not mutant p53 can repress transcription initiation in vitro by interfering with the binding of basal transcription factors to the TATA motif. Oncogene 8:1183-1193. [PubMed] [Google Scholar]
- 35.Robinson, M. J., S. A. Stippec, E. Goldsmith, M. A. White, and M. H. Cobb. 1998. A constitutively active and nuclear form of the MAP kinase ERK2 is sufficient for neurite outgrowth and cell transformation. Curr. Biol. 8:1141-1150. [DOI] [PubMed] [Google Scholar]
- 36.Ryan, K. M., and K. H. Vousden. 1998. Characterization of structural p53 mutants which show selective defects in apoptosis but not cell cycle arrest. Mol. Cell. Biol. 18:3692-3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sengupta, S., and B. Wasylyk. 2001. Ligand-dependent interaction of the glucocorticoid receptor with p53 enhances their degradation by Hdm2. Genes Dev. 15:2367-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharrocks, A. D. 2002. Complexities in ETS-domain transcription factor function and regulation: lessons from the TCF (ternary complex factor) subfamily. Biochem. Soc. Trans. 30:1-9. [DOI] [PubMed] [Google Scholar]
- 39.Stephen, C. W., P. Helminen, and D. P. Lane. 1995. Characterisation of epitopes on human p53 using phage-displayed peptide libraries: insights into antibody-peptide interactions. J. Mol. Biol. 248:58-78. [DOI] [PubMed] [Google Scholar]
- 40.Sun, Y., L. Wenger, J. L. Rutter, C. E. Brinckerhoff, and H. S. Cheung. 1999. p53 down-regulates human matrix metalloproteinase-1 (collagenase-1) gene expression. J. Biol. Chem. 274:11535-11540. [DOI] [PubMed] [Google Scholar]
- 41.Treisman, R. 1994. Ternary complex factors: growth factor regulated transcriptional activators. Curr. Opin. Genet. Dev. 4:96-101. [DOI] [PubMed] [Google Scholar]
- 42.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 43.Vousden, K. H., and X. Lu. 2002. Live or let die: the cell's response to p53. Nat. Rev. Cancer 2:594-604. [DOI] [PubMed] [Google Scholar]
- 44.Wasylyk, C., J. L. Imler, J. Perez-Mutul, and B. Wasylyk. 1987. The c-Ha-ras oncogene and a tumor promoter activate the polyoma virus enhancer. Cell 48:525-534. [DOI] [PubMed] [Google Scholar]
- 45.Wu, K., S. W. Jiang, and F. J. Couch. 2003. p53 mediates repression of the BRCA2 promoter and down-regulation of BRCA2 mRNA and protein levels in response to DNA damage. J. Biol. Chem. 278:15652-15660. [DOI] [PubMed] [Google Scholar]
- 46.Xue, Y., N. T. Ramaswamy, X. Hong, and J. C. Pelling. 2003. Association of JNK1 with p21waf1 and p53: modulation of JNK1 activity. Mol. Carcinog. 36:38-44. [DOI] [PubMed] [Google Scholar]
- 47.Yamabe, Y., A. Shimamoto, M. Goto, J. Yokota, M. Sugawara, and Y. Furuichi. 1998. Sp1-mediated transcription of the Werner helicase gene is modulated by Rb and p53. Mol. Cell. Biol. 18:6191-6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu, J., L. Zhang, P. M. Hwang, C. Rago, K. W. Kinzler, and B. Vogelstein. 1999. Identification and classification of p53-regulated genes. Proc. Natl. Acad. Sci. USA 96:14517-14522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhai, W., and L. Comai. 2000. Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol. Cell. Biol. 20:5930-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, W., and S. Chen. 2001. EGR-1, a UV-inducible gene in p53−/− mouse cells. Exp. Cell Res. 266:21-30. [DOI] [PubMed] [Google Scholar]
- 51.Zhao, R., K. Gish, M. Murphy, Y. Yin, D. Notterman, W. H. Hoffman, E. Tom, D. H. Mack, and A. J. Levine. 2000. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 14:981-993. [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng, H., C. Wasylyk, A. Ayadi, J. Abecassis, J. A. Schalken, H. Rogatsch, N. Wernert, S. M. Maira, M. C. Multon, and B. Wasylyk. 2003. The transcription factor Net regulates the angiogenic switch. Genes Dev. 17:2283-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]