Abstract
The mitogen-activated protein kinase (MAPK) signaling pathway regulates diverse biologic functions including cell growth, differentiation, proliferation, and apoptosis. The extracellular signal-regulated kinases (ERKs) constitute one branch of the MAPK pathway that has been implicated in the regulation of cardiac differentiated growth, although the downstream mechanisms whereby ERK signaling affects this process are not well characterized. Here we performed a yeast two-hybrid screen with ERK2 bait and a cardiac cDNA library to identify novel proteins involved in regulating ERK signaling in cardiomyocytes. This screen identified the LIM-only factor FHL2 as an ERK interacting protein in both yeast and mammalian cells. In vivo, FHL2 and ERK2 colocalized in the cytoplasm at the level of the Z-line, and interestingly, FHL2 interacted more efficiently with the activated form of ERK2 than with the dephosphorylated form. ERK2 also interacted with FHL1 and FHL3 but not with the muscle LIM protein. Moreover, at least two LIM domains in FHL2 were required to mediate efficient interaction with ERK2. The interaction between ERK2 and FHL2 did not influence ERK1/2 activation, nor was FHL2 directly phosphorylated by ERK2. However, FHL2 inhibited the ability of activated ERK2 to reside within the nucleus, thus blocking ERK-dependent transcriptional responsiveness of ELK-1, GATA4, and the atrial natriuretic factor promoter. Finally, FHL2 partially antagonized the cardiac hypertrophic response induced by activated MEK-1, GATA4, and phenylephrine agonist stimulation. Collectively, these results suggest that FHL2 serves a repressor function in cardiomyocytes through its ability to inhibit ERK1/2 transcriptional coupling.
Mitogen-activated protein kinases (MAPKs) and stress-activated protein kinases consist of an amplification cascade of successively acting kinases that culminate in the dual phosphorylation and activation of terminal effector kinases that subsequently phosphorylate diverse effector proteins. In mammalian cells, the MAPK signaling cascade is comprised of at least three major branches (named for the terminal effector kinases) including p38, c-Jun N-terminal kinases (JNKs), and extracellular signal-regulated kinases (ERKs) (reviewed in reference 38). In cardiomyocytes, the MAPK signaling cascade is initiated by both cell stretching and by diverse neuroendocrine factors mediated through G protein-coupled receptors and receptor tyrosine kinases (3, 32). Once activated, p38, JNKs, and ERKs each phosphorylate a wide array of intracellular targets, including diverse transcription factors resulting in the reprogramming of gene expression. The major upstream activators of ERK1/2 are two MAPK kinases, MEK1 and MEK2, which directly phosphorylate a dual acceptor motif in ERKs (Thr-Glu-Tyr).
In cardiomyocytes, MEK1/2-ERK1/2 have been implicated as important transducers of the hypertrophic growth response both in cell culture-based studies and within the intact heart. For example, ERK1/2 are activated in cultured cardiomyocytes by catecholamines, endothelin-1, angiotensin II, and stretching (2, 9, 28, 39, 41), whereas the adult heart shows ERK activation in response to pressure overload (29, 33). These correlative observations were mechanistically extended by adenovirus-mediated gene transfer of a dominant-negative MEK1 (dnMEK1) or dnRaf-1 cDNA, each of which blocked endothelin-1 and phenylephrine (PE)-induced myocyte hypertrophy in culture (35, 40). In vivo, transgenic mice expressing activated MEK1 within the heart demonstrated constitutive ERK1/2 activation (but not activation of p38 or JNKs) that was associated with a prominent hypertrophy response (4). One potential mechanism whereby ERK1/2 might promote hypertrophy was recently suggested by the observation that the cardiac expressed transcription factor GATA4 is directly phosphorylated by ERK1/2, resulting in augmented hypertrophic gene expression (19).
To further examine the downstream mechanisms whereby ERK signaling might regulate the cardiac growth response, a yeast two-hybrid screen was performed to identify ERK2 interacting proteins, resulting in the isolation of the LIM-only domain-containing factor FHL2. This LIM domain protein is largely restricted in its expression to the heart both throughout embryonic development and in the adult (6, 8). FHL2 is localized within both the cytoplasm and nucleus but can shuttle between these compartments as part of its function (26, 27). In the nucleus, FHL2 can serve as a transcriptional coactivator through its interaction with various factors such as the androgen receptor, activator protein-1 (AP-1), CREB, CREM, β-catenin in transformed cell-types, and factors in the wnt signaling cascade (12, 13, 24, 25, 37). However, FHL2 can also serve as a transcriptional corepressor of the promyelocytic leukemia zinc finger protein and β-catenin in muscle cells, suggesting that its ability to function as an activator or repressor may be cell type or promoter dependent (20, 21). Similarly, FHL2 gene-targeted mice showed greater cardiac hypertrophic growth following catecholamine infusion, suggesting that FHL2 can have a dominant repressor-like activity in the myocardium (17). The results we present here identify FHL2 as a modulator of the cardiomyocyte growth response through regulation of MEK1-ERK2 signaling.
MATERIALS AND METHODS
Two-hybrid screening and interaction assays.
Yeast two-hybrid screening was performed to identify ERK2 interacting proteins by using the Matchmaker System-3 from Clontech (Palo Alto, Calif.) according to the manufacturer's instructions. The ERK2 bait contained amino acids 12 to 358 cloned in frame with the Gal4 DNA binding domain (DBD) at the BamHI site in the pGBKT7 plasmid. An adult mouse heart cDNA library in the pGAD10 vector was custom made by Clontech. The yeast reporter strain AH109 was transformed with the ERK2 bait together with the mouse heart cDNA library, and two million transformants were screened. Positive colonies were subject to multiple rounds of additional selection in the appropriate media to verify specificity (of which two FHL2-containing clone were identified). The strengths of all interactions were quantified after retransformation into yeast by a liquid β-galactosidase assay with O-nitrophenyl β-d-galactopyranoside (Sigma, St. Louis, Mo.) as a substrate. The ERK2-Gal4 mammalian expression vector was generated by subcloning ERK2 into the pM1-Gal4 DBD-containing plasmid, and FHL2-VP16 was generated by subcloning FHL2 into a pVP16 expression plasmid. The muscle LIM protein (MLP)-VP16 expression vector was a gift from Brian L. Black (University of California, San Francisco). The MEK1 expression vector was described previously (4).
Mice.
Activated MEK1 transgenic mice under the control of the cardiac-specific α-myosin heavy chain (α-MHC) promoter have been previously described (4). Transgenic mice overexpressing ERK2 were constructed by subcloning the rat ERK2 cDNA by blunt-end ligation into the HindIII site of the 5.5-kb murine cardiac-specific α-MHC promoter (a gift of J. Robbins, Children's Hospital, University of Cincinnati, Cincinnati, Ohio). MEK1 transgenic mice were mated with ERK2 transgenic mice to create MEK1-ERK2 double transgenic mice as a means of producing substantially greater ERK2 activity in the heart. C57BL/6 mice were injected subcutaneously with either phosphate-buffered saline or PE (10 mg/kg) for 15 min, and after this their hearts were collected for generation of protein extracts.
Cell culture.
Primary neonatal rat cardiomyocytes were prepared from hearts of 1- to 2-day-old Sprague-Dawley rat pups as previously described (11). After differential separation from nonmyocytes, enriched cardiomyocytes were plated on 1% gelatin-coated 12-well plates for transfection assays or 6-cm-diameter dishes for all other experiments. Cells were grown in M199 medium containing 100 U of penicillin-streptomycin/ml and 2 mmol of l-glutamine/liter without serum for 24 h before transfection or infection. Both cardiomyocytes and 10T1/2 cells were transfected with Tfx-20 reagent (Promega, Madison, Wis.). Cultures were harvested 48 h after transfection, and luciferase assays were preformed as described previously (19). Adenoviral infections were performed as previously described at a multiplicity of infection of 10 to 100 PFU per ml (11). Where indicated, U0126 was added at a concentration of 20 μM 1 h prior to treatment with agonist or harvesting.
Adenoviral constructs and expression plasmids.
AdMEK1, Ad-dnMEK1, AdGATA4, and Adβgal have been previously described (4, 19). A Flag-FHL2 cDNA and rat Flag-ERK2 cDNA were each subcloned into the HindIII site of the pAC-CMVpLpA vector to generate AdFHL2 and AdERK2 as described previously (11). Expression vectors encoding the activated MEK1 and FHL2 and the reporter genes of atrial natriuretic factor (ANF)-luciferase (ANF-Luc), Gal4-Luc, Gal4-ELK1, and GATA-Luc have been previously described (16, 19, 34). The myocyte enhancer factor 2 (MEF2)-dependent luciferase reporter was a gift from Bruce E. Markham (Pfizer Pharmaceuticals, Ann Arbor, Mich.). The constructs encoding FHL1, FHL3, LIM0, LIM1, LIM2, LIM3, LIM4, LIM0-3, LIM1-3, LIM1-4, and pEF-RhoA-V14 were described previously (26). The constructs encoding MEK5 and ERK5 were a gift from J. Han (The Scripps Research Institute, La Jolla, Calif.).
Immunoblotting analysis, luciferase assays, and glutathione S-transferase (GST) pull downs.
The generation of protein extracts from cultured cardiomyocytes or heart tissue and their subsequent immunoblotting has been described previously (19). Antibodies included ERK1/2 pan, phospho-ERK1/2 (Biosource, Camarillo, Calif.), ERK2 (BD Bioscience Pharmingen, San Diego, Calif.), phospho-105 GATA4 (Biosource International, Hopkinton, Mass.), GATA4 (Santa Cruz, Santa Cruz, Calif.), and FHL2 (26). Cardiomyocyte cultures were washed 14 h after transfection and maintained in serum-free media. The cells were lysed, and luciferase activity was measured 48 h posttransfection as described previously (19).
The full-length GST-FHL2 construct along with the partial constructs GST-FHL2 (amino acids 1 to 162) and GST-FHL2 (amino acids 163 to 279) were described previously (25). GST-FHL2 proteins were purified by glutathione-agarose affinity chromatography (10). Heart extracts were made from wild-type FVB/N mice, ERK2 transgenic mice (FVB/N), or ERK2-MEK1 double transgenic mice (FVB/N) as described previously (19). Heart extracts were incubated with GST-FHL2 constructs bound to glutathione-Sepharose beads in GST binding buffer containing 40 mM HEPES (pH 7.2), 50 mM Na acetate (pH 7.0), 200 mM NaCl, 2 mM EDTA, 5 mM dithiothreitol, 0.5% Nonidet P-40, protease inhibitors, and 2 μg of bovine serum albumin/ml for 3 h at 4°C. Beads were washed extensively in the binding buffer and boiled in sodium dodecyl sulfate (SDS) Laemmli loading buffer to elute bound proteins for resolution on SDS-13% polyacrylamide gels. Proteins were transferred to a polyvinylidene difluoride membrane, and immunoblotting was performed with a 1:1,000 dilution of the monoclonal ERK2 antibody (BD Bioscience Pharmingen) as described previously (19).
Binding assay.
GST-ERK2 (Upstate Biotechnology, Charlottesville Va.) was incubated with radiolabeled FHL proteins synthesized in vitro with a transcription-translation-coupled reticulocyte lysate system (Promega) in the presence of [35S]methionine. Equal amounts of GST-ERK2 proteins were incubated with 20 μl of radiolabeled proteins in GST binding buffer (described above). Glutathione beads were washed three times in binding buffer and boiled in SDS sample buffer to elute complexed proteins for SDS-polyacrylamide gel electrophoresis (PAGE) on 13 and 19% gels.
Coimmunoprecipitation.
Neonatal rat cardiomyocytes were infected with or without adenoviruses expressing ERK2 and FHL2 for 24 h and then subjected to phorbol myristate acetate (PMA) stimulation (200 nM) for 5 min. Cells were harvested in lysis buffer as previously described (19) and then incubated with a monoclonal ERK2 antibody and protein A-Sepharose beads in GST binding buffer for 3 h at 4°C. The beads were washed extensively with binding buffer, and the proteins were resolved on an SDS-13% polyacrylamide gel for subsequent Western blotting with a polyclonal FHL2 antibody. The in vivo immunoprecipitation consisted of 500 μg of mouse heart protein extract derived from either PE-stimulated mice (15 min after a subcutaneous injection of PE at 10 mg/kg of body weight) or phosphate-buffered saline-injected mice incubated with ERK2 polyclonal antibody followed by addition of protein A-G agarose, each incubated for 4 h at 4°C. The precipitates were washed, and Western blot analysis for monoclonal FHL2 antibody was performed.
Immunohistochemistry and hypertrophy analysis.
Adult mouse cardiomyocytes were isolated and immunostained as previously described (5). Endogenous proteins were localized with monoclonal ERK2 antibody at a 1:200 dilution or polyclonal FHL2 antibody at a dilution of 1:100. ERK2 and FHL2 were also immunolocalized in neonatal rat cardiomyocytes following AdFHL2 and/or AdERK2 infection, with or without PMA stimulation, showing similar colocalization. Fluorescein isothiocyanate-conjugated anti-mouse and tetramethyl rhodamine isothiocyanate-conjugated anti-rabbit secondary antibodies (Sigma) were used for subsequent visualization at a 1:200 dilution. Immunostaining for phosphorylated ERK1/2 was also performed with an antibody from Biosource at a dilution of 1:200. For calculation of cardiomyocyte size, neonatal rat cardiomyocytes were infected with the indicated adenoviruses for 24 h and then treated with or without PE for 36 h, essentially as described previously (4, 11). Cardiomyocytes were visualized with a 1:400 dilution of a monoclonal antibody against α-actinin (Sigma). Surface area was determined with the image analysis software NIH 1.63. Cells from randomly selected fields in three independent experiments were examined, and the surface area was compared to that of the control (100 cells each).
In vitro phosphorylation assay.
Activated recombinant ERK2 protein (Upstate Biotechnologies) was incubated with full-length GST-FHL2, GST-GATA4, or myelin basic protein (MBP) as described previously (19). The generation of recombinant purified GST-FHL2 and GST-GATA4 protein has been described previously (10, 19, 25). Phosphorylation was assayed at 30°C for 30 min in 30 μl of kinase buffer (20 mM HEPES [pH 7.4], 20 mM MgCl2, 2 mM dithiothreitol, 20 mM β-glycerophosphate, 20 mM p-nitrophenyl phosphate, 10 mg of leupeptin/ml, 10 mg of aprotinin/ml, and 0.1 mM sodium vanidate) in the presence of 10 μM ATP-10 Ci of [γ-32P]ATP (10 Ci/mmol) with the various GST fusion proteins described. The reaction was terminated with Laemmli sample buffer. The proteins were resolved by SDS-13% PAGE and detected with a Storm 860 PhosphorImager.
Analysis of mRNA levels.
mRNA levels of ANF and β-MHC from unstimulated or PE-stimulated neonatal cardiomyocytes was performed by dot blotting essentially as described previously (4). The ANF and β-MHC signals were normalized to glyceraldehyde-3-phosphate-dehydrogenase levels. RNA was extracted with Trizol reagent (Gibco-BRL, Rockville, Md.) 48 h after stimulation. The RNA was quantified and blotted to nitrocellulose with a dot blot manifold (Bio-Rad, Hercules, Calif.).
RESULTS
Two-hybrid screening identifies FHL2 as an ERK-interacting factor.
MEK1-ERK1/2 signaling plays a prominent role in regulating cardiac hypertrophic growth, although only a few ERK regulatory effectors are known to serve as downstream mediators of this process. To identify additional downstream ERK effectors, a yeast two-hybrid screen was performed with ERK2 as bait (fused to the Gal4 DBD) in conjunction with a cardiac myocyte-specific cDNA library (fused to the Gal4 activation domain). One of the more interesting prey clones that was isolated corresponded to amino acids 26 to 279 of the cardiac-restricted LIM-only factor FHL2 (two independent FHL2 clones were isolated). To further validate the specificity of this interaction, a rescued FHL2 prey plasmid and the ERK2-containing bait plasmid were retransformed back into the appropriate yeast strain containing the interaction-dependent β-galactosidase and metabolic selection marker cassettes (Fig. 1A). The data demonstrate a specific interaction between ERK2 (amino acids 12 to 358) and FHL2 (amino acids 26 to 279) but not between either factor and the pairwise control containing the Gal4 activation domain only or the Gal4 DBD only (Fig. 1A).
FIG. 1.
Two-hybrid screening identifies FHL2 as an ERK-interacting protein. (A) Yeast two-hybrid assay with strain AH109, which contains an interaction-dependent β-galactosidase reporter. The bait consisted of the Gal4 DBD fused to ERK2, and the identified prey contained the Gal4 activation domain (AD) fused to the indicated portion of FHL2. (B) Mammalian two-hybrid assay performed with 10T1/2 cells in which a Gal4-dependent luciferase reporter construct was cotransfected with expression vectors encoding the Gal4DBD-ERK2, the transcriptional activating protein VP16 fused to FHL2, MLP, and MEK1 (n = 3). Luciferase activity was measured as relative light units (RLU) per microgram of protein.
To extend the observations made with yeast, the interacting regions from FHL2 and ERK2 were cloned into mammalian expression vectors to permit a two-hybrid analysis in 10T1/2 fibroblasts by using a Gal4-dependent luciferase reporter construct. Relative to their positions in the yeast screen, the bait and prey fusions were switched between FHL2 and ERK2 as a further measure of specificity. Neither the Gal4 DBD-ERK2 fusion nor the VP16 activation domain-FHL2 fusion showed activation of the Gal4-dependent luciferase reporter when transfected alone (Fig. 1B). However, cotransfection of the vectors encoding Gal4-ERK2 together with FHL2-VP16 resulted in a dramatic increase in transcriptional activity, indicating that FHL2 and ERK2 also interact in mammalian cells (Fig. 1B). Cotransfection of an activated MEK1 construct, which enhances ERK2 phosphorylation, significantly increased reporter activity, suggesting that activated ERK might interact more avidly with FHL2 (Fig. 1B). To determine if the observed interaction with ERK2 was promiscuous to any LIM domain-containing factor, a construct encoding the MLP-VP16 fusion was also assayed for activity with ERK2 bait (Fig. 1B). However, ERK2 did not interact with MLP, suggesting that ERK2 only interacts with a specific subclass of LIM domain-containing factors, such as FHL2.
Determinants of the ERK2-FHL2 interaction.
FHL2 is a member of a multigene family of LIM-only factors, of which FHL1 and FHL3 are close relatives. To further characterize the specificity of interaction, [35S]methionine in vitro-labeled FHL1-3 proteins were each incubated with bacterium-produced GST-ERK2 or GST for pull-down analysis. The data show that all three FHL proteins interacted with ERK2, although FHL1 and FHL2 appeared to interact the strongest (Fig. 2A). To more thoroughly characterize the ERK2-FHL2 interaction, a more-refined series of 35S-labeled FHL2 domain deletions were analyzed for interaction with GST-ERK2 (Fig. 2B). The FHL2 domains consisting of LIM0-3, LIM1-3, and LIM1-4 each interacted with GST-ERK2, although single LIM domains alone did not appreciably interact (LIM2 alone gave a very weak signal, potentially suggesting that this domain is more important for the observed interaction) (Fig. 2B). Collectively, these results suggest that more than one LIM domain from FHL-type proteins is needed for optimal interaction with ERK2.
FIG. 2.
FHL2 physically interacts with ERK2. (A) SDS-PAGE showing a GST pull-down assay with GST alone or GST-ERK2 in conjunction with 35S-labeled FHL1, FHL2, or FHL3. (B) A series of FHL2 deletion fragments were 35S-labeled and subjected to a GST-ERK2 pull-down assay followed by SDS-PAGE analysis. (C) Full-length GST-FHL2 and two different GST-FHL2 deletion fragments were used in a pull-down assay with heart protein extract (500 μg) from wild-type mice or transgenic mice overexpressing ERK2 in the heart followed by Western blotting for ERK1/2. A Coomassie-stained gel shows equivalent levels of each GST fusion protein in each reaction mixture. IP, immunoprecipitation.
The interaction between FHL2 and ERK2 was further evaluated with mouse heart protein extracts and three different GST-FHL2 fusion proteins. The fusion proteins were immobilized to glutathione-coupled Sepharose resin and incubated with heart protein extracts derived from wild-type mice or from mice containing a cardiac-specific ERK2 transgene as a means of increasing the sensitivity of the interaction assay. ERK2 transgenic mice contain approximately 10-fold-more ERK2 protein in the heart, a small portion of which is in the activated state (Fig. 2C and data not shown). The reaction mixtures were then washed, eluted, and subjected to Western blotting with ERK1/2 antibody. Lanes 2 to 4 show an interaction between ERK2 protein derived from the ERK2 transgenic heart extracts (Flag epitope tagged) and full-length FHL2, amino acids 1 to 162 of FHL2, and amino acids 163 to 279 of FHL2 (Fig. 2C). GST alone showed no interaction with ERK2 (lane 1). Equivalent amounts of GST fusion protein were present in each reaction mixture, as revealed by Coomassie brilliant blue protein staining (Fig. 2C, bottom panel). We failed to detect a significant interaction between FHL2 and endogenous ERK1/2 from wild-type hearts (lanes 5 to 7), potentially due to a sensitivity issue given the lower levels of ERK1/2 protein that are present in wild-type hearts compared with the ERK2 transgenic hearts. Alternatively, the lack of a detectable interaction between FHL2 and endogenous ERK1/2 may also partially reflect a need for ERK activation to enhance the interaction (see section below). Collectively, the results discussed above indicate that activated ERK2 interacts with both the N and C termini of FHL2, which optimally contains at least two LIM domains.
ERK2 activation status influences its interaction with FHL2.
Recombinant ERK2- and FHL2-encoding adenoviruses were generated to further examine the mechanisms of interaction between these two proteins in vivo (both proteins are Flag epitope tagged). Cultured neonatal rat cardiomyocytes were infected with AdERK2 and AdFHL2 as a means of efficiently overexpressing each factor to permit a more sensitive analysis of interaction (nearly 100% of the cells become infected at a multiplicity of infection of 50 PFU/ml). Twenty-four hours after adenoviral infection, cardiomyocytes were either left unstimulated in serum-free media or transiently stimulated with PMA to induce ERK phosphorylation. Protein extracts were generated and subjected to ERK2 immunoprecipitation followed by Western blotting for FHL2 protein (Fig. 3A, top panel). The data show a weak interaction between ERK2 and FHL2 when each factor was overexpressed in unstimulated cardiomyocytes (Fig. 3A, lane 1). The FHL2 generated by the recombinant adenovirus shows a slightly retarded migration due to the Flag epitope. Interestingly, activation of ERK2 with PMA enhanced the interaction between ERK2 and overexpressed FHL2 protein (Fig. 3A, lane 2). As an important control, equivalent levels of ERK2 and FHL2 protein were expressed in each of the extracts used (Fig. 3A, middle and lower panels). Similar results were observed in two additional independent experiments.
FIG. 3.
Activated ERK2 interacts with FHL2 in vivo. (A) Cardiomyocytes were infected with adenoviruses encoding FHL2 and ERK2, and protein extracts were generated with or without prior PMA stimulation (200 nM, 5 min). ERK2 was then immunoprecipitated from the extracts (500 μg) and subjected to Western blotting for FHL2. The lower panels show control Western blots without immunoprecipitation (IP) for total ERK2 or total FHL2 from the coinfected extracts. (B) GST pull-down assays were performed with GST-FHL2 (full length) or GST coupled to beads and followed by Western blotting with the indicated antibodies. The input was cardiac extract (50 μg) from either ERK2 transgenic mice (mild level of ERK activation, lanes 1, 3, and 5) or activated MEK1-ERK2 double transgenic mice, which have substantial ERK activation (lanes 2, 4, and 6). The amount of phosphorylated ERK1/2 protein was also examined from ERK2 transgenic hearts versus MEK1-ERK2 double transgenic hearts. (C) Cardiac protein extracts (500 μg) derived from wild-type unstimulated hearts or PE-stimulated mice (mice were given PE at 10 mg/kg for 15 min prior to sacrifice) were subject to ERK2 immunoprecipitation with a polyclonal antibody followed by Western blotting with an FHL2 monoclonal antibody. The positive control is from cultured neonatal cardiomyocyte protein extract. The bottom panel shows phospho-ERK1/2 Western blotting from the unstimulated or PE-stimulated heart extracts. (D) Confocal immunohistochemistry from adult cardiac myocytes showing colocalization of endogenous FHL2 (red) and endogenous ERK2 (green) in vivo. FHL2 was also enriched at the Z-lines in neonatal rat cardiomyocyte cultures. +, present; −, absent.
The results discussed above suggest that activated ERK interacts more efficiently with FHL2 in cultured cardiomyocytes as detected by immunoprecipitation. To more rigorously extend this observation, a GST-FHL2 pull-down assay was performed with heart extracts from ERK2 transgenic mice or ERK2-MEK1 double transgenic mice. The activated MEK1 transgene dramatically increased ERK2 protein phosphorylation in the heart (Fig. 3B, lower panel) but did not alter the expression of total ERK2 protein (Fig. 3B, lanes 5 and 6). These two extracts were then subject to pull-down analysis with GST only (lanes 3 and 4) or with GST-FHL2 (lanes 1 and 2). Consistent with the immunoprecipitation results described above, heart extracts containing activated ERK2 showed a dramatic increase in the ability to interact with FHL2 compared with the less activated ERK2 extract (lacking the MEK1 transgene) (Fig. 3B). These results indicate that FHL2 interacts more efficiently with activated ERK2 in the heart.
The two assays described above utilized overexpression of ERK2 or FHL2 to readily demonstrate an interaction in vivo. However, we also wanted to examine the ability of endogenous FHL2 to interact with endogenous ERK2 at physiologic levels. To this end, total ERK2 was immunoprecipitated from unstimulated wild-type heart extract or from PE-stimulated wild-type heart extract and analyzed by Western blotting for FHL2 (Fig. 3C). The data demonstrate that a small amount of endogenous FHL2 coprecipitates with ERK2 but that activation of endogenous ERK2 shows a much more robust interaction (Fig. 3C). These results not only confirm that activated ERK2 interacts more readily with FHL2, but they also demonstrate that these two proteins interact at physiologic levels.
Lastly, it was also of interest to examine the subcellular localization of ERK2 and FHL2 in adult cardiac myocytes by confocal microscopy. Adult mouse cardiac myocytes were isolated, fixed, and incubated with both monoclonal ERK2 antibody and polyclonal FHL2 antibody to permit immunolocalization. In each case, endogenous FHL2 and endogenous ERK2 showed colocalization at the level of the Z-line in adult myocytes (Fig. 3D). FHL2 also localized to Z-line structures in neonatal rat cardiomyocytes, although some amount of FHL2 was also observed in the nuclei of some myocytes as well as diffusely localized in the cytoplasm of others (Fig. 3D and data not shown). Despite a less variable distribution of FHL2 in cultured neonatal cardiomyocytes, a significant portion of FHL2 protein also colocalized with ERK2 protein in these cells (data not shown). Collectively, these results indicate that FHL2 colocalizes with ERK2, an observation that is consistent with the hypothesis that FHL2 might regulate ERK2.
ERK2 does not phosphorylate FHL2, nor does FHL2 inhibit ERK phosphorylation.
The interaction observed between ERK2 and FHL2 suggested three initial hypotheses: (i) that ERK2 might directly phosphorylate FHL2, thereby altering its coactivator or corepressor functions, (ii) that FHL2 might alter the ability of ERK2 to bind and phosphorylate substrates, and (iii) that FHL2 might alter ERK1/2 activation and phosphorylation. To test the first hypothesis, bacterium-purified GST-FHL2 was incubated with an activated form of recombinant ERK2 in an appropriate kinase reaction buffer with [32P]ATP. As a control, bacterium-purified GST alone, bacterium-purified GST-GATA4, or MBP was also incubated with activated ERK2. The data show essentially no phosphorylation of GST or GST-FHL2, whereas GATA4 and MBP were each robustly phosphorylated by ERK2 as previously observed (19) (Fig. 4A). These results indicate that ERK2 does not directly phosphorylate FHL2 in vitro nor does activated MEK1 (data not shown).
FIG.4.
ERK2 does not phosphorylate FHL2, nor does FHL2 inhibit ERK activation. (A) SDS-PAGE of an in vitro kinase assay with bacterium-purified GST-FHL2 or GST alone incubated with activated recombinant ERK2 (0.25 μg). Positive controls consisted of bacterium-purified GST-GATA4 and MBP. (B) SDS-PAGE of an in vitro kinase assay between recombinant ERK2 and GATA4 or MBP in the presence or absence of recombinant FHL2. (C) Western blot for phosphorylated p90rsk and MEK1 from cardiomyocytes infected with Adβgal, AdMEK1, or AdMEK1 with AdFHL2. (D) Western blot of phosphorylated ERK1/2 or total ERK1/2 from cardiomyocytes infected with the indicated adenoviruses and left unstimulated or acutely stimulated with PMA (200 nM, 5 min). +, present; −, absent.
To address the second proposed hypothesis, the ability of recombinant GST-ERK2 to phosphorylate GATA4 or MBP was assessed in the presence or absence of recombinant GST-FHL2. The data show that GST-ERK2 efficiently phosphorylated GATA4 and MBP in an in vitro kinase assay in the presence of saturating GST-FHL2 protein, suggesting that the observed interaction between ERK2 and FHL2 does not disrupt the ability of ERK2 to phosphorylate substrates in vitro (Fig. 4B). This observation was extended to a cellular context by using adenoviral-mediated gene transfer of activated MEK1 and FHL2 in cultured cardiomyocytes. Overexpression of activated MEK1 induced robust phosphorylation of the cytoplasmic kinase p90rsk (through ERK1/2) at the baseline or in the presence of overexpressed FHL2 (Fig. 4C). These results suggest that FHL2 overexpression does not block the ability of MEK1-ERK1/2 to phosphorylate a cytoplasmic substrate protein in a cellular context.
To address the third potential hypothesis, FHL2 was overexpressed in cultured cardiomyocytes by itself or in combination with ERK2. ERK2 was then activated by transient PMA stimulation, and the relative degree of phosphorylation was measured by Western blotting with an ERK1/2 phospho-specific antibody. The data show that FHL2 overexpression did not alter the phosphorylation status of endogenous ERK2 or overexpressed ERK2 compared to Adβgal-infected controls (Fig. 4D). Absolute ERK1/2 protein levels were also not affected by FHL2 overexpression at baseline or following ERK2 overexpression (Fig. 4D, bottom panel). Collectively, these results suggest that FHL2 does not directly influence ERK phosphorylation in cultured cardiomyocytes.
FHL2 inhibits nuclear translocation of ERK2 and its phosphorylation of target proteins.
That FHL2 did not inhibit ERK activation, its interaction with substrate proteins, or the ability of ERK to become phosphorylated, suggests a more complex regulatory role underlying the observed interaction between these two factors. However, two additional regulatory mechanisms suggest themselves: (i) that FHL2 antagonizes ERK1/2 translocation into the nucleus, thus altering its ability to activate various transcription factors, and (ii) that FHL2 functions within the nucleus itself to alter the effectiveness of ERK1/2 in phosphorylating target proteins. To directly address at least one of these potential mechanisms, immunocytochemistry was performed in adenovirus-infected neonatal cardiomyocytes to more carefully examine the ability of FHL2 to antagonize ERK nuclear occupancy. PMA stimulation produced noticeable translocation of a pool of ERK2 protein into the nucleus, consistent with previous observations in other cell types (18) (Fig. 5A). However, AdFHL2 significantly reduced the number of cells showing total ERK2 protein translocation to the nucleus following PMA stimulation (Fig. 5A). Quantitation of this immunocytochemical approach revealed approximately 80% of the cells with significant ERK2 translocation to the nucleus following PMA stimulation (5 min), which was reduced to nearly background levels in the presence of FHL2 overexpression (Fig. 5B). An even more dramatic effect was observed when a phospho-specific ERK1/2 antibody was used, since nearly all the ERK1/2 that translocates to the nucleus is phosphorylated (this reduces the background of the immunocytochemical approach). Here, both PMA and the hypertrophic agonist PE induced a robust phospho-ERK signal in the nucleus of ERK2-overexpressing cardiomyocytes (Fig. 5C). Overexpression of FHL2 did not block the ability of ERK2 to be phosphorylated in cardiomyocytes following PMA and PE stimulation, but it did dramatically attenuate the nuclear localization of phosphorylated ERK2 (Fig. 5C). As a control, the MEK1 inhibitor U0126 also reduced ERK2 phosphorylation and the percentage of cells showing a nuclear signal (Fig. 5C). These results suggest that FHL2 normally functions to partially restrain ERK2 translocation to the nucleus following agonist stimulation in cardiomyocytes.
FIG. 5.
FHL2 inhibits ERK translocation and nuclear activity. (A) Immunocytochemistry for ERK1/2 (pan ERK antibody) from cultured neonatal cardiomyocytes infected with the indicated adenoviruses at baseline or after PMA stimulation. The arrowheads show cells with predominantly cytoplasmic ERK protein, and the full arrows show cells with significant nuclear ERK protein accumulation. (B) Quantitation of the percentage of cells with nuclear ERK (n ≈ 200 cells each). (C) Immunocytochemistry with a phospho-ERK1/2 specific antibody from cardiomyocytes infected with Adβgal (control) or AdFHL2 and stimulated with the indicated agonist or the MEK1 inhibitor U0126. The arrowheads show cells with predominantly cytoplasmic ERK protein, and the full arrows show cells with significant nuclear ERK protein. (D) Western analysis of phosphorylated GATA4 at serine 105 or total GATA4 from cardiomyocyte protein extracts that were previously infected with AdGATA4 (all 4 lanes) and/or AdFHL2 (lanes 3 and 4) and stimulated with PE (lanes 2 and 3) at 50 μM for 16 h. +, present; −, absent.
To more directly examine the functional ramifications of inhibited ERK1/2 nuclear translocation in the presence of FHL2, and to better understand its potential biologic significance, the phosphorylation of GATA4 at serine 105 was directly monitored following agonist stimulation in cultured cardiomyocytes. Cardiomyocytes were first infected with AdGATA4 as a means of increasing the signal (sensitivity) of the GATA4-phospho-105 antiserum used for Western blotting (Fig. 5D). PE stimulation augmented GATA4 phosphorylation at serine 105 (lane 2), which was reduced by coinfection with AdFHL2 (lane 3). Also of note, overexpression of FHL2 alone without agonist stimulation reduced basal GATA4 phosphorylation at serine 105, presumably because basal levels of phosphorylation are present (compare lanes 1 and 4), whereas total GATA4 protein levels did not change (Fig. 5D). These results further support the proposed role for FHL2 as an inhibitor of ERK1/2 nuclear translocation and they suggest that FHL2 may limit ERK-dependent transcriptional activity.
FHL2 inhibits ERK-dependent transcriptional responsiveness.
The results presented above suggest that FHL2 might function, in part, as a repressor of ERK-induced gene expression in cardiomyocytes. To investigate this potential mechanism of action, an ERK-dependent transcriptional assay was performed in the presence or absence of FHL2. The transcriptional regulatory factor ELK-1 is a classic target for ERK-mediated alterations in gene expression and, hence, was examined first (34). As previously reported, cotransfection of Gal4ELK-1 with activated MEK1 dramatically induced Gal4-dependent luciferase reporter activity (Fig. 6A). However, cotransfection of an FHL2-encoding expression vector produced a dose-dependent inhibition of reporter activity in cardiomyocytes (Fig. 6A). As a control, Western blotting was performed for protein content of MEK1, FHL2, and Gal4ELK-1 from the transfected cells, indicating that the observed effect was not due to a secondary alteration in protein expression (Fig. 6A). These results indicate that FHL2 can repress an ERK-mediated effector function associated with gene expression.
FIG. 6.
FHL2 inhibits ERK-mediated transcriptional activation. (A) A Gal4-dependent luciferase reporter construct (1.0 μg) was cotransfected into cardiomyocytes with expression vectors encoding Gal4DBD-ELK-1 fusion (0.25 μg), activated MEK1 (0.25 μg), and/or FHL2 (0.1, 0.25, or 0.5 μg). (Bottom) Control Western blots were performed for protein levels of MEK1, FHL2, and Gal4ELK-1 from the aligned transfection reactions. (B) A GATA site-dependent luciferase reporter construct (1.0 μg) was cotransfected into cardiomyocytes with expression vectors encoding activated MEK1 (0.25 μg) and/or FHL2. (C) An ANF promoter (luciferase) construct (1.0 μg) was cotransfected in cardiomyocytes with expression vectors encoding activated MEK1 (0.25 μg) and/or FHL2. (D) Cotransfection experiment with the Gal4-luciferase reporter (1 μg) and a construct encoding Gal4cJun and/or FHL2 (0.25 μg) at baseline or after serum stimulation in cardiomyocytes. (E) Cotransfection experiment with the Gal4-luciferase reporter and constructs encoding Gal4DBD-ELK-1 (0.25 μg), MEK1 (0.25 μg), RhoA-V14 (0.25 μg), and FHL2 (0.50 μg). (F) Cotransfection experiments with a MEF2-dependent luciferase reporter construct (1.0 μg) and expression vectors encoding ERK5 (0.25 μg), MEK5 (0.25 μg), and FHL2 (0.5 μg). All data in each panel are expressed as relative light units (RLUs) per microgram of protein (n = 3 independent experiments). +, present; −, absent.
We have previously shown that MEK1-ERK1/2 signaling also enhances the transcriptional potency of the cardiac-expressed transcription factor GATA4 by direct phosphorylation at serine 105. As previously reported, MEK1-ERK1/2 signaling significantly augmented GATA transcriptional activity (19), which was inhibited by cotransfection of an FHL2-encoding expression vector in cardiomyocytes (Fig. 6B). These results extend the observations made with the Gal4ELK-1 reporter assay and together suggest that FHL2 can inhibit ERK-dependent transcriptional activity.
Finally, the observation that FHL2 inhibits ERK-dependent transcriptional responsiveness within the context of two synthetic reporter constructs was extended to a more physiological context through the use of the ANF promoter. The ANF promoter gene is up-regulated in response to hypertrophic stimuli such as agonist stimulation, cell stretching, or MEK1-ERK1/2 stimulation (1, 4). Here we observed that FHL2 overexpression significantly reduced MEK1-induced ANF promoter activity in cardiomyocytes (Fig. 6C). Collectively, these results indicate that FHL2 can antagonize the ability of MEK1-ERK1/2 to augment transcriptional responsiveness, suggesting a biologic function for the observed interaction between these two factors.
The observation that FHL2 functioned as a transcriptional repressor of ERK-sensitive gene expression is in contrast to other reports demonstrating a coactivator function for FHL2 (12, 13, 24, 25, 37). However, as previously reported, we observed that FHL2 cotransfection augmented expression of a cJun-dependent transcriptional assay (Fig. 6D), which is consistent with its reported ability to serve as a coactivator of AP-1 (24). Similarly, FHL2 also functioned as a coactivator of RhoA signaling (Fig. 6E), consistent with a previous report (26). Indeed, activated RhoA even blocked the repressor activity of FHL2 towards MEK1-dependent activation of Gal4ELK-1 (Fig. 6E). These latter two control experiments suggest that FHL2 can still function as an activator in conjunction with some cofactors and as a repressor with others.
Finally, we also evaluated whether FHL2 could serve as a transcriptional repressor of an ERK5-dependent transcriptional response through MEF2. ERK5 is activated by MEK5 and is partially related to ERK1/2 in domain structure (14). While MEK5 and ERK5 each significantly activated a MEF2-dependent reporter construct, neither was significantly affected by FHL2 cotransfection (Fig. 6F). These results suggest that FHL2 does not interact with ERK5 but is more restricted to the ERK1/2 subfamily.
FHL2 overexpression reduces MEK1-, GATA4-, and agonist-induced cardiomyocyte hypertrophy.
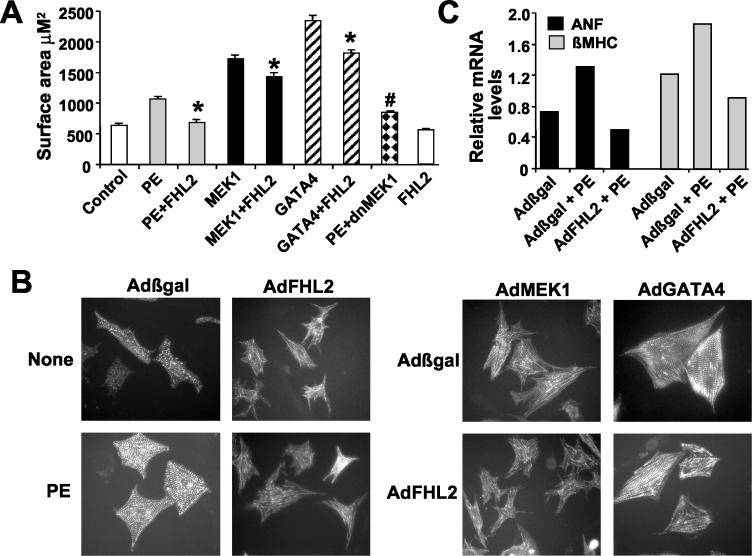
We have previously shown that MEK1-ERK1/2 signaling promotes cardiomyocyte hypertrophy in vitro and in vivo (4, 19). Similarly, the ability of ERK1/2 to activate GATA4 by direct phosphorylation is an important event in mediating a productive hypertrophic response (19). These prior results suggested that if FHL2 served as a biologically significant inhibitor of ERK1/2 signaling, overexpression of FHL2 might diminish the cardiac hypertrophic response to these upstream stimuli. To this end, cultured neonatal myocytes were infected with a prohypertrophic adenovirus expressing activated MEK1 alone or in combination with AdFHL2. Alternatively, the ability of PE stimulation to promote myocyte hypertrophy was compared between control Adβgal- and AdFHL2-infected cardiomyocytes. Finally, the ability of AdFHL2 to antagonize AdGATA4-mediated cardiomyocyte hypertrophy was also investigated. As a control, a dnMEK1-encoding adenovirus was also employed. The data demonstrate that AdFHL2 significantly reduced MEK1-, GATA4-, and PE-induced cardiomyocyte hypertrophy as assessed quantitatively by cell surface area measurement (Fig. 7A) and qualitatively by the relative degree of sarcomeric organization (Fig. 7B). AdFHL2 overexpression also diminished expression of the hypertrophy-associated genes ANF and β-MHC induced by PE stimulation (Fig. 7C). Collectively, these results provide additional support for the proposed role of FHL2 as a general repressor of ERK transcriptional coupling in neonatal cardiomyocytes.
FIG. 7.
FHL2 overexpression reduces cardiomyocyte hypertrophy due to MEK1, GATA4, and PE agonist stimulation. (A) Measurement of cell surface area of neonatal cardiomyocytes infected with the indicated adenoviruses in serum-free media for 48 h or stimulated with PE (50 μM) for the same period of time (n = 3 independent experiments, at least 200 cells measured in each experiment). *, P < 0.05 versus without AdFHL2; #, P < 0.05 versus PE. (B) Representative immunocytochemical analysis of α-actin-stained neonatal cardiomyocytes treated as indicated. (C) mRNA quantitation of ANF and β-MHC levels from cultured cardiomyocytes infected with the indicated adenovirus and stimulated with PE for 48 h.
DISCUSSION
Here we investigated the mechanisms whereby MEK1-ERK1/2 signaling might influence the growth response of mammalian cells by attempting to identify novel downstream effectors of ERK2 by yeast two-hybrid screening. While relatively few downstream effectors of MEK1-ERK1/2 signaling have been extensively characterized, previous work has suggested a few potential mechanisms whereby this pathway might regulate the growth response of cardiac myocytes. For example, ERK activation has been associated with p70 S6 kinase-2 activation in the regulation of protein translation and protein accumulation during the hypertrophic response (36). ERKs were also recently shown to regulate rRNA transcription by directly phosphorylating the transcription factor upstream binding factor (31). MEK1-ERK1/2 signaling has also been implicated in regulating the transcription of polymerase II-associated genes through direct phosphorylation of the transcriptional effectors Elk-1, Ets1, Sap1a, c-Myc, and STAT factors (reviewed in reference 14). ERK1/2 signaling has been associated with phosphorylation and activation of the cardiac-enriched transcription factor GATA4, which itself serves as an important mediator of the hypertrophic growth response (19, 22, 23). Finally, ERK1/2 appears to directly phosphorylate the N terminus of p300 and the C terminus of CBP to directly enhance the transcriptional potency of these coactivator proteins (15). Collectively, these observations support an emerging consensus whereby MEK1-ERK1/2 signaling plays a pivotal role in regulating the cardiomyocyte growth response through diverse downstream effectors.
Given the ability of ERK1/2 to mediate the hypertrophic growth response through direct phosphorylation of transcriptional regulatory proteins, the identification of FHL2 as an ERK2-interacting factor in the heart initially suggested a relatively straightforward hypothesis whereby the coactivator-like ability of FHL2 would be directly augmented through ERK-mediated phosphorylation. This initial hypothesis was also based on a number of previous publications showing that FHL2 functions as an important transcriptional coactivator when present in the nucleus. For example, FHL2 was shown to coactivate the androgen receptor, AP-1, β-catenin, CREB, CREM, and the transcriptional response downstream of wnt signaling (12, 13, 20, 24, 25, 37). However, our analysis showed that ERK2 did not directly phosphorylate FHL2, as is classically observed for positive transcriptional effectors of the MAPK pathway, nor did FHL2 augment the transcriptional potency of ERK. In dramatic contrast, FHL2 specifically attenuated the ability of ERK to function in a transcriptional activating role. This inhibitory activity was observed with both a GATA4- and ELK-1-dependent transcriptional reporter, each of which is specifically and potently activated by MEK1-ERK1/2 signaling. MEK1-ERK1/2 signaling also mediates the hypertrophic responsiveness of the ANF promoter (1), which was similarly blocked by FHL2. Collectively, these observations indicate that FHL2 serves as a specific inhibitor of MEK1-ERK1/2 signaling, consistent with the observation that FHL2 can also function as a transcriptional corepressor of the promyelocytic leukemia zinc finger protein and β-catenin (20, 21).
One likely mechanism whereby FHL2 serves as a corepressor is through diminishing the translocation of ERK into the nucleus or possibly by enhancing its export. To this end, overexpression of FHL2 significantly reduced ERK-mediated phosphorylation of GATA4 at serine 105 in the nucleus. FHL2 overexpression also consistently reduced the amount of ERK1/2 protein found within the nucleus following acute stimulation in cardiomyocytes. These results are somewhat intriguing given that FHL2 itself is partitioned between both the nucleus and cytoplasm of neonatal cardiomyocytes, although more is found in the cytoplasm (Fig. 5A). However, it is possible that the cytoplasmic pool of ERK, which is mostly associated with FHL2 at the level of the Z-line, is more difficult to mobilize than the ERK protein that may be associated with FHL2 in the nucleus. Alternatively, we cannot exclude the possibility that FHL2 within the nucleus might also directly inhibit the ability of ERK to interact with and phosphorylate select transcription factors or coactivators.
Finally, our results do not directly address the known ability of FHL2 to function as a coactivator of gene expression, independent of ERK1/2 signaling. For example, activation of Rho signaling in NIH 3T3 fibroblasts causes translocation of FHL2 into the nucleus where it functions as a transcriptional coactivator of the androgen receptor (26). Given these and other observations, it is likely that FHL2 can play dichotomous functions in regulating cardiomyocyte gene expression. FHL2 may serve as a coactivator of gene expression through specific interactions with discrete transcription factors, or it may serve an inhibitory role through its ability to diminish the effectiveness of ERK-mediated transcriptional coupling. Indeed, as will be discussed below, FHL2 gene-targeted mice show enhanced cardiac hypertrophic growth to catecholamine stimulation, suggesting a net repressor function for FHL2. Yet, pressure overload-induced hypertrophy was not enhanced in FHL2-null mice, suggesting that the repressor-like functions of FHL2 might be counterbalanced by coactivator-like functions. In our present study, we observed that FHL2 overexpression only partially inhibited the hypertrophic growth response of cultured neonatal cardiomyocytes, suggesting an interplay between both positive and negative regulatory effects. However, the fact that significant inhibition of cardiomyocyte growth was observed suggests that FHL2's ability to antagonize ERK-dependent transcriptional responsiveness may predominate over any simultaneous coactivator functions in cultured cardiomyocytes.
The gene encoding FHL2 has been disrupted by homologous recombination in the mouse (7, 17). FHL2 gene-targeted mice are viable, most likely due to redundancy with multiple other LIM-only family members that are also expressed in the heart, such as FHL1 and FHL3 (8, 30). If FHL2 truly functions as a negative regulator of MEK1-ERK1/2 signaling in the heart, one might predict that FHL2-null mice would have greater ERK activity following stimulation so that the hypertrophic growth response is enhanced. Indeed, FHL2-null mice were shown to have greater cardiac hypertrophic growth following catecholamine infusion (17). However, when FHL2-null mice were subjected to pressure overload stimulation by aortic banding, a hypertrophic response was observed that was indistinguishable from that of wild-type controls (7). The discrepancy between these two studies is potentially due to the nature of the stimulus, such that pressure overload produces a more global response which recruits diverse hypertrophic signaling pathways, although catecholamine infusion may evoke a more unitary response that is dependent on MEK1-ERK1/2 signaling. Further analysis of the cardiac hypertrophic potential of FHL2 gene-targeted mice is needed. Indeed, it would be interesting to cross MEK1 transgenic mice with FHL2-null mice as a means of more selectively assessing the functional role of this interaction in vivo. Alternatively, it may be necessary to generate and analyze gene-targeted mice for other members of the LIM-only family to more rigorously evaluate its coactivator properties versus its signaling inhibitory properties apart from potential compensatory effects.
Acknowledgments
This work was supported by the National Institute of Health and a Pew charitable trust Scholar Award to J.D.M. J.D.M. is also an Established Investigator of the American Heart Association. N.H.P was supported by a National Institutes of Health Individual Research Service Award F32 HL71427. O.F.B. was supported by a National Institutes of Health Individual Research Service Award F32 HL10336.
We thank Allen York for excellent technical assistance.
REFERENCES
- 1.Aoki, H., M. Richmond, S. Izumo, and J. Sadoshima. 2000. Specific role of the extracellular signal-regulated kinase pathway in angiotensin II-induced cardiac hypertrophy in vitro. Biochem. J. 347:275-284. [PMC free article] [PubMed] [Google Scholar]
- 2.Bogoyevitch, M. A., P. E. Glennon, M. B. Andersson, A. Clerk, A. Lazou, C. J. Marshall, P. J. Parker, and P. H. Sugden. 1994. Endothelin-1 and fibroblast growth factors stimulate the mitogen-activated protein kinase signaling cascade in cardiac myocytes. The potential role of the cascade in the integration of two signaling pathways leading to myocyte hypertrophy. J. Biol. Chem. 269:1110-1119. [PubMed] [Google Scholar]
- 3.Bueno, O. F., and J. D. Molkentin. 2002. Involvement of extracellular signal-regulated kinases 1/2 in cardiac hypertrophy and cell death. Circ. Res. 91:776-781. [DOI] [PubMed] [Google Scholar]
- 4.Bueno, O. F., L. J. De Windt, K. M. Tymitz, S. A. Witt, T. R. Kimball, R. Klevitsky, T. E. Hewett, S. P. Jones, D. J. Lefer, C. F. Peng, R. N. Kitsis, and J. D. Molkentin. 2000. The MEK1-ERK1/2 signaling pathway promotes compensated cardiac hypertrophy in transgenic mice. EMBO J. 19:6341-6350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cambon, N., and M. A. Sussman. 1997. Isolation and preparation of single mouse cardiomyocytes for confocal microscopy. Methods Cell Sci. 19:83-90. [Google Scholar]
- 6.Chan, K. K., S. K. Tsui, S. M. Lee, S. C. Luk, C. C. Liew, K. P. Fung, M. M. Waye, and C. Y. Lee. 1998. Molecular cloning and characterization of FHL2, a novel LIM domain protein preferentially expressed in human heart. Gene 210:345-350. [DOI] [PubMed] [Google Scholar]
- 7.Chu, P. H., W. M. Bardwell, Y. Gu, J. Ross, Jr., and J. Chen. 2000. FHL2 (SLIM3) is not essential for cardiac development and function. Mol. Cell. Biol. 20:7460-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu, P. H., P. Ruiz-Lozano, Q. Zhou, C. Cai, and J. Chen. 2000. Expression patterns of FHL/SLIM family members suggest important functional roles in skeletal muscle and cardiovascular system. Mech. Dev. 95:259-265. [DOI] [PubMed] [Google Scholar]
- 9.Clerk, A., M. A. Bogoyevitch, M. B. Anderson, and P. H. Sugden. 1994. Differential activation of protein kinase C isoforms by endothelin-1 and phenylephrine and subsequent stimulation of p42 and p44 mitogen-activated protein kinases in ventricular myocytes cultured from neonatal rat hearts. J. Biol. Chem. 269:32848-32857. [PubMed] [Google Scholar]
- 10.Dai, Y. S., P. Cserjesi, B. E. Markham, and J. D. Molkentin. 2002. The transcription factors GATA4 and dHAND physically interact to synergistically activate cardiac gene expression through a p300-dependent mechanism. J. Biol. Chem. 277:24390-24398. [DOI] [PubMed] [Google Scholar]
- 11.De Windt, L. J., H. W. Lim, S. Haq, T. Force, and J. D. Molkentin. 2000. Calcineurin promotes protein kinase C and c-Jun NH2-terminal kinase activation in the heart. Cross-talk between cardiac hypertrophic signaling pathways. J. Biol. Chem. 275:13571-13579. [DOI] [PubMed] [Google Scholar]
- 12.Du, X., P. Hublitz, T. Gunther, D. Wilhem, C. Englert, and R. Schüle. 2002. The LIM-only coactivator FHL2 modulates WT1 transcriptional activity during gonadal differentiation. Biochem. Biophys. Acta 19:93-101. [DOI] [PubMed] [Google Scholar]
- 13.Fimia, G. M., D. De Cesare, and P. Sassone-Corsi. 2000. A family of LIM-only transcriptional coactivators: tissue-specific expression and selective activation of CREB and CREM. Mol. Cell. Biol. 20:8613-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrington, T. P., and G. L. Johnson. 1999. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr. Opin. Cell Biol. 11:211-218. [DOI] [PubMed] [Google Scholar]
- 15.Gusterson, R., B. Brar, D. Faulkes, A. Giordano, J. Chrivia, and D. Latchman. 2002. The transcriptional co-activators CBP and p300 are activated via phenylephrine through the p42/p44 MAPK cascade. J. Biol. Chem. 277:2517-2524. [DOI] [PubMed] [Google Scholar]
- 16.Knowlton, K. U., E. Baracchini, R. S. Ross, A. N. Harris, S. A. Henderson, S. M. Evans, C. C. Glembotski, and K. R. Chien. 1991. Co-regulation of the atrial natriuretic factor and cardiac myosin light chain-2 genes during alpha-adrenergic stimulation of neonatal rat ventricular cells. Indentification of cis sequences within an embryonic and a constitutive contractile protein gene which mediate inducible expression. J. Biol. Chem. 266:7759-7768. [PubMed] [Google Scholar]
- 17.Kong, Y., J. M. Shelton, B. Rothermel, X. Li, J. A. Richardson, R. Bassel-Duby, and R. S. Williams. 2001. Cardiac-specific LIM protein FHL2 modifies the hypertrophic response to β-Adrenergic stimulation. Circulation 103:2731-2738. [DOI] [PubMed] [Google Scholar]
- 18.Lai, J. M., S. Wu, D. Y. Huang, and Z. F. Chang. 2002. Cytosolic retention of phosphorylated extracellular signal-regulated kinase and a Rho-associated kinase-mediated signal impair expression of p21Cip1/Waf1 in phorbol 12-myristate-13-acetate-induced apoptotic cells. Mol. Cell. Biol. 22:7581-7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang, Q., R. J. Wiese, O. F. Bueno, Y. S. Dai, B. E. Markham, and J. D. Molkentin. 2001. The transcription factor GATA4 is activated by extracellular signal-regulated kinase 1- and 2-mediated phosphorylation of serine 105 in cardiomyocytes. Mol. Cell. Biol. 21:7460-7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin, B., R. Schneider, S. Janetzky, Z. Waibler, P. Pandur, M. Kühl, J. Behrens, K. von der Mark, A. Starzinski-Powitz, and V. Wixler. 2002. The LIM-only protein FHL2 interacts with β-catenin and promotes differentiation of mouse myoblasts. J. Cell Biol. 159:113-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McLoughlin, P., E. Ehler, G. Carlile, J. D. Licht, and B. W. Schäfer. 2002. The LIM-only protein DRAL/FHL2 interacts with and is a corepressor for the promyelocytic leukemia zinc finger protein. J. Biol. Chem. 277:37045-37053. [DOI] [PubMed] [Google Scholar]
- 22.Molkentin, J. D. 2000. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. 275:38949-38952. [DOI] [PubMed] [Google Scholar]
- 23.Morimoto, T., K. Hasegawa, S. Kaburagi, T. Kakita, H. Wada, T. Yanazume, and S. Sasayama. 2000. Phosphorylation of GATA-4 is involved in alpha 1-adrenergic agonist-responsive transcription of the endothelin-1 gene in cardiac myocytes. J. Biol. Chem. 275:13721-13726. [DOI] [PubMed] [Google Scholar]
- 24.Morolon, A., and P. Sassone-Corsi. 2003. The LIM-only protein FHL2 is a serum-inducible transcriptional coactivator of AP-1. Proc. Natl. Acad. Sci. USA 100:3977-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller, J. M., U. Isele, E. Metzger, A. Rempel, M. Moser, A. Pscherer, T. Breyer, C. Holubarsch, R. Buettner, and R. Schüle. 2000. FHL2, a novel tissue-specific coactivator of the androgen receptor. EMBO J. 19:359-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller, J. M., E. Metzger, H. Greshik, A.-K. Bosserhoff, L. Mercep, R. Buettner, and R. Schüle. 2002. The transcriptional coactivator FHL2 transmits Rho signals from the cell membrane into the nucleus. EMBO J. 21:736-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng, E. K., K. K. Chan, C. H. Wong, S. K. Tsui, S. M. Ngai, S. M. Lee, M. Kotaka, C. Y. Lee, M. M. Waye, and K. P. Fung. 2002. Interaction of the heart-specific LIM domain protein, FHL2 with DNA-binding nuclear protein hNP220. J. Cell. Biochem. 84:556-566. [PubMed] [Google Scholar]
- 28.Post, G. R., D. Goldstein, D. J. Thuerauf, C. C. Glembotski, and J. H. Brown. 1996. Dissociation of p44 and p42 mitogen-activated protein kinase activation from receptor-induced hypertrophy in neonatal rat ventricular myocytes. J. Biol. Chem. 271:8452-8457. [DOI] [PubMed] [Google Scholar]
- 29.Rapacciuolo, A., G. Esposito, K. Caron, L. Mao, S. A. Thomas, and H. A. Rockman. 2001. Important role of endogenous norepinephrine and epinephrine in the development of in vivo pressure-overload cardiac hypertrophy. J. Am. Coll. Cardiol. 38:876-882. [DOI] [PubMed] [Google Scholar]
- 30.Retaux, S., and I. Bachy. 2002. A short history of LIM domain (1993-2002): from protein interaction to degradation. Mol. Neurobiol. 26:269-281. [DOI] [PubMed] [Google Scholar]
- 31.Stefanovsky, V. Y., G. Pelletier, R. Hannan, T. Gagnon-Kugler, L. I. Rothblum, and T. Moss. 2001. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell 8:1063-1073. [DOI] [PubMed] [Google Scholar]
- 32.Sugden, P. H., and A. Clerk. 1998. “Stress-responsive” mitogen-activated protein kinases (c-Jun N-terminal kinases and p38 mitogen-activated protein kinases) in the myocardium. Circ. Res. 24:345-352. [DOI] [PubMed] [Google Scholar]
- 33.Takeishi, Y., Q. Huang, J. I. Abe, M. Glassman, W. Che, J. D. Lee, H. Kawakatsu, E. G. Lawrence, B. D. Hoit, B. C. Berk, and R. A. Walsh. 2001. Src and multiple map kinase activation in cardiac hypertrophy and congestive heart failure under chronic pressure-overload: comparison with acute mechanical stretch. J. Mol. Cell. Cardiol. 33:1637-1648. [DOI] [PubMed] [Google Scholar]
- 34.Tian, J., and M. Karin. 1999. Stimulation of Elk1 transcriptional activity by mitogen-activated protein kinases is negatively regulated by protein phosphatase 2B (calcineurin). J. Biol. Chem. 274:15173-15180. [DOI] [PubMed] [Google Scholar]
- 35.Ueyama, T., S. Kawashima, T. Sakoda, Y. Rikitake, T. Ishida, M. Kawai, T. Yamashita, S. Ishido, H. Hotta, and M. Yokoyama. 2000. Requirement of activation of the extracellular signal-regulated kinase cascade in myocardial cell hypertrophy. J. Mol. Cell. Cardiol. 32:947-960. [DOI] [PubMed] [Google Scholar]
- 36.Wang, L., I. Gout, and C. G. Proud. 2001. Cross-talk between the ERK and p70 S6 kinase (S6K) signaling pathways. MEK-dependent activation of S6K2 in cardiomyocytes. J. Biol. Chem. 276:32670-32677. [DOI] [PubMed] [Google Scholar]
- 37.Wei, Y., C.-A. Renard, C. Labalette, Y. Wu, L. Levy, C. Neuvent, S. Prieur, and M.-A. Buendia. 2003. Identification of the LIM protein FHL2 as a coactivator of β-catenin. J. Biol. Chem. 278:5188-5194. [DOI] [PubMed] [Google Scholar]
- 38.Widmann, C., S. Gibson, M. B. Jarpe, and G. L. Johnson. 1999. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79:143-180. [DOI] [PubMed] [Google Scholar]
- 39.Yamazaki, T., K. Tobe, E. Hoh, K. Maemura, T. Kaida, I. Komuro, H. Tamemoto, T. Kadowaki, R. Nagai, and Y. Yazaki. 1993. Mechanical loading activates mitogen-activated protein kinase and S6 peptide kinase in cultured rat cardiac myocytes. J. Biol. Chem. 268:12069-12076. [PubMed] [Google Scholar]
- 40.Yue, T. L., J. L. Gu, C. Wang, A. D. Reith, J. C. Lee, R. C. Mirabile, R. Kreutz, Y. Wang, B. Maleeff, A. A. Parsons, and E. H. Ohlstein. 2000. Extracellular signal-regulated kinase plays an essential role in hypertrophic agonists, endothelin-1 and phenylephrine-induced cardiomyocyte hypertrophy. J. Biol. Chem. 275:37895-37901. [DOI] [PubMed] [Google Scholar]
- 41.Zou, Y., I. Komuro, T. Yamazaki, R. Aikawa, S. Kudoh, I. Shiojima, Y. Hiroi, T. Mizuno, and Y. Yazaki. 1996. Protein kinase C, but not tyrosine kinases or Ras, plays a critical role in angiotensin II-induced activation of Raf-1 kinase and extracellular signal-regulated protein kinases in cardiac myocytes. J. Biol. Chem. 271:33592-33597. [DOI] [PubMed] [Google Scholar]