Abstract
Metazoan NXF1-p15 heterodimers promote the nuclear export of bulk mRNA across nuclear pore complexes (NPCs). In vitro, NXF1-p15 forms a stable complex with the nucleoporin RanBP2/Nup358, a component of the cytoplasmic filaments of the NPC, suggesting a role for this nucleoporin in mRNA export. We show that depletion of RanBP2 from Drosophila cells inhibits proliferation and mRNA export. Concomitantly, the localization of NXF1 at the NPC is strongly reduced and a significant fraction of this normally nuclear protein is detected in the cytoplasm. Under the same conditions, the steady-state subcellular localization of other nuclear or cytoplasmic proteins and CRM1-mediated protein export are not detectably affected, indicating that the release of NXF1 into the cytoplasm and the inhibition of mRNA export are not due to a general defect in NPC function. The specific role of RanBP2 in the recruitment of NXF1 to the NPC is highlighted by the observation that depletion of CAN/Nup214 also inhibits cell proliferation and mRNA export but does not affect NXF1 localization. Our results indicate that RanBP2 provides a major binding site for NXF1 at the cytoplasmic filaments of the NPC, thereby restricting its diffusion in the cytoplasm after NPC translocation. In RanBP2-depleted cells, NXF1 diffuses freely through the cytoplasm. Consequently, the nuclear levels of the protein decrease and export of bulk mRNA is impaired.
Bidirectional macromolecular traffic between the nucleus and the cytoplasm is mediated by soluble transport receptors that shuttle through nuclear pore complexes (NPCs), large protein assemblies that form aqueous channels across the nuclear envelope. The three-dimensional architecture of the NPC is conserved and consists of three structural units. A ring-like central framework that embraces the central channel of the pore is positioned between two peripheral cytoplasmic and nucleoplasmic structures, the cytoplasmic ring from which eight cytoplasmic filaments emanate and the nuclear ring that anchors the nuclear basket (reviewed in references 24, 25, and 32).
The structural units of the NPC are composed of multiple copies of about 30 different polypeptides called nucleoporins. These proteins often contain clusters of phenylalanine-glycine (FG) dipeptide repeats (reviewed in references 25 and 32). The FG domains of nucleoporins interact with nuclear transport receptors, providing binding sites during translocation of receptor-cargo complexes through the central channel of the pore (references 22 and 23 and references therein).
Immunoelectron microscopy studies have shown that while most nucleoporins are detected on both sides of the central channel of the NPC, some are asymmetrically localized at either the nuclear or the cytoplasmic face of the pore. Among these are the components of the nuclear basket or the cytoplasmic filaments (reviewed in references 25 and 32). In vertebrates, a major component of the cytoplasmic filaments is the nucleoporin RanBP2 (also known as Nup358) (34, 36-38). Another nucleoporin localized to the cytoplasmic face of vertebrate NPCs is CAN (also known as Nup214) (18, 34). It has been suggested that RanBP2, CAN, and the additional asymmetrically localized nucleoporins act as platforms for the assembly or dissociation of receptor-cargo complexes before or after translocation through the central channel of the NPC (reviewed in references 24, 25, and 32).
The vast majority of nuclear transport receptors belong to the conserved family of RanGTP binding proteins called importins and exportins or karyopherins (reviewed in reference 5). However, NPC translocation can also be mediated by receptors that are structurally unrelated to the karyopherins. In particular, nuclear import of the GTPase Ran is facilitated by NTF2 homodimers, whereas export of bulk mRNA to the cytoplasm is mediated by a heterodimeric export receptor (NXF1-p15), which is related to NTF2 (reviewed in reference 5). The larger subunit of this heterodimeric receptor belongs to the conserved family of NXF proteins, which includes yeast Mex67p and metazoan NXF1.
NXF1 binds to FG-nucleoporin repeats via two distinct structural domains, the NTF2-like scaffold and the UBA-like domain, connected by a flexible linker. The NTF2-like scaffold results from the heterodimerization of the NTF2-like domain of NXF1 with p15 and has an overall structure similar to that of the NTF2 homodimer (11). The UBA-like domain is structurally related to ubiquitin-associated (UBA) domains (13). The NTF2-like scaffold and the UBA-like domain each feature a single hydrophobic pocket for the interaction with a phenylalanine of the nucleoporin FG-repeats, and both are required to promote translocation of mRNA export cargoes across the central channel of the NPC (3, 4, 11, 13, 20, 35).
NXF1 is a shuttling protein that localizes at steady state within the nucleoplasm and at the NPC (1, 2, 17). Nuclear import of human NXF1 is mediated by an N-terminal nuclear localization signal recognized by transportin-1 (also known as karyopherin β2A) (1, 30). Mutations that prevent binding of transportin-1 lead to the distribution of NXF1 throughout the nucleus and cytoplasm, indicating that nuclear import is impaired but not abolished (1, 2, 17). Similarly, the C-terminal half of NXF1 (comprising the NTF2-like scaffold and the UBA-like domain but not the nuclear localization signal) distributes between the nucleus and cytoplasm in human cells (1). Furthermore, the UBA-like domain, on its own, translocates across the NPC in both directions (1, 16, 27). Together, these observations indicate that NXF1 translocates through the NPC in both directions via interactions of its NPC binding domain with nucleoporins. Binding to transportin-1 at the cytoplasmic side of the NPC favors import, so that the protein is predominantly nucleoplasmic at steady state. Although it has been suggested that export of NXF1 would be facilitated by transportin-2 (also known as karyopherin β2B) (29), others have reported that transportin-2 acts as nuclear import receptor with similar specificity to that of transportin-1 (Ulrike Kutay, personal communication).
It has been shown before that NXF1-p15 dimers interact with RanBP2 both in HeLa and Drosophila melanogaster nuclear extracts (1, 9), suggesting a role for this nucleoporin in mRNA export. In this study we investigated, with Drosophila Schneider cells (S2 cells), the possible role of RanBP2 in the export of bulk mRNA. We show that RanBP2 is an essential nucleoporin that provides a major binding site for NXF1-p15 at the NPC. Partial depletion of RanBP2 leads to a significant decrease of the NPC-bound fraction of NXF1, an increase of its cytoplasmic levels and a partial inhibition of nuclear mRNA export. Under the same conditions, the steady-state subcellular localization of several nuclear or cytoplasmic proteins and CRM1-mediated protein export are not detectably affected. In particular, REF1 and Y14, two nuclear mRNA-binding proteins that interact with NXF1 and accompany cellular mRNAs to the cytoplasm (reviewed in reference 5), remained entirely nuclear in RanBP2-depleted cells. The release of NXF1 to the cytoplasm and the inhibition of mRNA export are therefore not the consequence of a general defect in NPC function, but rather they reveal a specific role for RanBP2 in the NXF1-mediated export pathway.
MATERIALS AND METHODS
Sequence analysis.
Human RanBP2 was used as a query sequence to search for orthologs in all completely sequenced eukaryotic genomes. All the orthologous sequences retrieved by homology searches except murine ranbp2 are predicted genes. Transcript borders were confirmed with GeneWise (http://www.sanger.ac.uk/Software/Wise2/) against the corresponding genomic region. The domain architecture of each protein was assigned with SMART (19). Since the domain organization varies considerably across species, it has been verified by scanning the genomic region of each protein with hidden Markov model profiles corresponding to the RanBD domain, the zinc finger motifs, and the domain homologous to cyclophilin A. The phylogenetic tree was built by using a bootstrap method (8) taking the alignment of the most conserved region of all the proteins (from the first to the second RanBD of the human RanBP2).
Cell culture and dsRNA interference.
RanBP2 and CAN cDNA fragments (corresponding to nucleotides 1 to 700 of the respective coding regions) were amplified by PCR from a random-primed S2 cDNA library. Primers were designed according to the predicted cDNAs. The identities of the amplified products were confirmed by sequencing. These cDNA fragments served as templates for the synthesis of double-stranded RNAs (dsRNAs). RNA interference (RNAi) was performed as described by Herold et al. (14).
FISH and indirect immunofluorescence.
Fluorescent in situ hybridization (FISH) with a Cy3-labeled oligo(dT) probe or with a digoxigenin-labeled hsp70 RNA probe was performed as described by Herold et al. (14). The nuclear envelope was stained with Alexa 488 wheat germ agglutinin (WGA) (1:1,500; Molecular Probes). Indirect immunofluorescence with specific antibodies was performed as described by Bachi et al. (1). Drosophila REF1 was detected with affinity purified anti-REF antibodies (KJ58, 1:10,000) and Cy5-coupled anti-rabbit secondary antibodies (1:300). Drosophila YPS was visualized with specific rat antisera and Cy5-coupled anti-rat secondary antibodies. TAP-tagged Drosophila Y14 and Drosophila PYM were detected with a polyclonal anti-protein A antibody (1:1,000; Sigma) and Cy5-coupled secondary anti-rabbit antibodies. Secondary antibodies were purchased from Jackson ImmunoResearch. Drosophila cells were mounted with fluoromount G (Southern Biotechnology Associates, Inc.). Images were taken with a confocal laser-scanning microscope (LSM 510; Zeiss).
Preparation of cell extracts for Western blotting and in vivo protein labeling.
Metabolic labeling and Western blot analysis were performed as described by Herold et al. (14). Rat antibodies raised against recombinant Drosophila NXF1 were described by Herold et al. (14). Mouse monoclonal antitubulin antibody E7 (Sigma) was diluted 1:10,000. Mouse monoclonal mAb414 (Babco) was diluted 1:3,000. Bound primary antibodies were detected with alkaline phosphatase-coupled anti-rat or anti-mouse (1:25,000) antibodies (Western Star kit from Tropix).
RNA isolation, RT-PCR, and Northern blots.
Total RNA was isolated with TRIzol reagent (Life Technologies). Isolation of cytoplasmic RNA, Northern blotting, and reverse transcription (RT)-PCR were performed as described by Herold et al. (14, 15).
Photobleaching experiments and image analysis.
Fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) were performed on a customized Zeiss LSM 510 essentially as described by Daigle et al. (6) with a 488-nm argon line for green fluorescent protein (GFP). To ensure reproducible bleaching frequencies, FLIP experiments were performed with a macro written in Visual BASIC for the LSM 2.8 SP2 software package (http://www.embl-heidelberg.de/ExternalInfo/ellenberg/homepage/macros.html). Average intensities from FRAP experiments were corrected for the total fluorescence lost during the photobleaching and the bleaching that occurred during scanning of recovery. Quantitative image analysis was performed with the LSM 2.8 SP2 software (Zeiss). Average intensities of regions of interest were exported to Microsoft Excel, the background was subtracted, and results were plotted. Other normalizations are described in the figure legends. For each experiment, at least 8 representative control or knockdown cells were measured. Each experiment was repeated in at least two independent knockdowns.
RESULTS
Purification of a multimeric complex involved in mRNA export by iTAP.
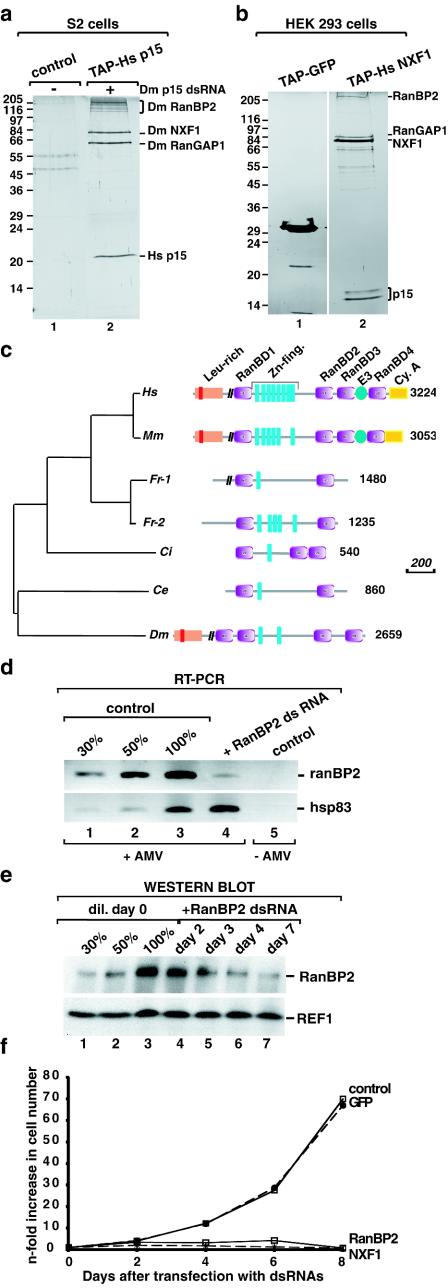
Using a combination of tandem affinity purification and RNAi (iTAP) in S2 cells, we have shown that human p15 (hsp15) fused to the tandem affinity purification (TAP) tag copurifies with three proteins: RanBP2, NXF1, and RanGAP1 (9). This tetrameric complex was purified from a polyclonal S2 cell line constitutively expressing TAP-tagged hp15, in which the expression of endogenous Drosophila p15 was depleted by RNAi. A typical purification is shown in Fig. 1a. When proteins bound to TAP-tagged hsp15 were purified from cells in which expression of RanBP2 was also silenced by RNAi, selection of RanGAP1 was strongly reduced, indicating that binding of NXF1-p15 dimers to RanBP2 was direct, whereas RanGAP1 was tethered to the complex via RanBP2 (data not shown). Similarly, TAP-tagged hsNXF1 transiently expressed in human embryonic kidney (HEK) 293 cells copurifies with RanBP2, RanGAP1, and hsp15 (Fig. 1b). Hence, this network of protein interactions is conserved.
FIG. 1.
RanBP2 forms a stable complex with NXF1-p15 dimers and is required for cell proliferation. (a) S2 cells expressing TAP-tagged hsp15 were depleted of endogenous Drosophila p15 by RNAi. Five days after addition of Drosophila p15 dsRNA, proteins bound to TAP-tagged hsp15 were purified and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Following silver staining, the selected proteins were identified by mass spectrometry as described by Forler et al. (9). Lane 1, proteins purified from lysates of cells expressing the TAP tag alone; lane 2, proteins bound to TAP-tagged hp15. (b) Proteins purified from lysates of HEK 293 cells transiently expressing TAP-tagged hsNXF1. Lane 1, proteins purified from lysates of cells expressing TAP-tagged GFP; lane 2, proteins bound to TAP-tagged hsNXF1. Proteins were analyzed as described for panel a. (c) Domain organization of RanBP2 orthologs. Abbreviations: Leu-rich, leucine-rich region with a TPR homology domain (in red); RanBD, RanBP1 homology domains; Zn-fing., zinc finger motifs; E3, SUMO1 E3 ligase activity; Cy. A, cyclophilin homologous region; Ce, C. elegans; Ci, C. intestinalis; Dm, D. melanogaster; Fr, F. rubripes; Hs, Homo sapiens; Mm, Mus musculus. The vertical black bars on the schematic representation of the proteins indicate that part of the sequence is not drawn to scale. Scale bar, 200 amino acids. (d) S2 cells growing in suspension were treated with dsRNAs specific for Drosophila RanBP2, Drosophila NXF1, and GFP. Aliquots of cells were collected on day 4 and analyzed by RT-PCR. The RanBP2 mRNA levels were reduced in cells treated with RanBP2 dsRNA (lane 4), whereas the levels of the unrelated hsp83 mRNA were not affected on day 4. In lanes 1 to 3, dilutions of the cDNA isolated from untreated cells were used in the PCR to determine the efficiency of the depletion. Lane 5 shows the control sample in which the reverse transcriptase was omitted. The PCR oligonucleotides amplified a fragment of RanBP2 coding sequence corresponding to nucleotides 1100 to 1370. (e) Cells from the same experiment as in panel d were analyzed by Western blotting with monoclonal antibody mAb414. In lanes 1 to 3, dilutions of the sample isolated on day 0 were loaded to assess the efficiency of the depletion. REF1 served as a loading control. (f) Cell numbers from the same experiment as in panel d were determined every 2 days up to 8 days after addition of dsRNAs. Results are given as the increase (n-fold) in cell numbers relative to the amount used for transfection on day 0.
Human RanBP2 contains an amino-terminal 700-residue leucine-rich region (a short part of which is similar to TPR repeats), four RanGTP binding domains related to the one in RanBP1 (RanBP1 homologous domains or RanBD), eight zinc finger motifs, and a carboxy terminus with homology to cyclophilin A (Fig. 1c). The protein contains several XFXFG repeats scattered along its C-terminal half (37, 38). We have identified orthologs in all the completely sequenced animal genomes but not in fungi (Fig. 1c). However, there are considerable differences in the domain architectures among the different species. The ortholog of RanBP2 in Caenorhabditis elegans and the two recently duplicated paralogs in Fugu rubipens are considerably shorter and lack structural domains at the N and C termini. The flexibility in the domain composition was also confirmed by the different number of zinc fingers in the human (8) and mouse (6) proteins. This difference is not due to alternative splicing patterns between the two species because the zinc finger variable region is encoded by a single exon (see also reference 7).
Strikingly, Drosophila RanBP2 architecture is more similar to the mammalian orthologs than to the other available chordate sequences (F. rubipens and Ciona intestinalis), which in turn resemble the C. elegans ortholog. A phylogenetic tree with the region shared between all sequences supports the expected taxonomy. It is thus likely that several independent domain losses occurred in F. rubipens, C. intestinalis, and C. elegans (Fig. 1c) but that domains have been gained by the mammalian genes. The additional domains in the mammalian proteins lead to novel functions, as in the case of the region of human RanBP2 which was recently shown to act as an E3 ligase in the sumoylation reaction (21).
In Drosophila cells, RanBP2 is an essential nucleoporin required for efficient export of bulk mRNA.
The presence of RanBP2 in a complex including NXF1-p15 dimers suggests a role for this protein in mRNA export. To address this possibility, we analyzed the effect of depleting RanBP2 on cell proliferation and export of bulk mRNA. S2 cells were treated with a dsRNA corresponding to the N terminus of Drosophila RanBP2. A dsRNA corresponding to GFP was used as a control. The efficiency of the depletion was tested by RT-PCR and Western blot analysis (Fig. 1d and e). Four days after the addition of RanBP2 dsRNA, the steady-state expression levels of the targeted mRNA were reduced below 30% of the levels detected in untreated cells (Fig. 1d, lane 4 versus lane 1), whereas the protein levels were below 50% of the wild-type levels (Fig. 1e, lane 6 versus lane 2).
Remarkably, cell proliferation was already inhibited 3 days after the addition of RanBP2 dsRNA (Fig. 1f). This inhibition was comparable to that observed when the essential mRNA export receptor NXF1 was depleted (Fig. 1f), despite the fact that levels of RanBP2 were reduced by less than 50% (Fig. 1e). This complete inhibition of cell proliferation indicates that the partial depletion of RanBP2 is not due to a low efficiency of the dsRNA but is likely to reflect a low turnover rate of the protein and the fact that the residual protein is not diluted over time because the cells stop dividing.
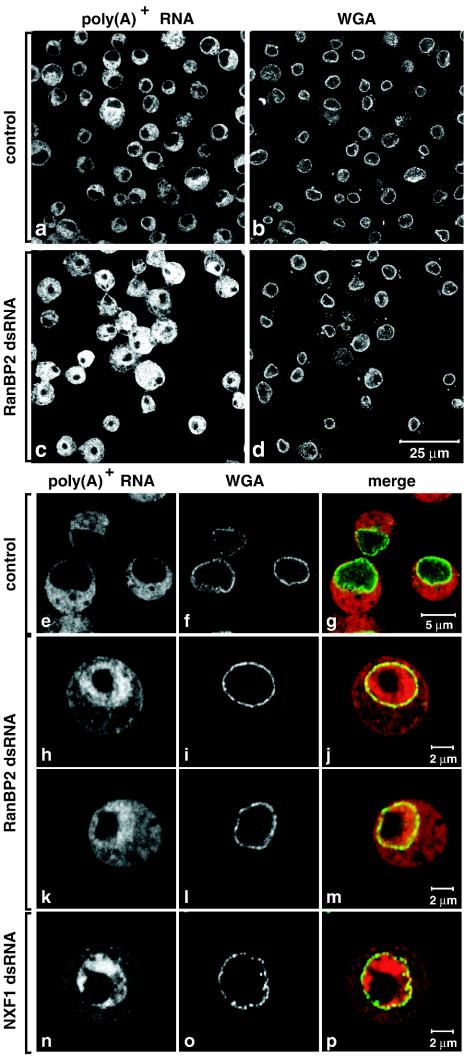
The intracellular distribution of bulk poly(A)+ RNA in control cells and in cells depleted of RanBP2 was investigated by FISH with an oligo(dT) probe. The NPCs were stained with fluorescently labeled WGA. In control cells the oligo(dT) signal was mainly cytoplasmic (Fig. 2a, b, and e to g). Following RanBP2 depletion a significant nuclear accumulation of poly(A)+ RNA was observed (Fig. 2c, d, and h to m). The oligo(dT) signal was widespread within the nucleoplasm but was excluded from the large nucleolus (Fig. 2h and k). The nuclear accumulation of poly(A)+ RNA correlated with RanBP2 depletion, as it was detected in about half of the cell population 3 days after the addition of RanBP2 dsRNA and in ca. 90% of the cell population on day 5, in several independent experiments (Fig. 2c and d and data not shown). This accumulation was, however, not as strong as that observed when NXF1 was depleted (Fig. 2h and k versus n).
FIG. 2.
Depletion of endogenous RanBP2 causes the accumulation of poly(A)+ RNA within the nucleus. (a to p) S2 cells were treated for 5 days with dsRNAs corresponding to GFP (control) Drosophila RanBP2 or Drosophila NXF1. poly(A)+ RNA was detected by FISH with an oligo(dT) probe (red). The nuclear envelope was stained with Alexa 488-WGA conjugates (green).
The partial inhibition of bulk mRNA export is likely to reflect the inefficient depletion of RanBP2. Since a stronger inhibition was not observed on days 7 and 8 (data not shown), we performed all experiments described below on days 5 and 6, when most cells accumulated poly(A)+ RNA within the nucleus. Notably, the mRNA export block observed in RanBP2-depleted cells is not a direct consequence of the inhibition of cell proliferation, as depletion of the essential protein eIF4G or Cdc37 also results in a strong inhibition of cell proliferation, which is not accompanied by a nuclear accumulation of poly(A)+ RNA (14, 15; A. Herold, unpublished data). Conversely, the inhibition of cell proliferation may not be a direct consequence of the mRNA export block but could reflect additional functions of this nucleoporin (26).
In contrast to the changes in the staining pattern of the oligo(dT) probe, the intensity of the WGA staining was not detectably affected in cells depleted of RanBP2 (Fig. 2d versus b). Similarly, nuclear envelope staining with the monoclonal antibody mAb414, which recognizes several nucleoporins, was not significantly affected in the RanBP2 knockdown (data not shown), suggesting that the integrity of the NPC is not severely compromised. Our results indicate that RanBP2 is an essential nucleoporin whose depletion leads to the inhibition of both mRNA export and cell proliferation.
RanBP2 functions in the mRNA export pathway.
It was recently shown that Drosophila cells depleted of essential mRNA export factors such as NXF1, p15, or UAP56 display strikingly similar mRNA expression profiles that represent a specific signature of the mRNA export pathway (15). In these cells, the total and cytoplasmic levels of most mRNAs are significantly reduced, whereas genes encoding export factors (i.e., NXF1, p15, and REF1) are upregulated as a result of a feedback loop whose underlying mechanism remains to be established (15).
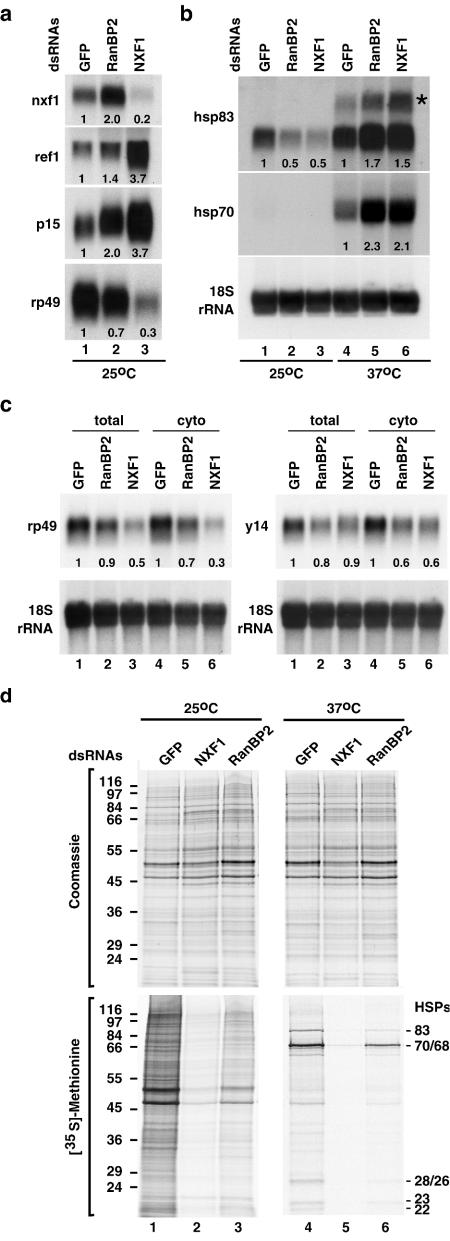
We expected that if RanBP2 were acting in the same pathway as NXF1 and p15, its depletion should lead to changes in mRNA expression levels similar to those observed in NXF1-depleted cells. Northern blot analysis of total RNA samples isolated from RanBP2-depleted cells revealed that the levels of nxf1, p15, and ref1 mRNAs were indeed increased (Fig. 3a). In contrast, the expression levels of rp49 mRNA (encoding ribosomal protein L32) or of hsp83 mRNA were reduced (Fig. 3a and b, lanes 1 to 3).
FIG. 3.
RanBP2 acts in the mRNA export pathway. (a and b) S2 cells were treated with GFP, NXF1, and RanBP2 dsRNAs. Six days later, cells were kept at 25°C or subjected to a 1-h heat shock at 37°C. Total RNA was isolated and analyzed by Northern blot (10 μg/lane) with probes specific for the indicated mRNAs and 18S rRNA. The asterisk indicates the position of the unspliced hsp83 precursor mRNA. Numbers below the lanes represent relative mRNA expression levels normalized to 18S rRNA and set arbitrarily to 1 in control cells. (c) S2 cells were treated with GFP, NXF1, and RanBP2 dsRNAs. Total and cytoplasmic RNA were isolated 4 days (NXF1) or 5 days (GFP and RanBP2) after addition of dsRNAs. Samples were analyzed by Northern blot as described for panel a. (d) S2 cells from the same experiment shown in panels a and b were pulse labeled with [35S]methionine for 1 h. Total lysates from equivalent numbers of cells were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Coomassie blue staining (upper panels) and fluorography (lower panels).
We also compared the steady-state expression levels of two mRNAs (rp49 and y14 mRNAs) that have been shown to be underrepresented in the cytoplasm of cells depleted of NXF1, p15, or UAP56 (15). Northern blot analysis revealed that rp49 and y14 mRNAs were underrepresented in the cytoplasm of cells depleted of RanBP2 (Fig. 3c, lanes 4 to 6). The reduction in rp49 or y14 mRNA levels was more pronounced in the cytoplasmic samples than in the total samples, as expected in cells in which mRNA export is inhibited (Fig. 3c, lanes 4 to 6 versus lanes 1 to 3). Overall, except for the nxf1 mRNA, the changes in mRNA expression levels in the RanBP2 knockdown exhibited the same trend as in the NXF1 knockdown but were less pronounced, in agreement with the observation that the inhibition of mRNA export is partial.
The reduction in mRNA levels observed in RanBP2-depleted cells is unlikely to reflect a general inhibition of transcription as hsp70 and hsp83 mRNAs were strongly induced after shifting the cells to 37°C for 1 h (Fig. 3b, lanes 4 to 6 versus lanes 1 to 3). Moreover, depletion of RanBP2 does not lead to a general block of pre-mRNA splicing, as no accumulation of unspliced hsp83 pre-mRNA was observed in depleted cells at temperatures below 37°C. In contrast, hsp83 pre-mRNA did accumulate in depleted cells as well as in control cells after inhibition of splicing by heat stress at 37°C (Fig. 3b, lanes 4 to 6).
The general inhibition of mRNA export in cells depleted of RanBP2 was confirmed indirectly by metabolic labeling. In cells depleted of RanBP2, the incorporation of [35S]methionine into newly synthesized proteins decreased significantly relative to the control cells (Fig. 3d, lane 3 versus lane 1). Moreover, the expression of HSP70 and HSP83 proteins following heat stress was also reduced (Fig. 3d, lane 6 versus lane 4) despite the fact that the corresponding mRNAs were strongly induced (Fig. 3b).
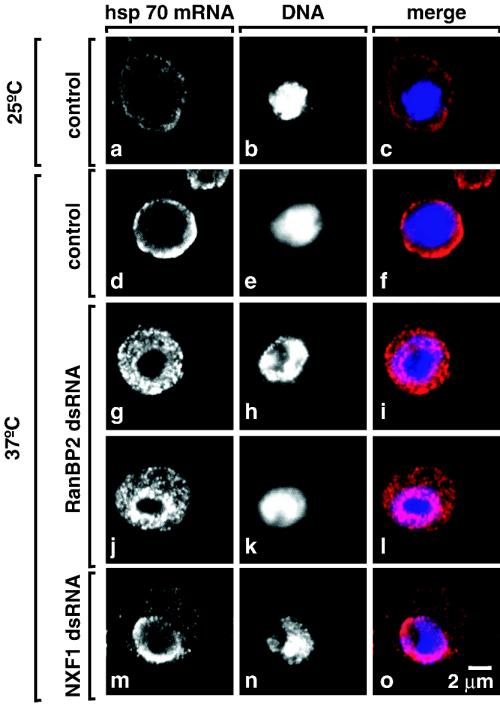
To determine whether the inhibition of heat shock protein synthesis in RanBP2-depleted cells was caused by a failure to export the heat shock mRNAs, we analyzed the intracellular distribution of hsp70 mRNA in control cells and cells depleted of RanBP2 after transcriptional induction at 37°C. In control cells, the hsp70 mRNA was detected mainly in the cytoplasm (Fig. 4d to f). In contrast, a clear nuclear accumulation of hsp70 mRNA was observed in cells depleted of RanBP2 (Fig. 4g and j). In agreement with the results obtained for poly(A)+ RNAs, the nuclear accumulation of hsp70 mRNA was not as strong as that observed when NXF1 was depleted (Fig. 4m).
FIG. 4.
hsp70 mRNA accumulates in the nucleus of cells depleted of RanBP2. (a to o) S2 cells were treated with GFP, NXF1, and RanBP2 dsRNAs. In panels d to o, cells were shifted to 37°C for 1 h, 4 days (NXF1), or 6 days (RanBP2) after the addition of the indicated dsRNAs. hsp70 mRNA was detected by FISH. DNA was stained with Hoechst 33342. The hsp70 signal is specific, as a decrease in intensity is observed when cells were not subjected to heat stress (panels a to c). Panels show representative examples of cells. In cells depleted of RanBP2, the inhibition of hsp70 mRNA export was almost complete (panel j, 10% of the cell population) or partial with an equal distribution of the hsp70 signal between the nucleus and the cytoplasm (panel g, 80% of the cell population).
The accumulation of poly(A)+ RNA and hsp70 mRNA within the nucleus of cells depleted of RanBP2 together with the similarities of the effects on mRNA expression levels and protein synthesis observed following RanBP2 or NXF1 depletions indicate that RanBP2 functions in the mRNA export pathway.
RanBP2 provides a major binding site for NXF1-p15 dimers at the NPC.
We next investigated the subcellular localization of NXF1 in cells in which expression of the ranBP2 gene was silenced. We expected that if NXF1-p15 is recruited to the NPC via the interaction with RanBP2, its depletion should result in the dissociation of NXF1-p15 from the NPC. To monitor the efficiency of RanBP2 depletion, cells were labeled with oligo(dT). To visualize the localization of the heterodimer, a polyclonal S2 cell line expressing GFP-NXF1 was used. GFP-NXF1 reflects the subcellular localization of endogenous NXF1 and promotes export of reporter mRNAs in human cells (1-4, 11). Moreover, NXF1 derivatives N-terminally fused to GFP restore mRNA export in S2 cells depleted of the endogenous protein (4), indicating that the GFP tag does not interfere with NXF1 export activity.
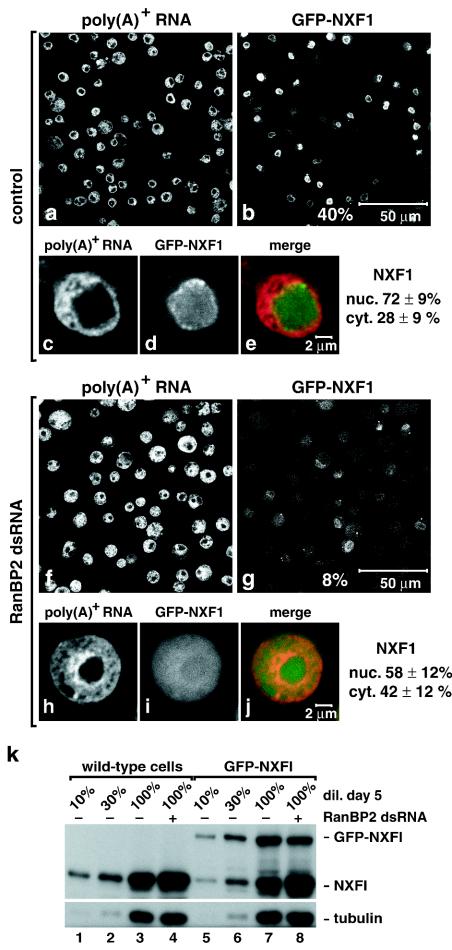
In the untreated cell population, GFP-NXF1 was detected within the nucleoplasm and at the nuclear rim in all cells expressing this protein at detectable levels (Fig. 5b and d). These represent ca. 40% of the cell population (Fig. 5b versus a). The nuclear localization of GFP-NXF1 was observed by confocal microscopy (Fig. 5b) and wide-field fluorescence microscopy (Fig. 5d), indicating that the protein is predominantly nuclear at steady state. After depletion of RanBP2, the intensity of the GFP signal detected in single optical sections was strongly reduced and only 8% of the cells exhibited clear nuclear staining (Fig. 5g versus f). Wide-field fluorescence microscopy revealed that the GFP signal was no longer localized at the nuclear rim and within the nucleoplasm but was dispersed throughout the cell with a significant fraction of the protein in the cytoplasm (Fig. 5i). The nuclear staining of the oligo(dT) probe indicated that RanBP2 was depleted efficiently (Fig. 5f and h).
FIG. 5.
RanBP2 provides a major binding site for NXF1 at the NPC. (a to j) S2 cells constitutively expressing GFP-NXF1 (green) were treated with RanBP2 dsRNA for 5 days as indicated. poly(A)+ RNA was detected by FISH with an oligo(dT) probe (red). All panels depict optical sections, except panels d and i, which show images recorded with the pinhole of the confocal microscope open. Control panels show untreated cells. The fraction of GFP-NXF1 in the nuclear and cytoplasmic compartments of control or knockdown cells is indicated. (k) Wild-type S2 cells or cells expressing GFP-NXF1 were treated with RanBP2 dsRNA for 5 days. Total cell lysates were analyzed by Western blotting with antibodies raised to recombinant NXF1. In lanes 1 to 3 and 5 to 7, dilutions of samples isolated on day 0 were loaded to assess the expression levels of the protein. Tubulin served as a loading control. Note that in agreement with the increased levels of nxf1 mRNA (Fig. 3a) the expression levels of endogenous NXF1 were increased in the RanBP2 knockdown (lanes 4 and 8 versus lanes 3 and 7).
The expression levels of GFP-NXF1 in the RanBP2 knockdown were similar to those detected in control cells, as judged by Western blot analysis (Fig. 5k, lane 8 versus lane 7), indicating that the detection of the GFP signal in the cytoplasm reflects a change in its subcellular localization. To estimate the shift in steady-state distribution of GFP-NXF1, we quantitated the fluorescence signal in the nucleus and cytoplasm of at least 18 representative control or depleted cells. The cytoplasmic fraction of NXF1 increased from 28% ± 9% to 42% ± 12% after depletion of RanBP2 (Fig. 5d versus i).
Similar results were obtained when the localization of a derivative of NXF1 having two UBA-like domains but no NTF2-like domain was investigated (Fig. S1 in the supplemental material [http://www.embl.de/ExternalInfo/izaurral/ranbp2]). Although GFP-NXF1-2× UBA does not interact with p15, it localizes at the nuclear rim (Fig. S1c and e in the supplemental material [http://www.embl.de/ExternalInfo/izaurral/ranbp2]) (4), and this nuclear rim staining was strongly reduced in RanBP2-depleted cells (Fig. S1h and j in the supplemental material [http://www.embl.de/ExternalInfo/izaurral/ranbp2]), indicating that NXF1-2× UBA binds to RanBP2 without p15. Thus, the interaction of NXF1-p15 with RanBP2 is mediated by NXF1. This conclusion is fully consistent with available data showing that NXF1, but not p15, binds directly to nucleoporin FG repeats and is responsible for the localization of the heterodimer at the NPC (1, 11, 13, 17, 35).
To summarize, in cells depleted of RanBP2, the association of NXF1 (or NXF1-2× UBA) with the NPC at steady state is strongly reduced and a larger fraction of the protein is detected in the cytoplasm.
The selectivity of the NPC is not altered in RanBP2-depleted cells.
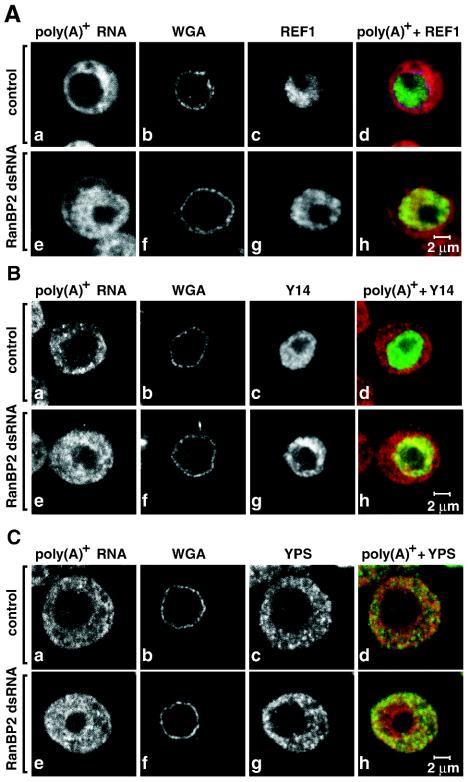
In the RanBP2 knockdown, NXF1 partially localizes to the cytoplasm. This observation led us to investigate whether the general selectivity of the NPC was altered so that many nuclear or cytoplasmic proteins redistributed between the two compartments. Moreover, since there is no RanBP1 homolog in Drosophila, RanBP2 is probably the only Ran binding protein that, together with RanGAP1, activates GTP hydrolysis by Ran. It was therefore of particular relevance to address whether the partial depletion of RanBP2 resulted in perturbations of the Ran system and, consequently, of nucleocytoplasmic transport. To this end, we analyzed the subcellular localization of proteins that reside either in the nucleoplasm or in the cytoplasm at steady state.
We found that in the RanBP2 knockdown, the nuclear localization of endogenous REF1 was unchanged, even in cells strongly accumulating poly(A)+ RNA within the nucleus (Fig. 6Ae to h versus a to d). Similarly, the distribution of Y14 was not changed in cells depleted of RanBP2 (Fig. 6Be to h versus a to d). These results indicate that not all nuclear proteins redistribute to the cytoplasm in the RanBP2 knockdown. Moreover, the cytoplasmic RNA-associated proteins YPS and Exuperantia were excluded from the nuclear compartment in cells depleted of RanBP2 (Fig. 6C and data not shown). We conclude that the selectivity of the NPC was not compromised following depletion of RanBP2. These results are consistent with those reported by Walther et al. (34), which show that nuclei assembled in vitro in Xenopus egg extracts depleted of RanBP2 lack cytoplasmic filaments but exclude nonimport cargoes and are still functional for protein import mediated by importin α/β or by transportin. Nevertheless, the possibility that RanBP2 depletion affects the rate of import or export of the analyzed proteins without changing their subcellular localizations at steady state cannot be ruled out (see below).
FIG. 6.
Depletion of RanBP2 does not alter the selectivity of the NPC. (A to C) S2 cells were fixed 5 days after the addition of RanBP2 dsRNA and labeled by FISH with an oligo(dT) probe [poly(A)+ RNA, red]. The nuclear envelope (WGA) was stained with Alexa 488-WGA conjugates. Cells shown in panel A were stained with antibodies that specifically recognize REF1 (green). Panel B shows the localization of TAP-tagged Drosophila Y14 (green). In panel C, cells were stained with antibodies raised to endogenous YPS (green). Control panels show untreated cells.
CRM1-mediated protein export is not detectably affected by partial depletion of RanBP2.
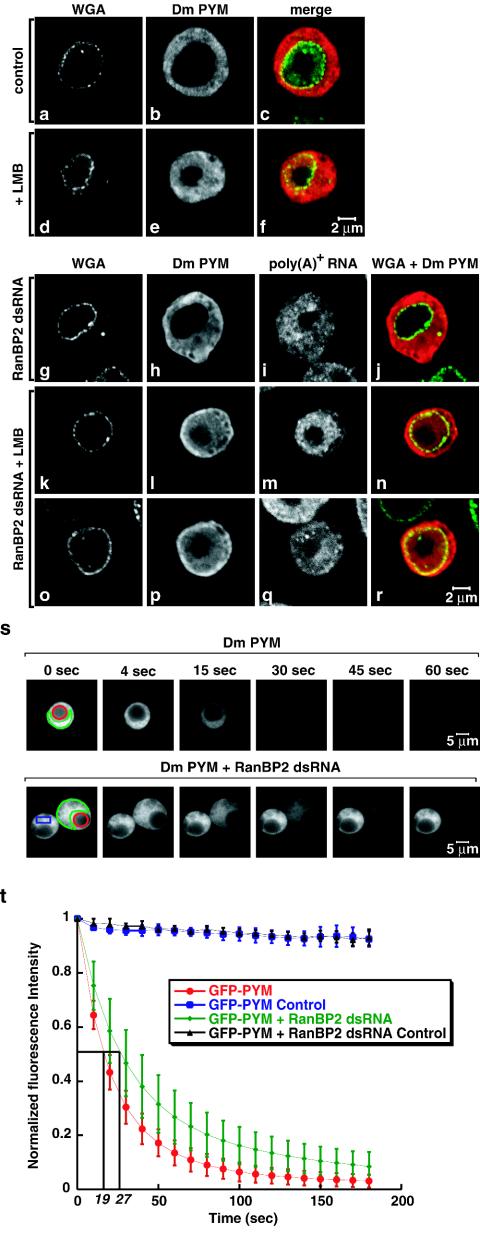
The results described above indicate that bulk protein import proceeds despite RanBP2 depletion so that no change in the steady-state subcellular distribution of the analyzed proteins (with the exception of NXF1) was detected. Under the same conditions (i.e., when the levels of RanBP2 were reduced to ca. 50% of the wild-type levels), bulk mRNA export was significantly impaired, so it was of interest to investigate whether other export pathways were affected. To this end, we analyzed the subcellular localization of a reporter protein whose export is mediated by CRM1. The PYM protein is the uncharacterized product of the wibg gene (9). At steady state, PYM was excluded from the nucleus (Fig. 7a to c). In cells treated with leptomycin B (LMB), PYM partially accumulated within the nucleoplasm, indicating that its nuclear exclusion is a consequence of being actively exported by CRM1 (Fig. 7d to f). LMB is a cytotoxic drug that binds to CRM1 and prevents its association with RanGTP and the export cargo (10).
FIG. 7.
CRM1-mediated export is not inhibited in RanBP2-depleted cells. (a to f) S2 cells expressing a TAP-tagged form of Drosophila PYM were treated with LMB (Sigma) as indicated. Three hours after the addition of LMB (final concentration, 20 ng/ml), cells were fixed and the tagged protein was visualized with an anti-protein A antibody (red). Cells were double-labeled by FISH with an oligo(dT) probe [poly(A)+ RNA]. The nuclear envelope (WGA) was stained with Alexa 488-WGA conjugates (green). (g to r) S2 cells expressing a TAP-tagged Drosophila PYM were treated with RanBP2 dsRNA for 5 days. One half of the cell population was treated with LMB (k to r). Cells were labeled as described for panels a to f. (s) Control and RanBP2-depleted cells expressing GFP-PYM were selected. A nucleoplasmic region (red circle) was photobleached repetitively 130 times every 1.6 s with a 40× PlanApochromat 1.4 NA oil immersion objective. After each bleaching, the depletion of the cytoplasmic fluorescence was monitored in a confocal section (region delimited in green). (t) Quantification of cytoplasmic fluorescence loss for GFP-PYM in untreated and RanBP2-depleted cells. The loss of the cytoplasmic signal was measured and normalized to the intensity of the cytoplasmic signal before bleaching (t = 0). Curves depict mean values (± standard deviations) of the results from 8 representative cells, including those shown in panel s. Control curves show that less than 10% of the cytoplasmic fluorescence was lost from a nonbleached neighboring cell (panel s, blue rectangle) due to image acquisition between bleach pulses and the fact that the bleach pulses did not affect areas outside the bleached regions. Note that the loss of cytoplasmic fluorescence in bleached cells due to the bleach pulses cannot be evaluated. However, this loss is comparable in control and depleted cells because at steady state the fraction of GFP-PYM in the cytoplasm is similar (81% ± 2% or 81% ± 5% in 8 representative control or depleted cells, respectively).
In cells depleted of RanBP2 [and consequently showing a nuclear accumulation of poly(A)+ RNA], PYM remained entirely cytoplasmic (Fig. 7g to j). This cytoplasmic localization was not due to the inhibition of its nuclear import because when RanBP2-depleted cells were treated with LMB, PYM accumulated within the nucleus, as in control cells (Fig. 7k to r). Thus, neither import nor CRM1-mediated export of PYM was severely compromised in the RanBP2 knockdown, indicating that the translocation of receptor-cargo complexes across the NPC is not apparently affected by the depletion. Similar results were obtained when the subcellular localization of the endogenous protein Extradenticle, which is exported by CRM1, was analyzed (data not shown).
The experiments described above showed that RanBP2 depletion did not affect the steady-state subcellular localization of several nuclear and cytoplasmic proteins but did not address whether import or export rates were affected. To investigate this issue, we analyzed the import of PYM in living cells by FLIP. Cells expressing GFP-PYM were repetitively bleached in the nucleoplasm (Fig. 7s), and loss of fluorescence in the cytoplasm was monitored over time (Fig. 7s). In both control and RanBP2-depleted cells, the fluorescent signal in the cytoplasm could be depleted to near background levels, showing that PYM import proceeded despite RanBP2 depletion. The half-time of depletion of the cytoplasmic signal was found to be 19 ± 5 s in control cells and 27 ± 8 s in RanBP2-depleted cells (Fig. 7t), indicating that the import rate of PYM was slightly reduced in RanBP2-depleted cells. We conclude that although nuclear import and export proceed in RanBP2-depleted cells, the transport rates are affected for at least a subset of receptor-cargo complexes. Nevertheless, these effects may not always alter the subcellular distribution of the cargo at steady state.
Nuclear import of NXF1 is not significantly inhibited by RanBP2 depletion.
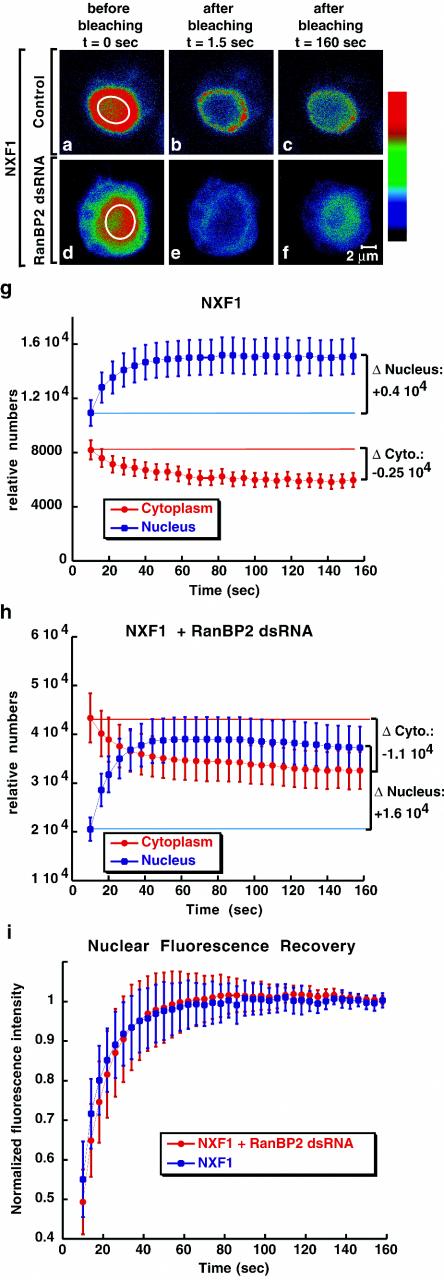
In contrast to the proteins analyzed above, the steady-state subcellular localization of NXF1 is significantly changed in RanBP2-depleted cells. This change could be due to a decrease of its import rate, an increased export, or both. To discriminate between these possibilities, we first analyzed the import of NXF1 in wild-type cells and cells treated with RanBP2 dsRNA by FRAP. In the experiment whose results are shown in Fig. 8, the nucleoplasmic GFP-NXF1 signal was bleached in control and knockdown cells, and the recovery of the nuclear fluorescence was measured over time. Immediately after bleaching a region of the nucleus (Fig. 8a and d), no discernible bleached zone could be seen (Fig. 8b and e). Instead, there was an immediate decrease in fluorescence intensity through the entire nucleus, indicating that NXF1 diffuses rapidly within the nucleus.
FIG. 8.
Import of NXF1 is not significantly affected in RanBP2 depleted cells. (a to f) Control and RanBP2-depleted cells expressing GFP-NXF1 were imaged immediately before photobleaching a defined zone in the nucleus (white circle) and every 1.5 s afterwards. Colors represent relative intensities of the GFP signal (red and blue correspond to high and low intensities, respectively). (g and h) Nucleoplasmic or cytoplasmic fluorescence intensities measured over time for each individual cell were multiplied by the nuclear or cytoplasmic volumes, respectively, and the relative number of molecules in each compartment was calculated. Curves depict mean values (± standard deviations) from measurements of 8 to 9 representative cells, including those shown in panel a. The nuclear recovery (Δ Nucleus) correlates with the loss of fluorescence in the cytoplasm (Δ Cytoplasm). (i) The recovery of the nucleoplasmic signal was measured over time and normalized to the intensity of the signal when recovery was completed (160 s). Curves depict mean values (± standard deviations) from measurements of 8 to 9 representative cells, including those shown in panels a and d.
After the bleach, the total nucleoplasmic fluorescence partially recovered, the total cytoplasmic fluorescence decreased by a similar amount (Fig. 8g and h), and the whole-cell fluorescence remained constant (data not shown), indicating that the recovery of the nuclear signal is due to the import of the cytoplasmic pool of the protein. To compare NXF1 import rates in control and depleted cells, the recovery of the nucleoplasmic signal was measured in representative cells and normalized to the intensity of the signal when recovery was complete (160 s) (Fig. 8i). The nucleoplasmic signal of NXF1 recovered with similar kinetics in control and RanBP2-depleted cells (with an error of less than 20% of the rate). Simulations indicated that a strong decrease in the NXF1 import rate (>20%), which would account for the change in the steady-state distribution of GFP-NXF1 in RanBP2-depleted cells, could not fit the rate of nuclear fluorescence recovery (Fig. S2 in the supplemental material [http://www.embl.de/ExternalInfo/izaurral/ranbp2]).
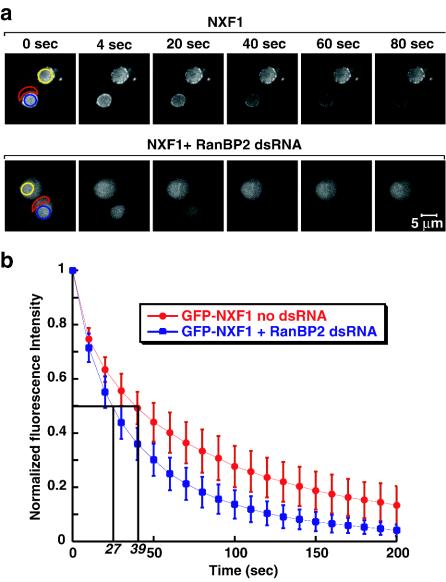
Depletion of RanBP2 affects the release of NXF1 into the cytoplasm.
Next, we investigated the relative export rates of GFP-NXF1 in control and depleted cells by FLIP. To compare the export rate of NXF1 quantitatively in these cells, the entire cytoplasmic signal was bleached in a single pulse at time zero (Fig. 9a). Subsequently, cells were repetitively bleached in the cytoplasm (Fig. 9a), and loss of fluorescence in the nucleus was monitored over time (Fig. 9a). In both control and RanBP2-depleted cells, the fluorescence signal in the nucleoplasm could be depleted to near background levels, showing that the protein was exported efficiently from the nucleus.
FIG. 9.
RanBP2 depletion increases the release of NXF1 in the cytoplasm. (a) Representative control and RanBP2-depleted cells expressing GFP-NXF1 were selected. A cytoplasmic region (delineated in red) was photobleached at time zero to reduce the cytoplasmic signal to background levels. Subsequently, the same region was repetitively bleached every 2.8 s. After each bleach, the depletion of the nuclear fluorescence was monitored in a confocal section (blue circle). (b) Quantitation of nuclear fluorescence loss with cytoplasmic FLIP for GFP-NXF1 in untreated and RanBP2-depleted cells. Curves depict mean values (± standard deviations) of the results from 8 representative cells, including those shown in panel a. Values are corrected for the loss of nuclear fluorescence from a nonbleached neighboring cell (panel a, yellow circle) due to image acquisition and to the bleach pulses.
To compare the export rates, the loss of the nuclear signal was measured in control or knockdown cells and normalized to the intensity of the nuclear signal before bleaching (t = 0) (Fig. 9b). The half-time of depletion of the nucleoplasmic signal, measured by FLIP in two independent experiments, was found to be 39 ± 11 s in control cells and 27 ± 7 s in depleted cells. Assuming that NXF1 export is limited by a single rate, these results indicate that, in RanBP2-depleted cells, NXF1 is released in the cytoplasm 1.4-fold more rapidly than in wild-type cells. Note that since the cytoplasmic fraction of the protein increased from 28 to 42% after RanBP2 depletion, but we could not detect a corresponding decrease of the import rate (Fig. 8i), a 1.5-fold stimulation of cytoplasmic release was expected.
In summary, in RanBP2-depleted cells, NXF1 shuttles as in wild-type cells, but a higher fraction of the protein is detected in the cytoplasm. Our results indicate that this is due mainly to an increased release of the protein into the cytoplasm.
The release of NXF1 in the cytoplasm is a specific effect of RanBP2 depletion.
The faster kinetics of NXF1 release in the cytoplasm could be a specific effect of RanBP2 depletion or alternatively due to a more general effect of depleting nucleoporins localized at the cytoplasmic face of the NPC. In particular, it was of interest to determine whether depletion of CAN (which also localizes at the cytoplasmic face of the NPC) could lead to a similar phenotype. CAN, like many nucleoporins possessing FG repeats, interacts with NXF1 and together with its yeast counterpart Nup159p has been implicated in mRNA export (1, 12, 17, 31, 35).
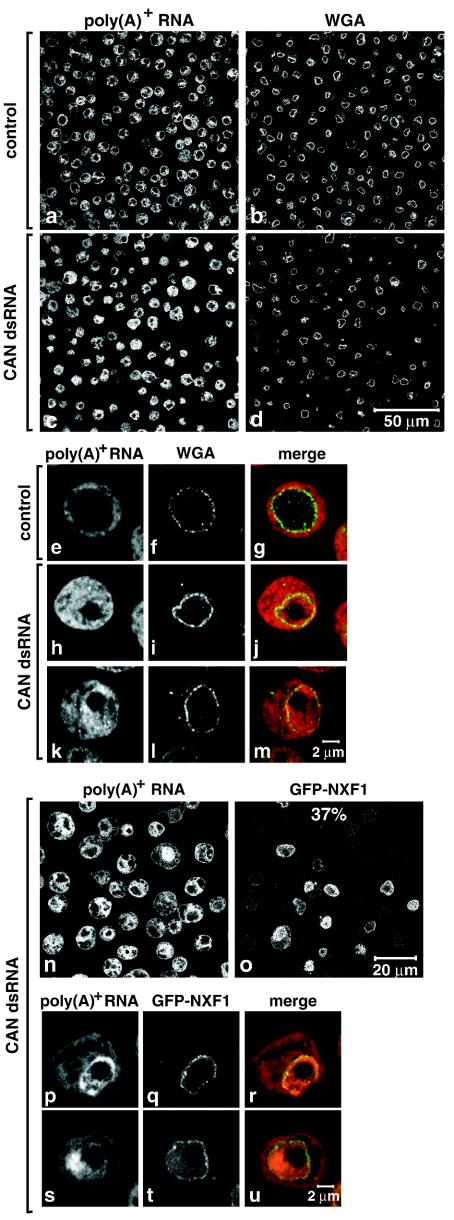
Depletion of CAN inhibited cell proliferation and led to a significant nuclear accumulation of poly(A)+ RNA (Fig. S3 in the supplemental material [http://www.embl.de/ExternalInfo/izaurral/ranbp2] and Fig. 10c, h, and k) (see also reference 31). These inhibitory effects on cell proliferation and mRNA export were comparable to the inhibitory effects observed when RanBP2 was depleted (Fig. S3 in the supplemental material [http://www.embl.de/ExternalInfo/izaurral/ranbp2] and Fig. 10 versus Fig. 2). As observed in the RanBP2 knockdown, depletion of CAN did not significantly affect NPC staining with fluorescently labeled WGA (Fig. 10d versus b).
FIG. 10.
Depletion of CAN inhibits mRNA export. (a to m) S2 cells were treated with a dsRNA corresponding to Drosophila CAN. poly(A)+ RNA was detected by FISH with an oligo(dT) probe (red). The nuclear envelope (WGA) was stained with Alexa 488-WGA conjugates (green). Control panels show untreated cells. (n to u) S2 cells constitutively expressing GFP-NXF1 (green) were treated with CAN dsRNA for 9 days. poly(A)+ RNA was detected by FISH (red).
In contrast to RanBP2 depletion, CAN depletion did not alter the steady-state subcellular localization of NXF1. Indeed, when cells expressing GFP-NXF1 were depleted of CAN, the GFP signal was detected at the nuclear rim and within the nucleus (Fig. 10o), even in cells exhibiting a strong nuclear accumulation of poly(A)+ RNA (Fig. 10p to u). Moreover, the fraction of the protein in the cytoplasm was not detectably changed despite the inhibition of mRNA export (Fig. 10p to u). These results highlight the specific role of RanBP2 in the recruitment of NXF1 to the NPC.
DISCUSSION
In this study we show that RanBP2 is an essential nucleoporin in Drosophila cells, playing a specific role in the export pathway of bulk mRNA. In RanBP2-depleted cells, the association of the mRNA export receptor NXF1 with the NPC is strongly reduced and a larger fraction of this otherwise predominantly nuclear protein is detected in the cytoplasm. The export of bulk mRNA is also partially inhibited. The observation that the steady-state subcellular localization of several nuclear and cytoplasmic proteins is not detectably affected in RanBP2-depleted cells makes unlikely the possibility that the release of NXF1 in the cytoplasm and the inhibition of mRNA export are due to a general defect in NPC function and suggests that the observed phenotypes are specifically due to the depletion of RanBP2. Moreover, the observation that depletion of CAN/Nup214, another nucleoporin localized at the cytoplasmic face of the NPC, also leads to the inhibition of cell proliferation and bulk mRNA export without affecting the subcellular localization of NXF1 provides further support for a specific role of RanBP2 in the recruitment of NXF1 to the NPC.
RanBP2 provides a major binding site for NXF1-p15 dimers at the cytoplasmic filaments of the NPC.
In this study we show that in cells partially depleted of RanBP2, shuttling of NXF1 is not inhibited, but the fraction of NXF1 associated with the NPC at steady-state decreases significantly. In addition, depletion of RanBP2 leads to an increased level of NXF1 in the cytoplasm, which is not caused by an import block. Based on these results, we suggest the following model. In wild-type cells, NXF1 shuttles across the central channel of the pore. The presence of high-affinity binding sites at the cytoplasmic filaments of the NPC may constrain the diffusion of NXF1 into the cytoplasm so that the efficiency of recycling is increased. In the absence of RanBP2, NXF1 molecules reaching the cytoplasmic side of the NPC may either reenter the nucleus or may diffuse freely into the cytoplasm. This diffusion would result in an increased number of molecules in the cytoplasm and a partial depletion of the nuclear pool so that the efficiency of mRNA export might be reduced.
RanBP2 functions in the mRNA export pathway.
The model described above implies that the role of RanBP2 in mRNA export is indirect, i.e., the primary effect of RanBP2 depletion would be to decrease the nuclear pool of NXF1. Note that the 1.5-fold decrease of the nucleoplasmic pool of NXF1 is likely to account for the observed nuclear accumulation of poly(A)+ RNA, as a similar accumulation is observed in S2 cells in which the levels of NXF1 are reduced to ca. 60% of the wild-type levels by RNAi (A. Herold and E. Izaurralde, unpublished data). Nonetheless, the possibility that the mRNA export block is not a direct consequence of the mislocalization of NXF1 cannot be ruled out.
Indeed, a more direct role of RanBP2 in mRNA export, such as facilitating the disassembly of mRNP export cargoes could be envisaged. However, in RanBP2-depleted cells shuttling mRNA-binding proteins, such as Y14 and REF1, do not accumulate in the cytoplasm, indicating that they dissociate from the mRNPs after export and are recycled back to the nucleus. Moreover, the possibility that the inhibition of mRNA export is an indirect consequence of a perturbation of the Ran system is also unlikely because transport processes mediated by receptors of the karyopherin family are less affected by the partial depletion.
The effects of RanBP2 depletion on the expression levels of the analyzed mRNAs parallel those observed following depletion of NXF1, p15, UAP56, or CAN (Fig. 3 and data not shown) (15). Particularly striking is the observation that RanBP2 depletion triggers the upregulation of nxf1, p15, and ref1 mRNAs. These mRNAs are upregulated when mRNA export is inhibited (e.g., in cells depleted of NXF1, p15, UAP56, RanBP2, or CAN) but not in cells in which proliferation has been inhibited by depletion of essential proteins that do not play a role in bulk mRNA export (e.g., eIF4G) (15). We therefore conclude that RanBP2 functions in the mRNA export pathway and that its depletion imposes a general block to this pathway which triggers a feedback loop, resulting in the upregulation of export factors.
Asymmetrically localized nucleoporins play a role in terminal steps of NPC translocation.
Current models of the mechanism of translocation of transport receptors through the NPC on their own or in association with cargoes propose that the central channel of the NPC would be occupied by a dense array of FG repeat nucleoporins (22, 23). Nuclear transport receptors generically interact with the FG repeat domains of nucleoporins and use these interactions to translocate cargoes across the central channel of the NPC at a high rate. These interactions must be transient in nature to allow fast translocation. However, in addition to transient generic interactions, transport receptors exhibit a high affinity for specific nucleoporins, and nucleoporin mutations affect specific transport pathways (reviewed in references 25 and 32). The basis for this selectivity is not known.
Previously, it has been shown that Nup153, a nucleoporin associated with the nuclear basket, provides high-affinity binding sites for importin β and is required for nuclear protein import mediated by this receptor (28, 33). In this study we show that RanBP2 provides major binding sites for NXF1-p15 at the NPC and is required for NXF1-mediated mRNA export. Together, these observations suggest that one function of the asymmetrically localized nucleoporins would be to restrict the diffusion of import receptors within the nucleoplasm or of export receptors within the cytoplasm after translocation so that the receptors are recycled efficiently. This would be a local effect, only affecting transport receptors in transit within the central channel of the NPC, consistent with our observation that RanBP2 depletion does not affect the import rate of the cytoplasmic pool of NXF1. Additionally, asymmetrically localized nucleoporins may also play a more active role in the disassembly of receptor-cargo complexes after translocation.
Acknowledgments
This study was supported by the European Molecular Biology Organization.
The technical support of Michaela Rode is gratefully acknowledged. We thank Iain W. Mattaj, David Thomas, and Tobias Walther for critical reading of the manuscript.
REFERENCES
- 1.Bachi, A., I. C. Braun, J. P. Rodrigues, N. Panté, K. Ribbeck, C. von Kobbe, U. Kutay, M. Wilm, D. Görlich, M. Carmo-Fonseca, and E. Izaurralde. 2000. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 6:136-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bear, J., W. Tan, A. S. Zolotukhin, C. Tabernero, E. A. Hudson, and B. K. Felber. 1999. Identification of novel import and export signals of human TAP, the protein that binds to the CTE element of the type D retrovirus mRNAs. Mol. Cell. Biol. 19:6306-6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, I. C., A. Herold, M. Rode, E. Conti, and E. Izaurralde. 2001. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J. Biol. Chem. 276:20536-20543. [DOI] [PubMed] [Google Scholar]
- 4.Braun, I. C., A. Herold, M. Rode, and E. Izaurralde. 2002. Nuclear export of messenger RNA by TAP/NXF1 requires two nucleoporin binding sites but not p15. Mol. Cell. Biol. 15:5405-5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conti, E., and E. Izaurralde. 2001. Nucleocytoplasmic transport enters the atomic age. Curr. Opin. Cell Biol. 3:310-319. [DOI] [PubMed] [Google Scholar]
- 6.Daigle, N., J. Beaudouin, L. Hartnell, G. Imreh, E. Hallberg, J. Lippincott-Schwartz, and J. Ellenberg. 2001. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154:71-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fauser, S., A. Aslanukov, R. Roepman, and P. A. Ferreira. 2001. Genomic organization, expression, and localization of murine Ran-binding protein 2 (RanBP2) gene. Mamm. Genome 12:406-415. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 9.Forler, D., T. Köcher, M. Rode, M. Gentzel, E. Izaurralde, and M. Wilm. 2003. An efficient protein-complex purification method for functional proteomics in higher eukaryotes. Nat. Biotechnol. 21:89-92. [DOI] [PubMed] [Google Scholar]
- 10.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 11.Fribourg, S., I. C. Braun, E. Izaurralde, and E. Conti. 2001. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol. Cell 8:645-656. [DOI] [PubMed] [Google Scholar]
- 12.Gorsch, L. C., T. C. Dockendorff, and C. N. Cole. 1995. A conditional allele of the novel repeat-containing yeast nucleoporin RAT7/NUP159 causes both rapid cessation of mRNA export and reversible clustering of nuclear pore complexes. J. Cell Biol. 129:939-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant, R. P., D. Neuhaus, and M. Stewart. 2003. Structural basis for the interaction between the Tap/NXF1 UBA domain and FG nucleoporins at 1A resolution. J. Mol. Biol. 326:849-858. [DOI] [PubMed] [Google Scholar]
- 14.Herold, A., T. Klimenko, and E. Izaurralde. 2001. NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA 7:1768-1780. [PMC free article] [PubMed] [Google Scholar]
- 15.Herold, A., L. Teixeira, and E. Izaurralde. 2003. Genome-wide analysis of nuclear mRNA export pathways in Drosophila. EMBO J. 22:2472-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang, Y., and B. R. Cullen. 1999. The human TAP protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 13:1126-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katahira, J., K. Strässer, A. Podtelejnikov, M. Mann, J. U. Jung, and E. Hurt. 1999. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 18:2593-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kraemer, D., R. W. Wozniak, G. Blobel, and A. Radu. 1994. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc. Natl. Acad. Sci. USA 91:1519-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Letunic, I., L. Goodstadt, N. J. Dickens, T. Doerks, J. Schultz, R. Mott, F. Ciccarelli, R. R. Copley, C. P. Ponting, and P. Bork. 2002. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30:242-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levesque, L., B. Guzik, T. Guan, J. Coyle, B. E. Black, D. Rekosh, M.-L. Hammarskjold, and B. M. Paschal. 2001. RNA export mediated by tap involves NXT1-dependent interactions with the nuclear pore complex. J. Biol. Chem. 276:44953-44962. [DOI] [PubMed] [Google Scholar]
- 21.Pichler, A., A. Gast, J. S. Seeler, A. Dejean, and F. Melchior. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109-120. [DOI] [PubMed] [Google Scholar]
- 22.Ribbeck, K., and D. Görlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rout, M. P., J. D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B. T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rout, M. P., and J. D. Aitchison. 2001. The nuclear pore complex as a transport machine. J. Biol. Chem. 276:16593-16596. [DOI] [PubMed] [Google Scholar]
- 25.Ryan, K. J., and S. R. Wente. 2000. The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 12:361-371. [DOI] [PubMed] [Google Scholar]
- 26.Salina, D., P. Enarson, J. B. Rattner, and B. Burke. 2003. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J. Cell Biol. 162:991-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt, I., and L. Gerace. 2001. In vitro analysis of nuclear transport mediated by the C-terminal shuttle domain of Tap. J. Biol. Chem. 276:42355-42363. [DOI] [PubMed] [Google Scholar]
- 28.Shah, S., S. Tugendreich, and D. J. Forbes. 1998. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J. Cell Biol. 141:31-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shamsher, M. K., J. Ploski, and A. Radu. 2002. Karyopherin beta 2B participates in mRNA export from the nucleus. Proc. Natl. Acad. Sci. USA 99:14195-14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Truant, R., Y. Kang, and B. R. Cullen. 1999. The human tap nuclear RNA export factor contains a novel transportin-dependent nuclear localization signal that lacks nuclear export signal function. J. Biol. Chem. 274:32167-32171. [DOI] [PubMed] [Google Scholar]
- 31.van Deursen, J., J. Boer, L. Kasper, and G. Grosveld. 1996. G2 arrest and impaired nucleocytoplasmic transport in mouse embryos lacking the proto-oncogene CAN/Nup214. EMBO J. 15:5574-5583. [PMC free article] [PubMed] [Google Scholar]
- 32.Vasu, S. K., and D. J. Forbes. 2001. Nuclear pores and nuclear assembly. Curr. Opin. Cell Biol. 3:363-375. [DOI] [PubMed] [Google Scholar]
- 33.Walther, T. C., M. Fornerod, H. Pickersgill, M. W. Goldberg, T. D. Allen, and I. W. Mattaj. 2001. The nucleoporin Nup153 is required for nuclear pore basket formation, nuclear pore complex anchoring and import of a subset of nuclear proteins. EMBO J. 20:5703-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walther, T. C., H. S. Pickersgill, V. C. Cordes, M. W. Goldberg, T. D. Allen, I. W. Mattaj, and M. Fornerod. 2002. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J. Cell Biol. 158:63-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiegand, H. L., G. A. Coburn, Y. Zeng, Y. Kang, H. P. Bogerd, and B. R. Cullen. 2002. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol. Cell. Biol. 22:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilken, N., J. L. Senecal, U. Scheer, and M. C. Dabauvalle. 1995. Localization of the Ran-GTP binding protein RanBP2 at the cytoplasmic side of the nuclear pore complex. Eur. J. Cell Biol. 68:211-219. [PubMed] [Google Scholar]
- 37.Wu, J., M. J. Matunis, D. Kraemer, G. Blobel, and E. Coutavas. 1995. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem. 270:14209-14213. [DOI] [PubMed] [Google Scholar]
- 38.Yokoyama, N., N. Hayashi, T. Seki, N. Pante, T. Ohba, K. Nishii, K. Kuma, T. Hayashida, T. Miyata, U. Aebi, et al. 1995. A giant nucleopore protein that binds Ran/TC4. Nature 376:184-188. [DOI] [PubMed] [Google Scholar]