Abstract
In this work, we report the implication of the pleckstrin homology (PH) domain-containing protein CKIP-1 in phosphatidylinositol 3-kinase (PI3-K)-regulated muscle differentiation. CKIP-1 is upregulated during muscle differentiation in C2C12 cells. We show that CKIP-1 binds to phosphatidylinositol 3-phosphate through its PH domain and localizes to the plasma membrane in a PI3-K-dependent manner. Activation of PI3-K by insulin or expression of an active form of PI3-K p110 induces a rapid translocation of CKIP-1 to the plasma membrane. Conversely, expression of the 3-phosphoinositide phosphatase myotubularin or PI3-K inhibition by LY294002, wortmannin, or mutant p85 abolishes CKIP-1 binding to the membrane. Upon induction of differentiation in low-serum medium, CKIP-1 overexpression in C2C12 myoblasts first promotes proliferation and then stimulates the expression of myogenin and cell fusion in a manner reminiscent of the dual positive effect of insulin-like growth factors on muscle cells. Interference with the PI3-K pathway impedes the effect of CKIP-1 on C2C12 cell differentiation. Finally, silencing of CKIP-1 by RNA interference abolishes proliferation and delays myogenin expression. Altogether, these data strongly implicate CKIP-1 as a new component of PI3-K signaling in muscle differentiation.
Upon activation, the phosphoinositide 3-kinase (PI3-K) family of enzymes produces 3′ phosphoinositide lipids (3-PIs), including phosphatidylinositol-3-phosphate (PtdIns-3P), PtdIns-3,4-biphosphate [PtdIns(3,4)P2], PtdIns(3,5)P2, and PtdIns(3,4,5)P3. These lipid products act as second messengers by interacting with the lipid binding domains of a variety of cellular proteins (16). Most proteins interacting with 3-PIs display a pleckstrin homology (PH), a FYVE, or a Phox homology (PX) domain. FYVE and PX domains bind preferentially PtdIns-3P (9, 27, 37), whereas PH domains have been shown to interact with PtdIns(3,4)P2 and PtdIns(3,4,5)P3 as well as PtdIns-3P (19). The binding of PH-, FYVE-, and PX-containing proteins to lipids affects their cellular localization, thereby mediating the regulation of diverse cellular activities, such as proliferation, differentiation, survival, cellular trafficking, and glucose metabolism. A negative feedback mechanism of PI3-K involves the 3-phosphoinositide phosphatases PTEN (phosphatase and tensin homolog deleted on chromosome 10) and myotubularin 1 (MTM1). PTEN dephosphorylates all 3-phosphorylated phosphoinositides in vitro and displays specificity for PtdIns(3,4)P2 and PtdIns(3,4,5)P3 in mammalian cells. Unlike PTEN, the lipid substrate of MTM1 appears to be restricted to PtdIns-3P (35).
Cellular responses triggered by PI3-K signaling are numerous and depend on the nature of the extracellular stimulus, the isoform of PI3-K activated, and the type of lipids produced.
Insulin-like growth factors (IGFs) are crucial extracellular signals involved in the activation of PI3-K. They are required for normal skeletal muscle development (18, 23) and potently stimulate myogenesis in cultured muscle cells (10, 11). The differentiation of myoblasts triggered by lowering serum concentration in the culture medium depends on autocrine production of IGF-II (12, 22, 29). Indeed, C2C12 myoblasts expressing an IGF-II antisense construct lose MyoD expression and fail to differentiate (22). The action of IGFs on myoblasts occurs through two phases: an initial proliferative response followed by a myogenic response characterized by expression of the muscle regulatory factor myogenin and of the cell cycle inhibitor p21. IGFs also promote muscle hypertrophy (7, 10). Recent studies have demonstrated that the PI3-K/AKT pathway mediates the stimulatory effects of IGFs on muscle differentiation. AKT expression is upregulated upon differentiation induced by IGFs with a corresponding increase in kinase activity (5, 30, 32). Inhibition of this pathway by the chemical inhibitor LY294002 (7, 32) or overexpression of dominant-negative kinases blocks muscle differentiation (15). Conversely, expression of active PI3-K or AKT induces myogenic differentiation (13, 14, 32). This pathway directly affects IGF-induced muscle differentiation (i.e., myogenin expression) through regulating transcriptional activation of the MEF2 proteins (30, 36).
In this paper, we report the identification of the PH domain-containing protein CKIP-1 (for “casein kinase 2-interacting protein-1”) (3) as a new PI3-K signaling component involved in muscle differentiation. In its N-terminal part, CKIP-1 contains a PH domain that confers on CKIP-1 the ability to bind to PtdIns-3P and to localize to the plasma membrane in a PI3-K-dependent manner. CKIP-1 expression is upregulated upon induction of C2C12 myoblast differentiation, and overexpression of CKIP-1 enhances myoblast differentiation. Interference with the PI3-K pathway impedes CKIP-1 effects on differentiation. Finally, the abolition of CKIP-1 expression by RNA interference delays myoblast differentiation. Altogether, our data strongly suggest the involvement of CKIP-1 in PI3-K-regulated muscle differentiation.
MATERIALS AND METHODS
Cell lines, culture conditions, and anti-CKIP-1 antibody.
C2C12 cells were maintained as myoblasts in growth medium (GM): 50% Dulbecco modified Eagle medium-50% HAM F12 (Gibco BRL), supplemented with 15% fetal calf serum (HyClone). Cells were differentiated in differentiation medium (DM): DMEM medium supplemented with 2% horse serum (Gibco BRL). Insulin (Sigma) was used at 25 μg/ml. LY294002 (Sigma) was used at 25 μM. In order to study CKIP-1 protein, a rabbit polyclonal antibody was raised against a peptide encoding 15 amino acids (241 to 255) in the carboxy-terminal part of mouse CKIP-1. This antibody detected denatured CKIP-1 in immunoblots as well as the native protein in immunoprecipitation and immunofluorescence experiments.
Plasmid constructs, siRNA, and transfections.
Full-length cDNA encoding M2 Flag-tagged CKIP-1 was subcloned into the KpnI-NotI sites of the pCDNA3 vector (Invitrogen). The cDNA encoding M2-CKIP-1 was obtained by PCR with the following oligonucleotide primers: 5′ GGTACCCCACCATGGACTACAAGGACGACGATGACAAGGAATTCAAGAAGAGCGGCTCC 3′ (sense primer with a KpnI site, a Kosack consensus sequence, an ATG codon followed by the M2-Flag sequence, an EcoRI site, and the first 15 bp of CKIP-1 excluding the ATG codon) and 5′ GCGGCCGCTCACATCAGGCTCTTCCGGT 3′ (antisense primer containing the last 20 bp of CKIP-1 cDNA followed by a NotI site). The mutant CKIP-1ΔPH (residues 147 to 409) was obtained by PCR with the 5′ primer AGCTATCTTGCCCACCCT preceded by an EcoRI site and the same antisense primer as above with the NotI site. The resulting PCR product was cloned in pcDNA3 in the EcoRI-NotI sites in place of the wild-type CKIP-1, i.e., after the M2 Flag. The cDNA encoding M2-CKIP-1 PH (residues 1 to 136) domain was obtained by PCR with the sense oligonucleotide primer used for cloning the full-length CKIP-1 cDNA (above). The antisense primer, containing the last 19 bp of PH domain cDNA followed by a stop codon and a NotI site, was the following 5′ GCGGCCGCTCAGATCCATGACTCCTTCTCT 3′. The resulting PCR product was cloned in pcDNA3, in the EcoRI-NotI sites. The construct containing the constitutively activated PI3-K molecule p110* was kindly provided by Anke Klippel (Atugen, Berlin, Germany). p110* is a chimeric protein in which the region between the two SH2 domains of p85, named iSH2 for inter-SH2, was covalently linked to its binding site at the N terminus of p110, using a flexible hinge region (17). The C-terminal of p110* was modified with a 10-amino-acid Myc epitope tag. The construct pLXSN-Δp85 and pECE-HA-AKT-1 have been previously described (26, 33). The Δp85 mutant is lacking the binding domain (iSH2) for p110. The expression constructs for wild-type and mutant MTM1 and pCMVTag3B-myc-2XFYVE were kindly provided by J. Laporte (IGBMC, Illkirch, France). The expression construct pEGFP-iPXp40 was generously given by C. D. Ellson (The Babraham Institute, Cambridge, United Kingdom). A small interfering RNA (siRNA) of the type AA(N19)UU (N, any nucleotide) was selected from the open reading frame of CKIP-1: AAATTCTGCGGGAAAGGGATTTT, nucleotides 85 to 107. Preannealed sense and antisense oligonucleotides were ordered from Genset. Cell transfections were performed with Lipofectamine Plus (for expression vectors) or Oligofectamine (for siRNA experiments) (Life Technologies, Gibco BRL) according to the manufacturer's instructions.
Cell growth measurement.
DNA synthesis was assayed by measuring BrdU incorporation by enzyme-linked immunosorbent assay (ELISA; cell proliferation ELISA, BrdU, chemiluminescence; Roche). BrdU incorporation was normalized to viable-cell number. Measurements of viable-cell numbers were realized by a colorimetric assay based on the cleavage of the tetrazolium salt WST-1 by a mitochondrial dehydrogenase in viable cells (cell proliferation reagent WST-1; Roche). Both BrdU incorporation measurements and WST-1 assays were carried out with the same wells.
Cell fractionation.
Cells were lysed in HES buffer (20 mM HEPES-NaOH [pH 7.4], 1 mM EDTA, 250 mM sucrose, protease inhibitors [Complete; Roche Molecular Biochemicals], 50 mM NaF, 10 mM β-glycerophosphate, and 6 mM Na3VO4) by trituration through a 25-gauge needle 25 times on ice. The nuclei were removed by centrifugation at 1,500 × g for 10 min at 4°C, and the P100 and S100 fractions were obtained by centrifugation at 100,000 × g for 30 min at 4°C. Supernatants (S100) were removed, and P100 fractions were resuspended in HES buffer containing 0.1% NP-40 and incubated at 4°C for 1 h. Protein concentration was measured (DC kit; Bio-Rad), and equal amounts of each fraction were analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and immunoblotting.
Northern analysis and real-time quantitative PCR.
Total cellular mRNA was isolated from cells grown on tissue culture dishes with Trizol (Gibco BRL). For Northern analysis, 20 μg of mRNA was loaded in each lane of a 1.5% agarose gel containing 5% formaldehyde and electrophoresed in a 1× MOPS (20 mM) buffer. RNA was transferred onto a nylon membrane (Hybond N; Amersham Pharmacia Biotech Inc.). Hybridization was performed overnight at 65°C in buffer containing 1% bovine serum albumin (BSA), 200 mM sodium phosphate buffer, 15% formamide, 1 mM EDTA, and 7% SDS. CKIP-1 probe was prepared from full-length CKIP-1 cDNA using a DNA labeling kit (Ready-To-Go; Amersham Pharmacia Biotech Inc.) to incorporate [32P]dCTP (3,000 Ci/mmol) (Amersham Pharmacia Biotech Inc.). Ethidium bromide staining was used to verify gel loading.
Real-time quantitative reverse transcription-PCR (RT-PCR) experiments were performed with a LightCycler system (Roche) according to the manufacturer's instructions. cDNA synthesis were carried out with random hexamer primers and superscript II RNase H (Gibco BRL). Amplifications were performed with the following primers: 5′-CTACAGGCCTTGCTCAGCTC-3′, nucleotides 394 to 414, and 5′-AGATTGTGGGCGTCTGTAGG-3′, nucleotides 573 to 593, for myogenin; 5′-GTCGATGTTGGTGCTTCTCA-3′, nucleotides 21 to 41, and 5′-AAGCAGCACTCTTCCACGAT-3′, nucleotides 196 to 215, for IGF-II; 5′-CGGTGGAACTTTGACTTCGT-3′, nucleotides 139 to 159, and 5′-GAGTGCAAGACAGCGACAAG-3′, nucleotides 329 to 349, for p21; 5′-TGAACTACCTGGACCGCTTC-3′, nucleotides 245 to 264, and 5′-CCACTTGAGCTTGTTCACCA-3′, nucleotides 431 to 450, for cyclin D1; and 5′-GCTGGTGAAAAGGACCTCT-3′, nucleotides 576 to 594, and 5′-CACAGGACTAGAACACCTGC-3′, nucleotides 805 to 824, for the control, hypoxanthine-guanine phosphoribosyltransferase.
Protein-lipid overlay assay.
Nitrocellulose-immobilized phospholipids from 100 to 1.6 pmol per spot were purchased from Echelon Biosciences Inc. (Salt Lake City, Utah). The membranes were blocked in 5% (wt/vol) fatty acid-free BSA in TBST (50 mM Tris-Cl [pH 7.5] 150 mM NaCl, and 0.1% [vol/vol] Tween 20) for 3 h. The membranes were then incubated overnight with gentle stirring in the same solution containing in vitro-translated proteins. Membranes were then washed six times over 30 min in TBST. Bound proteins were detected by autoradiography. CKIP-1 and CKIP-1ΔPH proteins were obtained in in vitro translation assays (TnT coupled reticulocyte lysate system; Promega, Madison, Wis.) using the T7 promoter in the pcDNA3 vector. The same reaction was performed for AKT-1 except that a PCR amplification of the AKT-1 cDNA plus the T7 promoter sequence was used as the template in the translation reaction.
Immunofluorescence analysis.
Immunofluorescence analysis was performed as follows. C2C12 cells were fixed for 5 min in 3.7% formalin-phosphate-buffered saline (PBS) prior to permeabilization for 5 min with 1% Triton in PBS. Cells were incubated with the primary antibodies diluted in PBS with 1% BSA for 1 h at 37°C or overnight at 4°C, then washed in PBS, and incubated for 30 min with biotinylated anti-rabbit or fluorescein-conjugated anti-mouse antibodies (Amersham). Biotinylated antibodies were revealed after a 30-min incubation with streptavidin-Texas red (Amersham). Primary antibodies were a rabbit polyclonal antibody directed against a peptide encoding 15 amino acids (241 to 255) in the carboxy-terminal part of CKIP-1, a mouse monoclonal antibody against myogenin (F5D) (PharMingen), a mouse monoclonal anti-c-Myc-tag antibody (9E10) (Eurogentec), a mouse monoclonal antihemagglutinin (anti-HA) tag antibody (HA-11) (Eurogentec), a mouse monoclonal anti-M2-Flag antibody (Sigma), and a mouse monoclonal anti-p85 antibody (Santa Cruz). The nuclei were stained with Hoechst reagent 33358 (1 μg/ml bisbenzemidine) (Sigma). Fluorescent images were visualized by confocal microscopy on a Zeiss LSM510 microscope or by regular microscopy on a Zeiss Axioplan 2 microscope. Images were captured with a Photometrics CoolSNAP fx camera and processed with Photoshop version 5.5 (Adobe Systems).
Western blot analysis.
Cells were lysed in buffer (100 mM Tris-Cl [pH 7], 5.2 mM EDTA, 100 mM NaCl, 1% Triton X-100, protease inhibitors [Complete, Roche Molecular Biochemicals], 10 mM β-glycerophosphate, 6 mM sodium vanadate, and 50 mM sodium fluoride). Total protein (50 μg; protein concentrations were determined with a DC kit; Bio-Rad) was separated by SDS-10% PAGE and transferred onto nitrocellulose membranes (Schleicher and Schuell). Membranes were blocked with TBS containing 5% skim milk and incubated overnight at 4°C with primary antibodies. Membranes were washed and incubated for 30 min with peroxidase-conjugated secondary antibodies (Amersham). After several washes in TBS, membranes were incubated with enhanced chemiluminescence reagents (Amersham). Primary antibodies for CKIP-1 and myogenin were those given above. A mouse monoclonal antibody against p21 (SC6246) (Santa Cruz) and a mouse monoclonal antibody against beta tubulin (Amersham) were also used.
RESULTS
Mouse CKIP-1 is upregulated during C2C12 myoblast differentiation.
In attempts to identify new phosphoinositide binding proteins involved in muscle differentiation, we found that the expression of the CKIP-1 mouse homologue (GenBank accession number AF168675) (3) was upregulated upon C2C12 myoblast differentiation. As seen in Fig. 1A, CKIP-1 was expressed at low levels in C2C12 myoblasts cultured in GM. Its expression was readily increased within 2 days after switching to low-serum DM at the time corresponding to the engagement of cells in differentiation, as judged by myogenin induction (Fig. 1A). In accordance with this observation, an immunofluorescence study showed a higher level of staining for CKIP-1 in myogenin-expressing cells before and after fusion into multinucleated myotubes than in proliferative myoblasts (Fig. 1B). Thereafter, the upregulation of CKIP-1 expression lasted up to 5 days in DM (Fig. 1A). The presence of additional bands suggested that CKIP-1 undergoes posttransduction modifications. The increase in CKIP-1 protein paralleled that of its mRNA, showing that the differentiation-dependent changes in CKIP-1 expression are most likely regulated at the transcriptional level (Fig. 1C). p21 expression was maximal 3 days after passage to DM, indicating an irreversible exit from the cell cycle (Fig. 1A).
FIG. 1.
Expression pattern of CKIP-1 in C2C12 cells (A) Immunoblots for endogenous CKIP-1, myogenin, and p21 obtained with whole-cell extracts of C2C12 myoblasts cultured in GM and DM for up to 5 days. Loading was verified by immunoblotting for beta-tubulin. (B) Immunofluorescence for endogenous CKIP-1 and myogenin in C2C12 myoblasts and myotubes. C2C12 cells were cultured in GM (myoblasts) or in DM for 5 days (myotubes), and CKIP-1 (red) and myogenin (green) were visualized with a rabbit polyclonal anti-CKIP-1 antibody and a mouse monoclonal antimyogenin antibody as primary antibodies. The nuclei were stained with Hoechst reagent 33358. Staining was visualized by confocal microscopy. Bar, 5 μm. (C) Northern blot analysis for CKIP-1 transcripts in C2C12 myoblasts (GM) and myotubes after 5 days in DM (DM 5). Equal loading was verified with ethidium bromide-stained ribosomal 28S and 18S RNAs (EtBr).
CKIP-1 is localized at the plasma membrane and in the nucleus in C2C12 cells.
Mouse and human CKIP-1 proteins share 90% homology (data not shown), and both proteins contain a conserved PH domain in their N-terminal part. PH domains usually function as membrane adapters or tethers, linking their host proteins to the membrane. In agreement, when overexpressed in C2C12 myoblasts, CKIP-1 was localized at the plasma membrane but also in the nucleus (Fig. 2A). A mutant CKIP-1 lacking its PH domain was predominantly located in the cytosol, indicating that the PH domain is responsible for both the membrane and nuclear localization of CKIP-1 (Fig. 2A). The intracellular localization of CKIP-1 and CKIP-1ΔPH was confirmed by cell fractionation experiments (Fig. 2B). Most of the wild-type CKIP-1 protein was detected in equal amounts in the nuclear and the P100 (membranes and insoluble cytoplasmic proteins) fractions. In agreement with the immunofluorescence data, a small fraction of CKIP-1 was observed in the S100 (soluble cytoplasmic proteins) fraction. The CKIP-1ΔPH mutant was found predominantly in this S100 fraction.
FIG. 2.
Intracellular localization of CKIP-1 (A) (Top) Schematic presentation of wild-type and mutant CKIP-1 expressed in C2C12 cells. The mutant CKIP-1ΔPH was obtained by deleting the first 146 amino acids of CKIP-1, corresponding to the PH domain. (Bottom) Immunolocalization of wild-type and mutant CKIP-1 proteins in growing C2C12 cells. C2C12 cells were transfected with pcDNA3 CKIP-1 (CKIP-1) or pcDNA3-CKIP-1ΔPH (CKIP-1ΔPH) and cultured in GM for a further 18 h. After fixation, cells were stained with a rabbit anti-CKIP-1 polyclonal antibody and visualized by confocal microscopy. Bar, 5 μm. (B) Distribution of wild-type and mutant CKIP-1 proteins in the nuclear (N), soluble (S100), and particulate (P100) fractions. Fractions were prepared from C2C12 cells expressing wild-type CKIP-1 or CKIP-ΔPH cultured in GM for 18 h following transfection. A 10-μg portion of each fraction was analyzed by immunoblotting using the anti-CKIP-1 polyclonal antibody.
Analysis of the amino acid sequence of CKIP-1 PH domain revealed a strong homology with the PH domain of AKT protein kinases. As shown in Fig. 3A, it shows 38 and 34% homology with mouse AKT-2 and AKT-1, respectively. Upon insulin stimulation, AKT is recruited at the plasma membrane and binds to PtdIns(3,4)P2 and PtdIns(3,4,5)P3 PI3-K products through its PH domain (1). Similarly to AKT-1, CKIP-1 rapidly translocated to the plasma membrane after insulin stimulation, whereas in the absence of any growth factor, CKIP-1 was almost exclusively nuclear (Fig. 3B). CKIP-1 was visualized at the membrane as early as 5 min after the addition of insulin (Fig. 3B) and a significant portion of CKIP-1 was concentrated at the plasma membrane after 10 min. This result was confirmed by cell fractionation experiments (Fig. 3C). Addition of insulin resulted in the redistribution of CKIP-1 from the nucleus to the membrane fraction (P100). Both AKT-1 and CKIP-1 accumulated primarily in membrane ruffles, which are known to form at the edges of the cells following PI3-K activation (34). By contrast, insulin had no effect on CKIP-1ΔPH localization (Fig. 3B), confirming the implication of the PH domain in CKIP-1 membrane localization.
FIG. 3.
Comparison between AKT-and CKIP-1ΔPH domains and subcellular localization after insulin stimulation. (A) Sequence comparison of the CKIP-1 PH domain with those of mouse AKT-2 and AKT-1. Conserved residues are shown in blue, and those predicted to bind 3-phosphoinositides with high affinity are shown in red. (B) C2C12 cells were transiently transfected with pcDNA3-CKIP-1 (CKIP-1) or pECE-HA-AKT-1 (AKT-1) or pcDNA3-CKIP-1ΔPH (CKIP-1ΔPH) plasmids. Eighteen hours after transfection, cells were serum starved by incubation in DMEM alone for 4 h (DMEM) and stimulated with insulin (Ins; 25 μg/ml) for 5 and 10 min. After fixation, cells were stained by indirect immunofluorescence using a rabbit anti-CKIP-1 polyclonal antibody and an anti-HA-tag monoclonal antibody to visualize CKIP-1 (red) and AKT-1 (green), respectively, and visualized by confocal microscopy. Arrows indicate membranous CKIP-1 and AKT-1. Bar, 5 μm. (C) C2C12 cells were transiently transfected with pcDNA3-CKIP-1. Eighteen hours after transfection, cells were serum starved by incubation in DMEM alone for 4 h and kept in DMEM alone (−) or stimulated with insulin (25 μg/ml) (+) for 10 min. Total cell extracts (T) and cellular fractions (N, S100, and P100) were then prepared and immunoblotted for CKIP-1.
CKIP-1 binds to phosphatidylinositol 3-phosphate.
We next investigated the binding of CKIP-1 to phosphoinositide lipids by performing a protein-lipid overlay assay. As expected, AKT-1 interacted with PtdIns(3,4,5)P3 and to a lesser extent with PtdIns(3,4)P2 (Fig. 4A). Surprisingly, CKIP-1 interacted with PtdIns-3P but not with PtdIns(3,4,5)P3 or any other phosphoinositide tested. Deletion of the PH domain completely abolished the interaction of CKIP-1 with PtdIns-3P (Fig. 4A).
FIG. 4.
CKIP-1 binds specifically to PtdIns-3P. (A) In vitro-translated CKIP-1, CKIP-1ΔPH, and AKT-1 proteins were analyzed by SDS-PAGE and autoradiography (top). One and two microliters of each were loaded on the gel. The ability of these proteins to bind a variety of phosphoinositides was analyzed in a protein-lipid overlay assay (bottom). The same amounts of different proteins were incubated with membranes shown in the lower panels. Dilutions of the indicated phosphoinositides are 100, 50, 25, 12.5, 6.3, 3.2, and 1.6 pmol/spot. (B) (Top) Colocalization of CKIP-1 PH and p40phox PX domains on the endosomes and at the plasma membrane. C2C12 cells were transiently transfected with pcDNA3-CKIP-1 PH and pEGFP-p40phox iPX expression vectors. Eighteen hours after transfection, cells were serum starved by incubation in DMEM alone for 4 h and stimulated with insulin (25 μg/ml) for 10 min. After fixation, C2C12 cells were stained by indirect fluorescence. The CKIP-1 PH domain was detected with the anti-M2-Flag antibody (red). The pEGFP-p40phox PX domain was visualized directly (green). Staining was visualized by confocal microscopy. Magnifications of areas outlined in white show the CKIP-1 PH and p40phox PX domain colocalization on the endosomes and at the plasma membrane. Bar, 5 μm. (Bottom) Insulin-induced CKIP-1 and 2XFYVE probe colocalization at the plasma membrane. C2C12 cells were transiently transfected with pcDNA3-CKIP-1 and pCMV-myc-2XFYVE expression vectors. Eighteen hours after transfection, cells were serum starved by incubation in DMEM alone for 4 h and stimulated with insulin (25 μg/ml) for 10 min. After fixation, cells were stained by indirect immunofluorescence using rabbit anti-CKIP-1 polyclonal and anti-Myc tag monoclonal antibodies to visualize CKIP-1 (red) and 2XFYVE (green), respectively. Staining was visualized by confocal microscopy. Bar, 5 μm. (C) C2C12 cells, transiently transfected with CKIP-1 in combination with pEGFP-wt-MTM1 (CKIP-1+MTM1) or pEGFP-MTM1 C375S (CKIP-1+MTM1C375S), were cultured in GM. Eighteen hours after transfection, cells were fixed and analyzed by indirect immunofluorescence for CKIP-1 (red). EGFP-MTM1 and -MTM1C375S (green) were visualized directly. Immunofluorescence analysis was performed with rabbit anti-CKIP-1 polyclonal antibody. Staining was visualized by confocal microscopy. Bar, 5 μm.
To confirm the binding of CKIP-1 PH domain to PtdIns-3P, we compared the intracellular localization of the isolated CKIP-1 PH domain with that of the PtdIns-3P binding p40phox PX domain (37). The CKIP-1 PH domain was found to localize mainly at the plasma membrane but also on cytoplasmic vesicles (Fig. 4B, top). This pattern of staining overlapped partially with that of the p40phox PX domain, indicating that CKIP-1 binds to plasma and endosomal membranes, where PtdIns-3P is present. We next examined the intracellular colocalization of the wild-type CKIP-1 protein and PtdIns-3P. C2C12 cells were cotransfected with CKIP-1 and the PtdIns-3P-specific probe 2XFYVE, which contains two FYVE domains from the receptor tyrosine kinase substrate Hrs (hepatocyte growth factor-regulated tyrosine kinase substrate) fused to a Myc epitope (27). Confocal microscopy revealed that in insulin-stimulated cells, CKIP-1 and the FYVE probe were colocalized at the plasma membrane (Fig. 4B, bottom) but not on the endosomes. In absence of insulin, 2XFYVE was detected only in the endosomes and not at the plasma membrane (data not shown). These results show that the CKIP-1 PH domain binds to PtdIns-3P in vivo and suggest that an interaction between CKIP-1 and another protein is probably necessary for plasma membrane PtdIns-3P targeting of CKIP-1. We then examined the effect of PtdIns-3P dephosphorylation by the 3-phosphoinositide-phosphatase MTM1 on CKIP-1 intracellular distribution. C2C12 cells were cotransfected with CKIP-1 and wild-type MTM1 or with inactive MTM1C375S mutant and kept in serum-containing medium. As shown in Fig. 4C, CKIP-1 was relocalized to the cytosol when coexpressed with wild-type MTM1, whereas the MTM1C375S mutant had no significant effects on CKIP-1 localization.
Altogether, these results show that the PH domain of CKIP-1 is necessary for its localization to the membrane via binding to D3-phosphorylated phosphatidylinositide PtdIns-3P. They also suggest that CKIP-1 localization is regulated by PI3-K.
CKIP-1 intracellular localization is regulated by PI3-K.
Since CKIP-1 binds to a PI3-K product, we next studied the influence of PI3-K activity on CKIP-1 intracellular localization. First, we investigated whether constitutive activation of PI3-K could modify CKIP-1 localization in nonstimulated cells. C2C12 myoblasts were cotransfected with CKIP-1 together with an active form of PI3-K p110*. p110* is composed of the p110 catalytic domain fused to the p85 iSH2 region mediating the interaction with and activation of p110 (17). As shown in Fig. 5A, and consistent with Fig. 3B, in the absence of growth factors, CKIP-1 was mainly localized in the nucleus. By contrast, in cells cotransfected with p110* (Fig. 5A), CKIP-1 was clearly associated with the membranes and only a slight nuclear staining could still be observed. p110* is therefore able to restore the membrane localization of CKIP-1 in serum-starved cells. This result demonstrates that activation of the PI3-K pathway promotes CKIP-1 translocation to the membranes.
FIG. 5.
Interference with the PI3-K pathway modifies CKIP-1 cellular localization. (A) C2C12 cells were transfected with pcDNA3-CKIP-1 alone (CKIP-1) or in combination with the Myc-tagged activated PI3-K p110* (CKIP-1+myc-p110*). Eighteen hours after transfection, cells were serum starved in DMEM alone for 4 h before fixation and immunofluorescence for CKIP-1 (red) with a rabbit anti-CKIP-1 polyclonal antibody and for PI3-K p110* (green) with an anti-c-Myc-tag monoclonal antibody. Staining was visualized by confocal microscopy. Bar, 5 μm. (B) C2C12 cells, transiently transfected with pcDNA3-CKIP-1 alone (CKIP-1) or in combination with pLXSN-Δp85 (CKIP-1 + Δp85), were cultured in GM. CKIP-1 transfected cells were also treated with LY294002 (25 μM) (CKIP-1+LY) for 4 h or wortmannin (300 nM) for 15 min (CKIP-1+W). Eighteen hours after transfection, cells were fixed and analyzed by indirect immunofluorescence for CKIP-1 (red) and cotransfected mutant p85 (green). Immunofluorescence was performed with a rabbit anti-CKIP-1 polyclonal antibody together with mouse anti-p85. Staining was visualized by confocal microscopy. Bar, 5 μm.
We then tested the effect of PI3-K inhibitors on CKIP-1 localization. C2C12 cells expressing CKIP-1 were kept in serum-containing medium and treated with the PI3-K chemical inhibitors LY294002 (25 μM) and wortmannin (300 nM) or cotransfected with a mutant p85 protein. The mutant form of the p85 regulatory subunit of PI3-K (Δp85) that fails to bind and activate the p110 catalytic subunit has been reported to act as a dominant negative mutant of this enzyme. We observed that treatment with LY294002 and wortmannin (Fig. 5B) or coexpression of Δp85 induced a nearly complete relocalization of CKIP-1 in the nucleus similar to that observed after serum starvation (Fig. 5A).
Altogether, these results demonstrate that the PI3-K pathway regulates CKIP-1 intracellular localization and strongly suggest that CKIP-1 may be a downstream component of this pathway. Indeed, activation of PI3-K, and therefore of PtdIns-3P production, promotes the binding of the CKIP-1 PH domain to the membrane, whereas inhibition of PI3-K or dephosphorylation of the D3 position of phosphoinositides by MTM1 abolishes CKIP-1 membrane binding.
CKIP-1 accelerates the proliferative phase that precedes C2C12 cell differentiation.
In C2C12 cells and primary myoblasts (25), differentiation occurs through two steps. Cells first exhibit a transient increase of proliferation, then withdraw from the cell cycle, and finally fuse to form multinucleated myotubes. The increase in CKIP-1 expression upon C2C12 cell differentiation led us to investigate for a possible functional role of CKIP-1 in this process. Expression vectors for wild-type and mutant CKIP-1 were transfected into C2C12 myoblasts. Cotransfection of a plasmid expressing green fluorescent protein (GFP) with the CKIP-1 expression plasmids allowed the estimation of the transfection efficacy to 60% by fluorescence-activated cell sorting quantification of GFP-positive cells (data not shown). Expression of the different CKIP-1 proteins in transfected cells was controlled by immunoblotting with an anti-CKIP-1 polyclonal antibody (Fig. 6A). Eighteen hours after transfection, cells were shifted to DM, and at various times afterwards, viable-cell numbers (WST-1 assay) were measured. Results are expressed as proliferation indexes, i.e., the ratio of the number of viable cells at the indicated times to the number of cells 24 h earlier. As shown in Fig. 6B, 24 h after DM addition, the proliferation index for CKIP-1-transfected cells was 1.8, versus 1.1 for control cells. The latter started to proliferate only between 24 and 48 h. Then the proliferation indexes of CKIP-1-expressing cells decreased progressively to reach 1 at 96 h, indicating that by this time, these cells were no longer proliferating. After 96 h in DM, the proliferation index of control cells was even lower than 1, showing that some cell death did occur between 72 and 96 h, since only viable cells are considered in WST-1 assays. CKIP-1ΔPH-expressing cells did not show any kinetic difference from control cells. BrdU incorporation reflected changes in proliferation indexes, thus indicating that CKIP-1 overexpression primarily affects cell growth and not survival (Fig. 6C).
FIG. 6.
CKIP-1 accelerates the initial peak of proliferation following DM addition and subsequently promotes cell fusion. (A) Immunoblot of wild-type and mutant CKIP-1 proteins transiently transfected in C2C12 cells. Cells were transfected with pcDNA3-CKIP-1, pcDNA3-CKIP-1ΔPH, or the empty pcDNA3 vector. Transfected cells were cultured for 18 h and cell lysates were analyzed by Western blotting with the anti-CKIP-1 polyclonal antibody. Fifty micrograms of total proteins was loaded. Due to a short exposure, endogenous CKIP-1 cannot be observed in exogenous CKIP-1 and CKIP-1ΔPH lanes. (B) Transfected cells were kept in GM for 18 h after transfection. Then they were trypsinized and replated at the same density (3,000 cells per well) in 96-well plates. Each condition was tested in triplicate. Twelve hours later, they were shifted into DM. At different times following addition of DM, viable-cell numbers were monitored in a WST-1 colorimetric assay. Results are expressed as proliferation indexes; i.e., ratio of the number of viable cells at a given time to the number of viable cells 24 h earlier. Data are means ± standard errors of the means for triplicates of one experiment. Two independent experiments were performed. (C) Relative BrdU incorporation and proliferation indexes in CKIP-1-overexpressing cells. The experimental procedure described for panel A was used, and BrdU was added at different times after addition of DM. Twenty-four hours later, BrdU incorporation was measured by ELISA and normalized to viable-cell numbers measured in a WST-1 assay. Results are expressed as the ratio of BrdU incorporation (gray symbols) and proliferation indexes (white symbols) in pcDNA3-CKIP-1-transfected cells versus empty-vector-transfected control cells. Data are means ± standard errors of the means for triplicates of one experiment. Two independent experiments were performed. (D) (Top) Morphological aspects of CKIP-1-overexpressing C2C12 myotubes. C2C12 cells were transiently transfected with pcDNA3-CKIP-1 (CKIP-1) or the empty pcDNA3 vector (V). Eighteen hours following transfection, cells were replated at the same density and shifted to DM. Seventy-two hours after addition of the DM, cells were fixed and analyzed by indirect immunofluorescence for CKIP-1 (red) using a rabbit anti-CKIP-1 polyclonal antibody. The nuclei were stained with Hoechst reagent 33358. Staining was visualized by regular microscopy. PC, phase-contrast image of the same field. Bar, 5 μm. (Bottom) Myotubes distribution according to the number of nuclei (left) and average number of nuclei per myotubes (right) in control empty pcDNA3 vector (black bars) and pcDNA3-CKIP-1 (white bars) transfected C2C12 cells 72 h after DM addition. One hundred different control and CKIP-1-overexpressing myotubes were examined for each condition. Numbers above the bars (right) indicate the percentage of differentiation, i.e., proportion of cells containing three or more nuclei. Only these cells were considered for the calculation of the average of nuclei per myotube.
CKIP-1 stimulates differentiation.
We next addressed the putative role of CKIP-1 in differentiation. Wild-type CKIP-1 and control vectors were transfected into C2C12 myoblasts. Eighteen hours after transfection, cells were shifted to DM for 72 h. As seen above, CKIP-1 overexpression did not noticeably affect the morphology of proliferating myoblasts. In contrast, after 72 h in DM, CKIP-1-overexpressing myoblasts formed thicker myotubes than control vector-transfected cells (Fig. 6D, top). This increase in size was paralleled by an increase in the number of nuclei per myotube (Fig. 6D, bottom). The majority of CKIP-1-overexpressing cells (85.1%) fused into multinucleated myotubes containing three or more nuclei, with an average of seven nuclei per cell. In contrast, only 47% of the control myotubes contained at least three or more nuclei, with an average of 3.8 nuclei per cell. CKIP-1 therefore appears to promote cell fusion.
In conclusion, upon induced differentiation, CKIP-1 overexpression in C2C12 myoblasts first promotes proliferation and then stimulates cell fusion.
CKIP-1 influences cell cycle and differentiation markers.
In order to dissect the early events leading to CKIP-1-stimulated proliferation and differentiation, we analyzed mRNA levels of the markers of differentiation myogenin and IGF-II as well as those of the cell cycle regulators p21 and cyclin D1 in control cells and wild-type- and mutant-CKIP-1-transfected cells. Eighteen hours after transfection, cells were shifted to DM, and at various times after, total mRNAs were extracted and analyzed by real-time quantitative RT-PCR. Levels of expression of CKIP-1 and CKIP-1ΔPH were equivalent to those shown in Fig. 6A. As shown in Fig. 7, 48 h after DM addition, myogenin mRNA levels were significantly increased (1.8-fold) in CKIP-1-expressing cells, whereas CKIP-1ΔPH had no effect, as determined by comparison to control cells transfected with empty pcDNA3 vector. No significant differences in IGF-II levels were observed between CKIP-1, CKIP-1ΔPH, and control cells, which all displayed the classical increase of IGF-II expression between 0 and 48 h after DM addition. There were no substantial differences in the level of p21 between control and CKIP-1ΔPH-transfected cells. CKIP-1 cells displayed a slightly lower level of p21 between 0 and 24 h. Cyclin D1 mRNA levels were twofold higher in CKIP-1-transfected cells than in controls at time zero. By 24 h after DM addition, cyclin D1 expression increased in control and CKIP-1ΔPH-transfected cells, while it remained constant in CKIP-1-expressing cells. At this time (24 h), cyclin D1 mRNA levels were thus equivalent in all the cells tested. As expected in differentiating cells, after 48 h in DM, the cyclin D1 mRNA level dramatically dropped in all conditions. These results suggest that CKIP-1 stimulates an initial proliferative phase via cyclin D1 and differentiation via myogenin expression. The inability of the CKIP-1ΔPH mutant to promote either cyclin D1 or myogenin expression suggests that membrane association and probably PtdIns-3P binding are required for CKIP-1 action.
FIG. 7.
CKIP-1 modulates accumulation of transcripts encoding myoblast proliferation and differentiation markers. Myogenin, IGF-II, cyclin D1, and p21 mRNA expression in C2C12 cells transfected with control empty pcDNA3 vector (V), pcDNA3-CKIP-1, and pcDNA3-CKIP-1ΔPH. Cells were first grown for 18 h in GM following transfection and then shifted to DM (time zero). At various time points after addition of DM, total mRNA was extracted, and the amount of mRNA for myogenin, IGF-II, cyclin D1, and p21 were quantified by real-time quantitative RT-PCR. An hypoxanthine-guanine phosphoribosyltransferase RT-PCR control reaction was carried out to normalize the amount of mRNA included in each RT-PCR. Each measure was performed in duplicate. Results are expressed as the increase relative to empty pCDNA3 vector-transfected cells at time zero (before the addition of DM). Data are means of duplicates of one representative experiment. Four independent experiments were performed.
Inhibition of CKIP-1-induced proliferation and myogenin increase by LY294002.
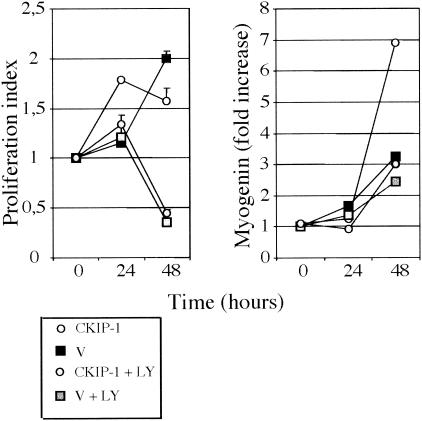
The PI3-K pathway is a key regulator of muscle differentiation. Our results suggest that CKIP-1 may be a component of this pathway. We therefore tested the influence of the PI3-K inhibitor LY294002 on CKIP-1-stimulated proliferation and myogenin expression. The previous experiments were repeated in the presence of the LY294002 inhibitor. As shown in Fig. 8 (left panel), LY294002 inhibited the transient increase in cell proliferation of CKIP-1-overexpressing and control cells. The respective MAP and p38 kinase inhibitors PD098059 and SB202190 had no effect on CKIP-1-induced proliferation (data not shown). LY294002 also abolished the effect of CKIP-1 on myogenin expression stimulation (Fig. 8, right panel).
FIG. 8.
Interference with the PI3-K pathway modifies CKIP-1-induced proliferation and CKIP-1-stimulated myogenin expression. C2C12 cells were transfected with pcDNA3-wt CKIP-1 alone or empty vector (V). Transfected cells were kept in GM for 18 h after transfection. After replating as for Fig. 6B and Fig. 7A, they were shifted into DM. Under some conditions, transfected cells were treated with LY294002 (25 μM) (LY). At different times following addition of DM, viable-cell number (left) was monitored in a WST-1 assay. Results are expressed as in Fig. 7A. Data are means ± standard errors of the means for triplicates of one experiment. Myogenin expression (right) was quantified by real-time quantitative RT-PCR as for Fig. 6A. Two (WST-1 assay) and three (myogenin) independent experiments were performed.
It thus appears that CKIP-1 affects both steps of differentiation in a PI3-K-dependent manner.
CKIP-1 silencing by siRNA affects cell proliferation and expression of the differentiation markers myogenin and p21.
Our results showed that CKIP-1 expression stimulates C2C12 differentiation. To determine whether CKIP-1 is required for this process to occur, we abolished its expression by siRNA. C2C12 cells were transfected with a 21-nucleotide siRNA duplex directed towards CKIP-1 mRNA or with the control sense RNA strand alone. After 48 h of culture in GM, the cells were trypsinized and replated at the same density in 96-well plates at 3,000 cells per well for DNA synthesis measurements and in 12-well plates at 30,000 cells per plate for protein expression analysis. Three hours later, they were shifted into DM. At various times following DM addition, BrdU incorporation was measured. Expression of endogenous CKIP-1 and differentiation markers myogenin and p21 was monitored in parallel by immunoblotting.
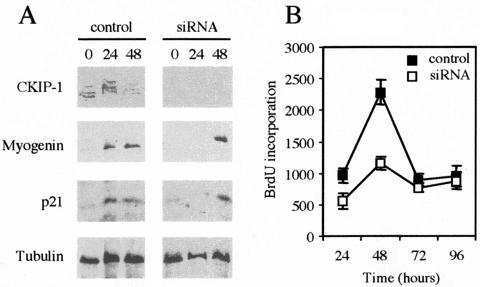
As expected, CKIP-1 expression was abolished in the siRNA-transfected cells (Fig. 9A). In these cells, myogenin and p21 expression was clearly delayed 24 h compared to expression in control cells, whereas expression of the control, beta-tubulin, was not affected (Fig. 9A). Consistently, 48 h after addition of DM, BrdU incorporation in siRNA cells was 50% lower than in control cells (Fig. 9B). No morphological changes between control and siRNA treated C2C12 myoblasts were observed (data not shown). The absence of CKIP-1 thus both reduces the initial proliferative phase and delays the second phase of differentiation, i.e., myogenin and p21 expression.
FIG. 9.
Silencing of CKIP-1 by siRNA abolishes the initial proliferative phase after DM addition and delays the expression of myogenin and p21. C2C12 cells were transfected with the double-stranded RNA specific for CKIP-1 (siRNA) or the sense strand alone (control). Forty-eight hours after transfection, cells were trypsinized and replated in 96- and 12-well plates. Twelve hours later, they were shifted into DM and at different time points, total protein extracts were prepared for CKIP-1, myogenin, p21, and tubulin expression analysis by immunoblotting and measure of BrdU incorporation. (A) Immunoblots for CKIP-1, myogenin, p21, and tubulin in control and CKIP-1 siRNA cells. (B) BrdU incorporation at different times following addition of DM in CKIP-1 siRNA cells and control cells. BrdU incorporation was normalized to viable-cell number. Data are means ± standard errors of the means for triplicates of one experiment. Two independent experiments were performed.
Altogether, these data demonstrate that CKIP-1 participates in the onset of the different PI3-K-mediated muscle differentiation processes.
DISCUSSION
In this work, we report the implication of the PH domain-containing protein CKIP-1 in PI3-K signaling. We have shown that CKIP-1, through its PH domain, interacts with PtdIns-3P in vitro. In agreement, we showed that the isolated CKIP-1 PH domain is located at the membrane but also on the endosomes and it localizes with the PtdIns-3P binding PX domain of p40phox (37). This confirmed that CKIP PH domain binds to PtdIns-3P in vivo. However, the entire CKIP-1 protein binds exclusively to plasma membrane PtdIns-3P. Indeed, it was found to be colocalized with the other PtdIns-3P-specific probe, 2XFYVE, at the plasma membrane. This result suggests that an interaction between CKIP-1 and another protein is necessary for the plasma membrane targeting of CKIP-1. It has been well established that, among the different products of PI3-K, PtdIns(3,4,5)P3 is the crucial second messenger in many signaling events (16). By contrast, it is generally considered that PtdIns-3P levels do not change upon cellular stimulation, and it has been shown that PtdIns-3P localizes mainly onto the endosomes where it interacts with FYVE finger proteins that regulate the endocytic pathway (28). However, a recent report clearly demonstrated that a plasma membrane-associated subpool of these lipids can be produced upon insulin stimulation in L6 cells and 3T3-L1 adipocytes (21). In agreement with our data, Maffucci et al. (21) also showed that the PtdIns-3P-specific 2XFYVE probe, localized only in endosomal structures in the absence of insulin, was translocated to the plasma membrane shortly after insulin stimulation. This translocation is specifically induced by insulin and not by other growth factors, such as epidermal growth factor and platelet-derived growth factor. These results strongly support our data and suggest that PtdIns-3P, like other products of PI3-K, can act as lipid second messengers in insulin signaling. Despite its binding specificity for PtdIns-3P, CKIP-1 was never found to be colocalized with the 2XFYVE probe at the endosomes. Another PtdIns-3P binding protein containing a PH domain, insulin receptor substrate 3 (20), was shown to localize at the plasma membrane but not in the PtdIns-3P-coated endosomes, suggesting the existence of a PtdIns-3P-dependent pathway different from that regulating vesicle trafficking.
An activated form of PI3-K promoted CKIP-1 association with membranes, whereas a reduction of PtdIns-3P by expression of wild-type MTM1 abolished CKIP-1 binding to the plasma membrane. Consistently, inhibition of PI3-K by coexpression of a dominant negative PI3-K Δp85 mutant or addition of LY294002 or wortmannin also prevented CKIP-1 binding to the membranes. These data clearly demonstrate that PI3-K regulates CKIP-1 cellular localization via the binding of its PH domain to PtdIns-3P. Given the essential role of MTM1 in muscle (4) and its effect on CKIP-1 localization, a potential connection between these two proteins in PI3-K signaling will have to be investigated.
CKIP-1 was found to be upregulated both at the mRNA and protein levels in differentiating C2C12 myoblasts, suggesting a positive role for CKIP-1 in the differentiation process. In C2C12 cells and primary myoblasts (25), differentiation occurs through two steps, an initial proliferative response followed by a myogenic response characterized by expression of myogenin and p21 and cell fusion. When overexpressed in C2C12 myoblasts, CKIP-1 accelerates the initial and transient increase in cell proliferation (as evidenced by the increase of cyclin D1 expression and proliferation index) and then promotes myogenin expression and cell fusion. Indeed, CKIP-1-expressing C2C12 cells formed thicker myotubes containing an average of twice as many nuclei as control vector-transfected cells. The inhibition of endogenous CKIP-1 by siRNA abolished the initial proliferative phase and delayed myogenin expression. These data clearly implicate CKIP-1 in the process of differentiation. The dual effect of CKIP-1 on both proliferation and differentiation of C2C12 cells is reminiscent of the known effects of IGF-I on muscle cells. Indeed, exogenous IGF potently stimulates myogenesis in cultured cells by first promoting myoblast proliferation and then stimulating the expression of differentiation markers (7). Moreover, IGF-I-treated myoblasts (10) form larger multinucleated myotubes than control cells.
Recent studies have demonstrated that the PI3-K pathway mediates the stimulatory effects of IGFs on muscle differentiation (7, 32). In addition, it is now well established that PI3-K is also involved in the hypertrophic response of muscle to IGF (24). Our data show that CKIP-1 is involved in the same steps as those controlled by the IGF/PI3-K pathway and suggest that CKIP-1 may mediate some of the effects of PI3-K in this process and therefore be a novel component of this pathway. Accordingly, the PI3-K inhibitor LY294002 consistently abolished the proliferative effect as well as the myogenin increase stimulated by CKIP-1. We can therefore conclude that CKIP-1 affects C2C12 cell differentiation in a PI3-K-dependent manner. Moreover, our results are in favor of a model in which in muscle differentiation, PI3-K signaling not only occurs through the well known PtdIns(3,4,5)P3 and AKT secondary intermediates but also can signal via PtdIns-3P and CKIP-1.
CKIP-1, with no identifiable catalytic domain, may be one scaffolding adapter. Indeed, CKIP-1 contains one PH domain that specifically binds PtdIns-3P and numerous PXXP motifs (3), signatures of the binding sites for SH3 domain-containing proteins. Several tyrosines within potential binding motifs for SH2-containing proteins are present in the CKIP-1 amino acid sequence (data not shown). CKIP-1 shuttles between the membrane and the nucleus. As a scaffolding protein, CKIP-1 may target signaling complexes between these two compartments in a PI3-K-dependent manner. CKIP-1 was isolated in a yeast two-hybrid screen with CK2α as bait (3). We have not detected CKIP-1 binding to CK2α in C2C12 cells. Moreover, when CKIP-1 and CK2α are coexpressed, no modifications of CK2α subcellular localization dependent on PI3-K activity could be observed. However, we cannot exclude the possibility that CKIP-1 is a regulator of CK2α in other cellular processes regulated by PI3-K. Indeed, several recent reports described a role for CK2 in PI3-K-regulated cell survival (31). Alternatively, one can imagine that CKIP-1 may serve as a signaling intermediate participating in the regulation of a transcription complex. Depending on its partner in AP-1 complexes, c-Jun regulates either muscle proliferation or differentiation (2, 8). CKIP-1 contains a leucine zipper motif (3) that was identified on the basis of its ability to interact with the leucine zipper of c-Jun (6). It is therefore conceivable that CKIP-1, by interacting with c-Jun or other components of AP-1 complexes, for example, may modulate their positive and/or negative effects on muscle differentiation.
In order to define the molecular mechanisms of CKIP-1 action in PI3-K-regulated differentiation, we are characterizing CKIP-1 membrane and nuclear partners.
Acknowledgments
We are grateful to Marc Billaud (CNRS UMR 5641, Lyon, France) for providing plasmids and for helpful discussions. We thank Anke Klippel, Atugen, for the PI3-K p110* mutant. We are grateful to Jocelyne Laporte (IGBMC, Strasbourg, France), Harald Stenmark (Norwegian Radium Hospital, Oslo, Norway), and Chris Ellson (The Babraham Institute, Cambridge, United Kingdom) for providing MTM-1, the 2XFYVE probe, and EGFP-ip40phox vectors, respectively.
This work was supported by the Association pour la Recherche sur le Cancer and the Association Française contre les Myopathies. L.V. is a Ph.D. fellow of La Ligue Nationale Contre le Cancer.
REFERENCES
- 1.Alessi, D. R., S. R. James, C. P. Downes, A. B. Holmes, P. R. Gaffney, C. B. Reese, and P. Cohen. 1997. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase B alpha. Curr. Biol. 7:261-269. [DOI] [PubMed] [Google Scholar]
- 2.Andreucci, J. J., D. Grant, D. M. Cox, L. K. Tomc, R. Prywes, D. J. Goldhamer, N. Rodrigues, P. A. Bedard, and J. C. McDermott. 2002. Composition and function of AP-1 transcription complexes during muscle cell differentiation. J. Biol. Chem. 277:16426-16432. [DOI] [PubMed] [Google Scholar]
- 3.Bosc, D. G., K. C. Graham, R. B. Saulnier, C. Zhang, D. Prober, R. D. Gietz, and D. W. Litchfield. 2000. Identification and characterization of CKIP-1, a novel pleckstrin homology domain-containing protein that interacts with protein kinase CK2. J. Biol. Chem. 275:14295-142306. [DOI] [PubMed] [Google Scholar]
- 4.Buj-Bello, A., V. Laugel, N. Messaddeq, H. Zahreddine, J. Laporte, J. F. Pellissier, and J. L. Mandel. 2002. The lipid phosphatase myotubularin is essential for skeletal muscle maintenance but not for myogenesis in mice. Proc. Natl. Acad. Sci. USA 99:15060-15065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calera, M. R., and P. F. Pilch. 1998. Induction of Akt-2 correlates with differentiation in Sol8 muscle cells. Biochem. Biophys. Res. Commun. 251:835-841. [DOI] [PubMed] [Google Scholar]
- 6.Chevray, P. M., and D. Nathans. 1992. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc. Natl. Acad. Sci. USA 89:5789-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coolican, S. A., D. S. Samuel, D. Z. Ewton, F. J. McWade, and J. R. Florini. 1997. The mitogenic and myogenic actions of insulin-like growth factors utilize distinct signaling pathways. J. Biol. Chem. 272:6653-6662. [DOI] [PubMed] [Google Scholar]
- 8.Daury, L., M. Busson, N. Tourkine, F. Casas, I. Cassar-Malek, C. Wrutbiak-Cabello, M. Castellazi, and M. Cabello. 2001. Opposing functions of ATF2 and Fos-like transcription factors in c-Jun-mediated myogenin expression and terminal differentiation in avian myoblasts. Oncogene 20:7998-8008. [DOI] [PubMed] [Google Scholar]
- 9.Ellson, C. D., S. Andrews, L. R. Stephens, and P. T. Hawkins. 2001. The PX domain: a new phosphoinositide-binding module. Nature 3:679-682. [DOI] [PubMed] [Google Scholar]
- 10.Engert, J. C., E. B. Berglund, and N. Rosenthal. 1996. Proliferation precedes differentiation in IGF-I-stimulated myogenesis. J. Cell Biol. 135:431-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewton, D. Z., S. L. Roof, K. A. Magri, F. J. McWade, and J. R. Florini. 1994. IGF-II is more active than IGF-I in stimulating L6A1 myogenesis: greater mitogenic actions of IGF-I delay differentiation. J. Cell Physiol. 161:277-284. [DOI] [PubMed] [Google Scholar]
- 12.Florini, J. R., K. A. Magri, D. Z. Ewton, P. L. James, K. Grindstaff, and P. S. Rotwein. 1991. “Spontaneous” differentiation of skeletal myoblasts is dependent upon autocrine secretion of insulin-like growth factor-II. J. Biol. Chem. 266:15917-15923. [PubMed] [Google Scholar]
- 13.Jiang, B. H., M. Aoki, J. Z. Zheng, J. Li, and P. K. Vogt. 1999. Myogenic signaling of phosphatidylinositol 3-kinase requires the serine-threonine kinase Akt/protein kinase B. Proc. Natl. Acad. Sci. USA 96:2077-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang, B. H., J. Z. Zheng, and P. K. Vogt. 1998. An essential role of phosphatidylinositol 3-kinase in myogenic differentiation. Proc. Natl. Acad. Sci. USA 95:14179-14183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaliman, P., F. Vinals, X. Testar, M. Palacin, and A. Zorzano. 1996. Phosphatidylinositol 3-kinase inhibitors block differentiation of skeletal muscle cells. J. Biol. Chem. 271:19146-19151. [DOI] [PubMed] [Google Scholar]
- 16.Katso, R., K. Okkenhaug, K. Ahmadi, S. White, J. Timms, and M. D. Waterfield. 2001. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu. Rev. Cell Dev. Biol. 17:615-675. [DOI] [PubMed] [Google Scholar]
- 17.Klippel, A., C. Reinhard, W. M. Kavanaugh, G. Apell, M. A. Escobedo, and L. T. Williams. 1996. Membrane localization of phosphatidylinositol 3-kinase is sufficient to activate multiple signal-transducing kinase pathways. Mol. Cell. Biol. 16:4117-4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, J. P., J. Baker, A. S. Perkins, E. J. Robertson, and A. Efstratiadis. 1993. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 75:59-72. [PubMed] [Google Scholar]
- 19.Maffucci, T., and M. Falasca. 2001. Specificity in pleckstrin homology (PH) domain membrane targeting: a role for a phosphoinositide-protein co-operative mechanism. FEBS Lett. 506:173-179. [DOI] [PubMed] [Google Scholar]
- 20.Maffucci, T., G. Razzini, A. Ingrosso, H. Chen, S. Iacobelli, S. Sciacchitano, M. J. Quon, and M. Falasca. 2003. Role of pleckstrin homology domain in regulating membrane targeting and metabolic function of insulin receptor substrate 3. Mol. Endocrinol. 17:1569-1579. [DOI] [PubMed] [Google Scholar]
- 21.Maffucci, T., A. Brancaccio, E. Piccolo, R. Stein, and M. Falasca. 2003. Insulin induces phosphatidylinositol-3-phosphate formation through TC10 activation. EMBO J. 22:4178-4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montarras, D., F. Aurade, T. Johnson, J. IIlan, F. Gros, and C. Pinset. 1996. Autonomous differentiation in the mouse myogenic cell line, C2, involves a mutual positive control between insulin-like growth factor II and MyoD, operating as early as at the myoblast stage. J. Cell Sci. 109:551-560. [DOI] [PubMed] [Google Scholar]
- 23.Powell-Braxton, L., P. Hollingshead, C. Warburton, M. Dowd, S. Pitts-Meek, D. Dalton, N. Gillett, and T. A. Stewart. 1993. IGF-I is required for normal embryonic growth in mice. Genes Dev. 7:2609-2617. [DOI] [PubMed] [Google Scholar]
- 24.Rommel, C., S. C. Bodine, B. A. Clarke, R. Rossman, L. Nunez, T. N. Stitt, G. D. Yancopoulos, and D. J. Glass. 2001. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 3:1009-1013. [DOI] [PubMed] [Google Scholar]
- 25.Sabourin, L. A., A. Girgis-Gabardo, P. Seale, A. Asakura, and M. A. Rudnicki. 1999. Reduced differentiation potential of primary MyoD−/− myogenic cells derived from adult skeletal muscle. J. Cell Biol. 144:631-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segouffin-Cariou, C., and M. Billaud. 2000. Transforming ability of MEN2A-RET requires activation of the phosphatidylinositol 3-kinase/AKT signaling pathway. J. Biol. Chem. 275:3568-3576. [DOI] [PubMed] [Google Scholar]
- 27.Stenmark, H., R. Aasland, and P. C. Driscoll. 2002. The phosphatidylinositol 3-phosphate-binding FYVE finger. FEBS Lett. 513:77-84. [DOI] [PubMed] [Google Scholar]
- 28.Stenmark, H., and D. J. Gillooly. 2001. Intracellular trafficking and turnover of phosphatidylinositol 3-phosphate. Semin. Cell Dev. Biol. 12:193-199. [DOI] [PubMed] [Google Scholar]
- 29.Stewart, C. E., and P. Rotwein. 1996. Insulin-like growth factor-II is an autocrine survival factor for differentiating myoblasts. J. Biol. Chem. 271:11330-11338. [DOI] [PubMed] [Google Scholar]
- 30.Tamir, Y., and E. Bengal. 2000. Phosphoinositide 3-kinase induces the transcriptional activity of MEF2 proteins during muscle differentiation. J. Biol. Chem. 275:34424-34432. [DOI] [PubMed] [Google Scholar]
- 31.Torres, J., J. Rodriguez, M. P. Myers, M. Valiente, J. D. Graves, N. K. Tonks, and R. Pulido. 2003. Phosphorylation-regulated cleavage of the tumor suppressor PTEN by caspase-3: implications for the control of protein stability and PTEN-protein interactions. J. Biol. Chem. 278:30652-30660. [DOI] [PubMed] [Google Scholar]
- 32.Tureckova, J., E. M. Wilson, J. L. Cappalonga, and P. Rotwein. 2001. Insulin-like growth factor-mediated muscle differentiation: collaboration between phosphatidylinositol 3-kinase-Akt-signaling pathways and myogenin. J. Biol. Chem. 276:39264-39270. [DOI] [PubMed] [Google Scholar]
- 33.Vandromme, M., A. Rochat, R. Meier, G. Carnac, D. Besser, B. A. Hemmings, A. Fernandez, and N. J. Lamb. 2001. Protein kinase B β/Akt2 plays a specific role in muscle differentiation. J. Biol. Chem. 276:8173-8179. [DOI] [PubMed] [Google Scholar]
- 34.Watton, S. J., and J. Downward. 1999. Akt/PKB localisation and 3′ phosphoinositide generation at sites of epithelial cell-matrix and cell-cell interaction. Curr Biol. 9:433-436. [DOI] [PubMed] [Google Scholar]
- 35.Wishart, M. J., and J. E. Dixon. 2002. PTEN and myotubularin phosphatases: from 3-phosphoinositide dephosphorylation to disease. Phosphatase and tensin homolog deleted on chromosome ten. Trends Cell Biol. 12:579-585. [DOI] [PubMed] [Google Scholar]
- 36.Xu, Q., and Z. Wu. 2000. The insulin-like growth factor-phosphatidylinositol 3-kinase-Akt signaling pathway regulates myogenin expression in normal myogenic cells but not in rhabdomyosarcoma-derived RD cells. J. Biol. Chem. 275:36750-36757. [DOI] [PubMed] [Google Scholar]
- 37.Zhan, Y., J. V. Virbasius, X. Song, D. Pomerleau, and G. W. Zhou. 2002. The p40phox and p47phox PX domains of NADPH oxidase target cell membranes via direct and indirect recruitment by phosphoinositides. J. Biol. Chem. 277:4512-4518. [DOI] [PubMed] [Google Scholar]