Abstract
Transforming growth factor β (TGF-β) superfamily members signal via complexes of activated Smads, comprising phosphorylated receptor-regulated Smads, such as Smad2 and Smad3, and Smad4. These complexes are recruited to DNA by specific transcription factors. The forkhead/winged-helix transcription factors, XFast-1/XFoxH1a and XFast-3/XFoxH1b, bind an activated Smad heterotrimer comprising two Smad2s and one Smad4. Here we identify a novel Smad2 interaction motif, the Fast/FoxH1 motif (FM), present in all known Fast/FoxH1 family members, N-terminal to the common Smad interaction motif (SIM). The FM is necessary and sufficient to bind active Smad2/Smad4 complexes. The FM differs from the SIM since it discriminates between Smad2 and Smad3, and moreover only binds phosphorylated Smad2 in the context of activated Smad complexes. It is the first Smad interaction motif with this property. Site-directed mutagenesis indicates that the binding site for the FM on a Smad2/Smad4 heterotrimer is a hydrophobic pocket that incorporates the Smad/Smad interface. We demonstrate that the presence of an FM and SIM in the Fast/FoxH1 proteins allows them to compete efficiently for activated Smad2/Smad4 complexes with transcription factors such as Mixer that only contain a SIM. This establishes a hierarchy of Smad-interacting transcription factors, determined by their affinity for active Smad complexes.
Members of the transforming growth factor β (TGF-β) superfamily signal to the nucleus through activation of the Smads (23). Ligand binding activates a heterotetrameric complex of two type II and two type I serine/threonine kinase receptors, which phosphorylate and activate receptor-regulated Smads (R-Smads) at two specific serines in their C-terminal SSXS motif. Type I receptors for TGF-β, Activin, and Nodal activate the R-Smads, Smad2 and Smad3, and the FYVE domain-containing protein SARA (Smad anchor for receptor activation) recruits these R-Smads to the receptors for phosphorylation. Once phosphorylated, the R-Smads form heteromeric complexes with the common-mediator Smad (Co-Smad) Smad4, which accumulate in the nucleus where they regulate transcription of TGF-β target genes.
Recruitment of Smads to promoter elements is achieved at least in part by interaction with transcription factors. In early Xenopus embryos, complexes of XSmad2 and the Xenopus Smad4, XSmad4β (12) are recruited to the distal element (DE) of the goosecoid promoter through the paired-like homeodomain transcription factors of the Mix family, Mixer, Milk and Bix3 (9, 32), and to the Activin-responsive element (ARE) of the Mix.2 promoter through the forkhead/winged-helix transcription factors of the Fast/FoxH1 family, XFast-1 and XFast-3 (4, 13), now referred to as XFoxH1a and XFoxH1b, respectively (Daniel Martínez, personal communication). In all cases the transcription factors interact with the C-terminal MH2 domain (Mad homology domain 2) of Smad2 and through this interaction bind an activated Smad2/Smad4 complex which is required for transcriptional activation.
The stoichiometry of Smad complexes in vitro has been somewhat controversial, with evidence existing for both dimers and trimers. Crystallographic evidence suggests that Smad complexes are trimers, interacting via their MH2 domains. The Smad4 MH2 domain monomer contains a core β-sandwich capped by a three-helix bundle at one end and a loop-helix region at the other (33). The Smad4 MH2 domains form a trimer which has 3 identical protein-protein interfaces comprising the three-helix bundle of one subunit packed against the loop-helix region of the adjacent subunit (33). The crystal structure of the phosphorylated Smad2 MH2 domain indicates that it too is a trimer, similar to Smad4 (42), except that it is strengthened by the phosphorylated C terminus of one monomer contacting the L3 loop/B8 β-strand pocket of the adjacent monomer. Heterotrimers of Smads may assemble in a similar manner (3, 31). However, there is also some biochemical evidence for dimeric Smad2/Smad4 complexes (41, 42), suggesting that Smads may be able to form dimers or trimers. In support of this view, Smad2 has recently been shown to form heterotrimers with Smad4 when complexed with XFoxH1a or XFoxH1b to form Activin-responsive factor (ARF) on the ARE, while the Smad3/Smad4 complexes that interact with the Smad-binding region of the c-Jun promoter are heterodimers (16).
The interactions of Smads with partner proteins are fundamental to the signaling pathway and critical for determining signaling specificity. It is therefore essential to understand these interactions at the molecular level. Members of the FoxH1 and Mix families interact with Smad2 through a highly conserved proline-rich Smad interaction motif (SIM) present in the C-terminal domain of these transcription factors, which binds the Smad2 MH2 domain (9, 32). The SIM can also interact with the MH2 domain of Smad3, and recent work has demonstrated that endogenous Smad3/Smad4-containing complexes that bind the c-Jun Smad-binding region or the Smad7 Smad binding element include SIM-containing transcription factors (16). The SIM has significant sequence similarity with the proline-rich rigid coil region of the SARA Smad-binding domain, which interacts with a shallow hydrophobic pocket in the MH2 domain of Smad2 and Smad3 (32, 40). The Mixer and the XFoxH1a SIMs have been shown to interact with a region of this same hydrophobic pocket (32, 43).
Mutation of the SIM in Mixer is sufficient to completely abolish interaction with Smad2 (9). However, we show here that XFoxH1a and XFoxH1b containing mutated SIMs still retain some ability to interact with Smad2/Smad4 complexes. This led us to the identification of a novel Smad2 interaction motif that is present within the C-terminal domain of all FoxH1 family members, N-terminal to the SIM in a region previously shown to be important for ARF formation (5). This motif, which we have called the Fast/FoxH1 motif (FM) is highly conserved. We demonstrate that the FM is required for the FoxH1 proteins to interact with active Smad2/Smad4 complexes. It is also sufficient to interact with activated Smad2/Smad4 complexes. In contrast to the SIM which binds both unphosphorylated and phosphorylated Smad2 and Smad3, the FM discriminates between these two R-Smads. Moreover, the FM only binds phosphorylated Smad2 in the context of activated Smad complexes. We identify a number of residues in the Smad2 MH2 domain with which the FM interacts, and consequently, a potential FM-binding pocket in the activated Smad2/Smad4 trimer. The presence of an FM and a SIM in the FoxH1s results in stronger binding to activated Smad2/Smad4 complexes than is achieved with a SIM alone. This establishes a hierarchy of Smad-interacting transcription factors, determined by their affinity for active Smad complexes, which has obvious implications for determining the specificity of ligand-induced transcriptional responses.
MATERIALS AND METHODS
Plasmids.
The following plasmids have been described: Mixer in pFTX9, pGEX-KG and pGEX-KG containing XSmad2C (9); XFast-1/XFoxH1a and XFast-3/XFoxH1b in pEF-HA and pEF-Flag (13); pSV7d-TβRI (8); pCMV5-HA-TβRII (39); pGal4 (1-95) and p(Gal4-OP)5-Luc (32); (ARE)3-Luc, (DE)4-Luc and pEFLacZ (28); and Pitx2 ASE-Luc (34). XSmad2C and its derivatives (9) were subcloned into pEF-Flag. Mixer was subcloned into pEF-HA, and XFoxH1b was subcloned into pFTX9. Mutations of full-length XSmad2 and XFoxH1a and XFoxH1b were made using standard PCR methods. The residues in XFoxH1a and XFoxH1b that were mutated to alanine are shown in Fig. 1D. Fusions were made of the Gal4 (1-95) DNA-binding domain with either the XFoxH1a or XFoxH1b SIM (amino acids 470 to 495 and 293 to 320, respectively), the XFoxH1a FM (amino acids 400 to 425), and the XFoxH1b FM (amino acids 219 to 244), and mutants thereof (the same as those made in the context of the full-length XFoxH1a and XFoxH1b). pEF-(HA)4 XSmad2 is an expression vector containing 4 HA tags upstream of XSmad2. All constructs were verified by sequencing.
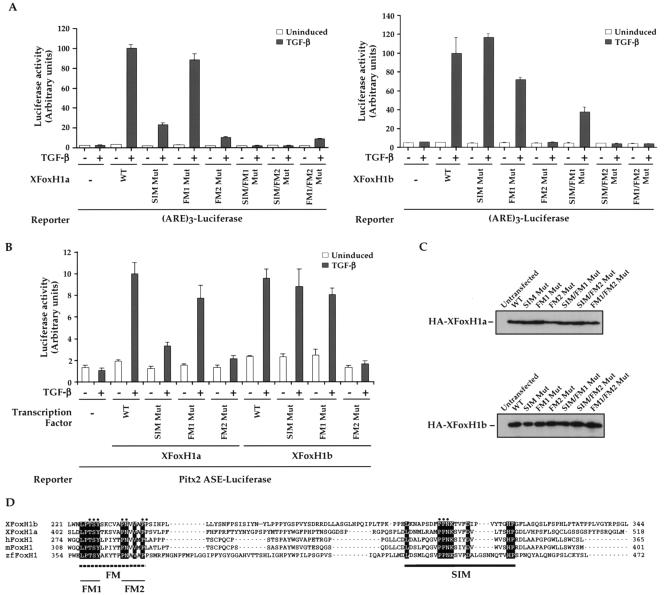
FIG. 1.
The FM is required in both XFoxH1a and XFoxH1b for mediating TGF-β-induced transcriptional activation through recruitment of active Smad2/Smad4 complexes. (A and B) NIH 3T3 cells were transfected with the (ARE)3-Luc reporter (A) or the Pitx2 ASE-Luc reporter (B) either alone or with plasmids expressing HA-tagged wild-type or mutant derivatives of XFoxH1a or XFoxH1b as indicated and treated with or without TGF-β. Luciferase was quantitated relative to β-galactosidase from the pEFLacZ internal control. The data are the means and standard deviations of three independent experiments. (C) Levels of expression of the mutants are shown by Western blot. (D) Alignment of the C-terminal regions of the five known FoxH1 family members: X, Xenopus; h, human; m, mouse; zf, zebrafish. Black dotted underlining indicates the Fast/FoxH1 motif (FM), with FM1 and FM2 denoted; black underlining indicates the Smad interaction motif (SIM). The dots indicate the residues that were mutated to alanines in the XFoxH1 mutants, and also in the Gal4-FM and Gal4-SIM mutants (see Fig. 3).
Transfections and reporter assays.
Maintenance of NIH 3T3, HaCaT and MDA-MB468 cells, transfection and reporter assays were as described (28) except that HaCaT cells were transfected using FuGENE (Roche). Smad2-null mouse embryo fibroblasts were cultured in Dulbecco's modified Eagle's medium (DMEM)/10% fetal bovine serum. TGF-β1 (Peprotech Ltd) was used at 2 ng/ml. For reporter assays, cells were induced with TGF-β for 8 h; for all other assays, inductions were for 1 h unless otherwise stated.
Band shift assays and glutathione S-transferase (GST) and peptide pulldowns.
Band shift probes corresponding to the Mixer binding site, c-Jun Smad-binding region or the ARE were generated as described (9, 16). Band shift assays were as previously described: those in Fig. 2 (9), those in Fig. 4A using in vitro translated Mixer, and GSTSmad2C (32), those in Fig. 4B and C using nuclear extracts (13), and those in Fig. 6B and C (16). GST and peptide pulldowns were performed as described previously (32). The wild-type and mutant Mixer SIM peptides have been previously described (9), and the XFoxH1a and XFoxH1b SIM peptides were as described (13). The XFoxH1a FM peptide was biotin-aminohexanoic acid-RQIKIWFQNRRMKWKKLLSLDLPTSYTKSVAPNVVAPPSVLP, where the last 26 amino acids correspond to residues 400 to 425 of XFoxH1a. The XFoxH1a mutant peptide was biotin-aminohexanoic acid-RQIKIWFQNRRMKWKKYSNLLDSTPLAPLVSVPKTLSPVAVP, where the last 26 amino acids are the scrambled version of residues 400 to 425 of XFoxH1a. In both cases, the first 16 amino acids are from α-helix 3 of Antennapedia (6). The antibodies used in band shifts and Western blots were anti-FLAG (M2; Sigma); anti-FLAG M2-peroxidase conjugate (FLAG-HRP; Sigma); anti-HA (Roche Diagnostics); anti-HA-peroxidase conjugate (HA-HRP; Roche Diagnostics); anti-Smad2/3 (Transduction Laboratories); anti-Smad4 (B8; Santa Cruz); anti-phosphorylated-Smad2 (P-Smad2; NEB or (7); anti-phosphorylated Smad3 (P-Smad3; a gift from Ed Leof); and anti-Smad3 (16).
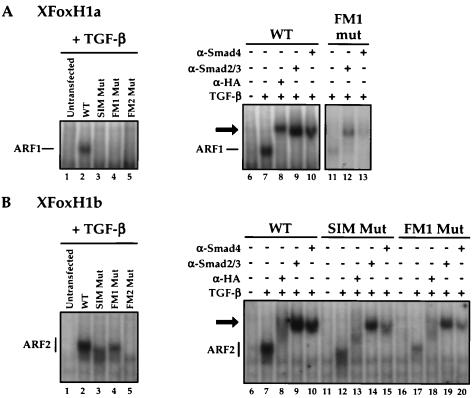
FIG. 2.
Mutation of the FM in XFoxH1a and XFoxH1b inhibits the ability of these transcription factors to form stable complexes with activated endogenous Smad2 and Smad4. NIH 3T3 cells were transfected with plasmids expressing HA-tagged wild-type or mutant derivatives of XFoxH1a (A) or XFoxH1b (B), and treated with TGF-β prior to making whole-cell extracts. Extracts were analyzed by band shift assay using the ARE probe. Antibody supershifts were used to demonstrate that the DNA-bound complexes contained all components (right panels). The Smad-containing complexes ARF1 (A) and ARF2 (B) are indicated, as are the supershifted complexes (black arrow). Note that as well as supershifting the ARF complexes, the anti-Smad2/3 antibody also stabilizes the ARF complexes, making it easier to detect weak complexes such as that formed by the XFoxH1a FM1 mutant.
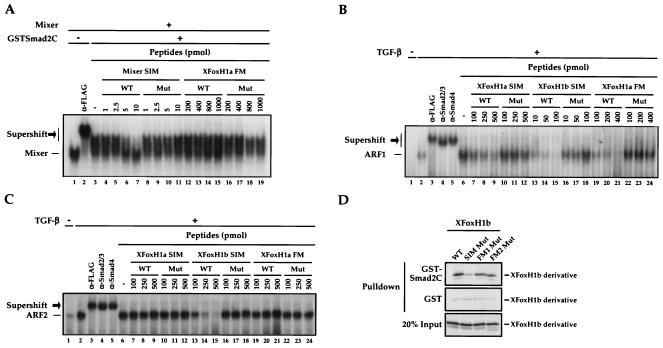
FIG. 4.
The SIM and FM do not bind Smad2 in an identical manner. (A) In vitro-translated FLAG-tagged Mixer was incubated with Mixer binding site probe alone or with anti-FLAG antibody or 5 ng of GSTSmad2C, as indicated. Different amounts (picomoles) of wild-type (WT) or mutant (Mut) Mixer SIM or XFoxH1a FM peptides were added to the 20-μl band shift reactions. Mixer complexed with the probe is shown, as is the ternary complex with anti-FLAG antibody or GSTSmad2C (black arrow). (B, C) NIH 3T3 cells were transfected with plasmids expressing FLAG-tagged XFoxH1a (B) or XFoxH1b (C), and treated with TGF-β prior to making nuclear extracts. Extracts were analyzed by band shift assay using the ARE probe. The Smad-containing complexes ARF1 (B) and ARF2 (C) are indicated, as are the supershifted complexes (black arrow). Wild-type or mutant XFoxH1a or XFoxH1b SIM or XFoxH1a FM peptides were used as in panel A. (D) 35S-labeled wild-type XFoxH1b and mutants thereof were generated in vitro in reticulocyte lysate and assayed for their ability to interact with GST and GSTSmad2C in a GST pulldown assay. Bound protein was visualized by SDS-PAGE and autoradiography; 20% input protein is shown.
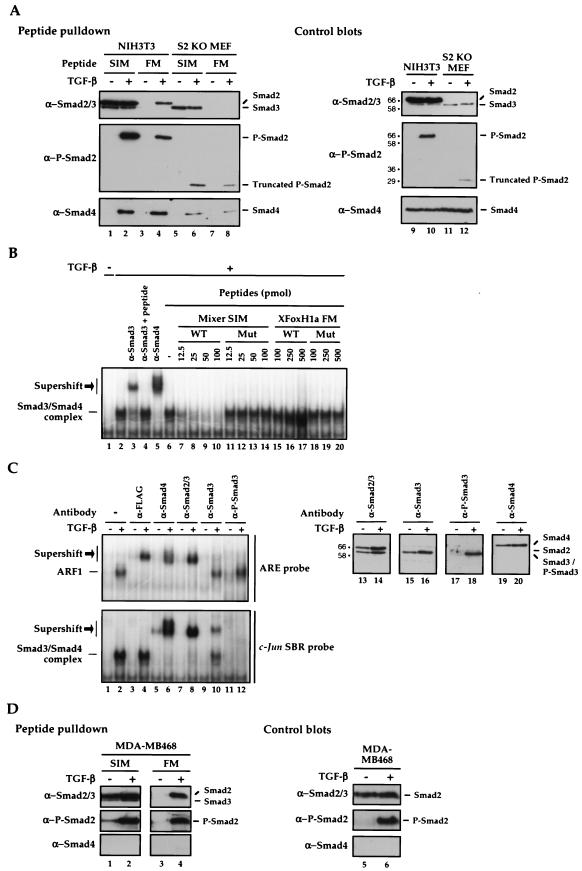
FIG.6.
The XFoxH1a FM interacts with phosphorylated Smad2 and through this interaction binds Smad4, but does not interact with Smad3. (A and D) The wild-type Mixer SIM and XFoxH1a FM peptides were immobilized on neutravadin beads and incubated with whole-cell extracts from uninduced or TGF-β-induced NIH 3T3 cells or Smad2-null mouse embryo fibroblasts (S2 KO MEF; A) or MDA-MB468 cells (D). Associated proteins were Western blotted for Smad2/3, phosphorylated Smad2 (P-Smad2), and Smad4 (left panels), and whole-cell extracts were blotted with the same antibodies (right panels). The numbers to the left of the control blots in panel A show the positions of molecular size markers (kilodaltons). (B) Nuclear extracts made from HaCaT cells treated with or without TGF-β for 1 h were analyzed by band shift using the c-Jun Smad-binding region (SBR) probe. The complexes were supershifted with the indicated antibodies to demonstrate that they contained Smad3 and Smad4. Different amounts (picomoles) of wild-type (WT) or mutant (Mut) Mixer SIM or XFoxH1a FM peptides were added to the 20-μl band shift reaction mixtures. The Smad3/Smad4 complex is indicated, as are the supershifted complexes (black arrow). (C) Nuclear extracts prepared from HaCaT cells transfected with a plasmid expressing FLAG-tagged XFoxH1a and treated with or without TGF-β for 1 h were analyzed by band shift using the ARE (upper panel) or the c-Jun Smad-binding region probe (lower panel). The complexes were incubated with antibodies as indicated to demonstrate that ARF1 contains XFoxH1a, Smad2 and Smad4, but not Smad3, while the Smad3/Smad4 complex on the c-Jun Smad-binding region contains Smad3 and 4. Right panels, Western blots of nuclear HaCaT extracts, treated with or without TGF-β, probed with the same antibodies to demonstrate antibody specificity. The numbers to the left show the positions of molecular size markers (kilodaltons).
Gel filtration analysis.
Untransfected or transfected NIH 3T3 cells treated with or without TGF-β were washed twice in cold PBS and lysed in buffer containing 50 mM Tris-HCl (pH 7.5), 400 mM NaCl, 1 mM EDTA (pH 8), 0.25% NP-40, 1 mM DTT, 25 mM NaF, 12.5 mM sodium β-glycerophosphate, and protease inhibitors. Extracts were sonicated and centrifuged at 13,000 rpm for 5 min at 4°C. The supernatant was diluted to contain 150 mM NaCl and 0.1% NP-40, centrifuged for 10 min at 4°C at 25,000 rpm and passed through a 0.2 μm filter. The supernatant was applied to a Superdex-200 gel filtration column (Amersham Pharmacia Biotech) preequilibrated with buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% NP-40, 1 mM EDTA, 1 mM dithiothreitol, 0.5 mM benzamidine, 25 mM NaF, 12.5 mM sodium β-glycerophosphate. The column was run at 0.5 ml/min, and 400 μl samples were collected, concentrated and analyzed by Western blot, probing with anti-Smad2/3 or anti-P-Smad2 antibodies. The column was calibrated with molecular mass standards (Bio-Rad).
Molecular modeling.
The model of the predicted Smad2/Smad4 MH2 domain heterotrimer was generated from the crystal structure of the phosphorylated Smad2 (42). From the PDB entry, a homotrimer was constructed based on the operations z, x, y; 1/2 + y, 1/2 - z, -x; and -y, 1/2 -z, x; (I 213 symmetry). A Smad4 subunit, taken from the Smad4 MH2 domain homotrimer (3, 30) was directly superimposed on one of the Smad2 subunits using the 3D-JIGSAW software (Cα RMSD: 1.33 Å; 45% identity) (1). The program PASS (2) was used to identify potential binding sites, based on the size, shape and extent of partially buried volumes across the protein surface.
RESULTS
Identification of the Fast/FoxH1 motif.
Mix family members that interact with Smad2 share only two regions of amino acid identity; the DNA-binding domain and the SIM (32). In Mixer, mutation of the two prolines in the central PPNK of the SIM to alanines abolishes interaction with Smad2 and, as a result, TGF-β-induced transcriptional activation mediated by Mixer/Smad2/Smad4 complexes (9). The SIM is also well conserved in all FoxH1 family members, which also interact with Smad2 (9, 32). We tested its importance in the Xenopus FoxH1s for their ability to confer TGF-β-induced transcriptional activation on the Mix.2 ARE through recruitment of endogenous active Smad complexes (9). Wild-type XFoxH1a and XFoxH1b mediated a 29- and 20-fold TGF-β-induced transcriptional activation, respectively (Fig. 1A). Mutation of the SIM in XFoxH1a decreased the induction to 11-fold, but did not eliminate it, and in XFoxH1b, this mutation had no effect (Fig. 1A). This suggested the presence of an additional motif in the XFoxH1s that could interact with Smad2 and/or Smad4 and thus compensate for the mutated SIM.
Sequence alignment of the 5 known FoxH1 family members (4, 19, 20, 29, 35, 38, 46) revealed a highly conserved region within their C-terminal domains, N-terminal to the SIM. This region, which we have termed the Fast/FoxH1 Motif (FM) is characterized by the sequences LPTSY and PN(V/A)V(A/M)P(L/P), which we refer to as FM1 and FM2, respectively (Fig. 1D).
We investigated the relative importance of each of these motifs in XFoxH1a and XFoxH1b for mediating TGF-β-induced transcription. This analysis was performed in NIH 3T3 cells which do not contain any endogenous FoxH1 family members that would interfere with the interpretation of the experiment. In the context of XFoxH1a, mutation of FM1 had a negligible effect on the ability of the transcription factor to activate TGF-β-induced transcription, but mutation of FM2 had a substantial effect (Fig. 1A, left panel). Mutation of both FM1 and FM2 resulted in a protein that retained only residual activity (4-fold activation), and mutation of either FM1 or FM2 in combination with a SIM mutation completely abolished activity (Fig. 1A, left panel). For XFoxH1b, mutation of FM1 had little effect on its ability to mediate TGF-β-induced transcriptional activation, whereas mutation of FM1 in combination with the SIM reduced the induction by TGF-β to 7-fold (Fig. 1A, right panel). Mutation of the XFoxH1b FM2 completely abolished TGF-β-induced transcription, as did combinations of the FM2 mutation with SIM or FM1 mutations (Fig. 1A, right panel). We also assayed the activity of the single mutants of both XFoxH1a and XFoxH1b on a naturally occurring Nodal responsive enhancer, the left side-specific enhancer of the mouse Pitx2 gene (Fig. 1B) (34). These mutants all behaved in exactly the same way as they did on the ARE3-luciferase reporter gene (Fig. 1A and B). All mutants were expressed at approximately equal levels (Fig. 1C).
These results demonstrate that the FM is required in both XFoxH1a and XFoxH1b for mediating TGF-β-induced transcriptional activation via the ARE and that FM2 is more important than FM1.
The FM is required to recruit active endogenous Smad complexes.
We next investigated the effect of mutation of the FM on the ability of XFoxH1a and XFoxH1b to form stable XFoxH1/Smad2/Smad4-DNA-bound complexes, ARF1 and ARF2, respectively, on the ARE (13). These ARF complexes are readily detected in a band shift assay using radiolabeled ARE probe and extracts from TGF-β-induced NIH 3T3 cells transfected with wild-type XFoxH1a or XFoxH1b, respectively (Fig. 2A and B). Mutations in the SIM or FM2 of XFoxH1a prevented ARF1 formation, but mutations in FM1 could allow very weak ARF1 formation (Fig. 2A). Mutation of the XFoxH1b FM2 completely abolished ARF2 formation, while mutation of the SIM or FM1 only partially inhibited formation of ARF2 (Fig. 2B). Supershift analysis demonstrated that the ARF complexes formed by wild-type XFoxH1a and XFoxH1b and the mutants all contained HA-tagged XFoxH1a or b, and endogenous Smad2 and Smad4 (Fig. 2A and B, right panels). However the mobility of ARF2 containing SIM-mutated XFoxH1b was greater than that of wild-type ARF2 for reasons we do not fully understand (Fig. 2B). In all cases the inhibition of ARF1/2 formation by the mutations reflected loss of XFoxH1a/b-Smad interaction, since none of the mutations affected the ability of the XFoxH1s to bind DNA alone (data not shown), as expected from the fact that their DNA-binding domains are distant from the FM and SIM region (13).
These results demonstrate that for XFoxH1a, binding of activated Smad2/Smad4 complexes to form a stable ARF1 complex requires both the SIM and FM. For XFoxH1b, formation of a stable ARF2 complex requires an intact FM, although mutation of the SIM can be tolerated. Note that whereas low affinity Smad-transcription factor interactions, such as that mediated by the XFoxH1a FM1 mutant, may be detected in a reporter assay (Fig. 1A), these weakly associated complexes do not readily withstand the electric field in the more stringent band shift assay.
The FM is sufficient for mediating TGF-β-induced transcriptional activation.
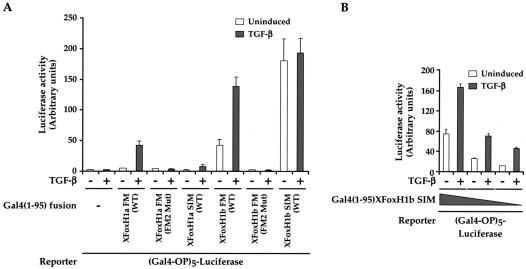
Having established that the FM is necessary for the interaction of the XFoxH1s with Smad2/Smad4 complexes, we determined if it is sufficient. The XFoxH1a and XFoxH1b SIM or FM (FM1 and FM2) coding regions, and mutants thereof, were fused to the Gal4 DNA-binding domain, and assessed for their ability to activate transcription of a Gal4 operator-driven reporter, which reflects their ability to bind activated endogenous Smads (32). All constructs were well expressed and bound DNA effectively (data not shown). Both the XFoxH1a and XFoxH1b FM fusions were sufficient to activate transcription in a TGF-β-inducible manner, the XFoxH1b FM being more active than the XFoxH1a FM (Fig. 3A). In both cases mutation of FM2, which is sufficient in the full-length XFoxH1s to prevent binding of the Smads, abolished this activity. By comparison, the XFoxH1a SIM fusion conferred a weak (4-fold) TGF-β-induced activation (Fig. 3A). The XFoxH1b SIM fusion conferred a very high basal level of transcription that was not increased upon TGF-β addition (Fig. 3A). This is due to its high affinity for Smad2 (13), enabling it to bind active Smad complexes that exist at low levels in uninduced cells, since titrating down the amount of the XFoxH1b SIM fusion until it was limiting allowed us to detect a fourfold TGF-β-dependent activation (Fig. 3B).
FIG. 3.
The FM is sufficient to confer TGF-β inducibility in vivo. (A) NIH 3T3 cells were transfected with the (Gal4-OP)5-Luc reporter in combination with plasmids expressing Gal4(1-95) fusions of the XFoxH1a or XFoxH1b SIM or FM and mutants thereof. (B) NIH 3T3 cells were transfected with decreasing amounts (25, 10, or 5 ng) of plasmid expressing Gal4(1-95)XFoxH1b SIM, with the (Gal4-OP)5-Luc reporter. TGF-β inductions and luciferase assays were as for Fig. 1, and the data are the means and standard deviations of three independent experiments.
Thus, the XFoxH1a and XFoxH1b FMs and SIMs are sufficient in vivo to interact with active Smad2/Smad4 complexes and thereby mediate TGF-β-induced transcriptional activation. The FM and SIM in XFoxH1b both appear to be more active than those in XFoxH1a.
The SIM and FM do not bind Smad2 via the same mechanism.
The assays described above indicate that the FM binds Smad2/Smad4 complexes. We next analyzed how the FM interacts with the Smads to determine whether it bound Smad2 using the same mechanism as the SIM. We have previously shown that the SIM efficiently binds the Smad2 MH2 domain expressed as a GST fusion protein (GSTSmad2C) and we have shown that the interaction of DNA-bound Mixer with GSTSmad2C is mediated solely through the SIM (9). Thus, a Mixer SIM peptide will efficiently compete the Mixer-GSTSmad2C interaction in a band shift assay (Fig. 4A, lanes 3 to 7) (9). A wild-type XFoxH1a FM peptide however had no effect on the Mixer-GSTSmad2C-DNA complex (Fig. 4A, lanes 12 to 15; see below and Discussion), indicating that the FM peptide could not compete with the SIM for binding to GSTSmad2C.
To ensure that the FM peptide was active, and could interact with Smad2 and/or Smad4 we assayed its ability to prevent ARF1 and ARF2 formation, comparing its effects with XFoxH1a and XFoxH1b SIM peptides. Both the XFoxH1a and XFoxH1b SIM peptides interfere with ARF1 complex formation, although higher concentrations of the lower affinity XFoxH1a SIM are required (13). ARF2 is a stronger complex whose formation can be prevented by the XFoxH1b SIM peptide, but not by the lower affinity XFoxH1a SIM peptide (13). The XFoxH1a FM peptide prevented ARF1 complex formation, but had no effect on the stronger ARF2 complex (Fig. 4B and C). Mutant SIM or FM peptides had no effect on any of the complexes (Fig. 4B and C).
Thus, the XFoxH1a FM peptide was not able to bind GSTSmad2C and compete the solely SIM-dependent Mixer-GSTSmad2C interaction. However, it could bind Smad2/Smad4 complexes and prevent ARF1 formation which is mediated through the combined activities of the SIM and FM in XFoxH1a. This result suggests that the FM does not interact with the Smads using the same mechanism as the SIM. We investigated this further by examining the interaction of GSTSmad2C with XFoxH1b, whose SIM and FM have a higher affinity for Smads than those of XFoxH1a (see above). XFoxH1b interacts efficiently with GSTSmad2C in a GST pulldown assay. This activity however is mediated solely through the SIM, since mutation of the SIM inhibited the interaction, but mutations in the FM had little effect (Fig. 4D). This result confirms that, unlike the SIM, the FM does not bind GSTSmad2C. The FM can however interact with Smad2/Smad4 complexes.
The presence of an FM and a SIM in the XFoxH1s allows them to compete effectively for activated Smads.
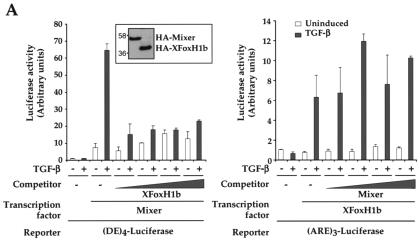
We investigated the potential biological relevance of the presence in the FoxH1s of two Smad interaction motifs (a SIM and an FM), hypothesizing that two Smad interaction motifs in the FoxH1s might result in stronger binding to Smad2/Smad4 complexes than is possible with just a SIM. To test this we investigated the ability of XFoxH1b to compete for activated Smads in transcription assays with Mixer, which only contains a SIM. In peptide competition assays, approximately equal amounts of Mixer SIM or XFoxH1b SIM peptide are required to inhibit formation of ARF complexes, suggesting that their SIMs have similar affinities for the Smads (Fig. 4B and C and data not shown). Both proteins were equally well expressed when the same amount of DNA was transfected (Fig. 5A, inset). The data show that even the lowest amount of XFoxH1b can compete efficiently with DNA-bound Mixer for activated Smads (Fig. 5A, left). Conversely, not even the highest levels of transfected Mixer can compete with DNA-bound XFoxH1b for activated Smads (Fig. 5A, right).
FIG. 5.
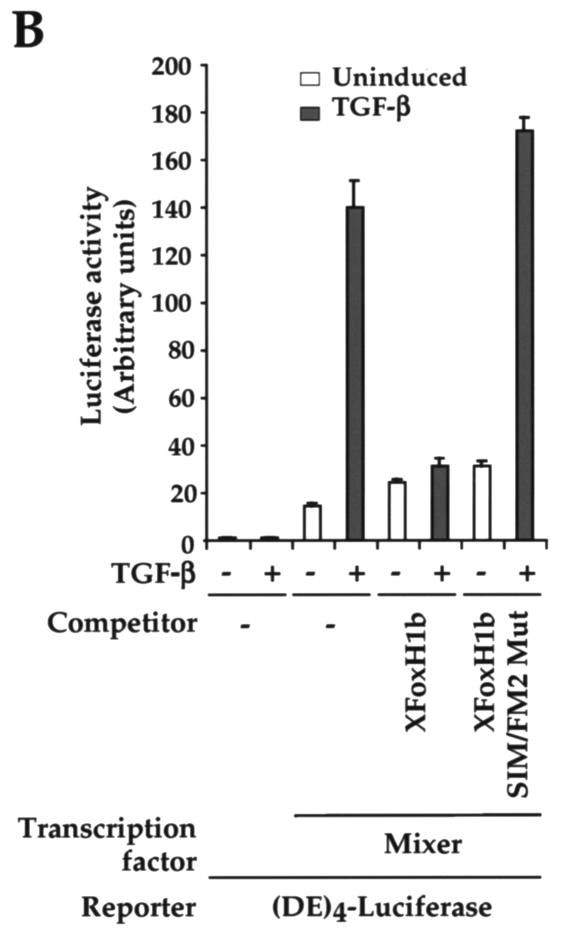
XFoxH1b competes with Mixer for activated Smads. (A) NIH 3T3 cells were transfected with the (DE)4-Luc reporter and 25 ng of plasmid expressing HA-tagged Mixer either alone or with 6.25, 12.5, 25, or 50 ng of plasmid expressing HA-tagged XFoxH1b or with (ARE)3-Luc reporter and 25 ng of plasmid expressing HA-tagged XFoxH1b either alone or with 6.25, 12.5, 25, or 50 ng of plasmid expressing HA-tagged Mixer. Both proteins were equally well expressed when the same amount of DNA was transfected, as detected in a Western blot probed with anti-HA (inset). (B) NIH 3T3 cells were transfected with the (DE)4-Luc reporter and 25 ng of plasmid expressing HA-tagged Mixer either alone or with 25 ng of plasmid expressing HA-tagged XFoxH1b or HA-tagged XFoxH1b SIM/FM2 mutant. TGF-β induction and luciferase assays were as for Fig. 1.
To prove that the ability of XFoxH1b to compete with Mixer for activated Smads depends on its containing an intact SIM and FM, and is not the consequence of nonspecific squelching of core transcription machinery, we also demonstrated that the XFoxH1b derivative with a mutated SIM and FM2 (XFoxH1b SIM/FM2 Mut) had no effect in this competition assay (Fig. 5B). Taken in conjunction with the mutational analysis shown in Fig. 1 and 2 for XFoxH1a and XFoxH1b and in (9) for Mixer, we conclude that the presence of a SIM and FM in the same molecule allows the FoxH1s to compete very efficiently for activated Smads with Mixer, which only contains a SIM.
The XFoxH1a FM peptide interacts with active Smad2, but not with active Smad3.
We then examined the interaction of the FM with endogenous Smad2 and/or Smad4 using peptide pulldown assays (32). As a control we used the well characterized Mixer SIM peptide (9, 32), which behaves the same as the XFoxH1a and XFoxH1b SIMs in these assays (data not shown). The FM and SIM peptides were immobilized on beads, incubated with whole-cell extracts from uninduced or TGF-β-induced NIH 3T3 cells, and Western blotted for endogenous unphosphorylated and phosphorylated Smad2, and Smad4. The FM peptide did not pull down any Smads from extracts from uninduced cells, but pulled down phosphorylated Smad2, and Smad4 from extracts made from TGF-β-induced cells (Fig. 6A). By comparison, the Mixer SIM peptide pulled down unphosphorylated Smad2 and Smad3, phosphorylated Smad2 and, through this interaction, Smad4 (Fig. 6A) (32). Mutant peptides did not pull down any of the Smads (data not shown) (32).
It appeared that the FM interacted with activated Smad2, but not with Smad3. To confirm this, we performed peptide pulldowns with extracts prepared from Smad2-null mouse embryonic fibroblasts (27). The FM peptide clearly did not interact at all with Smad3, but nevertheless pulled down Smad4 from extracts made from TGF-β-induced cells (Fig. 6A, lanes 7, 8). In contrast, the SIM peptide pulled down Smad3 from uninduced cell extracts, and Smad3 and Smad4 from TGF-β-induced cell extracts (lanes 5, 6). We investigated further why the FM pulled down Smad4 from extracts from TGF-β-induced cells when it could not interact with Smad3, and the cells contain no full-length Smad2. We demonstrated that these cells, which are homozygous for Smad2 disrupted in its first coding exon (exon 2) (11), actually express a truncated Smad2 that is phosphorylated upon TGF-β signaling. It is readily detected with an antibody directed against phosphorylated Smad2 in a Western blot (Fig. 6A), though not with the anti-Smad2/3 antibody (data not shown). We deduce from its size that this truncated Smad2 starts at Met 241, and encodes the entire MH2 domain and a small portion of linker. Thus, we conclude that the FM interacts with the C-terminal domain of Smad2 in response to TGF-β and through this interaction pulls down Smad4 (see also below). In addition, in contrast to the SIM, the FM discriminates between activated Smad2-containing complexes and activated Smad3-containing complexes.
We investigated this discrimination between Smad2 and Smad3 in more detail by testing whether the FM could interfere with the formation of an endogenous Smad3/Smad4-containing band shift complex that binds the c-Jun Smad-binding region (16). Consistent with the notion that the FM does not interact with Smad3, the FM peptide could not interfere with the formation of this complex, whereas a SIM peptide could, as previously reported (16) (Fig. 6B).
Our observation that the FM discriminates between activated Smad2/Smad4 and Smad3/Smad4 complexes prompted us to investigate whether this binding specificity was also a property of the FoxH1 transcription factors that contain this motif. We asked whether the FoxH1s discriminated between endogenous Smad2 and Smad3. FLAG-tagged XFoxH1a was therefore expressed in HaCaT cells, which contain approximately equal amounts of endogenous Smad2 and Smad3 (Fig. 6C; right-hand panels). The resulting ARF1 complex formed upon TGF-β stimulation only contains XFoxH1a, Smad2 and Smad4, as demonstrated by the fact that it was supershifted by a FLAG antibody, an anti-Smad4 antibody and an antibody that recognizes both Smad2 and Smad3, but was unaffected by an antibody that recognizes Smad3 only, or an antibody that recognizes phosphorylated Smad3 (P-Smad3; Fig. 6C). To confirm that had Smad3 been present in the ARF1 complex, it would have been detected with the Smad3 and P-Smad3 antibodies, we demonstrated that the Smad3 antibody supershifted the Smad3/Smad4-containing complex that binds the c-Jun Smad-binding region, and the anti-P-Smad3 antibody completely disrupted this complex (Fig. 6C). The specificity of the antibodies was confirmed by Western blotting (Fig. 6C, right-hand panels). Several studies have shown that FoxH1 family members can bind Smad3, but this has only been demonstrated for overexpressed Smad3, or recombinant purified Smad3 (18, 19, 24, 44, 45). Indeed, when Smad3 was overexpressed in HaCaT cells with XFoxH1a, the resulting ARF1 complex formed in response to TGF-β did contain Smad3 (data not shown).
Our results therefore indicate that XFoxH1a preferentially binds activated endogenous Smad2/Smad4 complexes in vivo. This preference for Smad2/Smad4 complexes over Smad3/Smad4 complexes is lost if Smad3 is artificially overexpressed.
The XFoxH1a FM peptide binds Smad4 indirectly through its interaction with activated Smad2.
The observation that the FM did not pull down Smad4 from uninduced cells suggested that it did not interact directly with Smad4. To confirm this, we performed peptide pulldowns using extracts from the Smad4-null cell line, MDA-MB468 where TGF-β stimulation leads to formation of activated homomeric Smad2 complexes (25) (Fig. 6D). The FM was still able to pull down phosphorylated Smad2 from extracts of TGF-β-induced MDA-MB468 cells, indicating that the FM makes no essential contacts with Smad4 and that the presence of Smad4 in the complex is not required for FM interaction.
Taken together with the results in the previous section, we conclude that the FM contacts the Smad2 subunit of an active Smad2/Smad4 complex; its ability to pull down Smad4 is a consequence of the Smad2-Smad4 interaction.
The FM interaction requires Smad2 phosphorylation.
The FM only interacts with activated Smad2. We considered three possible explanations for this. Firstly, TGF-β-induced phosphorylation might be important for releasing the MH2 domain from autoinhibition mediated by the MH1 domain (10). Secondly, it may induce essential conformational changes in the MH2 domain (42). Thirdly, Smad2 phosphorylation might be important because it mediates heteromeric complex formation, and the FM might recognize a binding pocket only present in the active Smad complex.
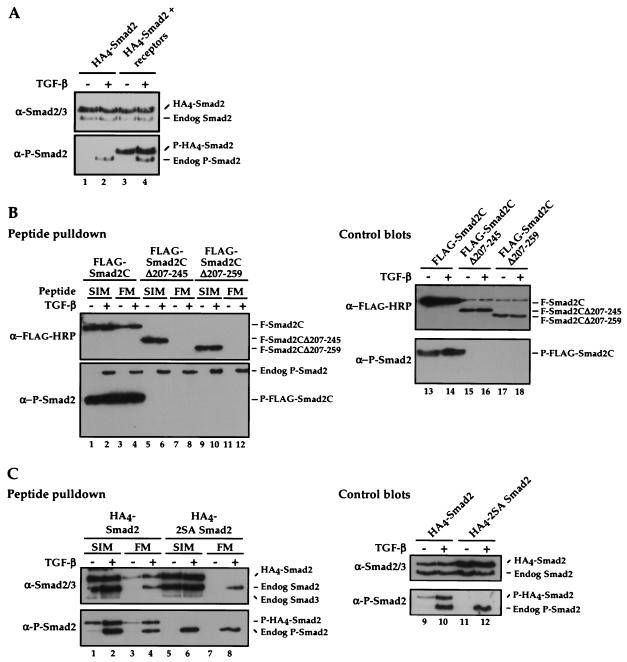
From the data in Fig. 6A, the FM clearly interacts with phosphorylated truncated Smad2 (amino acids 241 to 467), but we cannot tell from this experiment whether in the absence of the MH1 domain, phosphorylation was still necessary for the FM interaction, as we cannot detect this product in the absence of phosphorylation. We therefore tested in peptide pulldown assays whether the FM interacted with FLAG-tagged N-terminally truncated versions of Smad2, Smad2C (amino acids 198 to 467) and derivatives of it containing different amounts of linker. Transfected Smad2s are not efficiently phosphorylated by endogenous levels of receptors (Fig. 7A), so TGF-β type I and type II receptors were cotransfected in these experiments.
FIG. 7.
FM interaction requires Smad2 phosphorylation. (A) NIH 3T3 cells were transfected with HA4-tagged wild-type Smad2 with or without plasmids expressing TβRI and TβRII. Cells were either untreated or treated for 1 h with TGF-β. Whole-cell extracts were then blotted for Smad2/3 (upper panel) and phosphorylated Smad2 (P-Smad2; lower panel). (B and C) NIH 3T3 cells were transfected with FLAG-tagged Smad2C (amino acids 198 to 467), or linker-deletions (Smad2CΔ207-245 or Smad2CΔ207-259) (B) or wild-type or 2SA HA4-tagged Smad2 (C) and plasmids expressing TβRI and TβRII and incubated with or without TGF-β. Whole-cell extracts were incubated with immobilized wild-type Mixer SIM and XFoxH1a FM peptides and associated proteins were Western blotted for FLAG or Smad2/3 and P-Smad2 (left-hand panels). Whole-cell extracts were blotted with the same antibodies as a control (right-hand panels). Note that the band that appears in all lanes of the anti-FLAG-HRP control blot is a background band. In panels A and C, it is easy to distinguish the transfected Smad2s from endogenous Smad2 due to the large size of the 4 N-terminal HA tags on the transfected Smad2s.
The Smad2C was phosphorylated in the absence and presence of TGF-β as a result of receptor overexpression, thus we still could not distinguish between a requirement for phosphorylation versus a requirement for loss of autoinhibition by the MH1 domain (Fig. 7B, lanes 1 to 4). However, the Smad2Cs with further deletion of the linker (Smad2CΔ207-245 and Smad2CΔ207-259) were not phosphorylated, despite the overexpression of receptors, presumably because they lack sequences in the C-terminal portion of the linker downstream of M241, required to interact with the SARA Smad-binding domain for recruitment to the receptors (Fig. 7B) (40). These nonphosphorylated truncated Smad2s did not interact with the FM (Fig. 7B, lanes 7, 8, 11, 12). However as they contain an intact MH2 domain, they did interact with the SIM (Fig. 7B, lanes 5, 6, 9, 10). This indicates that phosphorylation of Smad2 is required for FM binding, and explains why the FM did not interact with unphosphorylated recombinant GSTSmad2C (Fig. 4A and D). This result was also confirmed by demonstrating that a full-length Smad2 in which the phosphorylated serines were mutated to alanines (2SA Smad2) also failed to interact with the FM, but interacted efficiently with the SIM (Fig. 7C).
FM binding requires Smad complex formation.
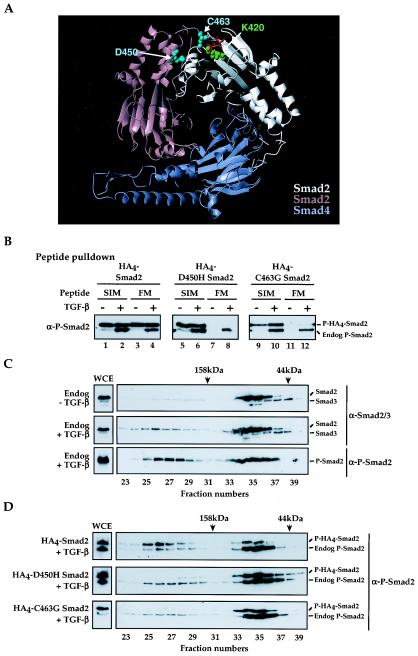
We next tested whether the FM interaction required Smad complex formation by investigating Smad mutants which cannot form complexes, but can be phosphorylated. We modeled the active Smad MH2 domain complex as a Smad2/Smad4 trimer with two Smad2s and one Smad4 (Fig. 8A), since a Smad2/Smad4 trimer associates in vivo with the XFoxH1s (16). We then mutated residues in full-length Smad2 required for complex formation. Three mutants were made in Smad2, K420A, D450H and C463G, that from the crystal structure were predicted to abolish contacts required for complex formation (Fig. 8A) (42).
FIG.8.
Smad2 phosphorylation and subsequent complex formation is necessary for FM interaction. (A) The predicted Smad2/Smad4 MH2 domain heterotrimer, highlighting interacting residues at the Smad2-Smad2 interface (cyan on the pink Smad2 subunit and green on the white subunit) and the phosphate groups (red) of the pink Smad2 subunit. Corresponding residues at the Smad2-Smad4 interfaces have been omitted for the sake of clarity. The two Smad2 subunits are not identical; the pink Smad2 subunit has no phosphoserine residues bound in its L3/B8 loop/strand pocket (42), while the white Smad2 subunit binds the phosphoserines from the pink Smad2 subunit. The Smad4 subunit (blue) binds the phosphoserines from the white Smad2 subunit. The figure was created using MOLMOL (17). (B) NIH 3T3 cells were transfected with plasmids expressing wild-type, D450H, or C463G HA4-tagged Smad2 and plasmids expressing TβRI and TβRII. Whole-cell extracts were made from uninduced or TGF-β-induced cells, incubated with immobilized wild-type Mixer SIM and XFoxH1a FM peptides, and associated proteins were Western blotted for P-Smad2. (C and D) Whole-cell extracts (WCE) were made from uninduced NIH 3T3 cells and cells induced with TGF-β for 40 min (C), as well as from TGF-β-induced NIH 3T3 cells transfected with wild-type, D450H or C463G HA4-tagged Smad2 and plasmids expressing TβRI and TβRII (D), and subjected to gel filtration analysis and Western blotting with antibodies against Smad2/3 (top two panels) and P-Smad2. Fraction numbers are indicated. The column was calibrated with standard molecular size markers; the centers of the elution peaks corresponding to 44 kDa and 158 kDa are indicated.
Mutation of K420 to alanine prevented phosphorylation of Smad2, thus precluding any further analysis of the FM interaction (Table 1). This was not unexpected as K420 is in the L3 loop, which is required for interaction with the type I receptor (21). The D450H and C463G Smad2 mutants, however, were both efficiently phosphorylated and, when assayed by peptide pulldown, did not interact with the XFoxH1a FM, but did interact with the Mixer SIM (Fig. 8B). We used gel filtration analysis to determine whether they formed Smad complexes upon TGF-β-induction. The assay was first validated using extracts from uninduced or TGF-β-induced NIH 3T3 cells to test for complex formation of endogenous Smad2. In the absence of TGF-β, Smad2 migrated predominantly in fractions corresponding to a molecular weight of above 44 kDa, consistent with it being monomeric (Fig. 8C). Upon TGF-β induction, a portion of the Smad2 could be detected in the fractions corresponding to molecular masses of above 158 kDa, indicating formation of higher order Smad complexes. When blotted for phosphorylated Smad2, it was clear that the Smad2 in these earlier fractions was phosphorylated, resulting in complex formation (Fig. 8C). After TGF-β stimulation, wild-type transfected Smad2 eluted in the same fractions as endogenous Smad2, indicating that it too forms higher-order Smad complexes (Fig. 8D). However, the D450H and C463G Smad2 mutants only eluted in fractions corresponding to monomeric Smad2, although the endogenous Smad2 clearly formed higher order complexes (Fig. 8D).
TABLE 1.
Investigation of the interaction between XFoxH1a FM and Smad2a
| Residuesb | Smad2c | Ability to be phosphorylated | Complex formation | FM interaction | SIM interaction |
|---|---|---|---|---|---|
| Wild type | + | + | + | + | |
| Interface residues | K420A | − | NTd | NT | NT |
| D450H | + | − | − | + | |
| K375A+D450H | − | NT | NT | NT | |
| K420A+D450H | − | NT | NT | NT | |
| K375A+K420A+D450H | − | NT | NT | NT | |
| C463G | + | − | − | + | |
| Residues located in potential | F356A | − | NT | − | − |
| hydrophobic grooves | V419A | + | NT | + | + |
| H331A | + | NT | + | + | |
| E326A | + | NT | + | + | |
| M327A | + | NT | + | + | |
| R334A | + | NT | + | + | |
| D352A | + | NT | + | + | |
| W274A | + | + | + | + | |
| Y340A | + | + | + | + | |
| Residues within the SIM-binding | W368A | + | + | − | − |
| hydrophobic pocket | N381A | + | NT | + | + |
| Y366A | + | NT | + | ± | |
| V373A | + | + | − | − | |
| Residue close to Smad/Smad interface | V431A | + | + | − | + |
All Smad2 mutants were tested for phosphorylation by Western blotting of whole-cell extracts with the anti-phosphorylated-Smad2 antibody. The ability of the mutants to form complexes was assayed by gel filtration analysis. Peptide pulldowns were used to determine whether the Smad2 mutants could interact with the Mixer SIM and the XFoxH1a FM.
Interface residues were as previously identified (42). SIM-binding hydrophobic pocket residues in the Smad2 MH2 domain were previously described (32).
Mutation of F356, a residue which is buried within Smad2, resulted in a protein that was not phosphorylated and could not interact with either the FM or the SIM, suggesting that this mutation caused a severe structural change in the Smad2. E326, M327 and H331 lie in a groove that forms in the vicinity of the phosphoserine-binding pocket upon complex formation. Although mutation of N381 has no effect on SIM binding under our experimental conditions, it is thought from modeling to be involved in forming a hydrogen bond with I304 of the Mixer SIM (32).
NT, not tested.
Thus, although D450H and C463G Smad2 were phosphorylated, they were unable to form Smad complexes. Therefore phosphorylation per se is not sufficient for the FM to interact with Smad2; the phosphorylation is required to induce complex formation, which in turn is necessary for FM-Smad2 interaction.
Determination of the FM binding site on Smad2/Smad4 trimers by site-directed mutagenesis.
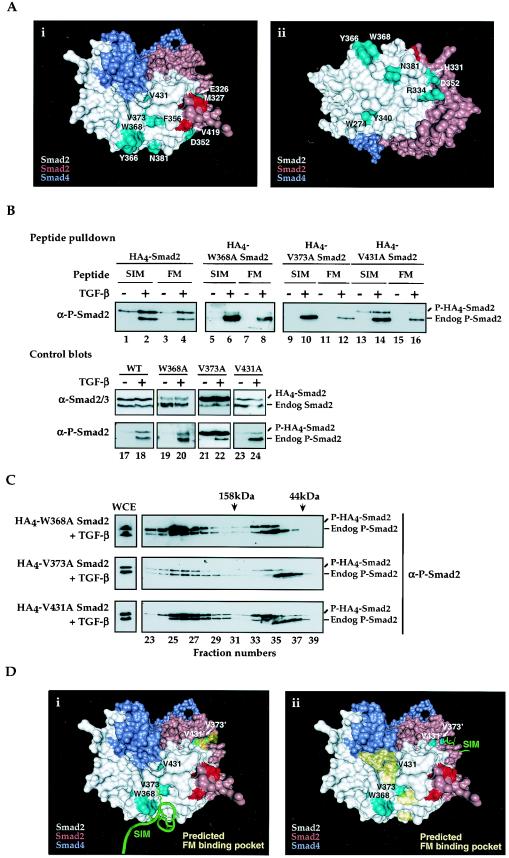
To determine where on the Smad2/Smad4 complex the FM binds, we examined the predicted Smad2/Smad4 MH2 domain heterotrimer structure using the program PASS (2) to identify potential hydrophobic grooves which might become available upon Smad2 phosphorylation and subsequent complex formation. Mutations were made in residues within these regions (Fig. 9Ai and ii) and tested for TGF-β-induced phosphorylation and, by peptide pulldown, for interaction with the FM and the SIM (Table 1).
FIG.9.
Residues required for FM binding to Smad2 are located in a hydrophobic pocket adjacent to the Smad trimer interface. (A) Solvent-accessible surface representations of the predicted Smad2/Smad4 MH2 domain trimer, highlighting mutated residues in cyan. Panels i and ii are identical but for a rotation about the horizontal axis. For orientation purposes, the phosphoserines from the pink subunit are colored red. The figure was created using VMD (15), together with the Surf accessible-surface generator (37). (B and C) NIH 3T3 cells were transfected with plasmids expressing wild-type or mutant (W368A, V373A, or V431A) HA4-tagged Smad2 and plasmids expressing TβRI and TβRII. (B) Whole-cell extracts were made from cells treated with or without TGF-β, incubated with immobilized wild-type Mixer SIM and XFoxH1a FM peptides, and Western blotted for P-Smad2. Whole-cell extracts were blotted with anti-P-Smad2 and anti-Smad2/3 antibodies as a control (lower panels). (C) Whole-cell extracts (WCE) were made from transfected NIH 3T3 cells induced with TGF-β for 40 min and separated by gel filtration. Fractions were concentrated and Western blotted for P-Smad2. Fraction numbers are indicated, and the column was calibrated as in Fig. 8. (D) Solvent-accessible surface representations of the predicted Smad2/Smad4 MH2 domain trimer, highlighting important Smad2 residues in cyan, and phosphoserines as in panel A in red. The Mixer SIM bound to the Smad2 MH2 domain (32) is shown in green, while the output from the PASS program is shown in yellow as an area indicating a possible binding groove for the FM fragment, consistent with the experimental observations. Panel i shows the SIM on one Smad2 subunit with the proposed FM-binding hydrophobic groove highlighted in yellow on the second Smad2 subunit, and panel ii gives the alternate arrangement. Residues on the pink Smad2 subunit are shown with primes. The FM and the SIM in XFoxH1a and XFoxH1b obviously cannot bind to the same Smad2 subunit simultaneously.
One obvious hydrophobic groove to test was that which binds the SIM. The FM and the SIM differ in their interaction with Smad2 since the SIM can bind monomeric Smad2, while the FM only binds complexed Smad2. However, these motifs are both proline-rich and contain a conserved Pro-Asn pair (Fig. 1D), so it was possible that they might interact with at least some of the same Smad2 residues. We tested the importance for FM binding of W368, which lies within a shallow hydrophobic pocket and is critical for the specific interaction of Smad2 with the SIM and with the SARA Smad-binding domain, and 3 other residues within this Smad2 hydrophobic pocket - N381, Y366 and V373 (Fig. 9A) (32). The resultant mutants were all phosphorylated efficiently (Fig. 9B; Table 1). Mutation of N381 had a negligible effect on the interaction of Smad2 with the SIM or the FM, and mutation of Y366 affected the SIM interaction, but was wild-type for FM interaction (Table 1). However, mutation of W368 or V373 to alanine resulted in proteins that did not bind the SIM or the FM, when compared with transfected wild-type Smad2 (Fig. 9B, lanes 1 to 12; and data not shown), suggesting that these residues are required for both SIM and FM binding. Importantly, these two mutants were efficiently expressed, were phosphorylated and formed higher order Smad complexes, indicating that their structures were not grossly disrupted (Fig. 9B and C).
Since the FM only interacts with Smad2 in active Smad complexes, we searched the Smad2/Smad4 trimer model for a hydrophobic groove in the vicinity of the SIM-interacting pocket that might also incorporate the trimer interface. Many of these predicted binding pockets were ruled out because mutation of potentially important residues in these pockets had no effect on SIM or FM binding (Table 1). However, we found one such groove that contained residues W368 and V373 of Smad2 and also, depending on the Smad2 subunit, incorporated the Smad2/Smad2 or Smad2/Smad4 interface (Fig. 9D). It is obviously not possible to mutate residues at the interface involved in stabilizing complex formation without affecting the ability of Smad2 to form complexes. However, we identified V431 as a residue that lay within this groove, adjacent to the trimer interface. Mutation of V431 to alanine resulted in a mutant Smad2 that was phosphorylated and formed Smad complexes efficiently (Fig. 9C). While the V431 mutant was able to interact with the SIM almost as efficiently as does wild-type Smad2 (Fig. 9B, compare lanes 1, 2 with 13, 14), it could not interact efficiently with the FM (compare lanes 3, 4 with 15, 16).
Taken together with our knowledge that W368 and V373 are required for FM binding, we propose a binding groove on the Smad trimer for the FM (Fig. 9D). The FM binding site is a hydrophobic groove that contains W368 and V373, and includes the Smad/Smad interface, adjacent to V431 (Fig. 9D). Since the XFoxH1s are monomeric and interact with a Smad complex containing two Smad2s (16), and given that the binding sites for the SIM and FM are partly overlapping, we propose that the SIM of XFoxH1a or XFoxH1b binds one Smad2 subunit, and the FM binds the other Smad2 subunit.
DISCUSSION
We have identified a new Smad2 interaction motif, the FM, present in all FoxH1 family members in their C-terminal domain, N-terminal to the previously identified SIM. The FM is composed of two highly conserved regions, which we refer to as FM1 and FM2. Dissection of these two submotifs in the context of both XFoxH1a and XFoxH1b has demonstrated the particular importance of the proline-rich FM2 for binding Smad2 and for mediating TGF-β-induced transcriptional activation. The FM does not interact with monomeric Smad2, even if it is phosphorylated, but will only interact with phosphorylated Smad2 in a homomeric complex or in a heteromeric complex with Smad4. It is the first Smad interaction motif with this property.
To guide our analysis of the interaction of the FM with the activated Smad complex, we constructed a model of a Smad2/Smad4 MH2 domain heterotrimer with a 2:1 stoichiometry, since it is a Smad trimer containing 2 Smad2s and 1 Smad4 that associates in vivo with the XFoxH1s (16). We identified a hydrophobic groove within the Smad2/Smad4 MH2 domain trimer that we propose is the FM binding pocket (Fig. 9D). It contains residues W368 and V373 that also interact with the SIM. For SIM binding, W368 is critical for determining specificity, allowing the SIM to interact only with Smad2 (and Smad3), and not with any of the other Smads (32). We propose that it plays a similar role in FM binding, although other properties of the FM allow it to distinguish between Smad2 and Smad3. However, whereas the SIM-binding pocket is distant from the trimer interface and involves residues N381 and Y339 of Smad2 (Fig. 9D; Table 1) (32, 40), the proposed FM-binding pocket extends instead towards the Smad interface. We have identified V431, located adjacent to the trimer interface within a hydrophobic groove that extends from residue W368, as a residue required for FM interaction. Our model predicts that the FM binds in this groove and runs along or across the trimer interface. Indeed, preliminary docking experiments using autodock software (22) indicate that the FM can dock within this identified groove, such that FM1 interacts with W368 and FM2 extends towards the interface (data not shown).
With the identification of the FM it is clear that the FoxH1 family members, which are monomeric, have two Smad2-interacting motifs and from recent studies of the stoichiometry of active Smad-transcription factor complexes on DNA they have been shown to interact with a Smad2-Smad2-Smad4 heterotrimer (16). We therefore propose that the SIM of the FoxH1 proteins binds to one Smad2 monomer and the FM to the other; they would be unable to bind the same Smad2 monomer as they require some of the same residues (W368 and V373) for binding. In contrast, Mixer, which binds DNA as a dimer (9) only contains one Smad interaction motif, the SIM.
The two Smad2 monomers in the heterotrimer are not identical, as only one has phosphoserines from the adjacent Smad2 subunit bound in its L3/B8 loop/strand pocket (the white subunit in Fig. 8A and 9D). Our results do not indicate which Smad2 subunit the FM binds, but one piece of evidence suggests that the FM may interact with the Smad2 subunit that does not bind phosphoserines (the pink subunit in Fig. 8A and 9D). We assume that because the FM interacts with activated homomeric Smad2 complexes in Smad4-null MDA-MB468 cells, it must recognize the Smad2/Smad2 interface. It may therefore interact with the subunit that forms the Smad2/Smad2 interface close to V431 (pink subunit in Fig. 9D), rather than the other (white) subunit that interacts with Smad4 at the interface close to residue V431 in Smad2.
We have demonstrated that the FM discriminates between active Smad2/Smad4 and Smad3/Smad4 complexes. This result was very surprising, since the MH2 domains of Smad2 and Smad3 are nearly identical (42). However, one striking difference between Smad2/Smad4 complexes and Smad3/Smad4 complexes is that the former are trimeric, but the latter have been shown, at least when bound to the c-Jun Smad-binding region, to be dimeric (16). Assuming the structure of the Smad3/Smad4 dimer is as recently proposed (42), it is tempting to speculate that the reason the FM does not bind Smad3/Smad4 complexes is because the R-Smad subunit it binds in the context of a Smad2/Smad4 trimer is missing (the pink subunit in Fig. 9D). It is also possible that the Smad3/Smad4 complexes interact with other components that prevent FM binding.
We propose that the presence of both an FM and a SIM in the FoxH1s has three important biological consequences. First, it allows the FoxH1s to distinguish between monomeric inactivated Smad2, and phosphorylated Smad2 in active complexes with Smad4. Second, it allows the FoxH1s to form very strong complexes with activated Smads. We have demonstrated that XFoxH1b can very effectively compete for activated Smad2/Smad4 complexes with the transcription factor Mixer, which only contains a SIM. This establishes a hierarchy between Smad-interacting transcription factors determined by their affinity for Smad complexes. This may be important for determining cell type-specific responses to TGF-β ligands, which would be sensitive to levels of active Smad2/Smad4 complexes in the nucleus, thus providing a mechanism for interpreting gradients of ligand activity. In cells therefore that coexpress these transcription factors, genes containing AREs in their promoters may be activated by TGF-β family members preferentially to genes containing Mixer binding sites, when levels of active Smads are limiting. Third, we have demonstrated that the FM discriminates between endogenous Smad3/Smad4 complexes and Smad2/Smad4 complexes, and moreover, XFoxH1a preferentially binds activated endogenous Smad2/Smad4 complexes at the ARE.
Previous results from others have shown that members of the FoxH1 family can form complexes with Smad3, but this has only been demonstrated with overexpressed Smad3, or with recombinant Smad3 in vitro (18, 19, 24, 44, 45), and we have confirmed this. The functional consequences of this interaction were controversial since in some cases incorporation of Smad3 into an ARF complex was shown to act positively (18, 44, 45), while in others it inhibited transcriptional activation (19, 24). In contrast, in our experiments performed with endogenous Smads, we demonstrate that the FM and XFoxH1a exhibit a high degree of selectivity in their binding to Smad2 versus Smad3. This property of the FoxH1 family members is potentially relevant in mouse and frog development, where FoxH1, Smad2 and Smad3 are coexpressed (4, 13, 14, 36, 38). In addition, it is clear that in some regions of the early mouse embryo, in particular the visceral endoderm, FoxH1 cooperates exclusively with Smad2 and Smad4 for the maintenance and amplification of Nodal transcription, as there is no Smad3 expressed in this region (26, 36). Also in Xenopus gastrula embryos the XFoxH1s must cooperate with Smad2 and Smad4 as there is little or no Smad3 expressed at this time (14).
In conclusion, we have identified a new Smad2-interacting motif in FoxH1 family members, which is the first such motif to recognize only active Smad complexes, and to distinguish between activated Smad2 and Smad3. Since a Smad interaction motif with this property will confer on transcription factors the ability to bind only activated Smad2-containing complexes in the nucleus, it will be very important to determine if the FM exists in any other Smad2-interacting transcription factors, and whether it is always found in association with a SIM.
Acknowledgments
We thank Hiroshi Hamada for the Pitx2 ASE-Luc reporter plasmid, Ed Leof for the anti-phosphorylated Smad3 antibody, Anita Roberts for the TβRII expression plasmid and the Smad2-null mouse embryo fibroblasts, Peter ten Dijke for the anti-phosphorylated Smad2 and anti-Smad3 antibodies and the TβRI expression plasmid, Annabel Borg for valuable help with the gel filtration analyses, Angelina Felici for advice on HaCaT transfections, and Nicola O'Reilly for peptide synthesis. We thank members of the lab, Richard Treisman, Mark Uden, and Frank Uhlmann for helpful comments on the manuscript.
The work was supported by Cancer Research UK.
REFERENCES
- 1.Bates, P. A., and M. J. Sternberg. 1999. Model building by comparison at CASP3: using expert knowledge and computer automation. Proteins Suppl. 3:47-54. [DOI] [PubMed] [Google Scholar]
- 2.Brady, G. P., Jr., and P. F. Stouten. 2000. Fast prediction and visualization of protein binding pockets with PASS. J. Comput. Aided Mol. Des. 14:383-401. [DOI] [PubMed] [Google Scholar]
- 3.Chacko, B. M., B. Qin, J. J. Correia, S. S. Lam, M. P. de Caestecker, and K. Lin. 2001. The L3 loop and C-terminal phosphorylation jointly define Smad protein trimerization. Nat. Struct. Biol. 8:248-253. [DOI] [PubMed] [Google Scholar]
- 4.Chen, X., M. J. Rubock, and M. Whitman. 1996. A transcriptional partner for MAD proteins in TGF-β signalling. Nature 383:691-696. [DOI] [PubMed] [Google Scholar]
- 5.Chen, X., E. Weisberg, V. Fridmacher, M. Watanabe, G. Naco, and M. Whitman. 1997. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature 389:85-89. [DOI] [PubMed] [Google Scholar]
- 6.Derossi, D., G. Chassaing, and A. Prochiantz. 1998. Trojan peptides: the penetratin system for intracellular delivery. Trends Cell Biol. 8:84-87. [PubMed] [Google Scholar]
- 7.Faure, S., M. A. Lee, T. Keller, P. ten Dijke, and M. Whitman. 2000. Endogenous patterns of TGFβ superfamily signaling during early Xenopus development. Development 127:2917-2931. [DOI] [PubMed] [Google Scholar]
- 8.Franzen, P., P. ten Dijke, H. Ichijo, H. Yamashita, P. Schulz, C. H. Heldin, and K. Miyazono. 1993. Cloning of a TGF β type I receptor that forms a heteromeric complex with the TGF β type II receptor. Cell 75:681-692. [DOI] [PubMed] [Google Scholar]
- 9.Germain, S., M. Howell, G. M. Esslemont, and C. S. Hill. 2000. Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 14:435-451. [PMC free article] [PubMed] [Google Scholar]
- 10.Hata, A., R. S. Lo, D. Wotton, G. Lagna, and J. Massagué. 1997. Mutations increasing autoinhibition inactivate tumour suppressors Smad2 and Smad4. Nature 388:82-87. [DOI] [PubMed] [Google Scholar]
- 11.Heyer, J., D. Escalante-Alcalde, M. Lia, E. Boettinger, W. Edelmann, C. L. Stewart, and R. Kucherlapati. 1999. Postgastrulation Smad2-deficient embryos show defects in embryo turning and anterior morphogenesis. Proc. Natl. Acad. Sci. USA 96:12595-12600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill, C. S. 2001. TGF-β signalling in early Xenopus development. Curr. Opin. Genet. Dev. 11:533-540. [DOI] [PubMed] [Google Scholar]
- 13.Howell, M., G. J. Inman, and C. S. Hill. 2002. A novel Xenopus Smad-interacting forkhead transcription factor (XFast-3) cooperates with XFast-1 in regulating gastrulation movements. Development 129:2823-2834. [DOI] [PubMed] [Google Scholar]
- 14.Howell, M., T. J. Mohun, and C. S. Hill. 2001. Xenopus Smad3 is specifically expressed in the chordoneural hinge, notochord and in the endocardium of the developing heart. Mech. Dev. 104:147-150. [DOI] [PubMed] [Google Scholar]
- 15.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33-38. [DOI] [PubMed] [Google Scholar]
- 16.Inman, G. J., and C. S. Hill. 2002. Stoichiometry of active Smad-transcription factor complexes on DNA. J. Biol. Chem. 277:51008-51016. [DOI] [PubMed] [Google Scholar]
- 17.Koradi, R., M. Billeter, and K. Wüthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14:51-55. [DOI] [PubMed] [Google Scholar]
- 18.Kretzschmar, M., J. Doody, I. Timokhina, and J. Massagué. 1999. A mechanism of repression of TGFβ/Smad signaling by oncogenic Ras. Genes Dev. 13:804-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labbé, E., C. Silvestri, P. A. Hoodless, J. L. Wrana, and L. Attisano. 1998. Smad2 and Smad3 positively and negatively regulate TGF β-dependent transcription through the forkhead DNA-binding protein FAST2. Mol. Cell 2:109-120. [DOI] [PubMed] [Google Scholar]
- 20.Liu, B., C. L. Dou, L. Prabhu, and E. Lai. 1999. FAST-2 is a mammalian winged-helix protein which mediates transforming growth factor β signals. Mol. Cell. Biol. 19:424-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo, R. S., Y. G. Chen, Y. Shi, N. P. Pavletich, and J. Massagué. 1998. The L3 loop: a structural motif determining specific interactions between SMAD proteins and TGF-β receptors. EMBO J. 17:996-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris, G. M., D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. Belew, and A. J. Olson. 1998. Automated Docking Using a Lamarckian Genetic Algorithm and an Empirical Binding Free Energy Function. J. Computational Chem. 19:1639-1662. [Google Scholar]
- 23.Moustakas, A., S. Souchelnytskyi, and C. H. Heldin. 2001. Smad regulation in TGF-β signal transduction. J. Cell Sci. 114:4359-4369. [DOI] [PubMed] [Google Scholar]
- 24.Nagarajan, R. P., J. Liu, and Y. Chen. 1999. Smad3 inhibits transforming growth factor-β and activin signaling by competing with Smad4 for FAST-2 binding. J. Biol. Chem. 274:31229-31235. [DOI] [PubMed] [Google Scholar]
- 25.Nicolás, F. J., and C. S. Hill. 2003. Attenuation of the TGF-β-Smad signaling pathway in pancreatic tumor cells confers resistance to TGF-β-induced growth arrest. Oncogene 22:3698-3711. [DOI] [PubMed] [Google Scholar]
- 26.Norris, D. P., J. Brennan, E. K. Bikoff, and E. J. Robertson. 2002. The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development. 129:3455-3468. [DOI] [PubMed] [Google Scholar]
- 27.Piek, E., W. J. Ju, J. Heyer, D. Escalante-Alcalde, C. L. Stewart, M. Weinstein, C. Deng, R. Kucherlapati, E. P. Bottinger, and A. B. Roberts. 2001. Functional characterization of transforming growth factor β signaling in Smad2- and Smad3-deficient fibroblasts. J. Biol. Chem. 276:19945-19953. [DOI] [PubMed] [Google Scholar]
- 28.Pierreux, C. E., F. J. Nicolás, and C. S. Hill. 2000. Transforming growth factor β-independent shuttling of Smad4 between the cytoplasm and nucleus. Mol. Cell. Biol. 20:9041-9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pogoda, H., L. Solnica-Krezel, W. Driever, and D. Meyer. 2000. The zebrafish forkhead transcription factor FoxH1/Fast1 is a modulator of nodal signaling required for organizer formation. Curr. Biol. 10:1041-1049. [DOI] [PubMed] [Google Scholar]
- 30.Qin, B., S. S. Lam, and K. Lin. 1999. Crystal structure of a transcriptionally active Smad4 fragment. Structure Fold Des. 7:1493-1503. [DOI] [PubMed] [Google Scholar]
- 31.Qin, B. Y., S. S. Lam, J. J. Correia, and K. Lin. 2002. Smad3 allostery links TGF-β receptor kinase activation to transcriptional control. Genes Dev. 16:1950-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Randall, R. A., S. Germain, G. J. Inman, P. A. Bates, and C. S. Hill. 2002. Different Smad2 partners bind a common hydrophobic pocket in Smad2 via a defined proline-rich motif. EMBO J. 21:145-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi, Y., A. Hata, R. S. Lo, J. Massagué, and N. P. Pavletich. 1997. A structural basis for mutational inactivation by the tumour suppressor Smad4. Nature 388:87-93. [DOI] [PubMed] [Google Scholar]
- 34.Shiratori, H., R. Sakuma, M. Watanabe, H. Hashiguchi, K. Mochida, Y. Sakai, J. Nishino, Y. Saijoh, M. Whitman, and H. Hamada. 2001. Two-step regulation of left-right asymmetric expression of Pitx2: initiation by nodal signaling and maintenance by Nkx2. Mol. Cell 7:137-149. [DOI] [PubMed] [Google Scholar]
- 35.Sirotkin, H. I., M. A. Gates, P. D. Kelly, A. F. Schier, and W. S. Talbot. 2000. fast1 is required for the development of dorsal axial structures in zebrafish. Curr. Biol. 10:1051-1054. [DOI] [PubMed] [Google Scholar]
- 36.Tremblay, K. D., P. A. Hoodless, E. K. Bikoff, and E. J. Robertson. 2000. Formation of the definitive endoderm in mouse is a Smad2-dependent process. Development 127:3079-3090. [DOI] [PubMed] [Google Scholar]
- 37.Varshney, A., F. P. J. Brooks, and W. V. Wright. 1994. Linearly scalable computation of smooth molecular surfaces. IEEE Comput. Graph. Appli. 14:19-25. [Google Scholar]
- 38.Weisberg, E., G. E. Winnier, X. Chen, C. L. Farnsworth, B. L. Hogan, and M. Whitman. 1998. A mouse homologue of FAST-1 transduces TGFβ superfamily signals and is expressed during early embryogenesis. Mech. Dev. 79:17-27. [DOI] [PubMed] [Google Scholar]
- 39.Wrana, J. L., L. Attisano, J. Cárcamo, A. Zentella, J. Doody, M. Laiho, X. F. Wang, and J. Massagué. 1992. TGFβ signals through a heteromeric protein kinase receptor complex. Cell 71:1003-1014. [DOI] [PubMed] [Google Scholar]
- 40.Wu, G., Y. G. Chen, B. Ozdamar, C. A. Gyuricza, P. A. Chong, J. L. Wrana, J. Massagué, and Y. Shi. 2000. Structural basis of Smad2 recognition by the Smad anchor for receptor activation. Science 287:92-97. [DOI] [PubMed] [Google Scholar]
- 41.Wu, J. W., R. Fairman, J. Penry, and Y. Shi. 2001. Formation of a Stable Heterodimer between Smad2 and Smad4. J. Biol. Chem. 276:20688-20694. [DOI] [PubMed] [Google Scholar]
- 42.Wu, J. W., M. Hu, J. Chai, J. Seoane, M. Huse, C. Li, D. J. Rigotti, S. Kyin, T. W. Muir, R. Fairman, J. Massagué, and Y. Shi. 2001. Crystal structure of a phosphorylated Smad2. Recognition of phosphoserine by the MH2 domain and insights on Smad function in TGF-β signaling. Mol. Cell 8:1277-1289. [DOI] [PubMed] [Google Scholar]
- 43.Xu, L., Y. Kang, S. Col, and J. Massagué. 2002. Smad2 nucleocytoplasmic shuttling by nucleoporins CAN/Nup214 and Nup153 feeds TGFβ signaling complexes in the cytoplasm and nucleus. Mol. Cell 10:271-282. [DOI] [PubMed] [Google Scholar]
- 44.Yagi, K., D. Goto, T. Hamamoto, S. Takenoshita, M. Kato, and K. Miyazono. 1999. Alternatively spliced variant of Smad2 lacking exon 3. Comparison with wild-type Smad2 and Smad3. J. Biol. Chem. 274:703-709. [DOI] [PubMed] [Google Scholar]
- 45.Yeo, C. Y., X. Chen, and M. Whitman. 1999. The role of FAST-1 and Smads in transcriptional regulation by activin during early Xenopus embryogenesis. J. Biol. Chem. 274:26584-26590. [DOI] [PubMed] [Google Scholar]
- 46.Zhou, S., L. Zawel, C. Lengauer, K. W. Kinzler, and B. Vogelstein. 1998. Characterization of human FAST-1, a TGF β and activin signal transducer. Mol. Cell 2:121-127. [DOI] [PubMed] [Google Scholar]