Abstract
Oct-1 is a sequence-specific DNA binding transcription factor that is believed to regulate a large group of tissue-specific and ubiquitous genes. Both Oct-1 and the related but tissue-restricted Oct-2 protein bind to a DNA sequence termed the octamer motif (5′-ATGCAAAT-3′) with equal affinity in vitro. To address the role of Oct-1 in vivo, an Oct-1-deficient mouse strain was generated by gene targeting. Oct-1-deficient embryos died during gestation, frequently appeared anemic, and suffered from a lack of Ter-119-positive erythroid precursor cells. This defect was cell intrinsic. Fibroblasts derived from these embryos displayed a dramatic decrease in Oct-1 DNA binding activity and a lack of octamer-dependent promoter activity in transient transfection assays. Interestingly, several endogenous genes thought to be regulated by Oct-1 showed no change in expression. When crossed to Oct-2+/− animals, transheterozygotes were recovered at a very low frequency. These findings suggest a critical role for Oct-1 during development and a stringent gene dosage effect with Oct-2 in mediating postnatal survival.
Gene expression is mediated by the interaction of trans-acting factors with cis-acting elements in promoters and enhancers. The octamer-binding proteins Oct-1 and Oct-2 control gene expression by interacting with the octamer element 5′-ATGCAAAT-3′ and related motifs (6, 50, 54, 55, 57). Their DNA binding domains are highly homologous, and both proteins bind DNA with equal affinity in vitro (29). Expression studies indicate that Oct-1 is ubiquitously expressed, whereas Oct-2 is restricted to B cells, macrophages, and other hematopoietic cells, as well as cells of the central nervous system.
Oct-1 and Oct-2 belong to the POU domain family of transcription factors, which was identified after the isolation of three mammalian transcription factors and a Caenorhabditis elegans developmental regulator: Pit-1, Oct-1, Oct-2 and unc-86 (16). The POU domain consists of a bipartite DNA-binding motif, the POU-specific and POU homeodomain, tethered by a linker domain of approximately 20 amino acids (60, 66). X-ray crystallography studies reveal that the POU subdomains of Oct-1 both contact the major groove but on opposite sides of the DNA (26), with the POU-specific domain contacting the 5′-ATGC subsite and the POU homeodomain binding the AAAT-3′ subsite. Such cooperative binding between adjacent subdomains resulting in the recognition of asymmetrical DNA motifs is characteristic of POU domain transcription factors.
With the exception of the ubiquitously expressed Oct-1, all of the known mouse POU domain factors are expressed in restricted temporal and spatial patterns during development (reviewed in reference 49). Gene targeting experiments have provided direct evidence that the POU domain proteins play a critical role in the determination of cell fates. For example, deficiency of the POU domain protein Pit-1 results in pituitary hypoplasia and dwarfism (33, 44). Mutation of Brn-3.1 leads to congenital auditory loss whereas Brn-3.2 knockout mice have defects in the terminal differentiation and survival of retinal ganglion cells (9). Disruption of Oct-3/4 results in loss of pluripotency of the inner cell mass in the developing embryo (39).
The canonical octamer was first described as a conserved motif present in immunoglobulin heavy- and light-chain promoters and enhancers (10, 37, 43). Point mutation of the octamer reduces the expression of an immunoglobulin transgene by over 20-fold, demonstrating the importance of this sequence in mediating immunoglobulin transcription (20). However, the same sequence occurs in the regulatory regions of other genes, most of which are not B-cell specific. Examples include the U2 and U6 snRNA and histone H2B genes (38, 55). Variants of the octamer sequence have been implicated in the regulation of an array of lymphoid-specific genes, such as CD20, CD21, CD36, interleukin-2, interleukin-4, and Pax-5 (5, 23, 27, 36, 45, 46, 53, 63, 69). Other tissue-specific genes thought to be regulated by Oct-1 include osteopontin, TIE1, Cdx-2, iNOS, and GADD45 (2, 11, 21, 22, 25, 30, 61, 67).
While Oct-2, with its lymphoid cell- and neuron-restricted expression, was first thought to be critical in mediating immunoglobulin expression, genetic evidence contradicted this assumption. Although Oct-2−/− animals die perinatally from unidentified causes, B-cell development is largely unimpaired and normal levels of immunoglobulin are produced (7). The discovery of OCA-B/Bob-1/OBF-1, a B-cell-specific cofactor that interacts with both Oct-1 and Oct-2 (12, 35, 58), provided a possible explanation for the B-cell-restricted activity of the octamer element. However, OCA-B mutation alone and in combination with Oct-2 generates B cells with normal immunoglobulin levels in mouse models (24, 40, 51).
To further investigate the importance of the octamer and its associated protein factors, we created an Oct-1 loss-of-function model by gene targeting. We demonstrate that Oct-1 is essential for embryonic viability and normal erythropoiesis in vivo and octamer-dependent transcription in transient transfection assays. Furthermore, Oct-1+/−;Oct-2+/− animals exhibit decreased postnatal viability, demonstrating a cooperative interaction between these two proteins.
MATERIALS AND METHODS
Inactivation of Oct-1.
An Oct-1 genomic clone containing exons 3 to 7 was isolated from a mouse 129/Sv phage library by standard techniques. A targeting construct was created by replacing exon 3 with a neomycin resistance cassette in the opposite transcriptional orientation and flanked by loxP sites. J1 embryonic stem (ES) cells (32) were electroporated at 240 V with 20 μg of NotI-linearized targeting construct and selected for resistance to G418 (200 μg/ml) and ganciclovir (0.2 μM). Homologous recombinants were identified by NheI digestion of genomic DNA and Southern blotting with a 1.2-kb 5′ external probe. Heterozygous clones showed a 7.2-kb band corresponding to the incorporation of the neomycin cassette. DNA from the positive clones was subsequently digested with HindIII and verified by Southern blotting by a 3′ probe.
Two different ES cell clones were injected into embryonic day 3.5 (E3.5) C57BL/6 blastocysts, and the resulting chimeras were backcrossed to C57BL/6 for germ line transmission. Heterozygous pups were detected in agouti offspring by Southern blotting and PCR of tail DNA. PCR for genotyping was performed with the following primers: 5′-TAGCACTTCTCCCCATCTTCTATC-3′ (common), 5′-GCAGAAGAGTTACACAGGATTTGA-3′ (wild-type allele), and 5′-CGAAGTTATTAGGTCCCGAATA-3′ (mutant allele).
Generation and analyses of Oct-1-deficient primary and immortalized MEFs.
Heterozygous Oct-1 mutant females and males were intercrossed. The morning that a vaginal plug was detected was designated day 0.5. At day 13.5 post coitum, embryos were isolated and prepared as described (18). Immortalized fibroblasts were obtained by a serial 3T3 protocol (64). Oct-1 was restored in Oct-1−/− immortalized mouse embryo fibroblast (MEF) lines after single-cell cloning by infection with retroviruses carrying human Oct-1 (pBabe-hOct-1).
Hematopoietic analyses.
For flow cytometry, single-cell suspensions from E12.5 to 13.5 fetal livers were prepared by passage through a 26-gauge needle. Enucleated red blood cells were lysed with PharmLyse (Pharmingen) for 15 min at 37°C. Cells were then incubated with anti-CD32/CD16 (FcγII/III receptor, 2.4G2, Pharmingen) for 5 min prior to the addition of primary antibodies. The following antibodies from Pharmingen were used according to the manufacturer's protocol: anti-TER-119-phycoerythrin (Ly-76), anti-Ly-6G-allophycocyanin (Gr-1, RB6-8C5), anti-CD11b-allophycocyanin (Mac-1), and anti-CD45.2-fluorescein isothiocyanate (Ly-5.2). Stained cells were analyzed on a FACSCalibur (Becton Dickinson) by gating on live cells. At least 20,000 events were collected per sample.
Adoptive transfers were performed by harvesting fetal livers at E12.5 (17). Recipient C57BL/6-CD45.1 animals were irradiated in two doses of 600 rad each, separated by 3 h, by a 137Cs source. Irradiated animals were maintained in autoclaved cages with trimethoprim-sulfamethoxazole. Bone marrow cells were analyzed by fluorescence-activated cell sorting 16 weeks after adoptive transfer.
For real-time reverse transcription PCR, RNA was prepared from E12 fetal livers enriched by affinity selection by magnetic microbeads conjugated with TER-119 antibody (Miltenyi). cDNA was generated by 1 μg of RNA and oligo(dT) primer (Invitrogen). SYBR green PCR (Qiagen) was performed at an annealing temperature of 50°C for 40 cycles with primer sets specific for β, α, ɛ, and βH1 globins. β-Actin (Ambion) was used in parallel to normalize for loading. Experiments were performed in triplicate.
Electrophoretic mobility shift assays.
Nuclear extract from the appropriate cell line was prepared from 15-cm dishes by a modified protocol (8). Concentration was quantified by the Bradford assay. Single-stranded DNA oligonucleotides were purified by polyacrylamide gel electrophoresis, resuspended to a final concentration of 100 μM, and annealed with its respective counterpart in equimolar concentration in 10 mM Tris, pH 8.0, 50 mM NaCl, and 1 mM EDTA. The sequences of the consensus and the mutant octamer oligonucleotides have been described previously (50). Annealed oligonucleotides were labeled with the Klenow fragment of DNA polymerase (Roche) and α-[32P]dATP and purified by gel electrophoresis. Mobility shift assays were performed essentially as described (4). After 15 min of incubation at room temperature, anti-Oct-1 antibody (Santa Cruz) was added to the samples where indicated. The gel was exposed and quantitated with a phosphoimager.
Oct-1 enrichment.
Latex beads were obtained from the laboratory of Hiroshi Handa (Frontier Collaborative Research Center, Tokyo, Japan). The beads were equilibrated in 0.6× buffer D (16 mM HEPES, pH 7.9, 1 mM EDTA, 16% glycerol, 1 mM dithiothreitol) containing 200 mM NaCl. Octamer-binding proteins were purified from nuclear extract as follows: 2.5 mg of nuclear extract was incubated with 17.5 μg of poly(dI · dC) and 10 μl of packed beads in a final volume of 500 μl in 0.6× buffer D with 1 mg of bovine serum albumin per ml for 20 min at room temperature. The beads were washed four times, in 0.6× buffer D with 200 and 300 mM NaCl successively, by centrifugation at 16,000 × g for 5 min. Bound proteins were eluted into two washes of 20 μl in 0.6× buffer D with 1.0 M NaCl at room temperature. The eluates were pooled, concentrated by trichloroacetic acid, and resolved on an SDS-10% polyacrylamide gel. For Western blotting, a mouse polyclonal antibody raised against the C terminus of Oct-1 (unpublished data) was supplemented with mouse monoclonal antibodies directed against the Oct-1 DNA binding domain (Calbiochem) and the C terminus (Santa Cruz). Detection was performed by chemiluminescence with an anti-mouse immunoglobulin-horseradish peroxidase secondary antibody (Amersham).
Triton-acetic acid-urea gel electrophoresis and mass spectroscopy.
Triton-acetic acid-urea gels and whole-cell extract were prepared essentially as described (34); 8 μg of extract derived from E11.5 embryos was loaded and electrophoresed for 24 h. The appropriate band was excised and digested with trypsin in 25 mM ammonium carbonate overnight. Tryptic peptides were eluted with acetonitrile. The peptides were dried and reconstituted with 8 μl of 0.1% trifluoroacetic acid, desalted by a C18 Ziptip (Millipore), and eluted with 4 μl of 50% acetonitrile-0.1% trifluoroacetic acid. Eluates were dried and reconstituted with 1 μl of α-cyano-4-hydroxycinnamic acid matrix, and half of the total volume was applied to separate matrix-assisted laser desorption ionization-time of flight (MALDI-TOF; Applied Biosystems model Voyager DESTR). Spectra were screened against the NCBI database by the Protein Prospector search engine (University of California-San Francisco).
Transfection.
A total of 105 immortalized fibroblasts were seeded into six-well plates. Plasmid constructs contained an H2B core promoter (pGL3-H2B-59+1) or a VH promoter-intronic enhancer combination (EVH186.2, gift of F. Alt) fused to a firefly luciferase gene; 10 ng of pCMV-Renilla luciferase was used per sample as an internal control for transfection efficiency. A promoterless vector (pGL3, Promega) served as a negative control. Transfections were performed with FuGene 6 (Roche) according to the manufacturer's protocol. Reporter activity was measured 36 h posttransfection by the dual luciferase assay (Promega). Experiments were performed in triplicate.
Northern hybridization.
Total RNA from primary MEFs was prepared by Trizol (Invitrogen); 10 μg of total RNA was used for Northern blotting according to standard procedures. Hybridization was carried out with γ-[32P]ATP-labeled oligonucleotides to the U2 and U6 snRNAs and H2B mRNA. Band intensity was quantitated with a PhosphoImager and ImageQuant software and normalized to β-actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Ambion).
RESULTS
Inactivation of the Oct-1 gene.
Oct-1 is located in a syntenic region on chromosome 1 in mice and humans (59). Sequencing the exon-intron boundaries of mouse exons 3 to 7 and comparison to known human sequences (59) showed that the genomic organization was conserved. Previous studies have shown that targeted disruption of exon 9 in the CD3ζ locus also affected Oct-1 transcription due to overlap of the 3′ regions of the two genes (28, 31, 41). To avoid disrupting the CD3 locus, a strategy was designed to inactivate Oct-1 by targeting the 5′ end. At least three isoforms of the Oct-1 protein resulting from alternative splicing have been identified in the mouse (19, 42, 47). The transcripts encoding these isoforms have a common 5′ sequence but each has a unique 3′ end distal to the POU domain. Our targeting strategy would thus minimize the production of all known alternatively spliced variants.
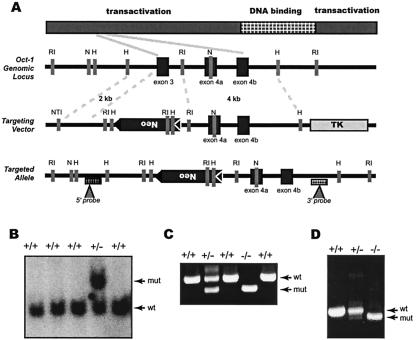
Exon 3 was replaced with a loxP-flanked neomycin gene in the antisense transcriptional orientation (Fig. 1A). Potential transcripts from the targeted allele that are spliced from exon 2 to exon 4 will harbor a frameshift mutation, resulting in premature termination and deletion of at least two-thirds of the encoded polypeptide, including the DNA binding domain. Clones with the mutation introduced on one allele were identified by NheI digest to detect a 7.5-kb fragment, indicating the incorporation of the neomycin cassette (Fig. 1B). Positive clones were confirmed by a second HindIII digest with a 3′ probe (not shown). Two independent clones were injected into C57BL/6 blastocysts to generate chimeric animals. Heterozygous Oct-1-deficient animals were viable and fertile (not shown).
FIG. 1.
Inactivation of Oct-1. (A) Schematic of the Oct-1 murine genomic locus and targeting vector. Exons are denoted by black boxes and positions of internal and external probes are indicated with triangles. Restriction enzymes: RI, EcoRI; H, HindIII; N, NheI; NT1, NotI. (B) Genomic blot by DNA derived from ES cells. The DNA was digested with NheI and probed with the 5′ external probe indicated in A. The endogenous allele is 6 kb and the targeted allele is 7.2 kb. Wt, wild-type allele; Mut, targeted allele. (C) Genotyping of tail DNA by PCR. PCR conditions and primers are described in Materials and Methods. Primers distinguished a 304-bp fragment of the targeted allele from a 424-bp fragment of the wild-type allele. (D) Reverse transcription-PCR of RNA from Oct-1 mutant mice. Primers amplified a region from exon 2 to the POU domain.
To characterize the consequences of the gene targeting to the structure of the mRNA, a reverse transcription PCR was performed. RNA derived from the heterozygous MEFs produced a doublet corresponding to the expected lengths of fragments from wild-type and mutant cDNAs. In contrast, RNA derived from homozygous MEFs produced a single band of reduced intensity (Fig. 1D). Sequencing verified that this band resulted from the splicing of exon 2 to exon 4a. These results showed that the mutant Oct-1 allele was transcribed and the mRNA was not completely subject to nonsense-mediated decay. However, splicing of exon 2 to exon 4a generates a nonsense mutation that upon translation results in a polypeptide truncated before the DNA binding domain.
Oct-1 is critical for embryonic survival.
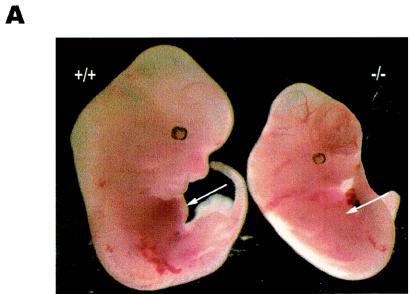
Oct-1+/− animals were intercrossed to generate homozygotes for analysis. On a mixed C57BL/6 × 129/Sv background, no viable Oct-1−/− animals were detected at birth among more than 300 offspring, indicating that the mutation is embryo lethal (Table 1). To assess the timing and the cause of death, timed matings were performed. The window of lethality on the mixed background was variable, beginning at ≈E12 and continuing close to birth (Table 1). At E13.5, the homozygous mutant embryos were already underrepresented, constituting only 17.7% of the entire population, compared with an expected frequency of 25% (Table 1). Of these, 10.5% did not show any gross histological abnormalities at the time of sacrifice, whereas 3.6% appeared runted (see, for example, Fig. 5A), and another 3.6% were dead. The window of embryonic lethality shifts much earlier with successive backcrosses to either C57BL/6 or 129/Sv (unpublished data). These observations indicate that Oct-1 is essential for embryonic development.
TABLE 1.
Analysis of embryos from Oct-1+/− crossesa
| Gestational age | Total no. of embryos | No. of resorbed embryos | No. genotyped | No. (%) of embryos
|
||||
|---|---|---|---|---|---|---|---|---|
| +/+ | +/− | −/− | −/− runted | −/− dead | ||||
| E13.5 | 211 | 20 | 191 | 47 (24.6) | 110 (57.6) | 20 (10.5) | 7 (3.6) | 7 (3.6) |
| E18.5 | 30 | 5 | 24 | 6 (25) | 17 (70.8) | 2 (8.3) | 1 (4.2) | 1 (4.2) |
| E20.5 (pups) | 380 | 12 | 368 | 145 (39.4) | 223 (60.6) | 0 (0) | 0 (0) | 0 (0) |
Genotypes of resorbed embryos were not available. Nonresorbed embryos and newborn pups were genotyped by PCR and Southern blotting with yolk sac and tail DNA as described in the text. The −/− group included only embryos which were alive and anatomically normal at the time of dissection, whereas the −/− runted group consisted of embryos which were visibly smaller and paler but alive.
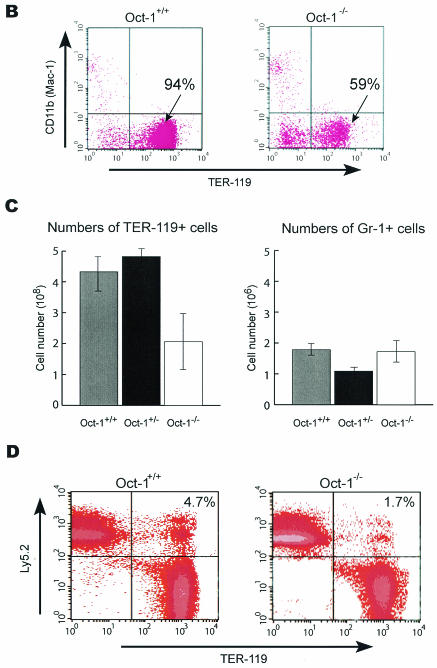
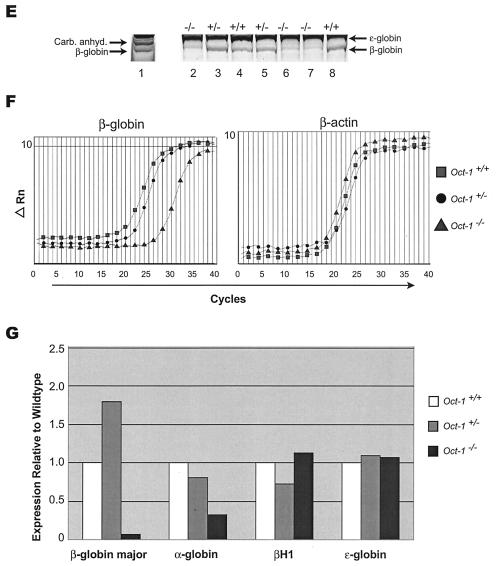
FIG. 5.
Oct-1-deficient embryos are runted and anemic and show reduced β-globin expression. (A) Comparison of an Oct-1-deficient embryo (right) with a wild-type littermate control (left) at E12.5. Oct-1 mutants appeared smaller with pale fetal livers that are reduced in size (arrows). (B and C) Flow cytometry analysis of TER-119-positive cells from E12.5 fetal livers. Oct-1 mutants exhibit a decreased proportion (B) and fewer total numbers (C) of TER-119-positive cells. Three wild-type and four homozygous Oct-1 mutant animals were used. Error bars denote standard deviations. (D) Fluorescence-activated cell sorting analysis of bone marrow from adoptive transfer of fetal livers. Irradiated recipient animals were injected with fetal liver cells from E12.5 Oct-1−/− embryos. At 16 weeks after transfer, bone marrow cells from the recipient mice were analyzed by staining with antibodies directed against TER-119 and CD45.2/Ly-5.2. (E) Separation of fetal and adult globins by Triton-acetic acid-urea gel electrophoresis of protein extract from E11.5 embryos. Lane 1: peripheral blood control derived from 8-week-old adult mouse; lanes 2 to 8, embryos with their genotypes indicated. Carb. Anhyd., carbonic anhydrase. (F) Real-time PCR amplification curves for β-globin and β-actin. Data are shown for one representative set of experiments. (G) Quantitation of globin RNA levels by SYBR green real-time PCR. RNA was purified from E12.5 TER-119-positive fetal liver cells.
Oct-1 protein levels are severely reduced in mutant cells.
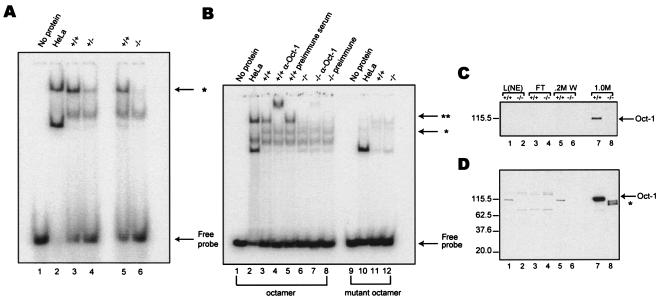
To examine Oct-1 protein activity in the mutant cells, electrophoretic mobility assays were performed with nuclear extract derived from immortalized MEFs. Oct-1/DNA complex formation was reduced by 50% in Oct-1+/− nuclear extract (Fig. 2A, lane 4) and was absent in Oct-1-deficient nuclear extracts (lane 6). The presence of Oct-1 in the protein-DNA complex was confirmed by supershift by a monoclonal antibody recognizing a C terminal epitope of the protein (Fig. 2B, lane 4). No supershift was detected when preimmune serum was used (lane 5). However, incubation of the Oct-1-deficient nuclear extract with an Oct-1-specific antibody revealed a slight amount of comigrating supershifted complex that was 40-fold lower in intensity compared to wild-type extract (Fig. 2B, lanes 6 and 7). The formation of both the Oct-1/DNA complex and the supershifted complex depended on a canonical octamer element as point mutation of the octamer element eliminated the protein-DNA complex (lanes 10 to 12).
FIG. 2.
Characterization of Oct-1 protein levels in mutant cells. (A) DNA binding activity is decreased in both heterozygous and homozygous MEF nuclear extracts. Lanes contained 5 μg of nuclear extract. HeLa extract was included as a positive control (lane 2). Lanes 3 and 5 contained wild-type extract, lane 4 had extract from heterozygous MEFs, and, lane 6 had extract from Oct-1-deficient MEFs. Oct-1/DNA complex is indicated by an asterisk. (B) Oct-1 DNA binding activity is dramatically reduced in mutant nuclear extract. EMSA of nuclear extract derived from wild-type (lanes 3 to 5, 11) and Oct-1-deficient immortalized cell lines (lanes 6 to 8, 12). Lanes contained 10 μg of total protein. **, supershifted complex formed upon addition of anti-Oct-1 antibody (lanes 4 and 7). Reactions in lanes 1 to 8 were incubated with an octamer oligonucleotide. and reactions in lanes 9 to 12 were incubated with a mutant. (C and D) A 1-s exposure and 2-min exposure, respectively, of a Western blot with a combination of three anti-Oct-1 antibodies (Materials and Methods). The load consisted of 2.5 μg of nuclear extract (lanes 1 and 2), representing 1% of the input sample. Truncated polypeptides are indicated by an asterisk (lane 8). Nuclear extract load (L[NE]), flowthrough (FT), wash (0.2 M W), and elution (1.0 M).
To investigate the expression of Oct-1 in the mutant cells, Western blotting experiments were performed. No Oct-1 protein was detected in repeated experiments with immortalized Oct-1-deficient MEF nuclear extracts (not shown). Because the Oct-1 signal from wild-type nuclear extract was relatively weak, latex microspheres (14) coupled to concatemerized octamer elements were used to enrich for octamer-binding proteins. Eluted samples were electrophoresed through SDS-10% PAGE gels and Western blotted by Oct-1 antibodies.
A combination of antibodies (recognizing the DNA-binding domain and C-terminal epitopes), did not detect Oct-1 in the mutant nuclear extract after enrichment with latex beads (Fig. 2C, lane 8). In contrast, a pronounced band was detected with the wild-type extract (lane 7). Upon prolonged exposure of the same blot, three truncated polypeptides were detected in the Oct-1−/− extract (Fig. 2D, lane 8). Based on their predicted molecular weights, these rare truncated products could arise from poorly utilized translation initiation sites downstream of exon 3 and/or possible rare splicing events joining exon 2 to downstream exons. Full-length Oct-1 was never detected even after prolonged exposure.
Serial dilution revealed that the sum of intensities of the truncated polypeptides was at least 30-fold less than the wild-type product (not shown). This value may represent an underestimation of the actual difference as some Oct-1 in the wild-type sample eluted during the washing steps (Fig. 2D, lane 5). Taken together, there is at least a 30- to 40-fold reduction in DNA binding activity. Thus, the engineered Oct-1 mutation represents a severely hypomorphic allele.
Oct-1 deficiency impairs octamer-dependent transcription.
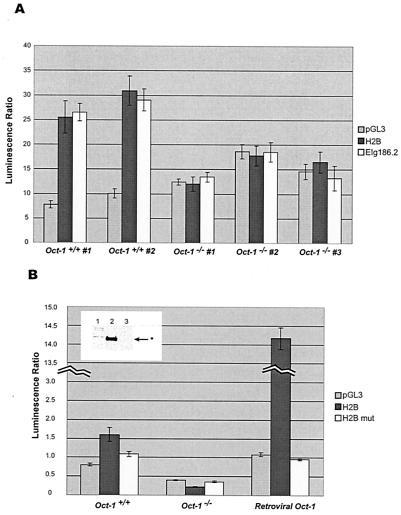
Transient transfection experiments were performed to examine the effect of Oct-1 deficiency on transcriptional regulation in vivo. A canonical octamer element is present in IgH variable region promoters (37, 43) and an inverted octamer element mediates the activity of the histone H2B promoter (55). Plasmid constructs containing these two promoters fused to a firefly luciferase reporter were transfected into wild-type and Oct-1-deficient immortalized fibroblasts. Transfection efficiency was normalized by cotransfecting plasmids carrying the Renilla luciferase gene under the control of a CMV promoter. The Eμ intronic enhancer was used in combination with the 186.2 VH promoter as the promoter alone gave weak signals in non-B-cell lines.
Figure 3A depicts results from three parallel transfections of immortalized fibroblast lines independently derived from wild-type and Oct-1-deficient mouse embryos. These promoters were inactive in Oct-1−/− cells. To assess whether this effect was mediated specifically through the octamer element, point mutations were introduced in the octamer sequence of the H2B promoter by altering 5′-ATGCAAAT-3′ to 5′-ATGCAAGC-3′. Promoters bearing mutant octamer elements reduced the reporter activity in wild-type cells to similar levels as negative controls. In contrast, reporter activity in Oct-1−/− cells was unaffected (Fig. 3B). These observations demonstrate that a reduction in cellular Oct-1 levels correspondingly decreases the transcriptional activity of transfected octamer-containing promoters.
FIG. 3.
Oct-1−/− immortalized cell lines fail to activate octamer-containing promoters. (A) H2B (black bars) and immunoglobulin VH (white bars) promoter constructs were transfected. pGL3 vector was used to establish a baseline of firefly luciferase activity (gray bars). (B) Point mutation in the octamer element abolishes promoter activity. Levels of exogenous Oct-1 delivered via a retroviral vector are shown in Western blot (inset). Lane 1 represents a wild-type cell line, lane 2 depicts a mutant cell line infected with retrovirus carrying pBABE-hOct-1, and lane 3 denotes the same line carrying an empty pBABE vector. All cell lines used in this experiments were single-cell cloned. *, Oct-1 protein.
To determine whether the defect in promoter activation could be rescued by ectopic expression of Oct-1, recombinant retroviruses containing human Oct-1 were introduced into the Oct-1-deficient fibroblast lines. Control retroviruses without Oct-1 had no effect on reporter activity (not shown). In contrast, expression of Oct-1 rescued the promoter activation defect (Fig. 3B). The octamer-dependent reporter activity in the rescued cell lines was approximately 10-fold higher than wild-type lines. Expression of Oct-1 in these cells was verified by Western blotting (Fig. 3B, inset). This result confirms that the defect in octamer-dependent promoter activity is due to the absence of Oct-1 and can be fully reversed upon its reintroduction.
Oct-1 is not uniquely required for the expression of putative target genes or cell cycle regulation in MEFs.
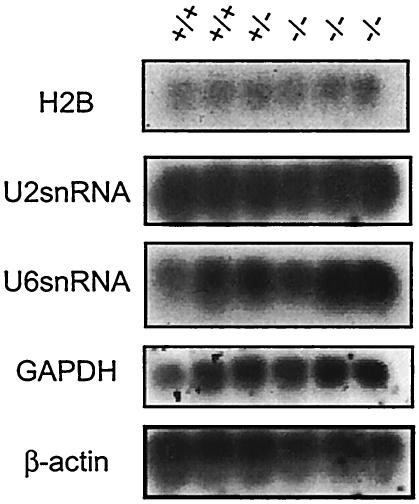
Oct-1 dependence has been proposed for a large number of ubiquitously expressed and tissue-specific genes. Putative target genes (histone H2B and U2 and U6 snRNA) were analyzed in primary MEFs by Northern analysis. β-actin and GAPDH were used as loading controls as Oct-1 has not been implicated in the regulation of either gene. These genes were expressed at significant and indistinguishable levels in Oct-1-deficient MEFs compared to wild-type and heterozygous controls (Fig. 4). These data suggest that Oct-1 is probably not essential for the expression of these genes. An initial analysis of the growth properties and cell cycle profile of Oct-1-deficient MEFs showed no differences between these cells and their wild-type counterparts (not shown).
FIG. 4.
Expression of putative Oct-1-dependent target genes is unaltered in the absence of Oct-1. Three independently isolated strains of Oct-1−/− primary MEFs were tested. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin were used as loading controls.
Oct-1-deficient embryos exhibit decreased erythropoiesis and a reduction in β-globin expression.
A subset of embryos appeared pale with small fetal livers even though the heart was beating at the time of dissection (Fig. 5A). We therefore tested whether Oct-1 deficiency resulted in reduced erythropoiesis. E12.5 fetal liver cells were tested for TER-119 expression because at later time points the embryos are frequently moribund. TER-119 reacts only with cells of the erythroid lineage and is expressed from the early proerythroblast through the mature erythrocyte stages. E12.5 fetal livers from Oct-1-deficient embryos contained a reduced percentage of TER-119-positive cells (Fig. 5B) as well as a significant decrease in the total number of TER-119-positive cells recovered (Fig. 5C). In contrast, the number of granulocyte precursors, as denoted by positive Gr-1 surface staining, was similar in both wild-type and mutant fetal liver populations. Oct-1 deficiency therefore results in a selective decrease in erythropoiesis and anemia in developing embryos.
To determine whether the decrease in TER-119-positive cells reflected a cell-intrinsic defect in the erythroid lineage due to Oct-1 loss, adoptive transfer experiments were performed. Oct-1−/− CD45.2/Ly-5.2 E12.5 embryos were injected into lethally irradiated CD45.1/Ly-5.1 C57BL/6 host animals as described in Materials and Methods. Erythroid cells from recipient bone marrow were stained with antibodies to TER-119 and CD45.2/Ly-5.2 and analyzed by fluorescence-activated cell sorting. As with the fetal livers, a clear reduction in Oct-1−/− TER-119 cells identified as CD45.2/Ly-5.2+ was evident (Fig. 5D). The analysis was conducted at 16 weeks posttransfer, and therefore it is unlikely that the observed differences were due to the rate of repopulation. Instead, the results likely reflect steady-state differences. These studies indicated that the erythroid phenotype of the Oct-1-deficient animals is cell autonomous.
To assess globin levels in the Oct-1-deficient animals, globin species derived from E11.5 embryos were fractionated on Triton X-100-acetic acid-urea-polyacrylamide gels (34). Coomassie blue staining revealed the presence of two closely spaced bands. The slow-migrating protein was consistently expressed at high levels in Oct-1−/−, Oct-1−/+, and Oct-1+/+ animals (Fig. 5E, lanes 2 to 8). The rapid-migrating band was reduced exclusively in Oct-1−/− animals. MALDI-TOF mass spectroscopy of tryptic fragments from this band matched 11 peptides to mouse adult β-globin, with 82% coverage and a mean error of −3.57 ppm. The slower-migrating band was identified as embryonic ɛ-globin by the same procedure. Both proteins were identified with high confidence intervals. These studies showed that the β-globin protein was selectively reduced in the Oct-1 homozygous mutant animals.
To distinguish whether the decrease in β-globin expression was due to a primary transcription defect or to secondary effects associated with anemia, real-time reverse transcription-PCR was performed with RNA isolated from purified TER-119-positive cells derived from E12.5 fetal livers. If the reduction in β-globin expression observed in the embryos reflected a decrease in the total number of red blood cells, β-globin levels on a per-cell basis would remain the same among wild-type, heterozygous, and homozygous preparations. Instead, there was a strong and specific reduction of β-globin mRNA levels in Oct-1−/− red blood cells relative to Oct-1+/+ and Oct-1+/− cells (Fig. 5F). Although α-globin mRNA appeared slightly reduced as well, no such difference was observed for the embryonic globin isoforms βH1 and ɛ (Fig. 5G). cDNA templates derived from the TER-119-negative fraction amplified approximately 32-fold less effectively for the globin messages (not shown). Thus, Oct-1 deficiency results in a specific molecular defect in the levels of β-globin mRNA.
Oct-1+/−; Oct-2+/− transheterozygous animals exhibit decreased postnatal viability.
To test whether Oct-1 and Oct-2 function redundantly during embryonic development, Oct-1+/− animals were crossed with Oct-2+/− animals. Oct-2 homozygotes die neonatally (7). Although genotypic analysis of E18.5 embryos revealed a ratio of transheterozygotes close to the expected Mendelian frequency, a large subset of the transheterozygotes died within 24 h of birth (Table 2). Histological examination did not reveal any gross anatomical abnormalities, and fluorescence-activated cell sorting analysis showed normal numbers of TER-119-positive cells (not shown). Surviving neonates became increasingly lethargic and runted, with lower body weights compared to littermate controls (not shown). A subset of these animals died prior to weaning. If the wild-type littermates were removed prior to weaning, some transheterozygotes animals could eventually attain normal body weights. Although the exact cause of death is unknown, these observations suggest that these two factors function together in a dose-dependent manner to ensure postnatal survival.
TABLE 2.
Analysis of embryos from Oct-1+/− × Oct-2+/− crossesa
| Age | Total no. | No. dead | No. genotyped | No. (%)
|
||||
|---|---|---|---|---|---|---|---|---|
| Wild type | Oct-1+/− | Oct-2+/− | Oct-1+/−; Oct-2+/− alive | Oct-1+/−; Oct-2+/− dead | ||||
| E18.5 | 17 | 0 | 17 | 4 (23.5) | 6 (35.3) | 1 (5.9) | 6 (35.3) | 0 (0) |
| Postnatal | 78 | 5 | 73 | 23 (31.5) | 11 (15.7) | 20 (27.4) | 5 (6.8) | 4 (5.5) |
| Week 3 | ||||||||
Embryos and pups were genotyped by PCR and Southern with yolk sac and tail DNA as described in the text. The category of dead Oct-1+/−; Oct-2+/− pups included animals that either died soon after birth due to unknown causes or until the time of weaning at 21 days.
DISCUSSION
We describe the phenotype of a targeted mutation of the transcription factor Oct-1. Mutant animals die as embryos with 100% penetrance, a finding in general accordance with those for another Oct-1 mutant generated by deleting the DNA binding POU domain (P. Matthias, unpublished data). Fibroblasts derived from these animals proliferate normally but are defective in their ability to express transfected reporter constructs driven by octamer-containing promoters. Mutant mice are also frequently growth retarded and anemic and are defective in erythropoiesis. Furthermore, erythropoietic precursor cells in Oct-1 mutant mice express β-globin at greatly reduced levels, indicating a decrease or delay in the expression of this protein. The defect in β-globin expression was observed with 100% penetrance, regardless of whether the embryos appeared runted or anemic.
In nuclear extracts from mutant fibroblasts, octamer binding activity was reduced by 40-fold relative to the wild-type level. By a combination of a novel DNA affinity chromatography technique and immunoblotting, three faint truncated polypeptides were detected in the mutant extract. Although the intensity and relative sizes of these products indicate that they are at least 30-fold reduced in expression and missing a large portion of the Oct-1 protein, it is possible that a small amount of residual activity is conferred by these polypeptides.
Transfection assays show that for two known Oct-1 target genes, IgH and H2B, reporter activity was severely impaired in Oct-1-deficient cells. In the case of H2B, the expression defect could be corrected by the introduction of retrovirally expressed human Oct-1. Yet surprisingly, previously implicated endogenous Oct-1 target genes such as histone H2B and U2 and U6 snRNAs are expressed normally. This apparent paradox may be explained in part by the configuration of endogenous genes. Endogenous promoters frequently have binding sites for multiple transcription factors, so the loss of Oct-1 may not be sufficient to abolish expression. For instance, in addition to the octamer located within the distal sequence element of the snRNA genes, there is also a functionally important stretch of DNA immediately upstream known as the SPH element that is thought to interact with Staf/SBF (reviewed in reference 15). The gene encoding the mouse U1 snRNA also contains an AP1 site and a putative SP1 site (13, 38).
Recent reports have demonstrated that in many IgH and some Igκ promoters, a conserved, pyrimidine-rich sequence, located downstream of the transcription initiation site and upstream of the ATG start codon, helps to mediate IgH promoter activity (3, 62). The transfection constructs used in these experiments contained only the minimal promoter without flanking sequences. Another possibility is that residual Oct-1 activity in mutant cells (1/30 to 1/40 of the wild-type level) is sufficient for full levels of expression for each of these genes. We deem this possibility unlikely. In other studies, for example, MEFs with this residual Oct-1 activity are severely impaired for herpes virus replication at normal multiplicities of infection, demonstrating a requirement in the infection process (T. Kristie, submitted for publication).
Oct-1 deficiency results in a decrease in erythropoiesis. Fluorescence-activated cell sorting analysis of fetal liver cells showed a significant decrease in the total numbers of erythroid cells in the mutant animals. Adoptive transfer experiments with Oct-1-deficient fetal livers further demonstrated that the defect resides within the hematopoietic compartment. In contrast to wild-type controls, recipient animals that received Oct-1-deficient fetal liver cells also showed enlarged spleens, consistent with extramedullary hematopoiesis secondary to anemia (not shown). We generated Oct-1 double-mutant ES cells by targeting the remaining Oct-1 allele by a similar targeting strategy and a puromycin selection cassette (unpublished). Oct-1-deficient ES cells are viable, consistent with our other findings (not shown). When these cells are differentiated in methylcellulose, a significant but incomplete impediment to erythroid differentiation was observed. These cells generated fewer erythroid CFU and increased numbers of undifferentiated erythroblasts (not shown).
β-Globin protein expression was examined in E11.5 Oct-1 mutant embryos. At this stage in development, β-globin expression has initiated in the fetal liver erythrocytes (65). Although no change was observed in the level of embryonic ɛ-globin, β-globin expression was strongly reduced in the Oct-1 homozygous embryos. Furthermore, the level of β-globin mRNA is selectively reduced on a per-cell basis in E12.5 embryos, indicating that the decrease in β-globin expression is a transcriptional defect. This defect may be due to direct transcriptional control by Oct-1. An octamer element within the second intron of the human β-globin gene was shown to be dispensable for the correct regulation of transgenes but necessary for their maximal expression (1, 48). Although this intronic octamer is not conserved in the mouse, two canonical octamer elements are present 700 bp and 1.2 kb upstream of the mouse β-globin promoter transcription start site and could potentially mediate a dependence on Oct-1.
Mice harboring mutations in β-globin have numerous features in common with the Oct-1-deficient mice, including embryonic lethality, decreased numbers of red blood cells, splenomegaly, and extramedullary hematopoiesis (52, 56, 68). Decreased β-globin expression was observed in Oct-1 mutants as early as E11.5, prior to the manifestation of anemia in the developing embryo. However, our data do not rule out the possibility that Oct-1 may indirectly regulate β-globin expression through the transcriptional control of transcription factors that control the expression of this gene. In addition, a developmental delay in switching to β-globin expression, or a delay in switching from primitive (yolk sac) to definitive (fetal liver) erythropoiesis, would also cause a decrease in β-globin expression in mutant cells relative to the wild type. At present these possibilities have not been distinguished.
Strikingly, the Oct-1+/−;Oct-2+/− transheterozygotes failed to thrive and survived to adulthood at greatly reduced frequencies, a finding in contrast to either of the single-heterozygous strains. The data suggest that Oct-1 and Oct-2 function in an interdependent fashion to regulate a critical physiological process during the late development and early life of the mouse. Single heterozygote animals are fully viable, presumably because sufficient total Oct protein remains in the cells to carry out this process. However, in the absence of two alleles, such as in the Oct-1+/−;Oct-2+/− transheterozygote (this study) or in Oct-2−/− animals (7), this Oct-dependent system is impaired and survival is compromised. In Oct-1 homozygotes, death occurs earlier during development, presumably because of a critical function in a tissue that lacks Oct-2 protein. The generation of double mutant animals or suppression of these proteins by RNA interference may reveal additional information about their function.
Acknowledgments
We thank P. Ernst, N. Nakayama, A. Dingwall, and G. Jones for discussion and advice, P. Matthias for communication of data prior to publication, W. Leonard and members of the Sharp lab for critical reading of the manuscript, H. Cargill for illustrations, D. Cook and M. Luo for help with MALDI mass spectroscopy, and M. Siafaca for assistance. The latex beads were a gift from H. Handa. Oct-2+/− mice were provided by L. Corcoran.
V.W. was supported by a Howard Hughes predoctoral fellowship and a David Koch graduate fellowship. D.T. was supported by fellowships from the Irvington Institute for Immunological Research and the Medical Foundation. This work was funded by U.S. Public Health Service grants PO1-CA42063 to P.A.S. and A140416 to J.C. and partially by Cancer Center Support grant P30-CA14051.
REFERENCES
- 1.Bharadwaj, R. R., C. D. Trainor, P. Pasceri, and J. Ellis. 2003. LCR-regulated transgene expression levels depend on the Oct-1 site in the AT-rich region of beta-globin intron-2. Blood 101:1603-1610. [DOI] [PubMed] [Google Scholar]
- 2.Boutet, S. C., T. Quertermous, and B. M. Fadel. 2001. Identification of an octamer element required for in vivo expression of the TIE1 gene in endothelial cells. Biochem. J. 360:23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casellas, R., M. Jankovic, G. Meyer, A. Gazumyan, Y. Luo, R. Roeder, and M. Nussenzweig. 2002. OcaB is required for normal transcription and V(D)J recombination of a subset of immunoglobulin kappa genes. Cell 110:575-585. [DOI] [PubMed] [Google Scholar]
- 4.Cepek, K. L., D. I. Chasman, and P. A. Sharp. 1996. Sequence-specific DNA binding of the B-cell-specific coactivator OCA-B. Genes Dev. 10:2079-2088. [DOI] [PubMed] [Google Scholar]
- 5.Christensen, S. M., B. K. Martin, S. S. Tan, and J. H. Weis. 1992. Identification of sites for distinct DNA binding proteins including Oct-1 and Oct-2 in the Cr2 gene. J. Immunol. 148:3610-3617. [PubMed] [Google Scholar]
- 6.Clerc, R. G., L. M. Corcoran, J. H. LeBowitz, D. Baltimore, and P. A. Sharp. 1988. The B-cell-specific Oct-2 protein contains POU box- and homeo box-type domains. Genes Dev. 2:1570-1581. [DOI] [PubMed] [Google Scholar]
- 7.Corcoran, L. M., M. Karvelas, G. J. Nossal, Z. S. Ye, T. Jacks, and D. Baltimore. 1993. Oct-2, although not required for early B-cell development, is critical for later B-cell maturation and for postnatal survival. Genes Dev. 7:570-582. [DOI] [PubMed] [Google Scholar]
- 8.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erkman, L., R. J. McEvilly, L. Luo, A. K. Ryan, F. Hooshmand, S. M. O'Connell, E. M. Keithley, D. H. Rapaport, A. F. Ryan, and M. G. Rosenfeld. 1996. Role of transcription factors Brn-3.1 and Brn-3.2 in auditory and visual system development. Nature 381:603-606. [DOI] [PubMed] [Google Scholar]
- 10.Falkner, F. G., and H. G. Zachau. 1984. Correct transcription of an immunoglobulin kappa gene requires an upstream fragment containing conserved sequence elements. Nature 310:71-74. [DOI] [PubMed] [Google Scholar]
- 11.Fan, W., S. Jin, T. Tong, H. Zhao, F. Fan, M. J. Antinore, B. Rajasekaran, M. Wu, and Q. Zhan. 2002. BRCA1 regulates GADD45 through its interactions with the OCT-1 and CAAT motifs. J. Biol. Chem. 277:8061-8067. [DOI] [PubMed] [Google Scholar]
- 12.Gstaiger, M., L. Knoepfel, O. Georgiev, W. Schaffner, and C. M. Hovens. 1995. A B-cell coactivator of octamer-binding transcription factors. Nature 373:360-362. [DOI] [PubMed] [Google Scholar]
- 13.Gunderson, S. I., J. T. Murphy, M. W. Knuth, T. H. Steinberg, J. H. Dahlberg, and R. R. Burgess. 1988. Binding of transcription factors to the promoter of the human U1 RNA gene studied by footprinting. J. Biol. Chem. 263:17603-17610. [PubMed] [Google Scholar]
- 14.Handa, H., Y. Yamaguchi, and T. Wada. 1999. Purification of DNA-binding proteins, p. 283-302. In P. Millner (ed.), High resolution chromatography: a practical approach. Oxford University Press, Oxford, England.
- 15.Hernandez, N. 2001. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem. 276:26733-26736. [DOI] [PubMed] [Google Scholar]
- 16.Herr, W., R. A. Sturm, R. G. Clerc, L. M. Corcoran, D. Baltimore, P. A. Sharp, H. A. Ingraham, M. G. Rosenfeld, M. Finney, G. Ruvkun, et al. 1988. The POU domain: a large conserved region in the mammalian pit-1, oct-1, oct-2, and Caenorhabditis elegans unc-86 gene products. Genes Dev 2:1513-1516. [DOI] [PubMed] [Google Scholar]
- 17.Horwitz, B. H., M. L. Scott, S. R. Cherry, R. T. Bronson, and D. Baltimore. 1997. Failure of lymphopoiesis after adoptive transfer of NF-kappaB-deficient fetal liver cells. Immunity 6:765-772. [DOI] [PubMed] [Google Scholar]
- 18.Humbert, P. O., R. Verona, J. M. Trimarchi, C. Rogers, S. Dandapani, and J. A. Lees. 2000. E2f3 is critical for normal cellular proliferation. Genes Dev. 14:690-703. [PMC free article] [PubMed] [Google Scholar]
- 19.Jaffe, J., M. Hochberg, J. Riss, T. Hasin, L. Reich, and R. Laskov. 1995. Cloning, sequencing and expression of two isoforms of the murine oct-1 transcription factor. Biochim. Biophys. Acta 1261:201-209. [DOI] [PubMed] [Google Scholar]
- 20.Jenuwein, T., and R. Grosschedl. 1991. Complex pattern of immunoglobulin mu gene expression in normal and transgenic mice: nonoverlapping regulatory sequences govern distinct tissue specificities. Genes Dev. 5:932-943. [DOI] [PubMed] [Google Scholar]
- 21.Jin, S., F. Fan, W. Fan, H. Zhao, T. Tong, P. Blanck, I. Alomo, B. Rajasekaran, and Q. Zhan. 2001. Transcription factors Oct-1 and NF-YA regulate the p53-independent induction of the GADD45 following DNA damage. Oncogene 20:2683-2690. [DOI] [PubMed] [Google Scholar]
- 22.Jin, T., and H. Li. 2001. Pou homeodomain protein OCT1 is implicated in the expression of the caudal-related homeobox gene Cdx-2. J. Biol. Chem. 276:14752-14758. [DOI] [PubMed] [Google Scholar]
- 23.Kamps, M. P., L. Corcoran, J. H. LeBowitz, and D. Baltimore. 1990. The promoter of the human interleukin-2 gene contains two octamer-binding sites and is partially activated by the expression of Oct-2. Mol. Cell. Biol. 10:5464-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, U., X. F. Qin, S. Gong, S. Stevens, Y. Luo, M. Nussenzweig, and R. G. Roeder. 1996. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature 383:542-547. [DOI] [PubMed] [Google Scholar]
- 25.Kim, Y. M., C. B. Ko, Y. P. Park, Y. J. Kim, and S. G. Paik. 1999. Octamer motif is required for the NF-kappaB-mediated induction of the inducible nitric oxide synthase gene expression in RAW 264.7 macrophages. Mol. Cell 9:99-109. [PubMed] [Google Scholar]
- 26.Klemm, J. D., M. A. Rould, R. Aurora, W. Herr, and C. O. Pabo. 1994. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell 77:21-32. [DOI] [PubMed] [Google Scholar]
- 27.Konig, H., P. Pfisterer, L. M. Corcoran, and T. Wirth. 1995. Identification of CD36 as the first gene dependent on the B-cell differentiation factor Oct-2. Genes Dev. 9:1598-1607. [DOI] [PubMed] [Google Scholar]
- 28.Koyasu, S., R. E. Hussey, L. K. Clayton, A. Lerner, R. Pedersen, P. Delany-Heiken, F. Chau, and E. L. Reinherz. 1994. Targeted disruption within the CD3 zeta/eta/phi/Oct-1 locus in mouse. EMBO J. 13:784-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.LeBowitz, J. H., T. Kobayashi, L. Staudt, D. Baltimore, and P. A. Sharp. 1988. Octamer-binding proteins from B or HeLa cells stimulate transcription of the immunoglobulin heavy-chain promoter in vitro. Genes Dev. 2:1227-1237. [DOI] [PubMed] [Google Scholar]
- 30.Lefort, K., J. P. Rouault, L. Tondereau, J. P. Magaud, and J. F. Dore. 2001. The specific activation of gadd45 following UVB radiation requires the POU family gene product N-oct3 in human melanoma cells. Oncogene 20:7375-7385. [DOI] [PubMed] [Google Scholar]
- 31.Lerner, A., L. D'Adamio, A. C. Diener, L. K. Clayton, and E. L. Reinherz. 1993. CD3 zeta/eta/theta locus is colinear with and transcribed antisense to the gene encoding the transcription factor Oct-1. J. Immunol. 151:3152-3162. [PubMed] [Google Scholar]
- 32.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 33.Li, S., E. B. Crenshaw, 3rd, E. J. Rawson, D. M. Simmons, L. W. Swanson, and M. G. Rosenfeld. 1990. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU domain gene pit-1. Nature 347:528-533. [DOI] [PubMed] [Google Scholar]
- 34.Lopez, R. A., S. Schoetz, K. DeAngelis, D. O'Neill, and A. Bank. 2002. Multiple hematopoietic defects and delayed globin switching in Ikaros null mice. Proc. Natl. Acad. Sci. USA 99:602-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo, Y., and R. G. Roeder. 1995. Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol. Cell. Biol. 15:4115-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mahmoud, M. S., and M. M. Kawano. 1996. Cloning and analysis of the human Pax-5 gene promoter. Biochem. Biophys. Res. Commun. 228:159-164. [DOI] [PubMed] [Google Scholar]
- 37.Mason, J. O., G. T. Williams, and M. S. Neuberger. 1985. Transcription cell type specificity is conferred by an immunoglobulin VH gene promoter that includes a functional consensus sequence. Cell 41:479-487. [DOI] [PubMed] [Google Scholar]
- 38.Murphy, J. T., R. R. Burgess, J. E. Dahlberg, and E. Lund. 1982. Transcription of a gene for human U1 small nuclear RNA. Cell 29:265-274. [DOI] [PubMed] [Google Scholar]
- 39.Nichols, J., B. Zevnik, K. Anastassiadis, H. Niwa, D. Klewe-Nebenius, I. Chambers, H. Scholer, and A. Smith. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95:379-391. [DOI] [PubMed] [Google Scholar]
- 40.Nielsen, P. J., O. Georgiev, B. Lorenz, and W. Schaffner. 1996. B lymphocytes are impaired in mice lacking the transcriptional coactivator Bob1/OCA-B/OBF1. Eur. J. Immunol. 26:3214-3218. [DOI] [PubMed] [Google Scholar]
- 41.Ohno, H., S. Goto, S. Taki, T. Shirasawa, H. Nakano, S. Miyatake, T. Aoe, Y. Ishida, H. Maeda, T. Shirai, and et al. 1994. Targeted disruption of the CD3 eta locus causes high lethality in mice: modulation of Oct-1 transcription on the opposite strand. EMBO J. 13:1157-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pankratova, E. V., I. E. Deyev, S. V. Zhenilo, and O. L. Polanovsky. 2001. Tissue-specific isoforms of the ubiquitous transcription factor Oct-1. Mol. Genet. Genomics 266:239-245. [DOI] [PubMed] [Google Scholar]
- 43.Parslow, T. G., D. L. Blair, W. J. Murphy, and D. K. Granner. 1984. Structure of the 5′ ends of immunoglobulin genes: a novel conserved sequence. Proc. Natl. Acad. Sci. USA 81:2650-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfaffle, R. W., G. E. DiMattia, J. S. Parks, M. R. Brown, J. M. Wit, M. Jansen, H. Van der Nat, J. L. Van den Brande, M. G. Rosenfeld, and H. A. Ingraham. 1992. Mutation of the POU-specific domain of Pit-1 and hypopituitarism without pituitary hypoplasia. Science 257:1118-1121. [DOI] [PubMed] [Google Scholar]
- 45.Pfeuffer, I., S. Klein-Hessling, A. Heinfling, S. Chuvpilo, C. Escher, T. Brabletz, B. Hentsch, H. Schwarzenbach, P. Matthias, and E. Serfling. 1994. Octamer factors exert a dual effect on the interleukin-2 and interleukin-4 promoters. J. Immunol. 153:5572-5585. [PubMed] [Google Scholar]
- 46.Pfisterer, P., J. Hess, and T. Wirth. 1997. Identification of target genes of the lymphoid-specific transcription factor Oct2. Immunobiology 198:217-226. [DOI] [PubMed] [Google Scholar]
- 47.Riss, J., and R. Laskov. 1999. Expression of novel alternatively spliced isoforms of the oct-1 transcription factor. Biochim. Biophys. Acta 1444:295-298. [DOI] [PubMed] [Google Scholar]
- 48.Rubin, J. E., P. Pasceri, X. Wu, P. Leboulch, and J. Ellis. 2000. Locus control region activity by 5′HS3 requires a functional interaction with beta-globin gene regulatory elements: expression of novel beta/gamma-globin hybrid transgenes. Blood 95:3242-3249. [PubMed] [Google Scholar]
- 49.Ryan, A. K., and M. G. Rosenfeld. 1997. POU domain family values: flexibility, partnerships, and developmental codes. Genes Dev. 11:1207-1225. [DOI] [PubMed] [Google Scholar]
- 50.Scheidereit, C., J. A. Cromlish, T. Gerster, K. Kawakami, C. G. Balmaceda, R. A. Currie, and R. G. Roeder. 1988. A human lymphoid-specific transcription factor that activates immunoglobulin genes is a homoeobox protein. Nature 336:551-557. [DOI] [PubMed] [Google Scholar]
- 51.Schubart, K., S. Massa, D. Schubart, L. M. Corcoran, A. G. Rolink, and P. Matthias. 2001. B-cell development and immunoglobulin gene transcription in the absence of Oct-2 and OBF-1. Nat. Immunol. 2:69-74. [DOI] [PubMed] [Google Scholar]
- 52.Shehee, W. R., P. Oliver, and O. Smithies. 1993. Lethal thalassemia after insertional disruption of the mouse major adult beta-globin gene. Proc. Natl. Acad. Sci. USA 90:3177-3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shore, P., W. Dietrich, and L. M. Corcoran. 2002. Oct-2 regulates CD36 gene expression via a consensus octamer, which excludes the co-activator OBF-1. Nucleic Acids Res. 30:1767-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh, H., R. Sen, D. Baltimore, and P. A. Sharp. 1986. A nuclear factor that binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature 319:154-158. [DOI] [PubMed] [Google Scholar]
- 55.Sive, H. L., and R. G. Roeder. 1986. Interaction of a common factor with conserved promoter and enhancer sequences in histone H2B, immunoglobulin, and U2 small nuclear RNA (snRNA) genes. Proc. Natl. Acad. Sci. USA 83:6382-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skow, L. C., B. A. Burkhart, F. M. Johnson, R. A. Popp, D. M. Popp, S. Z. Goldberg, W. F. Anderson, L. B. Barnett, and S. E. Lewis. 1983. A mouse model for beta-thalassemia. Cell 34:1043-1052. [DOI] [PubMed] [Google Scholar]
- 57.Staudt, L. M., H. Singh, R. Sen, T. Wirth, P. A. Sharp, and D. Baltimore. 1986. A lymphoid-specific protein binding to the octamer motif of immunoglobulin genes. Nature 323:640-643. [DOI] [PubMed] [Google Scholar]
- 58.Strubin, M., J. W. Newell, and P. Matthias. 1995. OBF-1, a novel B-cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell 80:497-506. [DOI] [PubMed] [Google Scholar]
- 59.Sturm, R. A., J. L. Cassady, G. Das, A. Romo, and G. A. Evans. 1993. Chromosomal structure and expression of the human OTF1 locus encoding the Oct-1 protein. Genomics 16:333-341. [DOI] [PubMed] [Google Scholar]
- 60.Sturm, R. A., and W. Herr. 1988. The POU domain is a bipartite DNA-binding structure. Nature 336:601-604. [DOI] [PubMed] [Google Scholar]
- 61.Takahashi, S., S. Saito, N. Ohtani, and T. Sakai. 2001. Involvement of the Oct-1 regulatory element of the gadd45 promoter in the p53-independent response to ultraviolet irradiation. Cancer Res. 61:1187-1195. [PubMed] [Google Scholar]
- 62.Tantin, D., and P. A. Sharp. 2002. A Mouse lymphoid cell line selected to have high immunoglobulin promoter activity. Mol. Cell. Biol. 22:1460-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thevenin, C., B. P. Lucas, E. J. Kozlow, and J. H. Kehrl. 1993. Cell type- and stage-specific expression of the CD20/B1 antigen correlates with the activity of a diverged octamer DNA motif present in its promoter. J. Biol. Chem. 268:5949-5956. [PubMed] [Google Scholar]
- 64.Todaro, G., and H. Green. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trimborn, T., J. Gribnau, F. Grosveld, and P. Fraser. 1999. Mechanisms of developmental control of transcription in the murine alpha- and beta-globin loci. Genes Dev. 13:112-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Verrijzer, C. P., M. J. Alkema, W. W. van Weperen, H. C. Van Leeuwen, M. J. Strating, and P. C. van der Vliet. 1992. The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J. 11:4993-5003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang, D., S. Yamamoto, N. Hijiya, E. N. Benveniste, and C. L. Gladson. 2000. Transcriptional regulation of the human osteopontin promoter: functional analysis and DNA-protein interactions. Oncogene 19:5801-5809. [DOI] [PubMed] [Google Scholar]
- 68.Yang, B., S. Kirby, J. Lewis, P. J. Detloff, N. Maeda, and O. Smithies. 1995. A mouse model for beta 0-thalassemia. Proc. Natl. Acad. Sci. USA 92:11608-11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, L., and G. J. Nabel. 1994. Positive and negative regulation of interleukin-2 gene expression: role of multiple regulatory sites. Cytokine 6:221-228. [DOI] [PubMed] [Google Scholar]