Abstract
The 3′ poly(A) tail of eukaryotic mRNAs and the poly(A) binding protein (PABP) play important roles in the regulation of translation. Recently, a human PABP-interacting protein, Paip2, which disrupts the PABP-poly(A) interaction and consequently inhibits translation, was described. To gain insight into the biological role of Paip2, we studied the Drosophila melanogaster Paip2 (dPaip2). dPaip2 is the bona fide human Paip2 homologue, as it interacts with dPABP, inhibits binding of dPABP to the mRNA poly(A) tail, and reduces translation of a reporter mRNA by ∼80% in an S2 cell-free translation extract. Ectopic overexpression of dPaip2 in Drosophila wings and wing discs results in a size reduction phenotype, which is due to a decrease in cell number. Clones of cells overexpressing dPaip2 in wing discs also contain fewer cells than controls. This phenotype can be explained by a primary effect on cell growth. Indeed, overexpression of dPaip2 in postreplicative tissues inhibits growth, inasmuch as it reduces ommatidia size in eyes and cell size in the larval fat body. We conclude that dPaip2 inhibits cell growth primarily by inhibiting protein synthesis.
Translation plays an important role in the regulation of gene expression and is implicated in the control of cell growth, proliferation, and differentiation. In eukaryotes, initiation is the rate-limiting step of translation in most circumstances and is a major target for regulation (reviewed in reference 18). The 5′ cap structure (m7GpppN, where m is a methyl group and N is any nucleotide) of the mRNA is recognized by the eukaryotic initiation factor 4F (eIF4F) complex. eIF4F is comprised of three subunits: (i) eIF4E, the cap binding protein; (ii) eIF4A, a bidirectional ATP-dependent RNA helicase; and (iii) eIF4G, a modular scaffolding protein, which possesses binding sites for eIF4E and eIF4A and recruits the 40S ribosomal subunit to the mRNA via its interaction with eIF3 (20). The 3′ poly(A) tail of the mRNA is bound by the poly(A) binding protein (PABP). PABP is a phylogenetically conserved protein that functions in mRNA stability and translation (48). PABP is an essential protein: in Saccharomyces cerevisiae, deletion of the PAB1 gene is lethal (49) and a P-element insertion in the Drosophila melanogaster PABP gene is embryonic lethal (53). PABP is an ∼630-amino-acid (aa) protein containing four RNA recognition motifs (RRMs) arranged in tandem and a proline-rich C-terminal domain (reviewed in reference 24). RRMs 1 and 2 are the major contributors to the poly(A) binding activity of PABP (10, 30). PABP directly interacts with eIF4G, leading to circularization of the mRNA by bridging the 5′ and 3′ extremities (closed-loop model) (40, 48). The closed-loop model explains the synergistic enhancement of translation by the 5′ cap structure and the 3′ poly(A) tail of the mRNA (15). By joining the 5′ and 3′ ends of the mRNA, circularization may facilitate recycling of ribosomes, initiation complex formation, or the 60S ribosome-joining step (24, 52).
Our group has identified two human proteins that interact directly with PABP: Paip1 and Paip2 (PABP-interacting proteins 1 and -2). Paip1 stimulates, while Paip2 represses, translation (8, 27). Paip2 inhibits translation by reducing the binding of PABP to the poly(A) tail and by competing with Paip1 for binding to PABP. Paip1 and Paip2 share two conserved PABP-interacting motifs (PAMs). PAM1 consists of a stretch of acidic amino acids in the middle of Paip2 (aa 22 to 75) and at the C terminus of Paip1 (aa 440 to 479), and it binds strongly to RRMs 2 and 3 and to RRMs 1 and 2 of PABP, respectively (25, 45). The second binding site, PAM2, also called the PABP C-terminal binding motif, resides in the C terminus of Paip2 (aa 106 to 120) (25) and the N terminus of Paip1 (aa 123 to 137) (45). PAM2 consists of a short stretch of 15 aa and binds to the C terminus of PABP (within aa 546 to 619) with a lower affinity (∼10- and ∼200-fold for Paip1 and Paip2, respectively) than that of the PAM1-PABP interaction (25, 45). PAM2 is also found in several additional proteins, including eukaryotic release factor 3 (eRF3), ataxin 2, and transducer of ErbB-2 (Tob) (11, 29). Thus, Paip2 and Paip1 might compete with some of these PAM2 binding partners to regulate PABP function.
To study the biological role of Paip2, the Drosophila homologue of the human Paip2 (hPaip2), dPaip2, was isolated and characterized. Its ability to interact with Drosophila PABP (dPABP), inhibit translation, and interfere with dPABP poly(A) binding activity was demonstrated. Importantly, dPaip2 inhibits growth in flies.
MATERIALS AND METHODS
Cloning of dPaip2.
Five partially sequenced expressed sequence tags (ESTs) (LD06786, LD15606, LD07934, LP05812, and GH10535) containing the dPaip2 cDNA were identified in the Berkeley Drosophila Genome Project database (46) based on their homology to hPaip2 (GenBank no. AF317675). These EST plasmids were obtained from Research Genetics and fully sequenced. By assembling the sequences of these ESTs, we determined the full-length dPaip2 cDNA sequence (Fig. 1A).
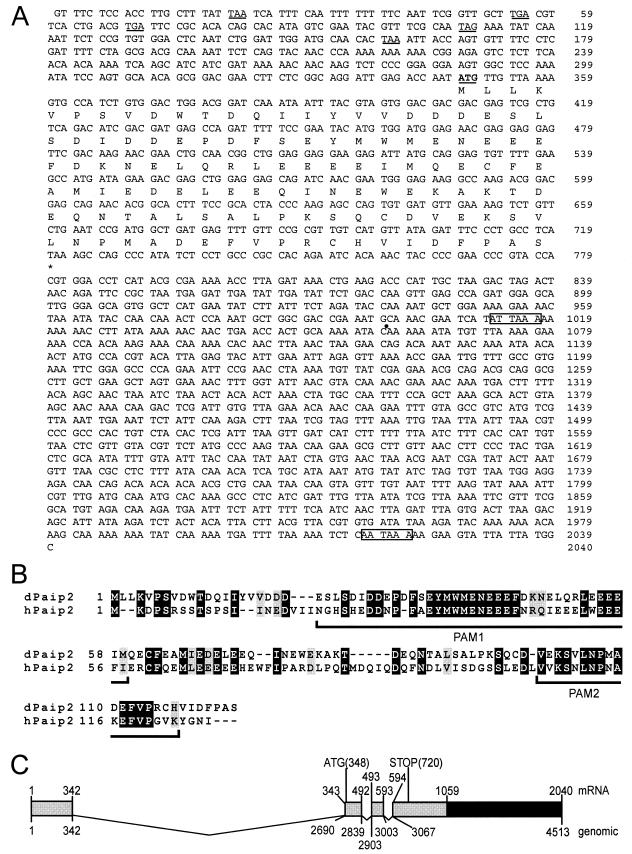
FIG.1.
Identification of dPaip2. (A) dPaip2 cDNA and amino acid sequences. The ATG initiation codon is underlined and in boldface. Five upstream in-frame stop codons are underlined. The stop codon is indicated by an asterisk. The two poly(A) signals are boxed. The dot indicates the first site of polyadenylation. (B) Sequence alignment of human and Drosophila Paip2 performed with PIMA multisequence alignment software (Baylor College of Medicine Search Launcher [http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html]). The solid and shaded boxes indicate conserved and similar residues, respectively. PAM1 and PAM2 are indicated. (C) Organization of dPaip2 gene. The numbers refer to the cDNA (top) and gene (bottom) sequences, respectively. The ATG and stop codons are indicated. The boxes and lines represent exons and introns, respectively. The shaded boxes correspond to the region shared by the two dPaip2 cDNAs, while the solid box highlights the extended 3′ UTR for the longest cDNA.
Plasmids.
For construction of pGST-dPaip2, the dPaip2 coding region and a short 3′ untranslated region (UTR) (i.e., nucleotides [nt] 348 to 771) was PCR amplified using LD06786 as a template with a forward and a reverse primer containing a BamHI or an XhoI restriction site, respectively. The resulting fragment was digested with BamHI/XhoI and ligated in pGEX6P1 (Amersham Pharmacia Biotech [APB]) digested with BamHI/XhoI. pBS-SKII-dPaip2 and pIND-1HA-dPaip2 were obtained by subcloning the BamHI/XhoI insert of pGST-dPaip2 into pBlueScript SKII(+) (Stratagene) and pIND-1HA (kindly provided by A. Brasey) digested with BamHI/XhoI, respectively. To construct pPacPL-HA-dPaip2, pIND-1HA-dPaip2 was digested with HindIII, blunt ended with Klenow fragment, and then digested with XbaI. The resulting insert was ligated into pPacPL (a kind gift of A. Rodriguez; pPacPL has an actin-5C promoter) digested with EcoRV and XbaI. pPacPL-dPaip2(ORF) and pPacPL-dPaip2(FL) were obtained by subcloning the BamHI/KpnI inserts of pBS-SKII-dPaip2 and LD06786, respectively, into pPacPL digested with BamHI/KpnI. To generate pUAS-HA-dPaip2, pIND-1HA-dPaip2 was digested with HindIII, blunt ended with Klenow, and then digested with XhoI. The resulting insert was ligated into pUAST (2) digested with BglII, blunt ended with Klenow, and then digested with XhoI.
The construct pBac-His-HMK-dPABP was prepared by PCR amplification of the dPABP coding region using pPA (GenBank no. L05109) (35) as a template with a forward and a reverse primer containing XhoI and NcoI restriction sites, respectively. The PCR fragment was digested with XhoI/NcoI and ligated in pBlueBac2C-His6-HMK (a kind gift of S. Morino) digested with XhoI/NcoI. To generate pGST-dPABP, pBac-His-HMK-dPABP was digested with NcoI, blunt ended with Klenow, and then digested with XhoI. The resulting insert was ligated in pGEX6P2 (APB), which had been digested with NotI, blunt ended with Klenow, and then digested with XhoI. pET-His-HMK-dPABP was obtained by subcloning the insert of pBac-His-HMK-dPABP digested with XhoI/NcoI and blunt ended with Klenow into pET-His-HMK (7) digested with XhoI and blunt ended with Klenow.
Protein expression and purification.
Fusion proteins were expressed in Escherichia coli BL21(DE3) as described previously (45). Glutathione S-transferase (GST) and GST-dPaip2 were purified on glutathione-Sepharose resin (APB) according to the manufacturer's recommendations. GST-dPABP and His-HMK-dPABP (where HMK is the phosphorylation site of heart muscle kinase) were found to be mainly insoluble, but a soluble form of the protein was purified according to the high-salt extraction procedure described previously (25) on glutathione-Sepharose and on Talon metal affinity resin (Clontech), respectively. To cleave the GST tag, GST-dPABP was digested with PreScission protease (APB) for 4 h to overnight at 4°C according to the manufacturer's instructions. Recombinant proteins were dialyzed against 1× phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4 · 7H2O, 1.4 mM KH2PO4, pH 7.4) or against buffer A (10 mM HEPES-KOH, pH 7.6, 100 mM KCl, 2 mM dithiothreitol) for in vitro translation assays.
GST pull-downs.
Purified GST or GST-dPaip2 (5 μg) was immobilized on glutathione-Sepharose (APB). dPABP (5 μg; with the GST tag cleaved) in 500 μl of pull-down buffer (20 mM HEPES-KOH, pH 7.5, 100 mM KCl, 1 mM dithiothreitol, 0.5 mM EDTA, 10% glycerol, and 0.5% NP-40) was added to the resin. After 2 h of incubation at 4°C, the resin was washed three times with 500 μl of pull-down buffer. Proteins were eluted with 40 μl of 1× Laemmli sample buffer (32). The samples were boiled for 5 min and resolved by SDS-12.5% PAGE, and the gel was stained with Coomassie blue R-250.
Antibodies and Western blotting.
To obtain an anti-dPaip2 antibody, a New Zealand White rabbit (rabbit 2929) was initially injected with 0.25 mg of dPaip2 peptide (aa 1 to 13; CMLLKVPSVDWTDQ) conjugated to keyhole limpet hemocyanin using the Imject Maleimide Activated Immunogen conjugation kit (Pierce). Additional injections were performed using 0.1 mg of dPaip2 peptide-keyhole limpet hemocyanin at 4-week intervals. Similarly, an anti-dPABP antibody was obtained by injection of 0.25 mg of GST-dPABP into a New Zealand White rabbit (rabbit 2482), followed by 0.125-mg injections at 4-week intervals. Western blotting was performed as previously described (45) with the following antibodies: rabbit crude anti-dPaip2, anti-dPABP, and anti-GST (M. Miron and N. Sonenberg, unpublished data) diluted at 1:1,000, 1:5,000, and 1:1000, respectively.
Cell culture and protein extracts.
Drosophila Schneider 2 (S2) cells (51) were grown at 27°C in Schneider's medium (Gibco-BRL) supplemented with 10% heat-inactivated fetal bovine serum (Gibco-BRL). The S2 cells (7 × 106) were seeded in a 60-mm-diameter dish 16 h prior to transfections. For transfection experiments, 2 μg of pPacPL, pPacPL-dPaip2(ORF), pPacPL-dPaip2(FL), or pPacPL-HA-dPaip2 was mixed with 16 μl of Enhancer and 25 μl of Effectene reagent (Qiagen) according to the manufacturer's instructions. Cells were harvested 48 h posttransfection for Western blot analysis. Briefly, S2 cells were scraped in 1× PBS, pelleted, rinsed twice with 1× PBS, and resuspended in 1× Passive lysis buffer (Promega) containing a protease inhibitor cocktail (Roche). This suspension was subjected to two freeze-thaw cycles, and cell debris was removed by a 10-min centrifugation at 16,000 × g. The protein concentrations of the supernatants were determined using the Bio-Rad protein assay.
Immunoprecipitation.
S2 cell extracts (300 μg) were diluted to 500 μl with 1× PBS supplemented with 0.2% NP-40 and incubated end over end with either rabbit polyclonal anti-dPaip2 (5 μl) or preimmune serum (5 μl) for 3 h at 4°C. A 50% slurry of protein A-Sepharose (20-μl bed volume; APB) was added to the mixture and incubated for an additional hour at 4°C. The mixture was centrifuged at 1,000 × g for 30 s, and the resin was washed three times with 500 μl of 1× PBS supplemented with 0.2% NP-40. Proteins were eluted with 1× Laemmli sample buffer (50 μl), and 25 μl was processed for Western blotting or far-Western analysis.
Far-Western analysis.
The far-Western analysis procedure was previously described (26, 45). 32P-labeled His-HMK-dPABP was used as a probe at a concentration of 250,000 cpm/ml.
Fly strains and analysis.
Transgenic flies were generated by microinjections of yw (yellow-white) embryos with pUAS-HA-dPaip2 for P-element-mediated germ line transformation according to standard protocols (2, 47). Six independent UAS-HA-dPaip2 lines (I to VI) were established. All genetic experiments described below were performed at 25°C.
For overexpression of dPaip2 in wings, UAS-HA-dPaip2 flies were crossed with MS1096-Gal4 flies, which drive expression of Gal4 throughout the wing disc (with the highest expression levels in the dorsal pouch) (6). The wings were dissected, mounted on a glass slide, and photographed, and their sizes, cell numbers, and cell densities were determined as previously described (38). Wing discs of 116-h-old (after egg deposition [AED]) third-instar larvae were also dissected, fixed, mounted, and measured as previously described (38).
For overexpression of dPaip2 in eyes, UAS-HA-dPaip2 flies were crossed with GMR-Gal4 flies, which drive the expression of Gal4 throughout the eye-imaginal disc (12). The flies were dehydrated gradually with increasing concentrations of ethanol and prepared by the hexamethyldisilazane method (59). The samples were then coated with gold, mounted, and observed with a scanning electron microscope. Ommatidia in the middle of the eye were measured using the Histogram function of Adobe Photoshop.
The flipase (FLP)-out technique (56) was used to induce clones of cells overexpressing dPaip2 and coexpressing the cell marker GFP-nls (green fluorescent protein fused to a nuclear localization signal) in different Drosophila tissues. The yw hs-FLP122/+; +; Act>CD2>Gal4 UAS-GFP-nls (where “>” represents the FLP recombinase target site) fly line (41) was used to generate those clones. The number of cells per clone in wing discs of 116-h-old (AED) third-instar larvae was determined as previously reported (38). The doubling time of cells in these clones was calculated as described previously (9). The larval fat bodies of 116-h-old (AED) third-instar larvae containing clones were dissected and analyzed by histology as previously described (38). The sizes of the GFP-positive cells and the GFP-negative cells contacting them were measured using the Histogram function of Adobe Photoshop. For each clone, the average size of the GFP-positive cells was expressed as a percentage of the average of their GFP-negative neighboring cells. The average cell size was obtained by averaging the cell size obtained from at least 20 clones for each genotype.
In vitro transcription.
Capped-luciferase (c+LucA−), capped-luciferase-poly(A) (c+LucA+), uncapped-luciferase (c−LucA−), and uncapped-luciferase-poly(A) (c−LucA+) mRNAs were prepared by in vitro transcription of pT3-Luc or pT3-Luc-pA plasmids (21) linearized with BamHI, using T3 RNA polymerase (MBI Fermentas) according to the manufacturer's instructions. Capped mRNAs were prepared in the presence of 1 mM cap analog [m7G(5′)ppp(5′)G] and 0.2 mM GTP.
S2 cell-free translation extracts and in vitro translation assays.
S2 cells were grown (see above) to a density of 10 × 106 to 15 × 106 cells/ml, and 200 ml was harvested by centrifugation at 3,000 × g. Cells were collected, and the translation extracts were prepared as described previously (3). Luciferase (Luc) mRNAs were used as reporter genes, and luciferase activity was assayed using the Luciferase assay kit (Promega) with a Lumat LB 9507 bioluminometer. Translation reaction mixtures (12 μl) containing 8 μl of S2 cell-free translation extracts, 20 mM HEPES-KOH, pH 7.6, 8 mM creatine phosphate, 0.5 mM spermidine, 0.2 mM GTP, 1 mM ATP, 20 μM amino acids, 150 mM potassium acetate, and 0.66 mM magnesium acetate were programmed with 50 ng of c+LucA+ mRNA. Buffer B (10 mM HEPES-KOH, pH 7.6) or various amounts of recombinant GST or GST-dPaip2 (diluted in buffer B) were added to the reaction mixtures. Following 60 min of incubation at 27°C, 3-μl aliquots were removed and added to 12 μl of 1× Reporter lysis buffer (Promega) and assayed for luciferase activity. The remaining 9 μl of translation reaction mixtures was used for RNase protection assays as described below.
RNase protection assays.
To isolate mRNA from in vitro translation mixtures, 50 μl of deproteinization buffer (20 mM Tris-HCl, pH 8.5, 100 mM NaCl, 2 mM EDTA, 1% SDS, 0.2 mg of yeast tRNA/ml) was added to 9 μl of translation samples. RNA was extracted with phenol-chloroform and was then ethanol precipitated. The RNA was resuspended in 30 μl of hybridization buffer and processed for RNase protection assays with RNase One (Promega) according to the manufacturer's instructions. The luciferase antisense probe was obtained by in vitro transcription of pGEM-Luc (Promega)-linearized EcoRV using T7 RNA polymerase (MBI Fermentas) in the presence of [α32P]GTP according to the manufacturer's instructions.
UV cross-linking experiments.
Capped-luciferase (c+LucA−) mRNA was polyadenylated and radiolabeled as follows: 5 μg of c+LucA− mRNA was incubated for 15 min at room temperature (in a 50-μl reaction mixture) in the presence of 0.1 mM ATP, 100 μCi of [α32P]ATP, 1× poly(A) polymerase buffer (U.S. Biochemicals), and 0.76 U of 1× poly(A) polymerase buffer. c+Luc− poly(A) tail-labeled mRNA (c+LucA+*) was purified on a ChromaSpin column TE-30 (Clontech) and scintillation counted. In vitro translation reaction mixtures (20 μl) were prepared as described above, but c+LucA+* mRNA (∼80 ng) was used instead of the c+LucA+ mRNA. The reaction mixtures were incubated for 15 min at 27°C and then transferred onto a glass plate, placed on ice, and irradiated at 4°C for 30 min with a UV lamp (254 nm) at a distance of ∼1 cm. The mixtures were then nuclease treated for 20 min at 37°C in the presence of 5 mM CaCl2, 0.15 μg of RNase A/μl, and 0.15 U of micrococcal nuclease (S7 nuclease; Roche)/μl. Half of each of the reaction mixtures (10 μl) was analyzed by autoradiography and Western blotting, while the rest was subjected to immunoprecipitation. For immunoprecipitations, the mixtures were diluted to 150 μl with 1× PBS-0.1% SDS and incubated for 30 min at 30°C. Then, 150 μl of 1× PBS-0.5% NP-40 was added to the samples with either rabbit polyclonal anti-dPABP (5 μl) or preimmune serum (5 μl) prebound to protein A-Sepharose (20-μl bed volume; APB). Following washing of the resin, the proteins were eluted as described above.
Nucleotide sequence accession number.
The complete assembled sequence of the dPaip2 cDNA has been deposited in GenBank under accession no. AY333418.
RESULTS
Cloning of dPaip2.
The dPaip2 cDNA was identified based on sequence homology with hPaip2 by searching the Berkeley Drosophila Genome Project database (46). dPaip2 is a single gene (GenBank accession no. AE003698; Gadfly no. CG12358), while two different genes exist in humans (GenBank no. AF317675 and AB032981). The complete assembled sequence of the dPaip2 cDNA is shown in Fig. 1A. The dPaip2 mRNA (2,040 nt) contains two polyadenylation signals (58) at positions 1012 and 2017. Two different mRNAs, which are likely generated by alternative polyadenylations, are expressed in flies, as Northern blot analysis detects two RNA species of the expected sizes (∼1,000 and ∼2,000 nt; data not shown). Both dPaip2 mRNAs encode a 124-aa protein with a predicted molecular mass of 14.6 kDa. dPaip2 shows 40% identity and 48% similarity to hPaip2 at the amino acid level (Fig. 1B). The two PAMs are highly conserved. Interestingly, PAM1 contains a stretch of 11 aa (aa 34 to 44 in dPaip2) that is 100% conserved between Drosophila and humans (Fig. 1B), consistent with its importance for PABP binding. Alignment of dPaip2 cDNA and genomic DNA sequences reveal that the dPaip2 gene is composed of four exons and three introns and that the gene is localized to chromosome 3R in cytological region 87D6 to 87D7 (Fig. 1C) (1, 13).
dPaip2 interacts with dPABP in vitro and in transfected S2 cells.
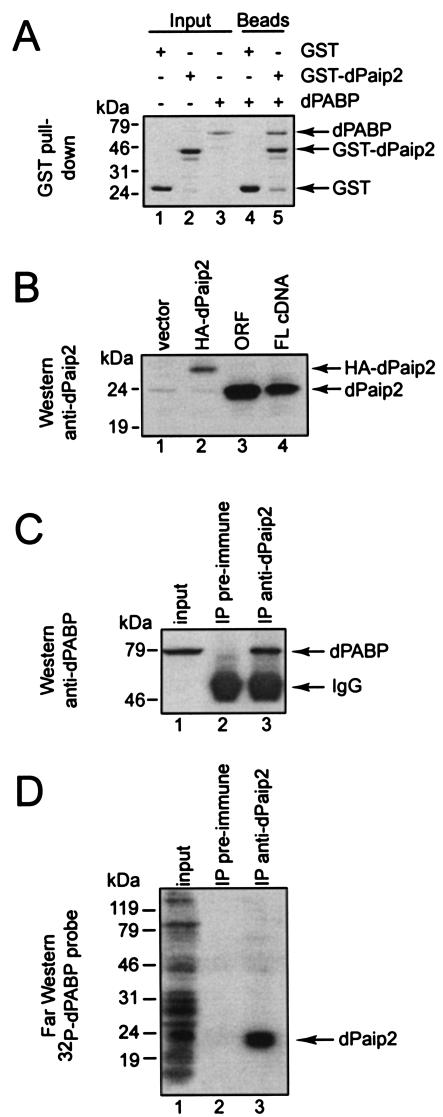
To ensure that the isolated dPaip2 is a functional hPaip2 homologue, its ability to interact with dPABP was examined. dPaip2 was expressed as a GST fusion protein for GST pull-down experiments (Fig. 2A). The recombinant proteins used in this experiment were largely intact (lanes 1 to 3, input). GST-dPaip2, but not GST, interacted with dPABP (compare lanes 4 and 5), demonstrating that dPaip2 is a PABP-interacting protein. This interaction is independent of RNA, since treatment of the proteins with a combination of RNase A and micrococcal nuclease did not diminish the binding (data not shown).
FIG. 2.
dPaip2 interacts with dPABP in vitro and in transfected S2 cells. (A) GST and GST-dPaip2 (5 μg) were immobilized on glutathione-Sepharose beads for GST pull-down analysis and incubated with dPABP (5 μg). Bound proteins were eluted in Laemmli sample buffer and resolved by SDS-12.5% PAGE. Recombinant proteins (1 μg; input) were run as controls. The gel was stained with Coomassie blue R-250. +, present; −, absent. (B) S2 cells were transfected with pPacPL (vector), pPacPL-HA-dPaip2, pPacPL-dPaip2(ORF) (containing the open reading frame only), and pPacPL-dPaip2(FL) (containing the full-length cDNA) as indicated. Cell extracts (50 μg) were subjected to Western blotting with an anti-dPaip2 serum. (C and D) Western blot analysis with anti-dPABP antibody (C) and far-Western analysis with 32P-His-HMK-dPABP probe (D) following immunopre-cipitation (IP) with anti-dPaip2 antibody on protein extracts (300 μg) prepared from S2 cells transfected with pPacPL-dPaip2(ORF). After immunoprecipitation, the proteins were resuspended in Laemmli sample buffer, resolved by SDS-12.5% PAGE, and subjected to Western and far-Western blotting as described in Materials and Methods. The input was 10% (30 μg) of the crude extract. The positions of the molecular mass markers are shown on the left. Proteins are identified on the right. IgG, heavy chain of immunoglobulin G.
To investigate the interaction of dPaip2 and dPABP in Drosophila cells, antibodies were raised against dPaip2 and dPABP in rabbits. S2 cells were transfected with plasmids encoding dPaip2 or hemagglutinin (HA)-tagged dPaip2. dPaip2 is expressed at very low levels in S2 cells (Fig. 2B, lane 1); however, Western blotting using dPaip2 antiserum detected a polypeptide migrating at ∼24 kDa in extracts transfected with dPaip2 constructs expressing just the open reading frame or full-length cDNA (lanes 3 and 4, respectively). The protein detected in extracts transfected with HA-dPaip2 migrated more slowly, as expected (lane 2). The apparent migration of dPaip2 on SDS-polyacrylamide gels differs substantially from the predicted molecular mass (14.6 kDa), but an anomalous migration on SDS-polyacrylamide gel electrophoresis (PAGE) was also observed for hPaip2 (27), probably due to the very acidic nature of hPaip2 (pI, 3.9) and dPaip2 (pI, 3.6). To demonstrate that exogenous dPaip2 is able to interact with dPABP, coimmunoprecipitations were performed (Fig. 2C). dPABP coprecipitated with dPaip2 antiserum but not with preimmune serum (lanes 2 and 3). We also performed far-Western analysis using 32P-dPABP as a probe (Fig. 2D). dPaip2 was detected following immunoprecipitation with anti-dPaip2 serum but not with preimmune serum (lanes 2 and 3). These results confirm that dPaip2 interacts with dPABP in Drosophila cells.
Overexpression of dPaip2 in Drosophila wings.
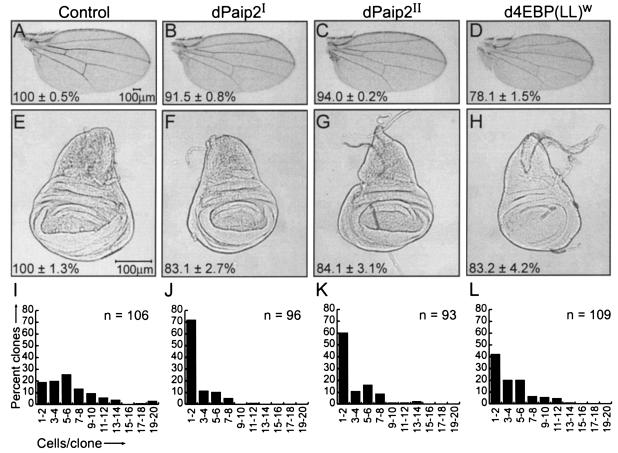
Cell growth is strongly dependent on the cellular translation capacity (for reviews, see references 22 and 57). It was therefore of interest to determine whether dPaip2 affects growth. To this end, we generated transgenic fly lines for ectopic overexpression of dPaip2 using the UAS-Gal4 system (2). Two independent UAS lines, UAS-HA-dPaip2I and UAS-HA-dPaip2II (hereafter referred to as dPaip2I and dPaip2II) were used to overexpress dPaip2 in different Drosophila tissues. First, to investigate whether dPaip2 inhibits growth in proliferating cells, dPaip2 was overexpressed in the wing-imaginal disc using the MS1096-Gal4 driver (6). Overexpression of the dPaip2I or dPaip2II transgene in male wings had no effect on the wing patterning but caused a statistically significant reduction of the overall wing size (91.5% ± 0.8% and 94.0% ± 0.2% of control wings, respectively) (Fig. 3A to C and Table 1). As a control for the Gal4 driver expression, we overexpressed the Drosophila 4E-BP from the UAS-d4E-BP(LL)w line [hereafter referred to as d4E-BP(LL)w] (38) and obtained a wing size reduction (78.1% ± 1.5% of the control wings) (Fig. 3D), in agreement with a previous report (38). We also examined wing-imaginal discs from third-instar larvae (Fig. 3E to H). Wing discs from larvae expressing the dPaip2I, dPaip2II, and d4E-BP(LL)w transgenes were significantly smaller than the control discs (83.1% ± 2.7%, 84.1% ± 3.1%, and 83.2% ± 4.2%, respectively).
FIG. 3.
Effects of ectopic expression of dPaip2 in wing tissues. Adult wings (A to D) or wing-imaginal discs (E to H) from third-instar larvae from control, dPaip2I, dPaip2II, and d4E-BP(LL)w transgenes upon crossing with MS1096-Gal4 flies. The values in the bottom left corners represent the average (± standard error of the mean) total wing (A to D) or wing disc (E to H) area measured for at least six wings (P < 0.0001) or at least six wing discs (P ≤ 0.0004) of each genotype. Clones were generated in wing discs using the FLP-out technique, and the numbers of cells per clone expressing GFP alone (I), GFP and dPaip2I (J), GFP and dPaip2II (K), and GFP and d4E-BP(LL)w (L) were determined. The total number of clones counted (n) is indicated in the top right corner of each panel. (A to H) The genotypes are MS1096-Gal4/+; +; + (control) (A and E), MS1096-Gal4/+; UAS-HA-dPaip2I/+; + (B and F), MS1096-Gal4/+; +; UAS-HA-Paip2II/+ (C and G), and MS1096-Gal4/+; +; UAS-d4E-BP(LL)w/+ (D and H). (I to L) All genotypes are yw hsFLP122; +; Act>CD2>Gal4; UAS-GFP-nls in addition to the following transgenes: +; UAS-HA-dPaip2I+ (J), +; +; UAS-HA-dPaip2II (K), and +; +; UAS-d4E-BP(LL)w (L).
TABLE 1.
Effects of dPaip2 overexpression on wing cell number and sizea
| Genotype | Total size of wing (106 μm2)b | % of control (total wing)c | Size of area 1 (105 μm2)d | % of control (area 1)e | Cell density in area 1 (10−2 cells/μm2)f | Cell size in area 1 (μm2/cell)g | No. of cells in area 1h | % of control (no. of cells)i |
|---|---|---|---|---|---|---|---|---|
| MS1096-Gal4/+ | 1.28 ± 0.01 | 100 ± 0.5 | 2.46 ± 0.02 | 100 ± 0.7 | 1.44 ± 0.01 | 69.5 ± 0.6 | 3,535 ± 48 | 100.0 ± 1.4 |
| MS1096-Gal4/UAS-HA-dPaip2I | 1.17 ± 0.01j | 91.5 ± 0.8 | 2.23 ± 0.02j | 90.8 ± 0.9 | 1.38 ± 0.02k | 72.4 ± 1.2 | 3,083 ± 46j | 87.2 ± 1.5 |
| MS1096-Gal4/UAS-HA-dPaip2II | 1.20 ± 0.01j | 94.0 ± 0.2 | 2.31 ± 0.01j | 94.0 ± 0.5 | 1.38 ± 0.02k | 72.3 ± 0.6 | 3,193 ± 35j | 90.3 ± 1.1 |
| MS1096-Gal4/UAS-d4EBP(LL)w | 1.00 ± 0.02j | 78.1 ± 1.5 | 1.89 ± 0.04j | 77.1 ± 1.7 | 1.55 ± 0.02k | 64.6 ± 0.8 | 2,930 ± 83j | 82.9 ± 2.8 |
| MS1096-Gal4/UAS-HA-dPaip2I/UAS-HA-dPaip2II | 1.09 ± 0.02j | 85.3 ± 2.0 | 2.06 ± 0.04j | 83.7 ± 0.8 | 1.41 ± 0.02l | 71.1 ± 0.7 | 2,892 ± 48j | 81.8 ± 1.7 |
| MS1096-Gal4/UAS-HA-dPaip2I/UAS-dPABP | 1.25 ± 0.01k | 98.1 ± 0.6 | 2.52 ± 0.02l | 102.5 ± 0.9 | 1.45 ± 0.02l | 69.2 ± 0.9 | 3,522 ± 113l | 99.6 ± 3.2 |
| MS1096-Gal4/P(10970) | 1.20 ± 0.02j | 94.0 ± 1.0 | 2.31 ± 0.02j | 94.2 ± 0.7 | 1.45 ± 0.02l | 69.2 ± 0.7 | 3,271 ± 70j | 92.5 ± 2.1 |
| MS1096-Gal4/P(10970)/UAS-HA-dPaip2II | 1.17 ± 0.01j | 91.3 ± 0.6 | 2.25 ± 0.02j | 91.5 ± 0.9 | 1.42 ± 0.02l | 70.4 ± 1.0 | 3,208 ± 51j | 90.8 ± 1.6 |
At least six wings were analyzed for each genotype. Values are ± standard error of the mean.
Measured using the Histogram function of Adobe Photoshop and converting the number of pixels to square micrometers.
Generated by comparing total size of wing with control value.
Area 1 is bound by the ACV LII, and LIV veins, and the wing margin and the size of this area were assessed as described in note b.
Generated by comparing size of area 1 with control value.
Assessed by counting the number of bristles (each representing a single cell) in a small area of 22,500 μm2 just distal to the wing margin. This small area is comprised within area 1.
Reciprocal of the cell density.
Estimation of cell number in area 1 generated by multiplying size of area 1 by cell density.
Generated by comparing number of cells in area 1 with control value.
P < 0.0001.
P < 0.05.
P ≥ 0.05.
Morphometric analysis revealed that wings from adult flies expressing the dPaip2I, dPaip2II, and d4E-BP(LL)w transgenes contained significantly (P < 0.0001) fewer cells than control wings (87.2% ± 1.5%, 90.3% ± 1.1%, and 82.9% ± 2.8%, respectively) (Table 1). The reduction in cell numbers seen with the dPaip2I, and dPaip2II alleles was accompanied by a minor decrease in cell density, and as a result, a minor increase in cell size (Table 1). However, fluorescence-activated cell sorter analysis (mean forward light scatter) of dissociated wing-imaginal disc cells of clones coexpressing dPaip2 and GFP induced by the FLP-out technique (56) failed to detect a size difference compared to normal cells (data not shown). The reduction in cell numbers could be caused by either apoptosis or inhibition of proliferation or growth. Wing-imaginal discs overexpressing dPaip2I and dPaip2II (using the MS1096-Gal4 driver) were stained with acridine orange and showed no significant increase in the number of apoptotic cells compared to control discs (data not shown), suggesting that apoptosis is not the primary cause of cell number reduction.
To examine the possible role of dPaip2 in cell proliferation or growth, clones of cells expressing GFP alone (control) or coexpressing GFP and dPaip2 were induced in the wing-imaginal disc 72 h after egg deposition and analyzed 44 h after induction (Fig. 3 I to L). Control clones contained on average five or six cells per clone (26% of the clones), while the dPaip2I- and dPaip2II-expressing clones contained one or two cells (72 and 60%, respectively). d4E-BP(LL)w clones also contained fewer cells than the control clones, as previously reported (38). In addition, approximately the same number of clones were recovered from control and dPaip2-overexpressing wing discs. Pyknotic nuclei, which are characteristic of apoptotic cells (55), were not observed in the dPaip2-overexpressing clones, excluding apoptosis as a significant cause of cell number reduction. The reduction in the number of cells per clone could result from a defect in cell division or growth. In general, cell growth is dominant over proliferation, as cells need to reach a critical size for cell division to occur (42, 50). Consequently, reducing the growth rate usually causes retardation of cell division (for a review, see reference 22). In agreement with these observations, dPaip2 clones are smaller than the control, as they consist of fewer cells of the same size. Also, inhibition of proliferation usually leads to an increase in cell size (22), which is not the case with dPaip2 overexpression. In fact, dPaip2 cells take more time than control cells to reach the critical size necessary for cell division (doubling time, ∼17 and ∼44 h for control and dPaip2 clones, respectively). Thus, the cell number reduction phenotype is most likely a consequence of inhibition of cell growth. This is supported by the growth inhibition phenotypes observed in nonproliferating cells of the eye and of the larval fat body (see below).
Epistatic interaction between dPaip2 and dPABP.
To provide further evidence that the wing size reduction observed upon dPaip2 overexpression was due to a difference in dPaip2 levels, we cooverexpressed dPaip2I and dPaip2II (hereafter referred to as double-dPaip2 flies) under the control of the MS1096-Gal4 driver. The wings of double-dPaip2 flies are smaller than the wings of flies with dPaip2I and dPaip2II alone (85.3% ± 2.0%, 91.5% ± 0.8%, and 94.05 ± 0.2% of control wings, respectively) (Table 1), and this size reduction is additive. To investigate if the phenotype of dPaip2 overexpression was mediated through its interaction with dPABP, we cooverexpressed dPABP (from the UAS-dPABP allele) (53) together with dPaip2I. This reversed the wing size reduction phenotype of dPaip2I (98.1% ± 0.6% versus 91.5% ± 0.8% for wings overexpressing dPaip2I alone) (Table 1). In addition, coexpression of dPABP rescued the cell number reduction phenotype of dPaip2I alone (3,522 ± 113 cells versus 3,083 ± 46 cells, compared to 3,535 ± 48 cells for control wings) (Table 1). To further highlight the dPaip2 and dPABP interaction, we overexpressed dPaip2II in flies heterozygous for a loss-of-function allele of dPABP [P(10970)] (53). Alone, animals heterozygous for this allele of dPABP displayed a wing size reduction, but this phenotype was slightly increased by overexpression of dPaip2II (94.0% ± 1.0% and 91.3% ± 0.6% of control wings, respectively) (Table 1). Similarly to the dPaip2 overexpression phenotype, the wings of dPABP-heterozygous animals also had fewer cells than control wings, and the size of these cells was not altered (Table 1). Together, these experiments show an epistatic interaction between dPaip2 and dPABP. Furthermore, they show that a reduced amount of PABP [as in P(10970)] causes a size reduction phenotype, highlighting the importance of PABP in growth. These results also provide in vivo evidence to support the in vitro dPaip2 and dPABP interaction data (Fig. 2).
Overexpression of dPaip2 in Drosophila eyes.
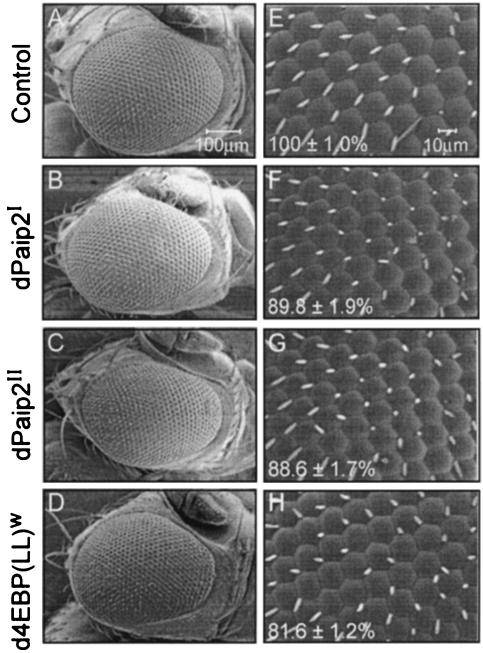
To support the observation that dPaip2 affects growth, dPaip2 expression was targeted to the eye-imaginal disc using the GMR-Gal4 driver (12). This driver allows the expression of transgenes late in eye development, during the differentiation of postmitotic cells (14). Thus, it is ideal for the investigation of the effects of dPaip2 on cell growth without interference from proliferation effects. Upon overexpression of the dPaip2I, dPaip2II, and d4E-BP(LL)w transgenes in eyes, no gross abnormalities in the overall shape or patterning of the eye was observed (Fig. 4A to D). The sizes of individual ommatidia in a flat region in the middle of the eye were measured (Fig. 4E to H). Ommatidia overexpressing the dPaip2I, dPaip2II, and d4E-BP(LL)w transgenes were smaller than the control (89.8% ± 1.9%, 88.6% ± 1.7%, and 81.6% ± 1.2%, respectively) (Table 2). The total number of ommatidia per eye was not significantly changed by overexpression of the dPaip2I and dPaip2II transgenes (687 ± 19 and 703 ± 10, respectively, compared to 707 ± 13 for control eyes) (Table 2). These results demonstrate that in the absence of proliferation, such as in differentiating eye cells, dPaip2 inhibits growth.
FIG. 4.
Effects of ectopic expression of dPaip2 in eyes. Scanning electron micrographs of eyes (A to D) and ommatidia (E to H) from control, dPaip2I, dPaip2II, and d4E-BP(LL)w transgenes upon crossing with GMR-Gal4 flies. Magnifications are ×170 for eyes and ×270 for ommatidia. The values in the bottom left corners of panels E to H represent the average (± standard errors of the mean) ommatidia sizes measured for at least 20 ommatidia (from four male eyes) of each genotype (P < 0.0001). The genotypes are +; GMR-Gal4/+; + (control) (A and E), +; GMR-Gal4/UAS-HA-dPaip2I; + (B and F), +; GMR-Gal4/+; UAS-HA-Paip2II/+ (C and G), and +; GMR-Gal4/+; UAS-4E-BP(LL)w/+ (D and H).
TABLE 2.
Effect of dPaip2 overexpression on eye and ommatidium size and numbera
| Genotype | Total size of eye (105 μm2)b | % of control (total eye)c | Size of ommatidia (μm2)b | % of control (ommatidia)d | No. of ommatidia per eyee |
|---|---|---|---|---|---|
| GMR-Gal4/+ | 1.26 ± 0.05 | 100 ± 4.2 | 253 ± 3 | 100 ± 1.0 | 707 ± 13 |
| GMR-Gal4/UAS-HA-dPaip2I | 1.15 ± 0.06f | 91.1 ± 5.5 | 228 ± 4i | 89.8 ± 1.9 | 687 ± 19j |
| GMR-Gal4/UAS-HA-dPaip2II | 1.17 ± 0.05g | 92.8 ± 3.8 | 224 ± 4i | 88.6 ± 1.7 | 703 ± 10k |
| GMR-Gal4/UAS-d4EBP(LL)w | 1.18 ± 0.06h | 93.7 ± 5.2 | 207 ± 3i | 81.6 ± 1.2 | 745 ± 10l |
Four male eyes and 20 ommatidia were analyzed for each genotype. Values are ± standard errors of the mean.
Measured using the Histogram function of Adobe Photoshop and converting the number of pixels to square micrometers.
Generated by comparing total eye size with control value.
Generated by comparing ommatidium size with control value.
Assessed by counting the number of ommatidia per eye.
P = 0.22.
P = 0.24.
P = 0.36.
P < 0.0001.
P = 0.42.
P = 0.86.
P = 0.06.
Overexpression of dPaip2 in Drosophila larval fat body.
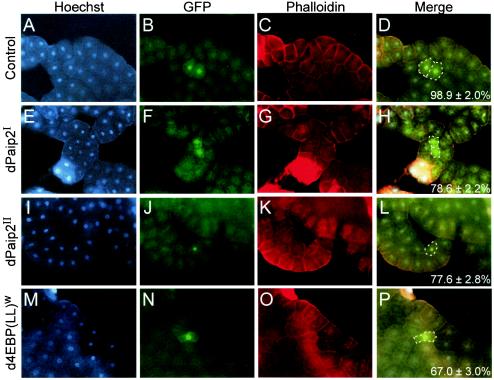
To further support the conclusion that dPaip2 primarily inhibits growth, we induced clones of cells overexpressing dPaip2 in another postmitotic tissue, the larval fat body. Polyploid fat body cells undergo successive rounds of DNA synthesis without mitosis and are not subjected to apoptosis (4). The sizes of fat body cells expressing GFP alone are not different from those of neighboring cells that do not express GFP (average size, 98.9% ± 2.0% of that of the GFP-negative cells) (Fig. 5A to D). However, fat body cells which coexpress GFP with the dPaip2I, dPaip2II, or d4E-BP(LL)w transgene are significantly smaller than neighboring cells that do not express GFP and the transgene (average size, 78.6% ± 2.2%, 77.6% ± 2.8%, and 67.0% ± 3.0% of that of the GFP-negative cells, respectively) (Fig. 5E to P). These results provide compelling evidence that dPaip2 inhibits the growth of nonproliferating fat body cells. Taken together, our results clearly demonstrate that dPaip2 plays a critical role in cell growth in several Drosophila tissues.
FIG. 5.
Effect of ectopic expression of dPaip2 in larval fat body. Clones were generated in larval fat bodies using the FLP-out technique, and cells expressing GFP alone (A to D), GFP and dPaip2I (E to H), GFP and dPaip2II (I to L), and GFP and d4E-BP(LL)w (M to P) were examined using a fluorescence microscope. The values in the bottom right corners of panels D, H, L, and P represent the average (± standard errors of the mean) cell sizes of GFP-positive cells (versus their GFP-negative neighboring cells) measured for at least 20 clones of each genotype (P < 0.0001). (A, E, I, and M) Staining of nuclei with Hoechst 33258. (B, F, J, and N) Expression of GFP. (C, G, K, and O) Staining of actin with phalloidin conjugated to rhodamine. (D, H, L, and P) Merged images (Hoechst, GFP, and phalloidin); the contours of GFP-expressing cells are delineated by white dashed lines. All the genotypes are yw hsFLP122; +; Act>CD2>Gal4 UAS-GFP-nls in addition to the following transgenes: +; UAS-HA-dPaip2I; + (E to H), +; +; UAS-HA-dPaip2II (I to L), and +; +; UAS-d4E-BP(LL)w (M to P).
dPaip2 represses translation and inhibits dPABP-poly(A) tail interaction in a Drosophila cell-free translation system.
To gain insight into the molecular mechanisms by which dPaip2 inhibits cell growth, its ability to repress translation and to interfere with the binding of PABP to the poly(A) tail of an mRNA was examined. We used a cell-free translation system from Drosophila S2 cells and luciferase as the reporter mRNA. The S2 cell-free translation system was optimized for salt and mRNA concentrations to reproduce the 5′-3′ synergy that is observed in vivo for translation of an mRNA that is both capped and polyadenylated (16). The synergy was calculated as the fraction of translation of a capped-polyadenylated mRNA divided by the sum of the translations of noncapped-polyadenylated and capped-nonpolyadenylated mRNAs [c+A+/(c+A− + c−A+)]. Under optimized conditions, we observed a 2.9-fold synergy between the cap and the poly(A) tail. The c+ LucA+ mRNA was translated ∼4- and ∼15-fold better than c+ LucA− and c−LucA− mRNAs, respectively (data not shown). We confirmed by RNase protection assays that the difference observed in the translation of these mRNAs cannot be attributed to differences in mRNA stability (data not shown). Thus, this S2 cell-free translation system is cap and poly(A)-tail dependent.
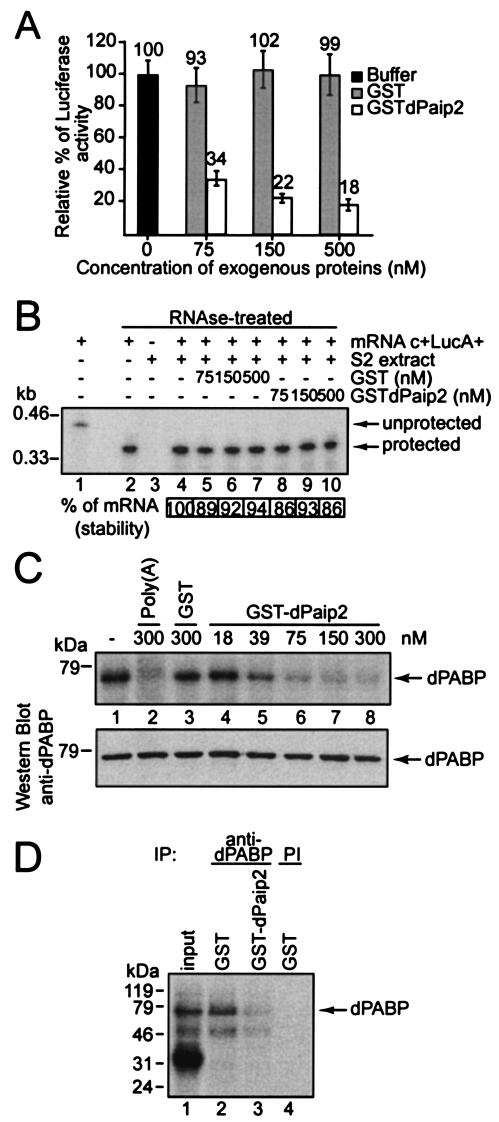
To examine the effect of dPaip2 in the S2 in vitro translation system, extracts were programmed with 50 ng of c+LucA+ mRNA in the presence of exogenous GST or GST-dPaip2. The addition of GST-dPaip2 resulted in a dramatic inhibition of translation of the reporter mRNA, whereas GST had no effect (Fig. 6A). At the highest concentration of GST-dPaip2 tested, 500 nM, translation was impaired by ∼80%. To exclude the possibility that GST-dPaip2 affects the stability of the reporter mRNA, RNase protection assays were performed on cell-free translation mixtures after 60 min of translation (Fig. 6B). There was no significant difference between the levels of luciferase mRNA in the presence of GST or of GST-dPaip2, as determined by quantification of the protected fragment (Fig. 6B, compare lanes 5 to 7 to lanes 8 to 10). As the difference in translation was not due to a difference in reporter mRNA stability, dPaip2 is a bona fide inhibitor of translation.
FIG. 6.
dPaip2 represses translation and inhibits dPABP-poly(A) tail interaction in a novel Drosophila cell-free translation system. (A) Capped luciferase mRNA with a 98-nt poly(A) tail (c+LucA+; 50 ng) was translated in S2 cell extracts in the presence of increasing amounts of GST or GST-dPaip2. The results are shown as relative luciferase activity (± standard errors of the mean) and are the averages of three independent experiments. (B) Stability of c+LucA+ mRNA after 60 min in translation reaction mixtures as analyzed by RNase protection assays. The percent stability of the protected fragment (relative to the buffer control in lane 4) was determined using a phosphorimager (BAS-2000-TR; Fujix). The contents of each sample are indicated above the lanes (+, present; −, absent). Lane 1 is probe alone without RNase treatment. Protected and unprotected fragments,as well as molecular size markers, are indicated on the right and the left, respectively. (C) Capped-luciferase mRNA labeled in the poly(A) tail (c+LucA+*) was incubated in S2 cell-free translation extracts in the presence of exogenous cold poly(A) RNA (Sigma), GST, or increasing concentrations of GST-dPaip2 and subjected to UV cross-linking followed by autoradiography (top). The membrane shown was then subjected to Western blot analysis with anti-dPABP antibody (bottom). (D) Immunoprecipitation (IP) with anti-dPABP antibody or preimmune (PI) serum of UV-cross-linking mixtures containing either GST or GST-dPaip2. The immunoprecipitates were resolved by SDS-PAGE and visualized by autoradiography; 30% of a GST-containing cross-linking mixture (input) was run as a control.
To determine whether dPaip2 interferes with the binding of PABP to the poly(A) tail of an mRNA, UV cross-linking experiments were performed (Fig. 6C). A labeled poly(A) tail was added to the c+LucA− mRNA using the poly(A) polymerase and [α32P]-ATP (generating c+LucA+* mRNA). The c+LucA+* mRNA was incubated with S2 cell-free translation extracts in the presence of buffer alone (lane 1), cold poly(A) as a competitor (lane 2), GST (lane 3), or increasing amounts of GST-dPaip2 (lanes 4 to 8), and the mixtures were UV irradiated. Cold poly(A) and increasing amounts of GST-dPaip2, but not GST, diminished the cross-linking of an ∼70-kDa band to the labeled poly(A) tail (Fig. 6C, top). Western blot analysis demonstrated that the radiolabeled protein comigrated with dPABP (Fig. 6C, bottom). To confirm the identity of the radiolabeled protein as dPABP, immunoprecipitation with anti-dPABP antibody was performed (Fig. 6D). Mixtures containing exogenous GST, but not those containing GST-dPaip2, precipitated a significant amount of the radiolabeled dPABP (compare lanes 2 and 3). Preimmune serum failed to precipitate the labeled proteins (lane 4). A UV-cross-linked mixture containing GST was run on the gel as a control without immunoprecipitation (lane 1, input). These results demonstrate that dPaip2 inhibits the binding of dPABP to the poly(A) tail of an mRNA. The results also support the conclusion that Paip2 inhibits translation by disrupting the interaction between PABP and the mRNA poly(A) tail, thereby preventing the formation of the mRNA closed loop, which is important for efficient translation initiation (27). They also prove the usefulness of the S2 cell-free translation system for studying proteins implicated in translation in Drosophila.
Taken together, these experiments demonstrate that dPaip2 is the bona fide homologue of the human Paip2 because it possesses all its known characteristics: (i) it interacts with dPABP, (ii) it represses translation, and (iii) it interferes with the ability of dPABP to bind poly(A). Thus, dPaip2 inhibits cell growth by repressing translation.
DISCUSSION
We have shown that dPaip2 reduces growth without altering patterning in several Drosophila tissues, including the larval fat body, eyes, wings, and wing-imaginal discs. dPaip2 strongly inhibits translation in vitro, as was shown for hPaip2 (27). Thus, dPaip2 most likely inhibits growth by repressing translation. Translation is a major target of growth control, as cells need to increase their protein content before they can divide in order to ensure daughter cell survival (reviewed in reference 22). Deregulation of translation has often been associated with growth defects. For example, in Drosophila, a collection of mutations in genes encoding ribosomal proteins (known as Minute mutations) have low overall growth rates and are delayed in development (reviewed in reference 33). Interference with the formation of the eIF4F complex at the 5′ end of the mRNA by the translation suppressor d4E-BP results in a reduced-growth phenotype (38). Mutations in the translation initiator factors deIF4E and deIF4A caused a more dramatic larval growth arrest phenotype (17, 31), which is similar to that seen upon amino acid starvation (4, 5). It is well established that nutrient starvation causes inhibition of translation by affecting discrete translational-control pathways (reviewed in references 28 and 37).
A number of signaling pathways have been implicated in the promotion of cell growth. Nutrient availability plays a key role in growth, and the insulin-signaling pathway coordinates cellular metabolism with nutritional conditions (5). The insulin-signaling pathway promotes translation via stimulation of S6K and inactivation of the translational repressor 4E-BP (reviewed in reference 37). Ectopic overexpression in Drosophila of positive components of the insulin-signaling pathway, for example, dInr or dPI3K, or mutations in negative regulators, such as dPTEN and dTSC1/2, cause dramatic increases in cell size and, to a lesser extent, increases in cell numbers (reviewed in references 34 and 37). Moreover, mutations in these same positive signaling components, or ectopic overexpression of the negative regulators, such as a highly active version of d4E-BP (38), primarily reduce cell size and have significantly weaker effects on cell numbers (with the exception of dS6K, which affects only cell size) (39). In addition, increased insulin signaling stimulates transition through the G1/S phases of the cell cycle, but the overall doubling time of these cells is unchanged due to a compensatory lengthening of the G2/M phases. Hence, this pathway seems primarily to stimulate mass accumulation, creating an imbalance between growth and proliferation signals, which results in an alteration of cell size. The reasons for this imbalance remain unclear. dMyc and dRas also control cell growth (reviewed in reference 22). Ectopic overexpression of dMyc or activated dRas (dRasV12) promotes growth and results in increased cell size and numbers. Conversely, loss of the dMyc gene inhibits growth and results in fewer and smaller cells (23). Interestingly, dRas appears to upregulate dMyc at a posttranscriptional level. Similar to the insulin-signaling pathway, overexpression of dMyc and dRas affects growth in an unbalanced fashion, as they also shorten the G1 phase of the cell cycle and the G2 phase is lengthened to compensate. However, these proteins have weaker effects on cell size than components of the insulin-signaling pathway.
In contrast to the insulin-signaling pathway and to dMyc and dRas, cooverexpression of dCdk4 and dCyclin D promotes growth and accelerates cell cycling (9). This results in a balanced, proportional cell growth in which cell size remains unchanged while cell numbers increase. Consistent with this finding, deletion of the Cdk4 gene represses growth without altering cell size and leads to a decrease in cell numbers (36). Interestingly, the phenotypes observed upon loss of the Cdk4 gene are similar to the phenotype of dPaip2 overexpression. In dividing cells of the wing discs, dPaip2 decreases cell numbers without reducing cell size, while in nonproliferating cells of the larval fat body and of the eye, dPaip2 overexpression decreases cell size. These tissue-specific effects are consistent with an inhibition of growth. In proliferating cells, the reduced translation capacity of the dPaip2-overexpressing cells likely affects all phases of the cell cycle equally. The reduced growth of these cells results in longer cell doubling times, but growth remains coordinated with proliferation and cell size is not affected. Consistent with a primary effect on growth, the nonproliferating cells of the eye and the larval fat body are reduced in size owing to impairment in translation. It is unclear at present why d4E-BP overexpression creates an imbalance between growth and proliferation signals (see above), which leads to an alteration in cell size, while dPaip2 does not. The different sensitivities of some mRNAs to these translational inhibitors might be responsible for the different phenotypes.
What is the molecular mechanism by which dPaip2 mediates its effect on cell growth? A likely possibility is that dPaip2 reduces growth by inhibiting the interaction of dPABP with the poly(A) tail, thus disrupting the mRNA 5′-3′ loop and inhibiting translation (24, 27). eIF4F disproportionately stimulates the translation of mRNAs containing extensive secondary structures in their 5′ UTRs, which mainly encode growth factors and their receptors, cyclins, and other mitogens (reviewed in references 19 and 54). For example, the level of cyclin D1 increases when eIF4E is overexpressed (43, 44). Inasmuch as PABP stimulates the translation of a subset of mRNAs by activating eIF4F, dPaip2 inhibition of translation might disproportionately affect the same subset of mRNAs. Cyclin D and Cdk4 are interesting candidates. It would be important to link dPaip2 and cyclin D/Cdk4 translation. In addition, the translation of some mRNAs might be especially sensitive to the level of dPABP or dPaip2.
The phenotypes observed upon overexpression of dPaip2 in different tissues are less dramatic than would have been expected from the in vitro translation experiments (∼80% inhibition of translation). It is conceivable that dPaip2 levels in vivo are tightly regulated to prevent deleterious interference with PABP function (PABP’s gene is essential). There is one Drosophila homologue of Paip1 (dPaip1; GenBank no. AE003804; Gadfly no. CG8963) that has not been studied yet. Since Paip1 stimulates translation, it is possible that it counteracts the effects of overexpression of the repressor dPaip2 (8, 27). dPaip2, like Paip1, eRF3, ataxin-2, and Tob (11, 29, 45), interacts with the C-terminal domain of PABP through its conserved PAM2 site (G. Roy and N. Sonenberg, unpublished data). The different PAM2-containing proteins might compete with Paip2 to modulate the activity of PABP and consequently attenuate the effects of dPaip2 overexpression. Furthermore, since hPaip2 is a phosphoprotein (A. Kahvejian, B. Raught, A.-C. Gingras, and N. Sonenberg, unpublished data), it is possible that dPaip2 is also a phosphoprotein and that its activity is controlled by its phosphorylation state.
In conclusion, we have demonstrated that dPaip2 is an inhibitor of cell growth, most likely because of its ability to repress translation. This study also highlights the importance of regulating PABP function in translation and growth. This system should serve as a basis to identify regulators of dPaip2 activity by screening for genetic interacting partners.
Acknowledgments
We thank C. Lister and C. Binda for technical assistance and A. Kahvejian, Y. Svitkin, H. Imataka, J. Berlanga, A. Brasey, J. Dostie, and A. Brueschke for helpful discussions. We are grateful to S. J. Leevers, T. Radimerski, and A. Brueschke for critical reading of the manuscript. We are indebted to V. Evdokimova for her advice. We thank C. DeMaria for preparation of the pBac-His-HMK-dPABP, pGST-dPABP, and pET-His-HMK-dPABP plasmids. We thank C. M. Schuster and the Bloomington Stock Center for providing the UAS-dPABP and P(10970) fly lines, respectively.
This research was supported by grants from the National Institutes of Health (1 RO1 GM66157-01) to N.S. and from the Canadian Institute of Health Research (CIHR) to N.S. and P.L. N.S. is a CIHR Distinguished Scientist, a Howard Hughes Medical Institute International Scholar, and a James McGill Professor. P.L. is a CIHR Investigator. G.R. and K.K. were recipients of doctoral studentships from the CIHR. M.M. was the recipient of a doctoral studentship from the Cancer Research Society. G.R. was the recipient of a McGill Major Studentship and now holds a McGill Faculty of Medicine internal studentship.
REFERENCES
- 1.Adams, M. D., S. E. Celniker, R. A. Holt, C. A. Evans, J. D. Gocayne, P. G. Amanatides, S. E. Scherer, P. W. Li, R. A. Hoskins, R. F. Galle, R. A. George, S. E. Lewis, S. Richards, M. Ashburner, S. N. Henderson, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185-2195. [DOI] [PubMed] [Google Scholar]
- 2.Brand, A. H., and N. Perrimon. 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118:401-415. [DOI] [PubMed] [Google Scholar]
- 3.Brasey, A., M. Lopez-Lastra, T. Ohlmann, N. Beerens, B. Berkhout, J. L. Darlix, and N. Sonenberg. 2003. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 77:3939-3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britton, J. S., and B. A. Edgar. 1998. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development 125:2149-2158. [DOI] [PubMed] [Google Scholar]
- 5.Britton, J. S., W. K. Lockwood, L. Li, S. M. Cohen, and B. A. Edgar. 2002. Drosophila's insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2:239-249. [DOI] [PubMed] [Google Scholar]
- 6.Capdevila, J., and I. Guerrero. 1994. Targeted expression of the signaling molecule decapentaplegic induces pattern duplications and growth alterations in Drosophila wings. EMBO J. 13:4459-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, B. P., and T. Hai. 1994. Expression vectors for affinity purification and radiolabeling of proteins using Escherichia coli as host. Gene 139:73-75. [DOI] [PubMed] [Google Scholar]
- 8.Craig, A. W., A. Haghighat, A. T. Yu, and N. Sonenberg. 1998. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature 392:520-523. [DOI] [PubMed] [Google Scholar]
- 9.Datar, S. A., H. W. Jacobs, A. F. de la Cruz, C. F. Lehner, and B. A. Edgar. 2000. The Drosophila cyclin D-Cdk4 complex promotes cellular growth. EMBO J. 19:4543-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deardorff, J. A., and A. B. Sachs. 1997. Differential effects of aromatic and charged residue substitutions in the RNA binding domains of the yeast poly(A)-binding protein. J. Mol. Biol. 269:67-81. [DOI] [PubMed] [Google Scholar]
- 11.Deo, R. C., N. Sonenberg, and S. K. Burley. 2001. X-ray structure of the human hyperplastic discs protein: an ortholog of the C-terminal domain of poly(A)-binding protein. Proc. Natl. Acad. Sci. USA 98:4414-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis, M. C., E. M. O'Neill, and G. M. Rubin. 1993. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development 119:855-865. [DOI] [PubMed] [Google Scholar]
- 13.FlyBase Consortium. 1999. The FlyBase database of the Drosophila Genome Projects and community literature. Nucleic Acids Res. 27:85-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freeman, M. 1996. Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87:651-660. [DOI] [PubMed] [Google Scholar]
- 15.Gallie, D. R. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 5:2108-2116. [DOI] [PubMed] [Google Scholar]
- 16.Gallie, D. R. 1998. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene 216:1-11. [DOI] [PubMed] [Google Scholar]
- 17.Galloni, M., and B. A. Edgar. 1999. Cell-autonomous and non-autonomous growth-defective mutants of Drosophila melanogaster. Development 126:2365-2375. [DOI] [PubMed] [Google Scholar]
- 18.Gingras, A.-C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 19.Gray, N. K., and M. Wickens. 1998. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 14:399-458. [DOI] [PubMed] [Google Scholar]
- 20.Hershey, J. W. B., and W. C. Merrick. 2000. Pathway and mechanism of initiation of protein synthesis, p. 33-88. In N. Sonenberg, J. W. B. Hershey, and M. B. Matthews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Iizuka, N., L. Najita, A. Franzusoff, and P. Sarnow. 1994. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell. Biol. 14:7322-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston, L. A., and P. Gallant. 2002. Control of growth and organ size in Drosophila. Bioessays 24:54-64. [DOI] [PubMed] [Google Scholar]
- 23.Johnston, L. A., D. A. Prober, B. A. Edgar, R. N. Eisenman, and P. Gallant. 1999. Drosophila myc regulates cellular growth during development. Cell 98:779-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kahvejian, A., G. Roy, and N. Sonenberg. 2001. The mRNA closed loop model: the function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harbor Symp. Quant. Biol. 66:293-300. [DOI] [PubMed] [Google Scholar]
- 25.Khaleghpour, K., A. Kahvejian, G. De Crescenzo, G. Roy, Y. V. Svitkin, H. Imataka, M. O'Connor-McCourt, and N. Sonenberg. 2001. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol. Cell. Biol. 21:5200-5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khaleghpour, K., S. Pyronnet, A. C. Gingras, and N. Sonenberg. 1999. Translational homeostasis: eukaryotic translation initiation factor 4E control of 4E-binding protein 1 and p70 S6 kinase activities. Mol. Cell. Biol. 19:4302-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khaleghpour, K., Y. V. Svitkin, A. W. Craig, C. T. DeMaria, R. C. Deo, S. K. Burley, and N. Sonenberg. 2001. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol. Cell 7:205-216. [DOI] [PubMed] [Google Scholar]
- 28.Kimball, S. R., and L. S. Jefferson. 2000. Regulation of translation initiation in mammalian cells by amino acids, p. 561-579. In N. Sonenberg, J. W. B. Hershey, and M. B. Matthews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Kozlov, G., J. F. Trempe, K. Khaleghpour, A. Kahvejian, I. Ekiel, and K. Gehring. 2001. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc. Natl. Acad. Sci. USA 98:4409-4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn, U., and T. Pieler. 1996. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J. Mol. Biol. 256:20-30. [DOI] [PubMed] [Google Scholar]
- 31.Lachance, P. E., M. Miron, B. Raught, N. Sonenberg, and P. Lasko. 2002. Phosphorylation of eukaryotic translation initiation factor 4E is critical for growth. Mol. Cell. Biol. 22:1656-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 33.Lambertsson, A. 1998. The minute genes in Drosophila and their molecular functions. Adv. Genet. 38:69-134. [DOI] [PubMed] [Google Scholar]
- 34.Lasko, P. 2002. Diabetic flies? Using Drosophila melanogaster to understand the causes of monogenic and genetically complex diseases. Clin. Genet. 62:358-367. [DOI] [PubMed] [Google Scholar]
- 35.Lefrère, V., A. Vincent, and F. Amalric. 1990. Drosophila melanogaster poly(A)-binding protein: cDNA cloning reveals an unusually long 3′-untranslated region of the mRNA, also present in other eukaryotic species. Gene 96:219-225. [DOI] [PubMed] [Google Scholar]
- 36.Meyer, C. A., H. W. Jacobs, S. A. Datar, W. Du, B. A. Edgar, and C. F. Lehner. 2000. Drosophila Cdk4 is required for normal growth and is dispensable for cell cycle progression. EMBO J. 19:4533-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miron, M., and N. Sonenberg. 2001. Regulation of translation via TOR signalling: insights from Drosophila melanogaster. J. Nutr. 131:2988S-2993S. [DOI] [PubMed] [Google Scholar]
- 38.Miron, M., J. Verdu, P. E. Lachance, M. J. Birnbaum, P. F. Lasko, and N. Sonenberg. 2001. The translational inhibitor 4E-BP is an effector of PI(3)K/Akt signalling and cell growth in Drosophila. Nat. Cell Biol. 3:596-601. [DOI] [PubMed] [Google Scholar]
- 39.Montagne, J., M. J. Stewart, H. Stocker, E. Hafen, S. C. Kozma, and G. Thomas. 1999. Drosophila S6 kinase: a regulator of cell size. Science 285:2126-2129. [DOI] [PubMed] [Google Scholar]
- 40.Munroe, D., and A. Jacobson. 1990. mRNA poly(A) tail, a 3′ enhancer of translational initiation. Mol. Cell. Biol. 10:3441-3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neufeld, T. P., A. F. de la Cruz, L. A. Johnston, and B. A. Edgar. 1998. Coordination of growth and cell division in the Drosophila wing. Cell 93:1183-1193. [DOI] [PubMed] [Google Scholar]
- 42.Polymenis, M., and E. V. Schmidt. 1999. Coordination of cell growth with cell division. Curr. Opin. Genet. Dev. 9:76-80. [DOI] [PubMed] [Google Scholar]
- 43.Rosenwald, I. B., A. Lazaris-Karatzas, N. Sonenberg, and E. V. Schmidt. 1993. Elevated levels of cyclin D1 protein in response to increased expression of eukaryotic initiation factor 4E. Mol. Cell. Biol. 13:7358-7363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rousseau, D., R. Kaspar, I. Rosenwald, L. Gehrke, and N. Sonenberg. 1996. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl. Acad. Sci. USA 93:1065-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roy, G., G. De Crescenzo, K. Khaleghpour, A. Kahvejian, M. O'Connor-McCourt, and N. Sonenberg. 2002. Paip1 interacts with poly(A) binding protein through two independent binding motifs. Mol. Cell. Biol. 22:3769-3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rubin, G. M., L. Hong, P. Brokstein, M. Evans-Holm, E. Frise, M. Stapleton, and D. A. Harvey. 2000. A Drosophila complementary DNA resource. Science 287:2222-2224. [DOI] [PubMed] [Google Scholar]
- 47.Rubin, G. M., and A. C. Spradling. 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218:348-353. [DOI] [PubMed] [Google Scholar]
- 48.Sachs, A. 2000. Physical and functional interactions between the mRNA cap structure and the poly(A) tail, p. 447-466. In N. Sonenberg, J. W. B. Hershey, and M. B. Matthews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Sachs, A. B., R. W. Davis, and R. D. Kornberg. 1987. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 7:3268-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saucedo, L. J., and B. A. Edgar. 2002. Why size matters: altering cell size. Curr. Opin. Genet. Dev. 12:565-571. [DOI] [PubMed] [Google Scholar]
- 51.Schneider, I. 1972. Cell lines derived from late embryonic stages of Drosophila melanogaster. J. Embryol. Exp. Morphol. 27:353-365. [PubMed] [Google Scholar]
- 52.Searfoss, A., T. E. Dever, and R. Wickner. 2001. Linking the 3′ poly(A) tail to the subunit joining step of translation initiation: relations of Pab1p, eukaryotic translation initiation factor 5B (Fun12p), and Ski2p-Slh1p. Mol. Cell. Biol. 21:4900-4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sigrist, S. J., P. R. Thiel, D. F. Reiff, P. E. Lachance, P. Lasko, and C. M. Schuster. 2000. Postsynaptic translation affects the efficacy and morphology of neuromuscular junctions. Nature 405:1062-1065. [DOI] [PubMed] [Google Scholar]
- 54.Sonenberg, N., and A. C. Gingras. 1998. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 10:268-275. [DOI] [PubMed] [Google Scholar]
- 55.Soriano, E., J. A. Del Rio, and C. Auladell. 1993. Characterization of the phenotype and birthdates of pyknotic dead cells in the nervous system by a combination of DNA staining and immunohistochemistry for 5′-bromodeoxyuridine and neural antigens. J. Histochem. Cytochem. 41:819-827. [DOI] [PubMed] [Google Scholar]
- 56.Struhl, G., and K. Basler. 1993. Organizing activity of wingless protein in Drosophila. Cell 72:527-540. [DOI] [PubMed] [Google Scholar]
- 57.Weinkove, D., and S. J. Leevers. 2000. The genetic control of organ growth: insights from Drosophila. Curr. Opin. Genet. Dev. 10:75-80. [DOI] [PubMed] [Google Scholar]
- 58.Wickens, M. 1990. How the messenger got its tail: addition of poly(A) in the nucleus. Trends Biochem. Sci. 15:277-281. [DOI] [PubMed] [Google Scholar]
- 59.Wolff, T. 2000. Histological techniques for the Drosophila eye. Part II: adult, p. 229-243. In W. Sullivan, M. Ashburner, and R. S. Hawley (ed.), Drosophila protocols. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.