Abstract
During tumorigenesis, tumor suppressor and cancer-related genes are commonly silenced by aberrant DNA methylation in their promoter regions. Recently, we reported that zebularine [1-(β-d-ribofuranosyl)-1,2-dihydropyrimidin-2-one] acts as an inhibitor of DNA methylation and exhibits chemical stability and minimal cytotoxicity both in vitro and in vivo. Here we show that continuous application of zebularine to T24 cells induces and maintains p16 gene expression and sustains demethylation of the 5′ region for over 40 days, preventing remethylation. In addition, continuous zebularine treatment effectively and globally demethylated various hypermethylated regions, especially CpG-poor regions. The drug caused a complete depletion of extractable DNA methyltransferase 1 (DNMT1) and partial depletion of DNMT3a and DNMT3b3. Last, sequential treatment with 5-aza-2′-deoxycytidine followed by zebularine hindered the remethylation of the p16 5′ region and gene resilencing, suggesting the possible combination use of both drugs as a potential anticancer regimen.
The abnormal de novo methylation of promoter CpG islands in numerous tumor suppressor and other cancer-related genes has been shown to be associated with their silencing during carcinogenesis (3, 18, 19, 30). This frequent alteration in human cancer cells may represent an alterative mechanism to mutations and chromosomal deletions for gene inactivation during tumorigenesis. The prevalence of aberrant methylation in cancer has encouraged the search for therapeutic agents which can inhibit methylation and which may thus be utilized to reverse this effect by reactivating genes which have become abnormally silenced.
5-Azacytidine (5-Aza-CR) and its deoxy analog, 5-aza-2′-deoxycytidine (5-Aza-CdR), are two of the most well-known DNA methylation inhibitors (15, 20). Both drugs are nucleoside analogs which have been widely used for studying the role of DNA methylation in biological processes as well as for clinical treatment of patients with acute myeloid leukemia and myelodysplastic syndrome (28, 34, 42). 5-Aza-CR and 5-Aza-CdR are potent mechanism-based inhibitors of DNA methyltransferases (DNMTs) and function by substituting for cytosine residues during DNA replication and forming covalent bonds with the DNMT, which ultimately leads to the inhibition of the DNMT's activity (8, 12, 31, 39). Unfortunately, these drugs are both unstable in aqueous solutions and toxic (4, 7, 39), and these characteristics have complicated their clinical use; hence there is the need for an effective, stable, and minimally toxic inhibitor of DNA methylation.
Previously, we characterized zebularine [1-(β-d-ribofuranosyl)-1,2-dihydropyrimidin-2-one) as a novel inhibitor of DNA methylation which is stable and minimally toxic both in vitro and in vivo (6). Zebularine is a cytidine analog containing a 2-(1H)-pyrimidinone ring that was originally developed as a cytidine deaminase inhibitor because it lacks an amino group on position 4 of the ring (22, 24). Studies with synthetic oligonucleotides containing the 2-(1H)-pyrimidinone ring have demonstrated the formation of a tight complex with bacterial methyltransferases in vitro (17), and this was further corroborated by a recent study demonstrating the crystallization of a bacterial DNA methyltransferase with the 2-(1H)-pyrimidinone ring forming a covalent bond at the active site (43). In a previous study, we have shown that zebularine can be orally administered to cause reactivation and demethylation of a silenced and hypermethylated p16 gene in human bladder tumor cells grown in nude mice (6). Nonetheless, one of the major challenges with the usage and application of nucleoside analogs as inhibitors of DNA methylation is the problem of remethylation of genes that were demethylated after treatment with these agents, which eventually results in their resilencing (5). This phenomenon of remethylation following cessation of drug treatment makes the clinical application of these drugs quite limited.
Here we demonstrate that the single treatment of T24 bladder carcinoma cells with zebularine resulted in a rapid induction of the p16 gene, followed by its resilencing and the remethylation of its 5′ region. We therefore examined the possibility of exploiting zebularine's stability to achieve an effective demethylation of aberrantly silenced genes and to maintain their expression over extended time periods and found that zebularine can effectively sustain demethylation of the p16 5′ region and prevent gene resilencing when administered in a continuous fashion to cultured cancer cells. Continuous zebularine treatment also caused demethylation of the entire p16 gene locus, which was most pronounced in CpG-depleted regions. Furthermore, zebularine induced a complete depletion of extractable DNMT1, but not DNMT3a and -3b, proteins in T24 cells. Last, we found that sequential treatment of T24 cells with an initial dose of 5-Aza-CdR followed by a sustained low dose of zebularine hindered the remethylation of the p16 5′ region and the resilencing of p16 gene expression. Our results suggest new strategies for cancer therapy using prolonged and continuous zebularine treatment, as well as a combination therapeutic regimen of 5-Aza-CdR and zebularine.
MATERIALS AND METHODS
Cell lines.
The T24 human bladder transitional carcinoma-derived cell line was obtained from the American Type Culture Collection (Manassas, Va.) and cultured in McCoy's 5A medium supplemented with 10% heat-inactivated fetal calf serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml (Gibco/Life Technologies, Inc., Palo Alto, Calif.). Normal LD419 human bladder fibroblasts were generated in our laboratory and cultured as previously described (41).
Drug treatments.
For kinetic studies, T24 cells were plated (3 × 105 cells/100-mm-diameter dish) and treated 24 h later with 5 × 10−4 M zebularine. The medium was changed 48 h after drug treatment, and DNA and RNA were harvested at various time points for methylation and reverse transcription-PCR (RT-PCR) analyses, respectively.
For the continuous drug treatment, T24 cells were plated (3 × 105 cells/100-mm dish) and treated 24 h later with 10−4 M zebularine. The medium was changed every 3 days, along with fresh zebularine treatment, for up to 40 days. Cells were treated continuously for 40 days with either 10−4 M zebularine or with an increasing concentration of zebularine from 10−4 to 4 × 10−4 M (an incremental increase of 5 × 10−5 M every 6 days). DNA and RNA were harvested at various time points for methylation and RT-PCR analyses, respectively. Protein lysates were collected at various time points for Western blot analysis of the DNMT protein levels.
For sequential drug treatment, T24 cells were plated (3 × 105 cells/100-mm dish) and treated 24 h later with either (i) 5 × 10−5 M zebularine continuously for 30 days, (ii) 10−6 M 5-Aza-CdR for 24 h, or (iii) 10−6 M 5-Aza-CdR for 24 h followed by 10−4 M zebularine for up to 30 continuous days. DNA and RNA were harvested at various time points for methylation and RT-PCR analyses, respectively.
Zebularine, synthesized as previously described (2), was dissolved in phosphate-buffered saline (PBS).
Nucleic acid isolation.
RNA was collected and extracted from cultured T24 and LD419 cells with the RNeasy Protect minikit (Qiagen, Valencia, Calif.) according to the manufacturer's recommended protocol. DNA was collected as previously described (13).
RT-PCR analysis.
Total RNA (5 μg) extracted from cultured cells was reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Invitrogen, Carlsbad, Calif.) and random hexamers (Amersham-Pharmacia, Piscataway, N.J.) in a total volume of 25 μl. The reverse transcription was performed as previously described (13). cDNA was amplified with primers specific for either p16 or GAPDH. The RT-PCR conditions were as follows: for p16, 94°C for 3 min, 28 cycles of 94°C for 1 min, 56°C for 30 s, and 72°C for 40 s, and a final extension step at 72°C for 5 min; for GAPDH, 94°C for 1 min, 19 cycles of 94°C for 1 min, 58°C for 30 s, and 72°C for 45 s, and a final extension step at 72°C for 2 min. The primer sequences are as follows: p16 sense, 5′-AGC CTT CGG CTG ACT GGC TGG-3′; p16 antisense, 5′-CTG CCC ATC ATC ATG ACC TGG A-3′; GAPDH sense, 5′-CAG CCG AGC CAC ATC GCT CAG ACA-3′; GAPDH antisense, 5′-TGA GGC TGT TGT CAT ACT TCT C-3′. RT-PCR amplification of each of the expressed genes was performed with 200 ng of cDNA, 10% dimethyl sulfoxide, 100 μM deoxynucleoside triphosphates, Taq DNA polymerase (Sigma), and 1 μM primers. The RT-PCR conditions, primers, and sequences for DNMT1, -3a, and -3b were as previously described (38). All reactions were analyzed in the linear range of amplification. PCR products were resolved on 2% agarose gels and subsequently transferred to a nylon membrane (Zetaprobe; Bio-Rad, Richmond, Calif.) under alkaline conditions. All blots were hybridized with a γ-32P-labeled internal oligonucleotide probe for p16 as previously described (13, 38).
Quantitative RT-PCR analysis.
The quantitation of mRNA levels was carried out by a real-time fluorescence detection method as described previously (9, 16). Briefly, after RNA isolation, cDNA was prepared from each sample as described above. The specific cDNA of interest (p16) and reference cDNA (GAPDH) were amplified by PCR separately using an oligonucleotide probe with a 5′ fluorescent reporter dye and a 3′ quencher dye (27). The 5′-to-3′ nuclease activity of Taq DNA polymerase cleaved the probe and released the reporter, whose fluorescence could be detected by the laser detector of the ABI Prism 7700 sequence detection system (Perkin-Elmer Corp., Foster City, Calif.). All of the samples were normalized to the reference GAPDH gene. The experiment was performed in duplicate.
Western blot analysis of DNMT protein levels.
Cells were rinsed twice with ice-cold PBS and lysed by the addition of radioimmunoprecipitation assay buffer (PBS, 0.1% sodium dodecyl sulfate [SDS], 0.5% Nonidet P-40, 0.5% sodium deoxycholate). Cells were scraped off dishes and placed on ice for 30 min. The mixture was centrifuged at 12,000 × g for 30 min at 4°C, and the supernatant was used for Western blot analysis. Approximately 60 μg of total protein extract was loaded onto 4 to 15% gradient Tris-HCl gels (Bio-Rad, Hercules, Calif.), electrophoresed in Tris-glycine-SDS running buffer (25 mM Tris, 192 mM glycine, 0.1% SDS; pH 8.3), and transferred to polyvinylidene difluoride membranes in Tris-glycine buffer (25 mM Tris, 192 mM glycine; pH 8.2) overnight at 4°C. The membranes were hybridized with antibodies against human DNMT1 (1:1,000 dilution; New England Biolabs, Beverly, Mass.), human DNMT3b (T-16; 1:500 dilution; Santa Cruz Biotechnology, Santa Cruz, Calif.), and PCNA (1:4,000 dilution, Santa Cruz Biotechnology) in Tris-buffered saline-Tween (TBS-T) buffer (0.1 M Tris, 1.5 M NaCl, 1% Tween 20) with 5% nonfat dry milk overnight at 4°C. The human DNMT3a was kindly provided by Guo-Liang Xu (Shanghai, People's Republic of China). The membranes were washed three times with TBS-T buffer at room temperature and incubated with secondary antibodies as follows: anti-mouse immunoglobulin G (IgG)-horseradish peroxidase (HRP) (1:3,000 dilution for PCNA; Santa Cruz Biotechnology), anti-rabbit IgG-HRP (1:2,000 dilution for DNMT1; Santa Cruz Biotechnology), and anti-goat IgG-HRP (1:10,000 dilution for DNMT3b; Calbiochem, San Diego, Calif.). All were incubated with the membrane for 1 h at room temperature. Afterwards, the membranes were washed five times with TBS-T at room temperature. Proteins were detected with the ECL chemiluminescence detection kit (Amersham-Pharmacia) and by exposure to Kodak (Rochester, N.Y.) X-Omat AR film. Autoradiograms from two independent Western gels were analyzed by scanning densitometry using a model GS-710 imaging densitometer (Bio-Rad).
Hemimethylation assay.
Hemimethylation analysis was performed as previously described (25, 32, 41). Undigested and HpaII-digested DNA from T24 cells was subjected to bisulfite modification. HpaII digests unmethylated DNA but does not cut a fully or hemimethylated configuration of its CCGG target sequence. Bisulfite-treated DNA was then amplified by PCR using primers that flanked the first HpaII sites in p16 intron 1 and p16 exon 2. The CpG site targeted by the intron 1 single-nucleotide primer extension (SNuPE) primer is also located in an HpaII site, so this primer was also used for hemimethylation analysis (41). The equations used to determine hemimethylation levels were as described previously (25). The H/F ratio represents the ratio of the percentage of hemimethylated (H) molecules to the percentage of fully methylated (F) molecules.
Quantitation of DNA methylation levels by Ms-SNuPE.
The mean cytosine methylation levels of CpG sites in the fragment were determined by treatment of genomic DNA (4 μg) with sodium bisulfite as described by Frommer et al. (11). Methylation analysis was performed using the methylation-sensitive SNuPE (Ms-SNuPE) assay (14). The bisulfite PCR and the qualitative Ms-SNuPE assay for the p16 promoter 5′ region (6) and p16 regions 1 to 8 (41) were performed as previously described. The bisulfite PCR primers for other loci are as follows: for p53 Alu, 5′-TGG GTT TAA TTA TTG TAT AGT TGA A-3′ (sense) and 5′-CTC AAC TCA CTA CAA ACT CCA-3′ (antisense); for D4Z4, 5′-GGG TTG AGG GTT GGG TTT AT-3′ (sense) and 5′-AAC TTA CAC CCT TCC CTA CA-3′ (antisense); for M4-4, 5′-ATG GTT TGA GGG TTT AGA TTA GGT-3′ (sense) and 5′-ACA TCA AAA TAA ACT TCC TCT TAC CA-3′ (antisense). The PCR conditions for p53 Alu were 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 1 min, annealing at 52°C for 45 s, and then extension at 72°C for 45 s. The D4Z4 PCR was performed in the same manner, except the annealing temperature was 58°C. The PCR conditions for M4-4 were 95°C for 2 min, followed by 42 cycles of denaturation at 95°C for 1 min, annealing at 56°C for 30 s, and then extension at 72°C for 1 min. A final extension step at 72°C for 10 min followed each PCR program. The Ms-SNuPE conditions for p53 Alu and D4Z4 were the same as those described previously (41). The Ms-SNuPE conditions for M4-4 were 95°C for 1 min, 46°C for 30 s, and 72°C for 20 s. The SNuPE primers for the p53 Alu are as follows: 5′-GTT AAG GGT TTT TTT TGT TTG GTT GGG-3′, 5′-TTT GGG AGG TTA AGG TAG G-3′, 5′-GTT TTT ATT GAA AAA TAT AAA AAA AAA TTA GT-3′, and 5′-GAA GGA GAA TGG TGT GAA TTT GGG-3′. The D4Z4 SNuPE primers are as follows: 5′-TGA GGG TTG GGT TTA TAG T-3′, 5′-GTG GTT TAG GGA GTG GG-3′, 5′-TAT ATT TTT AGG TTT AGT TTT GTA A-3′, and 5′-GAA AGG TTG GTT ATG T-3′. The M4-4 SNuPE primers are as follows: 5′-GGG TTT AGA TTA GGT TTT TT-3′, 5′-GTA ATA AGG ATT ATT TGA ATA G-3′, and 5′-TAA TAA TGT GGA TTT GTT TAA ATT-3′.
Bisulfite genomic sequencing.
The bisulfite genomic sequencing was used to determine methylation levels in individual molecules of DNA. The region of interest, the p16 5′ region, was amplified by PCR using DNA that had undergone sodium bisulfite conversion, as described above (11). The PCR-amplified region of the promoter was 530 bp, with 28 CpG dinucleotides, including CpGs both upstream of the transcriptional start site and in the first exon. PCR conditions have been previously described (33). Primers for the p16 5′ region were as follows: 5′-GGT GGG GTT TTT ATA ATT AGG AAA GAA TAG TTT TG-3′ (sense) and 5′-TCT AAT AAC CAA CCA ACC CCT CC-3′. PCR products were then cloned in into the pCR2.1 vector, followed by chemical transformation and plating, all performed with the TOPO-TA cloning kit (Invitrogen) by following the manufacturer's instructions. After positive clone identification, plasmid purification was performed with the Qiagen plasmid minikit. Sequencing was then performed at Laragen, Inc. (Los Angeles, Calif.)
Determination of population doublings and cell growth.
Cells were counted with a Z1 Coulter particle counter (Beckman Coulter Corporation, Hialeah, Fla.) at various time points. Untreated cells were analyzed under similar conditions as a control. The average cell number from two plates was determined, and the mean cell numbers were plotted to define population doublings. Initial drug treatment was started 24 h after seeding.
RESULTS
Kinetics of p16 mRNA induction and demethylation of the 5′ region by zebularine.
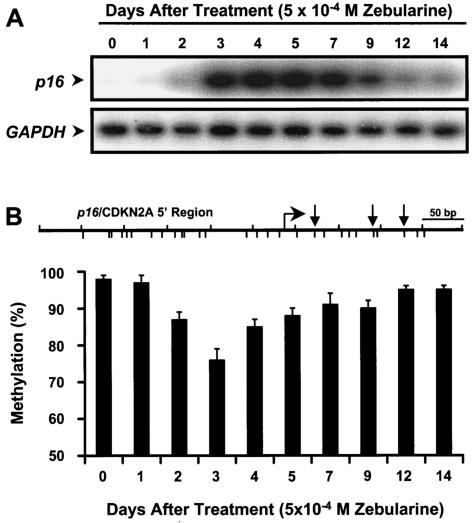
The kinetics of p16 mRNA induction and the demethylation of 5′ region of p16 by 5-Aza-CdR have been studied in detail (5), so it was of obvious interest to evaluate these parameters in T24 cells treated with zebularine. Treatment of T24 cells with 5 × 10−4 M zebularine for 48 h induced a slight expression of p16 by day 2, which increased dramatically up to day 5 and began to diminish thereafter (Fig. 1A). This resilencing of the p16 gene was not surprising, since the same phenomenon was previously demonstrated with 5-Aza-CdR and was presumably due to the remethylation of the 5′ region of p16 gene (5, 41). The methylation of the 5′ region of p16 decreased from 97% at day 0 to 75% at day 3 and then slowly increased thereafter, and this paralleled the decrease in p16 expression (Fig. 1B). Thus, remethylation is a common problem that is observed with zebularine as well as other inhibitors of DNA methylation and is a potential complication in the clinical applications of these drugs.
FIG. 1.
Kinetics of p16 mRNA induction and demethylation of the 5′ region by zebularine. T24 cells were exposed to zebularine (5 × 10−4 M) for 48 h. DNA and RNA were isolated at 24-h intervals after drug addition. (A) Expression levels of p16 mRNA at each time point were determined by RT-PCR analysis. GAPDH mRNA expression levels were measured to control for relative cDNA input. (B) Demethylation of the p16 5′ region by zebularine at each time point was quantified by Ms-SNuPE analysis. Methylation percentages represent the averages of three individual CpG sites in the p16 5′ regions as assayed by Ms-SNuPE analysis from two independent experiments. Error bars, standard deviations of four determinations. At the top is a partial map of the 5′ region of p16 gene analyzed. Tick marks below the line, CpG sites; bent arrow, transcription start site; straight arrows, CpG sites analyzed by Ms-SNuPE analysis. Marks above the line indicate 50-bp lengths of DNA.
Continuous treatment with zebularine sustains the expression and demethylation of the p16 gene.
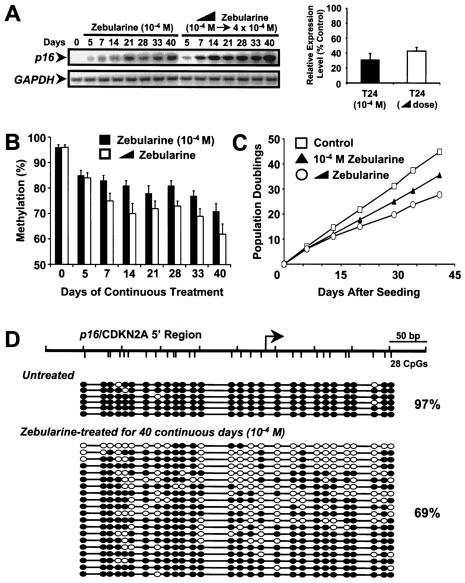
To circumvent the problems of remethylation, we took advantage of zebularine's stability and minimal cytoxicity (1, 6, 21) to investigate the effects of continuous drug treatment on p16 expression in T24 cells. Continuous zebularine treatment (10−4 M) for up to 40 days led to an induction of p16 expression at day 5, which increased over time (Fig. 2A, left). Treatment with increasing concentrations of zebularine (from 10−4 to 4 × 10−4 M) for 40 days led to an even greater level of p16 gene expression (Fig. 2A, left). We compared the extent of p16 reactivation from these two regimens to that in a normal fibroblast cell line (LD419), which expresses a p16 which is unmethylated in the 5′ region. Reactivation by zebularine led to p16 expression levels 31 (10−4 M) to 43% (increasing doses) of those seen in LD419 controls (Fig. 2A, right). This finding is not surprising since the methylation of the 5′ region with the increasing doses was reduced to approximately 63% over the same period (Fig. 2B). T24 cells were found to be growth suppressed by drug treatment, which is likely due to the reactivation of the p16 gene (Fig. 2C). Bisulfite genomic sequencing of DNA obtained from continuously treated cells showed substantial demethylation of the p16 5′ region, especially around the transcription start site (Fig. 2D). The percentage of methylation from all molecules (69%) was relatively consistent with the result obtained from the Ms-SNuPE analysis (71%) (Fig. 2B and D).
FIG. 2.
Effects of continuous zebularine treatment on p16 mRNA expression, demethylation of the 5′ region of the p16 gene, and cellular growth. T24 cells were treated with either zebularine (10−4 M) or an increasing dose of zebularine from 10−4 to 4 × 10−4 M over a period of 40 continuous days. For the increasing dose of zebularine, the drug was added in increments of 5 × 10−5 M every 6 days starting from 10−4 M. DNA and RNA were isolated at the indicated time points. (A, left) Expression levels of p16 mRNA at the indicated time point were measured by RT-PCR analysis. GAPDH mRNA expression levels were measured to control for relative cDNA input. (A, right) Relative expression levels of p16 mRNA (normalized to the reference GAPDH mRNA) for the indicated drug regimens were measured by quantitative real-time RT-PCR analysis. The levels are reported as percentages of expression of normal LD419 cells. Results represent averages and ranges of two separate determinations. (B) The methylation level of the 5′ region of the p16 gene for the indicated time points after either treatment was measured by Ms-SNuPE analysis. Methylation percentages represent the averages for three individual CpG sites in the 5′ regions of the p16 gene as assayed by Ms-SNuPE from two independent experiments. Error bars, standard deviations from four determinations. (C) Levels of cellular growth after treatment with the indicated drug regimens were plotted as population doublings against time. Initial drug treatment was started 24 h after seeding. (D) Bisulfite genomic sequencing of the 5′ region of the p16 gene in T24 cells. Cells were either untreated or treated with (10−4 M) zebularine continuously for 40 days. DNAs were then isolated and treated with sodium bisulfite, followed by the cloning and sequencing of individual molecules. Each horizontal line with a string of circles represents the methylation profile of one molecule. White circles, unmethylated CpG sites; black circles, methylated CpG sites. Total methylation of all molecules is noted to the right of the molecules as a percentage. Distances between CpGs are roughly to scale. (Top diagram) Schematic of the 5′ region analyzed indicating CpG density distributions. Tick marks above the line, 50-bp lengths of DNA; tick marks below the line, CpG sites; bent arrow, transcriptional start site.
Continuous zebularine treatment causes variable demethylation of the entire p16 gene locus.
We then assessed the effects of continuous zebularine treatment for 40 days on the methylation levels of several other regions within the p16 gene locus (41) (Fig. 3A). Regions 1 and 2 are CpG-poor sequences, region 3 is a CpG island containing Alu repetitive elements located upstream of the p16 promoter, region 4 is the CpG island of the promoter, regions 5 to 7 include three individual CpGs residing in intron 1 of p16, and region 8 is the CpG island of the second exon. All regions analyzed showed measurable demethylation; however, the CpG sites in regions 5 and 6, which are located in CpG-depleted areas, showed preferential demethylation (Fig. 3A).
FIG. 3.
Effects of continuous zebularine treatment on methylation levels of the p16 locus. (A) Methylation levels of regions within the p16 gene locus in T24 bladder cancer cells before and after continuous treatment with zebularine as measured by Ms-SNuPE analysis. (Top) Eight regions of various CpG densities were identified upstream and downstream of the p16 promoter region (region 4) (41). Regions 1 and 2 are CpG-poor sequences (three CpGs analyzed in region 1 and four CpGs in region 2), region 3 is a CpG island containing an Alu repetitive element located upstream of the p16 promoter (three CpGs analyzed), regions 5 to 7 consist of three individual CpG dinucleotides residing in intron 1 of p16, and region 8 is the CpG island of the second exon (three CpGs analyzed). Bent arrow, transcription start site for the p16 gene; vertical arrows, specific CpG sequences analyzed by Ms-SNuPE. (Bottom) Methylation values for each region were measured by Ms-SNuPE in T24 cells, before and after continuous treatment with zebularine for 40 days. T24 cells were treated with 10−4 M zebularine for 40 continuous days and DNA and RNA were harvested immediately afterwards. (B) Levels of fully methylated (F), hemimethylated (H), and unmethylated (U) DNA at the first HpaII site of p16 intron 1 (region 5) after continuous treatment with 10−4 M zebularine. (C) Levels of fully methylated, hemimethylated, and unmethylated DNA at the first HpaII site of p16 exon 2 (region 8) after continuous treatment with 10−4 M zebularine. The ratio of H to F (H:F) represents the ratio of the percentage of hemimethylated molecules to the percentage of fully methylated molecules.
To better understand this preferential demethylating effect of zebularine, we performed hemimethylation assays to determine the distribution of fully methylated, hemimethylated, and unmethylated sites (25) at both the CpG-depleted region 5 (p16 intron 1) and the CpG-rich region 8 (p16 exon 2) (Fig. 3B and C). In region 5, the levels of fully methylated sites substantially decreased after continuous treatment with zebularine and unmethylated sites increased dramatically over the 40-day time course (Fig. 3B). Methylation of the CpG site in region 5 is therefore poorly maintained after zebularine treatment, as most fully methylated CpGs were converted to the unmethylated state by day 40. The percentage of hemimethylated sites increased slowly over the course of continuous zebularine treatment but eventually decreased to starting levels (Fig. 3B). Zebularine appeared to preferentially target this CpG-depleted region, presumably by inhibiting the DNMT(s) responsible for maintaining its modification. The ratio of hemimethylated to fully methylated sites rose dramatically by day 21 and slowly decreased thereafter, indicating the effectiveness of the drug in this region. In contrast, most of the DNA molecules in the CpG island of p16 exon 2 remained fully methylated throughout the course of treatment, with a transitory increase in hemimethylated molecules and a slow increase in unmethylated molecules after 40 days (Fig. 3C). The ratio of hemimethylated to fully methylated sites remained low throughout the treatment, possibly due to the recruitment of another DNMT(s) to this region, which may methylate these hemimethylated molecules. Our results suggest that zebularine seemingly targets CpG-depleted regions more efficiently than CpG-rich regions.
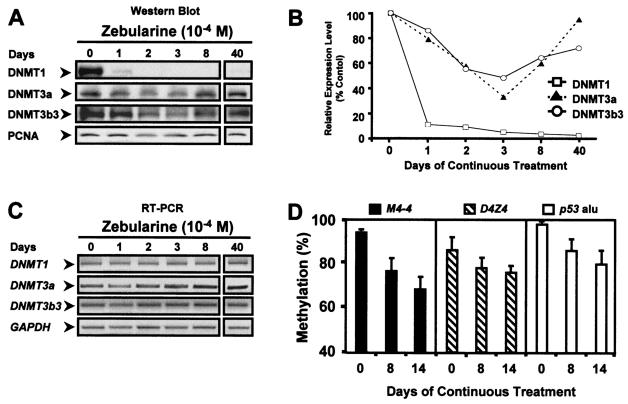
Zebularine selectively depletes DNMT1.
Previous work has shown that DNMTs work cooperatively to facilitate and maintain DNA methylation patterns in mammalian cells (25, 35). Since zebularine appears to target CpG-depleted regions, we next assessed the levels of DNMT1, -3a, and -3b in T24 bladder cancer cells before and during drug treatment. Western blot analysis showed a drastic depletion of extractable DNMT1 by day 1 of treatment in T24 cells, and virtually no extractable DNMT1 protein was present in cells growing in the presence of the drug, even after 40 days (Fig. 4A and B). DNMT3a and -3b3 were also affected in T24 cells, and this was most pronounced after 3 days of continuous zebularine treatment, yet both proteins gradually recovered thereafter (Fig. 4A and B). T24 cells express DNMT3b3 almost exclusively, and the human DNMT3b isoform, DNMT3b3, was recently shown to have reduced catalytic activity (40, 41a), possibly explaining the partial depletion of the enzyme by zebularine. DNMT3a, unlike DNMT1 or DNMT3b, is expressed throughout the cell cycle (37) and might therefore interact with zebularine only when it is incorporated into DNA during the S phase. The levels of DNMT mRNA transcripts were found to be unaffected by drug treatment, as detected by semiquantitative RT-PCR (Fig. 4C), supporting the idea that the depletion in DNMT protein levels was due to trapping of enzymes to the zebularine-substituted DNA rather than an inhibition of transcription or cell proliferation. Moreover, the sustained methylation of CpG-rich regions may be the result of the less-efficient depletion of DNMT3b3, since a recent study showed that DNMT1 and DNMT3b cooperate to methylate CpG islands in cancer cells (35).
FIG. 4.
Effects of continuous zebularine treatment on methylation status of various loci and on DNMT protein levels in T24 cells. T24 bladder cancer cells were treated with 10−4 M zebularine for up to 40 continuous days. DNA, RNA, and protein lysates were harvested at the indicated days. (A) Levels of DNMT1, -3a, and -3b3 proteins after continuous zebularine treatment in T24 cells. PCNA was used as a control for cell proliferation in the Western blot analysis. (B) Relative protein expression levels of DNMT1, -3a, and -3b3 (normalized to PCNA) were measured and quantitated by scanning densitometry. The initial protein expression level at day 0 was set as 100% for each DNMT, and the levels of protein expression from other days of treatment were compared to this level. The percentages represent the averages of two independent experiments. (C) Semiquantitative RT-PCR of DNMT mRNA levels in T24 cells under untreated or treated conditions. GAPDH mRNA levels were measured to control for the quantity and integrity of the input RNA. (D) Methylation levels of various hypermethylated loci in T24 cells before and after continuous zebularine treatment. M4-4 (GC percentage, 56.5%), D4Z4 (GC percentage, 73.3%), and p53 Alu (GC percentage, 55.7%) are hypermethylated loci found in T24 bladder cancer cells. The methylation status for each locus was quantitated by Ms-SNuPE analysis. The methylation percentages represent averages of two separate determinations from two independent experiments. Error bars, standard deviations of four determinations.
We next assessed the effectiveness of continuous zebularine on the methylation status of other methylated loci in T24 cells. The methylation levels of the D4Z4 subtelomeric repeat (a DNMT3b target sequence; CpG island; chromosome 4q35; GC percentage, 73.3%) (23), an Alu element in the p53 gene (repetitive sequence, CG-rich region; chromosome 17p13; GC percentage, 55.7%), and the M4-4 sequence (single-copy sequence; CpG island; chromosome 16q22; GC percentage, 56.5%) were all substantially affected in treated T24 cells, although D4Z4 was least affected (Fig. 4D). Our results indicated that continuous zebularine treatment can effectively and globally demethylate various methylated regions throughout the human genome.
Sequential treatment of T24 cells with 5-Aza-CdR followed by zebularine.
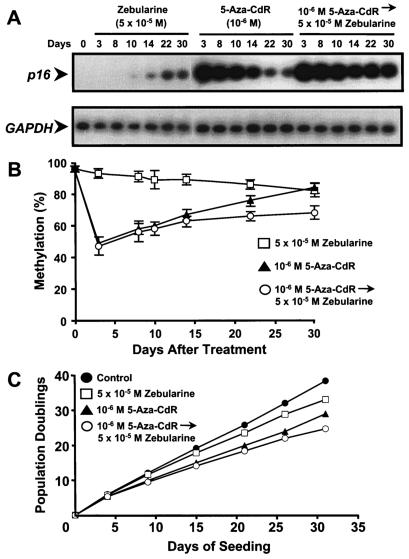
We next tested whether an initial treatment of 5-Aza-CdR followed by a continuous low dose of zebularine (5 × 10−5 M) in T24 cells could maintain p16 expression and hinder or prevent the remethylation of the p16 5′ region. Continuous zebularine treatment (5 × 10−5 M) resulted in a slow increase of p16 induction beginning at day 10 (Fig. 5A). Treatment with 10−6 M 5-Aza-CdR caused substantial p16 gene expression by day 3, which gradually diminished over time, consistent with the results as shown previously in our laboratory (5). Sequential treatment of 10−6 M 5-Aza-CdR followed continuously with 5 × 10−5 M zebularine resulted in robust p16 gene expression by day 3, which was well maintained throughout the treatment, as opposed to that from 5-Aza-CdR alone (Fig. 5A).
FIG. 5.
Sequential treatment of 5-Aza-CdR followed by zebularine and its effect on p16 mRNA expression, demethylation of the 5′ region of the p16 gene, and cellular growth suppression. T24 cells were treated with either (i) 5 × 10−5 M zebularine continuously for 30 days, (ii) 10−6 M 5-Aza-CdR for 24 h, or (iii) 10−6 M 5-Aza-CdR for 24 h followed sequentially by 5 × 10−5 M zebularine continuously of up to 30 days. DNA and RNA were harvested at the indicated time points for methylation and RT-PCR analyses, respectively. (A) The expression levels of p16 mRNA were determined for each of the three separate regimens at the indicated time points by RT-PCR analysis. GAPDH mRNA expression levels were measured to control for relative cDNA input. (B) The methylation status of the p16 5′ region was determined for each regimen at the indicated time points by Ms-SNuPE analysis. Methylation percentages represent the averages for three individual CpG sites in the p16 5′ regions as measured by Ms-SNuPE from two independent experiments. Error bars, standard deviations of four determinations. (C) Levels of cellular growth after treatment with the indicated drug regimens were plotted as population doublings against time. Cell counts were taken at the indicated time points to calculate population doublings. Initial drug treatment was started 24 h after seeding.
The methylation of the p16 5′ region decreased slowly over a period of 30 days (97 to 85%) after continuous zebularine (5 × 10−5 M) treatment (Fig. 5B). Treatment with 10−6 M 5-Aza-CdR alone resulted in an appreciable drop of the methylation of p16 5′ region, followed by a gradual remethylation over time. In contrast, sequential treatment with 10−6 M 5-Aza-CdR followed by 5 × 10−5 M zebularine produced a drastic decline of the methylation of p16 5′ region, followed by minimal remethylation, which was considerably slower than that following treatment with 5-Aza-CdR alone. Remethylation was therefore hindered by treatment of T24 cells with 5-Aza-CdR followed by continuous zebularine. These results indicated a potential clinical regimen combining both drugs, perhaps using 5-Aza-CdR as an initial loading drug and the less toxic zebularine as a maintenance drug.
We then analyzed the growth of the T24 cells under the various drug regimens and found that the cells treated sequentially with 5-Aza-CdR followed by zebularine were the most growth suppressed compared to the control cells and cells subjected to other drug regimens (Fig. 5C). Thus the sequential treatment which caused the most sustained expression of p16 resulted in the slowest growth rate of the treated cells (Fig. 5A and C).
DISCUSSION
Zebularine is a novel inhibitor of DNA methylation which is stable and minimally toxic (1, 6, 21). Transient treatments with methylation inhibitors are commonly followed by resilencing of genes, which is most likely due to the occurrence of remethylation (5, 10, 25, 41). Zebularine's stability and minimal cytotoxicity allowed us to grow cells in the continuous presence of the drug, and this led to the induction and maintenance of p16 expression in T24 cells, which thereby circumvented the problem of remethylation. The global demethylating effects of zebularine suggested the effectiveness of this methodology for applying the drug, especially for CpG-poor regions. Moreover, our previous work with nude mice showed that high doses of zebularine were not highly toxic to the mice (6). These observations suggest possible therapeutic strategies and clinical benefits in the continuous application of zebularine as a cancer therapy.
Continuous treatment of T24 bladder cancer cells with zebularine resulted in a complete depletion of DNMT1, and, surprisingly, these cells were still growing even after the depletion of DNMT1 throughout the 40-day time course. This appears to contrast with results of a recent study which showed that an intra-S-phase arrest can be triggered by the reduction in DNMT1 resulting from the use of an antisense oligonucleotide inhibitor (29). The authors, however, also mentioned that depletion of DNMT1 by 5-Aza-CdR did not induce this arrest, presumably due to the fact that DNMT1 is trapped only after the replication fork has formed and to the inability to prevent de novo synthesis of DNMT1. In our treatment of T24 cells with zebularine, which presumably has a mechanism akin to 5-Aza-CdR, we did not observe a distinct intra-S-phase arrest, as demonstrated with the antisense oligonucleotide, suggesting different mechanisms of action for these inhibitors. Perhaps, continuous zebularine treatment can offer an alternative means to study methylation effects in the absence of DNMT1 alone. This will be the focus of future studies.
Interestingly, with the apparently complete depletion of DNMT1, T24 cells still retained substantial methylation of the p16 CpG islands (p16 5′ region and exon 2), D4Z4, M4-4, and an Alu element in p53. However, DNMT3a and -3b3 were only partially affected by continuous zebularine treatment, suggesting that these enzymes may play important roles in the methylation of these four loci, especially of D4Z4, which is a specific DNMT3b target sequence. It is also intriguing that zebularine appears to preferentially target CpG-depleted regions (such as region 5 of p16 locus) over CpG-rich regions (such as region 8 of p16 locus). Since zebularine selectively depleted DNMT1, this suggests that the CpG-depleted regions are largely maintained by this enzyme, as we previously found with mouse embryonic stem cells (25). DNMT1 may therefore not be the only enzyme required to maintain methylation of CpG-rich regions, as both DNMT3a and -3b3 were still largely present after drug treatment. As indicated by the hemimethylation data, DNMT3a and -3b, which are presumably targeted to CpG-rich regions, may function to randomly methylate hemimethylated molecules generated by zebularine (26), whereas, in CpG-poor regions, there are more hemimethylated molecules than fully methylated molecules throughout the period of zebularine treatment, suggesting that perhaps these regions are basically maintained by DNMT1 alone.
Our observations also support recent findings that DNMT1 works cooperatively with DNMT3a and DNMT3b to maintain methylation of CpG-rich regions, such as CpG islands and repetitive elements (25, 35). On the other hand, another recent study from Robert et al. (36) indicated that DNMT1 alone was necessary and sufficient to maintain global methylation and aberrant CpG island methylation in human cancer cells. The discrepancy between these results may arise from the differences in methodologies used to deplete cellular DNMT1 levels. Rhee et al. (35) used a gene targeting approach, and the study by Robert et al. used an antisense approach (36), while we used a mechanism-based inhibitor of DNA methylation. Each study may affect unknown variables and may therefore include unaccounted biases. These models need to be investigated further in order to elucidate the mechanism behind the maintenance of aberrant CpG islands in human cancer cells.
Our results strongly suggest that continuous zebularine treatment is an effective strategy in sustaining demethylation of various loci and preventing their remethylation. When given sequentially after 5-Aza-CdR in a continuous fashion, zebularine can substantially hinder the rate of remethylation, implying that this is a powerful combination in treating cancer caused by aberrant epigenetic mechanisms. These findings suggest clinical potential as a cancer therapy as well as combination therapy with other drugs, such as 5-Aza-CdR, histone deacetylase inhibitors, and chemotherapies.
Acknowledgments
We thank Gerry Coetzee (Universty of Southern California) and Peter Laird (University of Southern California) for the use of the scanning densitometer and the TaqMan real-time PCR system, respectively.
This work was supported by NIH grants 1RO1 CA 82422 and 1RO1 CA 83867. Daniel Weisenberger was supported by NIH Training Grant in Basic Research in Oncology T32 CA09659.
REFERENCES
- 1.Barchi, J. J., S. Musser, and V. E. Marquez. 1992. The decomposition of 1-(beta-D-ribofuranosyl)-1,2-dihydropyrimidin-2-one (zebularine) in alkali: mechanism and products. J. Org. Chem. 57:536-541. [Google Scholar]
- 2.Barchi, J. J., Jr., D. A. Cooney, Z. Hao, Z. H. Weinberg, C. Taft, V. E. Marquez, and H. Ford, Jr. 1995. Improved synthesis of zebularine [1-(beta-D-ribofuranosyl)-dihydropyrimidin-2-one] nucleotides as inhibitors of human deoxycytidylate deaminase. J. Enzyme Inhib. 9:147-162. [DOI] [PubMed] [Google Scholar]
- 3.Baylin, S. B., M. Esteller, M. R. Rountree, K. E. Bachman, K. Schuebel, and J. G. Herman. 2001. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum. Mol. Genet. 10:687-692. [DOI] [PubMed] [Google Scholar]
- 4.Beisler, J. A. 1978. Isolation, characterization, and properties of a labile hydrolysis product of the antitumor nucleoside, 5-azacytidine. J. Med. Chem. 21:204-208. [DOI] [PubMed] [Google Scholar]
- 5.Bender, C. M., M. L. Gonzalgo, F. A. Gonzales, C. T. Nguyen, K. D. Robertson, and P. A. Jones. 1999. Roles of cell division and gene transcription in the methylation of CpG islands. Mol. Cell. Biol. 19:6690-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, J. C., C. B. Matsen, F. A. Gonzales, W. Ye, S. Greer, V. E. Marquez, P. A. Jones, and E. U. Selker. 2003. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J. Natl. Cancer Inst. 95:399-409. [DOI] [PubMed] [Google Scholar]
- 7.Constantinides, P. G., P. A. Jones, and W. Gevers. 1977. Functional striated muscle cells from non-myoblast precursors following 5-azacytidine treatment. Nature 267:364-366. [DOI] [PubMed] [Google Scholar]
- 8.Constantinides, P. G., S. M. Taylor, and P. A. Jones. 1978. Phenotypic conversion of cultured mouse embryo cells by aza pyrimidine nucleosides. Dev. Biol. 66:57-71. [DOI] [PubMed] [Google Scholar]
- 9.Eads, C. A., K. D. Danenberg, K. Kawakami, L. B. Saltz, P. V. Danenberg, and P. W. Laird. 1999. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res. 59:2302-2306. [PubMed] [Google Scholar]
- 10.Flatau, E., F. A. Gonzales, L. A. Michalowsky, and P. A. Jones. 1984. DNA methylation in 5-aza-2′-deoxycytidine-resistant variants of C3H 10T1/2 C18 cells. Mol. Cell Biol. 4:2098-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frommer, M., L. E. McDonald, D. S. Millar, C. M. Collis, F. Watt, G. W. Grigg, P. L. Molloy, and C. L. Paul. 1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA 89:1827-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabbara, S., and A. S. Bhagwat. 1995. The mechanism of inhibition of DNA (cytosine-5-)-methyltransferases by 5-azacytosine is likely to involve methyl transfer to the inhibitor. Biochem. J. 307(Pt. 1):87-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-Zulueta, M., C. M. Bender, A. S. Yang, T. Nguyen, R. W. Beart, J. M. Van Tornout, and P. A. Jones. 1995. Methylation of the 5′ CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res. 55:4531-4535. [PubMed] [Google Scholar]
- 14.Gonzalgo, M. L., and P. A. Jones. 1997. Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE). Nuleic Acids Res. 25:2529-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris, M. 1982. Induction of thymidine kinase in enzyme-deficient Chinese hamster cells. Cell 29:483-492. [DOI] [PubMed] [Google Scholar]
- 16.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 17.Hurd, P. J., A. J. Whitmarsh, G. S. Baldwin, S. M. Kelly, J. P. Waltho, N. C. Price, B. A. Connolly, and D. P. Hornby. 1999. Mechanism-based inhibition of C5-cytosine DNA methyltransferases by 2-H pyrimidinone. J. Mol. Biol. 286:389-401. [DOI] [PubMed] [Google Scholar]
- 18.Jones, P. A., and S. B. Baylin. 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3:415-428. [DOI] [PubMed] [Google Scholar]
- 19.Jones, P. A., and P. W. Laird. 1999. Cancer epigenetics comes of age. Nat. Genet. 21:163-167. [DOI] [PubMed] [Google Scholar]
- 20.Jones, P. A., and S. M. Taylor. 1980. Cellular differentiation, cytidine analogs and DNA methylation. Cell 20:85-93. [DOI] [PubMed] [Google Scholar]
- 21.Kelley, J. A., J. S. Driscoll, J. J. McCormack, J. S. Roth, and V. E. Marquez. 1986. Furanose-pyranose isomerization of reduced pyrimidine and cyclic urea ribosides. J. Med. Chem. 29:2351-2358. [DOI] [PubMed] [Google Scholar]
- 22.Kim, C. H., V. E. Marquez, D. T. Mao, D. R. Haines, and J. J. McCormack. 1986. Synthesis of pyrimidin-2-one nucleosides as acid-stable inhibitors of cytidine deaminase. J. Med. Chem. 29:1374-1380. [DOI] [PubMed] [Google Scholar]
- 23.Kondo, T., M. P. Bobek, R. Kuick, B. Lamb, X. Zhu, A. Narayan, D. Bourc'his, E. Viegas-Pequignot, M. Ehrlich, and S. M. Hanash. 2000. Whole-genome methylation scan in ICF syndrome: hypomethylation of non-satellite DNA repeats D4Z4 and NBL2. Hum. Mol. Genet. 9:597-604. [DOI] [PubMed] [Google Scholar]
- 24.Laliberte, J., V. E. Marquez, and R. L. Momparler. 1992. Potent inhibitors for the deamination of cytosine arabinoside and 5-aza-2′-deoxycytidine by human cytidine deaminase. Cancer Chemother. Pharmacol. 30:7-11. [DOI] [PubMed] [Google Scholar]
- 25.Liang, G., M. F. Chan, Y. Tomigahara, Y. C. Tsai, F. A. Gonzales, E. Li, P. W. Laird, and P. A. Jones. 2002. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 22:480-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, I. G., L. Han, A. Taghva, L. E. O'Brien, and C. L. Hsieh. 2002. Murine de novo methyltransferase Dnmt3a demonstrates strand asymmetry and site preference in the methylation of DNA in vitro. Mol. Cell. Biol. 22:704-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak, K. J., S. J. Flood, J. Marmaro, W. Giusti, and K. Deetz. 1995. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 4:357-362. [DOI] [PubMed] [Google Scholar]
- 28.Lubbert, M. 2000. DNA methylation inhibitors in the treatment of leukemias, myelodysplastic syndromes and hemoglobinopathies: clinical results and possible mechanisms of action. Curr. Top. Microbiol. Immunol. 249:135-164. [DOI] [PubMed] [Google Scholar]
- 29.Milutinovic, S., Q. Zhuang, A. Niveleau, and M. Szyf. 2003. Epigenomic stress response: knock-down of DNA methyltransferase 1 triggers an intra S-phase arrest of DNA replication and induction of stress response genes. J. Biol. Chem. 278:14985-14995. [DOI] [PubMed] [Google Scholar]
- 30.Momparler, R. L., and V. Bovenzi. 2000. DNA methylation and cancer. J. Cell Physiol. 183:145-154. [DOI] [PubMed] [Google Scholar]
- 31.Momparler, R. L., and D. Derse. 1979. Kinetics of phosphorylation of 5-aza-2′-deoxyycytidine by deoxycytidine kinase. Biochem. Pharmacol. 28:1443-1444. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen, C. T., D. J. Weisenberger, M. Velicescu, F. A. Gonzales, J. C. Lin, G. Liang, and P. A. Jones. 2002. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 62:6456-6461. [PubMed] [Google Scholar]
- 33.Pao, M. M., G. Liang, Y. C. Tsai, Z. Xiong, P. W. Laird, and P. A. Jones. 2000. DNA methylator and mismatch repair phenotypes are not mutually exclusive in colorectal cancer cell lines. Oncogene 19:943-952. [DOI] [PubMed] [Google Scholar]
- 34.Pinto, A., and V. Zagonel. 1993. 5-Aza-2′-deoxycytidine (Decitabine) and 5-azacytidine in the treatment of acute myeloid leukemias and myelodysplastic syndromes: past, present and future trends. Leukemia 7(Suppl. 1):51-60. [PubMed] [Google Scholar]
- 35.Rhee, I., K. E. Bachman, B. H. Park, K. W. Jair, R. W. Yen, K. E. Schuebel, H. Cui, A. P. Feinberg, C. Lengauer, K. W. Kinzler, S. B. Baylin, and B. Vogelstein. 2002. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416:552-556. [DOI] [PubMed] [Google Scholar]
- 36.Robert, M. F., S. Morin, N. Beaulieu, F. Gauthier, I. C. Chute, A. Barsalou, and A. R. MacLeod. 2003. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat. Genet. 33:61-65. [DOI] [PubMed] [Google Scholar]
- 37.Robertson, K. D., K. Keyomarsi, F. A. Gonzales, M. Velicescu, and P. A. Jones. 2000. Differential mRNA expression of the human DNA methyltransferases (DNMTs) 1, 3a and 3b during the G0/G1 to S phase transition in normal and tumor cells. Nuleic Acids Res. 28:2108-2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson, K. D., E. Uzvolgyi, G. Liang, C. Talmadge, J. Sumegi, F. A. Gonzales, and P. A. Jones. 1999. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nuleic Acids Res. 27:2291-2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santi, D. V., A. Norment, and C. E. Garrett. 1984. Covalent bond formation between a DNA-cytosine methyltransferase and DNA containing 5-azacytosine. Proc. Natl. Acad. Sci. USA 81:6993-6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soejima, K., W. Fang, and B. J. Rollins. 2003. DNA methyltransferase 3b contributes to oncogenic transformation induced by SV40T antigen and activated Ras. Oncogene 22:4723-4733. [DOI] [PubMed] [Google Scholar]
- 41.Velicescu, M., D. J. Weisenberger, F. A. Gonzales, Y. C. Tsai, C. T. Nguyen, and P. A. Jones. 2002. Cell division is required for de novo methylation of CpG islands in bladder cancer cells. Cancer Res. 62:2378-2384. [PubMed] [Google Scholar]
- 41a.Weisenberger, D. J., et al. Mol. Cancer Res., in press.
- 42.Wijermans, P., M. Lubbert, G. Verhoef, A. Bosly, C. Ravoet, M. Andre, and A. Ferrant. 2000. Low-dose 5-aza-2′-deoxycytidine, a DNA hypomethylating agent, for the treatment of high-risk myelodysplastic syndrome: a multicenter phase II study in elderly patients. J. Clin. Oncol. 18:956-962. [DOI] [PubMed] [Google Scholar]
- 43.Zhou, L., X. Cheng, B. A. Connolly, M. J. Dickman, P. J. Hurd, and D. P. Hornby. 2002. Zebularine: a novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J. Mol. Biol. 321:591-599. [DOI] [PMC free article] [PubMed] [Google Scholar]