Abstract
The p53 protein acts a tumor suppressor by inducing cell cycle arrest and apoptosis in response to DNA damage or oncogene activation. Recently, it has been proposed that phosphorylation of serine 15 in human p53 by ATM (mutated in ataxia telangiectasia) kinase induces p53 activity by interfering with the Mdm2-p53 complex formation and inhibiting Mdm2-mediated destabilization of p53. Serine 18 in murine p53 has been implicated in mediating an ATM- and ataxia telangiectasia-related kinase-dependent growth arrest. To explore further the physiological significance of phosphorylation of p53 on Ser18, we generated mice bearing a serine-to-alanine mutation in p53. Analysis of apoptosis in thymocytes and splenocytes following DNA damage revealed that phosphorylation of serine 18 was required for robust p53-mediated apoptosis. Surprisingly, p53Ser18 phosphorylation did not alter the proliferation rate of embryonic fibroblasts or the p53-mediated G1 arrest induced by DNA damage. In addition, endogenous basal levels and DNA damage-induced levels of p53 were not affected by p53Ser18 phosphorylation. p53Ala18 mice developed normally and were not susceptible to spontaneous tumorigenesis, and the reduced apoptotic function of p53Ala18 did not rescue the embryo-lethal phenotype of Mdm2-null mice. These results indicate that phosphorylation of the ATM target site on p53 specifically regulates p53 apoptotic function and further reveal that phosphorylation of p53 serine 18 is not required for p53-mediated tumor suppression.
The p53 tumor suppressor is a transcription factor that functions to regulate cell proliferation and apoptosis. The importance of p53 in suppressing cancer is highlighted by the very high incidence of spontaneous tumor formation in p53-deficient mice and in mice bearing genetic alterations in other members of the p53 pathway (14, 27, 30, 42). The critical role of p53 in tumor suppression is further underscored by the high frequency of p53 mutations in human cancer, and malignancies that retain a wild-type p53 gene often have acquired other mutations that affect one or more of the pathways known to govern p53 function (53). Therefore, it is likely that inactivation of p53 is a common mechanistic step in the development of most human cancers.
Cellular p53 activity is greatly increased following treatment of cells with DNA-damaging agents such as ionizing radiation and adriamycin (62). Induction of p53 activity can also occur in response to inappropriate growth-stimulatory signals, such as expression of an activated oncogene or exposure of the cells to other forms of stress (37, 38, 50, 69). Increased p53 activity resulting from DNA damage blocks the proliferation of cells in the G1 phase of the cell cycle by inducing the expression of genes such as the cyclin-dependent kinase inhibitor p21 (6, 13, 16, 20). In addition, increased p53 activity in the cell can initiate apoptosis by inducting the expression of proapoptotic BCL-2 family members involved in mitochondrion-associated cell death and by transactivating the expression of death receptor-associated genes (7, 41, 52, 66). Both p53-mediated inhibition of cell cycling and induction of apoptosis appear to be highly dependent upon the cell context. Induction of p53 has also been proposed to regulate passage of cells through the G2 phase of the cell cycle, chromosomal stability, and mitosis, although the precise molecular mechanisms used by p53 to perform these regulatory functions are less clear.
Given the multiple roles of p53 in controlling cell cycling and cell death, proper regulation of p53 activity is obviously crucial to permit cells to undergo normal growth and to respond appropriately to stress. Regulation of cellular p53 activity has been reported to occur at the level of p53 transcription and translation (19, 49, 63). However, it is clear that Mdm2-mediated alteration of p53 protein stability is a major pathway used by many cells to govern p53 function. The Mdm2 oncoprotein binds to the amino-terminal portion of the p53 protein (11, 48), induces p53 monoubiquitination and nuclear export (35, 39), and targets p53 for proteosomal degradation (23, 34). The importance of Mdm2 as a negative regulator of p53 activity in cell growth and development has been demonstrated in mice by rescue of the embryo-lethal phenotype of Mdm2-deficient mice by deletion of p53 (28, 45).
Numerous in vitro studies have determined that p53 is subject to posttranslational modification. Phosphorylation occurs in response to DNA damage on human p53 serine residues 15 and 20 (mouse Ser18 and Ser23, respectively) and has been found in several in vitro studies to upregulate p53 activity (15, 18, 36, 43, 60) and interfere with Mdm2-mediated degradation of p53 (55).
The ATM (mutated in ataxia telangiectasia) kinase has been demonstrated to phosphorylate human p53 serine 15 (3, 8, 33, 51). Furthermore, both human p53Ser15 phosphorylation (8, 46, 56) and total p53 protein levels (31, 32, 46) are reduced in ATM-null cells following ionizing radiation treatment. These observations suggest that ATM-mediated phosphorylation of this residue increases the stability of p53. Expression of inactive forms of the ataxia telangiectasia-related kinase ATR also interferes with p53Ser15 phosphorylation (47, 58, 65). In addition, mouse embryonic fibroblasts (MEFs) deficient in both ATM and ATR fail to undergo G1 arrest in response to DNA damage and display greatly reduced phosphorylation of p53Ser18, the mouse homologue of human p53Ser15 (5). These data suggest that ATM and ATR functions may depend, in part, upon the ability of these related kinases to phosphorylate p53 at this serine residue.
Recently mouse p53Ser18 was mutated in embryonic stem (ES) cells, and the p53 apoptotic response and cell cycle arrest following DNA damage of these cells were delayed (10). However, p53-mediated pathways of cell cycle inhibition and apoptosis are not fully functional in ES cells (1). Thus, the relationship between p53Ser18 phosphorylation and p53 stability and function remains unclear.
In order to directly address the role of p53Ser15 phosphorylation in regulating p53 levels and p53 function in a genetically defined in vivo system, we used gene targeting in mouse ES cells to generate mice bearing p53 alleles that contain a p53Ser18 to Ala18 substitution. Our results indicate that the p53-dependent apoptosis induced by ionizing radiation is significantly reduced in the thymocytes and splenocytes of p53Ala18/Ala18 mice. Therefore, phosphorylation of p53Ser18 regulates p53-dependent apoptosis. However, phosphorylation of this residue does not alter the stability of the p53 protein. Wild-type mice and p53Ala18 mice display similar basal p53 levels and similar induced levels of p53 following DNA damage. Surprisingly, phosphorylation of p53Ser18 does not affect the rate of cell proliferation of embryonic fibroblasts. In addition, loss of Ser18 does not alter the p53-dependent G1 arrest of cell growth following DNA damage. Thus, the G1 arrest induced by ATM and ATR following DNA damage is not mediated by p53Ser18 phosphorylation. Furthermore, the reduced apoptotic activity of p53Ala18 does not rescue the embryonic lethality of Mdm2-null mice and does not alter p53-mediated tumor suppression. These results indicate that phosphorylation of the ATM/ATR target serine 18 regulates p53 apoptotic functions and not p53-mediated cell growth arrest and that phosphorylation of this site is not required for p53 suppression of spontaneous tumorigenesis.
MATERIALS AND METHODS
Generation of targeted ES cells.
The targeting vector was generated with a portion of the p53 gene (129SVBrd-strain) spanning a BamHI fragment 9.9 kb from intron 1 and intron 6 of the p53 gene. Serine 18 in exon 2 was mutated by oligonucleotide site-directed mutagenesis (QuickChange kit; Stratagene) of a subfragment of the targeting vector. The oligonucleotides used were HKS1Ala18 (5′-GGATATCAGCCTCGAACTCCCTCTGGCCCAGGAGACATTTTCAGG-3′) and HKS2BAla (5′-CCTGAAAATGTCTCCTGGGCCAGAGGGAGTTCGAGGCTGATATCC-3′). The subfragment was DNA sequenced to confirm the mutation and subcloned back into the targeting vector. A neor cassette gene flanked by loxP sites was introduced in an AvrII site, resulting in loss of the AvrII site. An MC1-thymidine kinase gene was added to the 3′ end of the targeting vector.
Homologous recombination and generation of p53Ala18 mice.
PC3 ES cells (26) were electroporated with the targeting vector and drug selected with G418 and 1-(2′-deoxy-2′-fluoro-β-d-arabinofuranosyl-5-iodouracil (FIAU). A total of 600 ES clones were screened, and two correctly targeted clones were identified by Southern analysis of EcoRI-digested genomic DNA with 5′ and 3′ probes external to the targeting vector. The presence of the alanine mutation in the ES clones was confirmed by PCR with oligonucleotide primers to intron1 forward (1F; 5′-GACAAGTTATGCATCATAC-3′) and intron 3 reverse (3R; 5′-CAAAGCCCAAGTCCCTTTC-3′) and subsequent digestion of the PCR fragments with XhoI. The wild-type PCR product was digested to yield two fragments, 365 bp and 70 bp. Cells were microinjected into embryonic day 3.5 blastocysts (C57BL/6 strain), and the embryos were surgically implanted into pseudopregnant foster mice by standard procedures. Excision of the neor cassette in F1 offspring of male chimeric mice was confirmed by digestion with AvrII and Southern analysis with a p53 exon 7 probe.
Research involving mice complied with all relevant federal and institutional policies, as well as guidelines established by the Institutional Animal Care and Use Committee at the University of Massachusetts Medical School.
Cell culture, proliferation, and immortalization assays.
Murine embryonic fibroblasts (MEFs) were generated from day 13.5 embryos from the various genotypes and maintained in Dulbecco's modified Eagle's medium supplement with 10% fetal bovine serum, penicillin, and streptomycin. Cells were maintained at subconfluency, and low-passage MEFs (passages 2 to 4) were used for all subsequent assays. Growth curves were performed with triplicate plating of wild-type, p53−/−, p53Ala18/+ and p53Ala18/Ala18 homozygous MEFs. Cells were plated (5 × 105 cells/10-cm dish) and counted every 24 h with a Beckman Coulter counter. Proliferation assays were performed with two independent lines of each genotype. 3T3 assays were performed with multiple plates of each genotype to examine rates of spontaneous immortalization as described previously (22).
DNA damage and cell cycle analysis.
Replicative DNA synthesis was quantified by bivariate flow cytometry. In brief, cells were irradiated with 2, 5, or 8 Gy. Cells were irradiated with a cesium 137 source (Gammacell 40). Cells were pulsed with bromodeoxyuridine (60 μM) from15 to 18 h postirradiation and harvested by trypsinization and fixation in 70% ethanol. Fixed cells were washed and incubated in 4 N HCl for 30 min at room temperature. The HCl was neutralized by addition of 0.1 M borax. Cells were incubated with a fluorescein isothiocyanate-conjugated antibromodeoxyuridine monoclonal antibody (Becton Dickinson), washed, and resuspended in phosphate-buffered saline containing 50 μg of RNase per ml and 20 μg of propidium iodide per ml. Fluorescence from propidium iodide complexes and fluorescein isothiocyanate-conjugated bromodeoxyuridine was measured by flow cytometry. Similar numbers of cells were analyzed for each sample, and cell cycle distribution was determined with FlowJo software.
Protein analysis and antibodies.
Following treatment, cells were lysed in EBC buffer (120 mM NaCl, 50 mM Tris-HCl, pH 8.0, 1.0% NP-40, 0.1% sodium dodecyl sulfate, 1 mM EDTA) with a cocktail of protease and phosphatase inhibitors; 100 μg of cell extract were run on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel. The p53 antibody (Ab-7 kit [Oncogene Research Products], diluted 1:1,000), the phospho-p53 (Ser 15) (Cell Signaling 9284S, diluted 1:1,000), the Mdm2 antibody (Santa Cruz sc-812, diluted 1:500), the p21 (Santa Cruz sc-397, diluted 1:500), the α-tubulin antibody (Sigma, diluted 1:10,000), and the Puma antibody (Abcham Limited, diluted 1:1,000) were obtained from the indicated suppliers. Immune complexes were detected by enhanced chemiluminescence (ECL kit, Amersham).
Thymocyte and splenocyte studies.
Thymocytes and splenic cells were removed from 4- to 6-week-old mice. Thymuses were removed and ground up with a syringe tip. Cells were washed with phosphate-buffered saline-1% serum solution, and resuspended at 106 cells/ml in phosphate-buffered saline-1% serum. For T-cell population profiling, whole animals were irradiated with 8 Gy. Thymocytes were recovered from untreated and irradiated animals and stained with CD4-phycoerythrin and CD8-fluorescein isothiocyanate antibodies (Pharmingen). Thymocytes were then washed and fixed in 1% paraformaldehyde and subjected to fluorescence-activated cell sorting analysis. For sub-G1 DNA analysis, whole animals were irradiated with 8 Gy. Thymocytes were removed 8 h postirradiation from irradiated and control unirradiated animals. Thymocytes were fixed in 70% ethanol. Sub-G1 DNA content was determined by fluorescence-activated cell sorting analysis.
Viability analysis was performed on in vitro-cultured thymocytes and thymocytes derived from animals irradiated in vivo. For viability analysis, thymocytes were stained with the annexin V-fluorescein isothiocyanate apoptosis kit (Oncogene Research Products). Viability was determined as the population negative for annexin V/7-amino actinomycin D (7AAD) relative to unirradiated control animals. Spleens were dissected and recovered as described (57). Splenocytes were plated (2 × 105) in RPMI supplemented with 10% serum. Cells were irradiated with 8 Gy. Viable cells were counted by trypan blue exclusion 7 days post-ionizing radiation treatment.
RESULTS
Generation of p53Ala18 mice.
To analyze the effect of p53 serine 18 phosphorylation on p53 functions, a gene targeting experiment was performed in PC3 ES cells (129Sv-Brd strain). A gene replacement vector containing strain 129 mouse p53 genomic DNA was generated that encoded a serine to alanine substitution at amino acid position 18 (Fig. 1A, D) in exon 2 of the coding sequences. This substitution also deleted an XhoI restriction endonuclease site in exon 2 DNA sequences without altering the other amino acids encoded in exon 2. This vector also contained a neomycin drug resistance marker flanked by loxP sites inserted in intron 4 of the p53 gene and a herpes simplex virus thymidine kinase gene downstream of the 3′ region of homology. Following positive (G418) and negative (FIAU) drug selection of the ES cells, 600 surviving clones were screened by Southern analysis, and 3 clones were identified that had undergone targeting of the p53 allele.
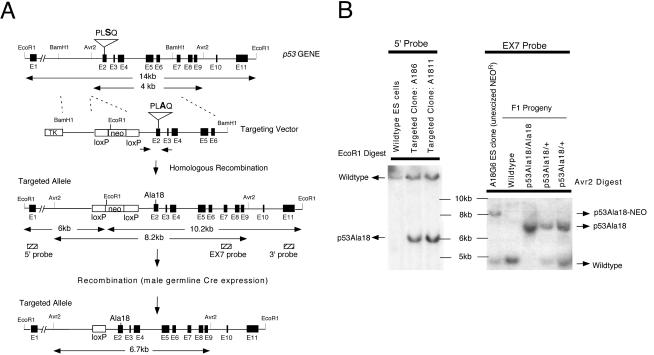
FIG. 1.
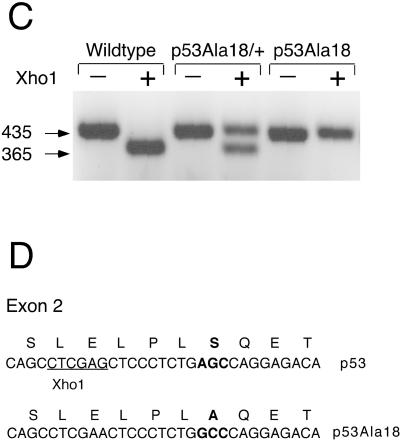
Mutation of serine 18 in the p53 gene. (A) The targeting vector encodes a serine to alanine missense mutation in exon 2. An XhoI restriction site was also deleted in this exon as a diagnostic marker. A floxed neor gene was introduced in intron 1 to permit positive selection of ES cells. Homologous recombination of the targeting vector in ES cells is illustrated. The neo gene will be excised in the sperm of chimeric mice due to expression of a protomine-Cre transgene present in the PC3 ES cells. The p53Ala18 allele is shown at the bottom. Probes used for Southern blot analysis are shown (hatched boxes). (B) Southern blot analysis of targeted ES cell DNA with a 5′ external probe (left panel). EcoRI digestionof ES cell DNA indicates the presence of the mutant allele (unexcised neor gene, 6 kb) and the wild-type allele (14 kb). Southern blot analysis of F1 progeny with EX7 probe (right panel). An AvrII digest distinguishes the wild-type allele (4 kb), unexcised mutant allele (8.2 kb), and excised mutant allele (6.7 kb). (C) The presence of the mutation was determined by PCR amplification of exon 2 and loss of the XhoI restriction site. (D) Diagram of the mutation generated in exon 2 of p53. Serine18 was mutated to an alanine. An adjacent XhoI restriction site was deleted for diagnostic purposes.
Detailed analysis of these three clones with 5′ and 3′ probes external to the targeting vector confirmed proper targeting of the locus (Fig. 1B). Two of these clones (A18-6 and A18-11) were used to perform blastocyst injection experiments, and both clones gave rise to multiple high-degree chimeras. Male chimeras were crossed with C57BL/6 mice to obtain agouti F1 offspring. Analysis of genomic DNA isolated from tail biopsies of these mice confirmed that a subset of the mice had inherited the targeted allele and had excised the neomycin drug selection marker due to expression of the protamine-Cre transgene (endogenous in PC3 ES cells, Fig. 1B). The presence of the mutation encoding the Ala18 substitution was confirmed in a subset of F1 mice by DNA sequencing and PCR analysis (Fig. 1C). Inheritance of the p53Ala18 allele was subsequently followed in the colony by use of a PCR strategy that used the newly deleted XhoI site in exon 2 adjacent to the Ala18 mutation. Mice heterozygous for the p53Ala18 allele were intercrossed, and mice homozygous for the mutant allele were recovered at the expected frequency of 25%. Mice heterozygous or homozygous for the p53Ala18 allele developed normally. Large cohorts of p53Ala18/+ and p53Ala18/Ala18 mice were generated for further analysis.
In vivo analysis of apoptosis in p53Ala18/Ala18 cells.
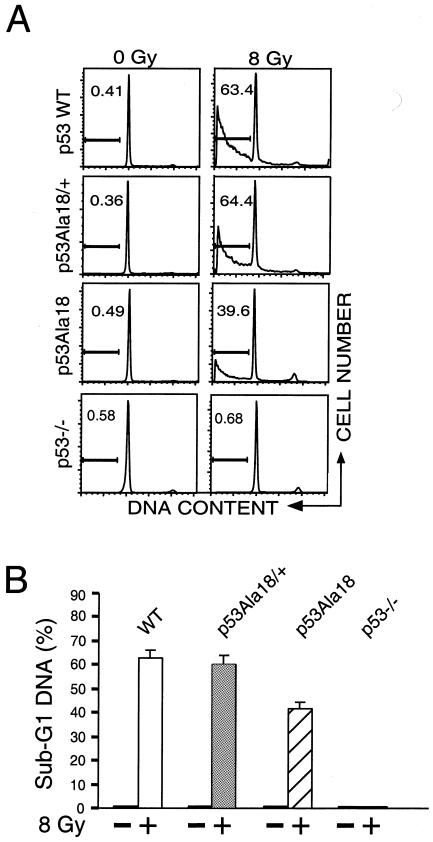
Mouse thymocytes are induced to undergo p53-dependent apoptosis following ionizing radiation treatment (12, 40). To determine the contribution of p53 ser18 phosphorylation to p53 apoptosis, we compared apoptosis in thymocytes of p53Ala18/Ala18 mice and in wild-type mice and p53-null mice following exposure to DNA damage. Thymocytes were recovered from nontreated mice and from treated mice at 8 h after whole-body exposure of the mice to 8 Gy of ionizing radiation. The thymocytes were analyzed for apoptosis by analyzing sub-G1 DNA content (Fig. 2A). Following DNA damage, a similar increase in the percentage of apoptotic cells was observed in wild-type thymocytes and in thymocytes bearing one copy of the p53Ala18 allele, suggesting that the presence of the p53Ala18 allele was not exerting a dominant negative effect on wild-type p53 (Fig. 2A). However, mice homozygous for p53Ala18 displayed significant reductions in the percentage of thymocytes undergoing apoptosis following DNA damage (Fig. 2B), indicating that phosphorylation of p53 serine 18 does alter p53-mediated apoptosis in vivo.
FIG. 2.
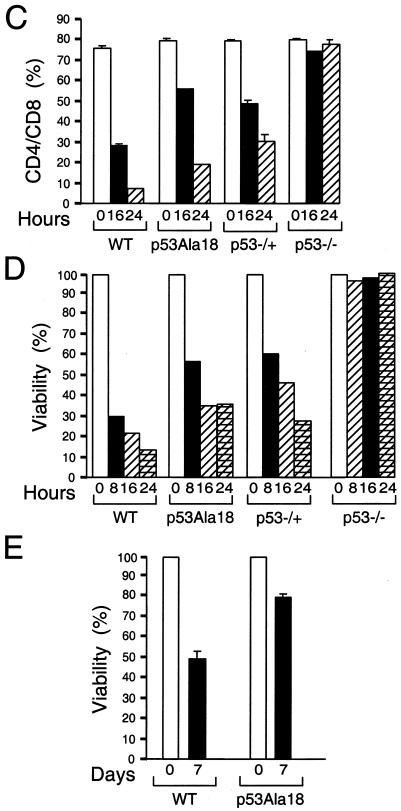
p53-mediated apoptosis is defective in p53Ala18 mice. (A) Analysis of sub-G1 content of thymocytes removed from irradiated and unirradiated animals (8 Gy, 8 h posttreatment). The y axis depicts cell number, and the x axis represents DNA content. The percentage of sub-G1 cells is given for each sample. WT, wild type. (B) Histograph of sub-G1 DNA from irradiated and nonirradiated mice. Two independent animals were analyzed in triplicate for each genotype. (C) Time course of CD4+ CD8+ profile of mice irradiated (8 Gy) in vivo and nonirradiated mice. The graph depicts double-positive cells. (D) Viability of thymocytes over time in response to ionizing radiation. Thymocytes were removed from mice, irradiated (8 Gy), and harvested at various times (0, 8, 16, and 24 h). Cells were stained with annexin V-fluorescein isothiocyanate, anti-CD4+, and 7AAD. Values given are the averages of the viable population (annexin V-fluorescein isothiocyanate and 7AAD negative) and are normalized to the number of viable cells in the untreated animal for each genotype. (E) Apoptosis in splenocytes is reduced in p53Ala18 mice. Splenocytes were prepared and plated (2 × 105 cells). Cells were irradiated with 8 Gy. Cell viability was determined 7 days post-ionizing radiation. The x axis represents time (days), and the y axis represents viability relative to unirradiated control cells.
CD4/CD8 thymocytes undergo p53-dependent apoptosis following ionizing radiation damage (40). Mice were irradiated in vivo with 8 Gy, and a time course of CD4/CD8 staining was performed post-ionizing radiation with multiple p53Ala18/Ala18, wild-type, and p53-deficient mice. A significant depletion of CD4/CD8 thymocytes was observed in p53Ala18/Ala18 mice (Fig. 2C). p53Ala18/Ala18 mice most closely resembled the phenotype of p53-heterozygous mice. The fraction of surviving thymocytes was also measured at multiple time points postirradiation by staining cells for annexin V and 7AAD (Fig. 2D). Annexin V (an early marker for apoptosis)-negative and 7AAD (a DNA intercalator)-negative populations were determined and compared to untreated thymocytes. The reduced viability of thymocytes in p53Ala18/ala18 mice confirmed that these cells were compromised in p53-dependent apoptosis.
To investigate if p53 apoptosis is altered in a different cell context, splenocytes were also harvested and analyzed for the percentage of cells undergoing p53-mediated apoptosis following whole-body ionizing radiation treatment (Fig. 2F). Similar to the results obtained with thymocytes, the p53Ala18/Ala18 splenocytes showed increased viability relative to wild-type splenocytes, indicating that p53-mediated apoptosis was also compromised in these cells.
Analysis of the role of p53 serine 18 phosphorylation in p53 stability.
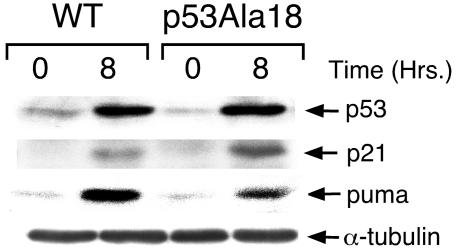
Phosphorylation of serine 18 has been proposed to inhibit Mdm2-p53 complex formation (55), leading to the stabilization of p53 levels by preventing Mdm2 from targeting p53 for ubiquitination and proteosomal degradation. Therefore, reduction in p53Ala18/Ala18 thymocyte apoptosis might reflect differences in the stabilization of p53 levels by phosphorylation. To explore this possibility, we examined the basal level of p53 and DNA damage-induced levels of p53 in wild-type and p53Ala18/Ala18 thymocytes. As expected, p53 was strongly induced in these cells 8 h after whole-body irradiation of mice (Fig. 3). However, no difference was detected between the basal p53 levels or the p53 levels induced by ionizing radiation in wild-type and p53Ala18/Ala18 thymocytes. In addition, while expression of p53-responsive genes such as p21 is upregulated in thymocytes following ionizing radiation, no defect was observed in the p21 induction in p53Ala18/Ala18 thymocytes. In contrast, induction of the proapoptotic gene PUMA was decreased in p53Ala18/Ala18 cells relative to p53 wild-type cells. PUMA has recently been shown to be the principal mediator of p53-dependent death in thymocytes (25).
FIG. 3.
Mutation of p53Ser18 does not affect basal or ionizing radiation-induced levels of p53 protein. Western analysis of extracts from irradiated (20 Gy) and nonirradiated thymocytes. Extracts were prepared at 0 h (nonirradiated) or 8 h after ionizing radiation treatment. α-Tubulin was used as a control for the Western blot. WT, wild type.
Analysis of the growth characteristics of p53Ala18/Ala18 embryonic fibroblasts.
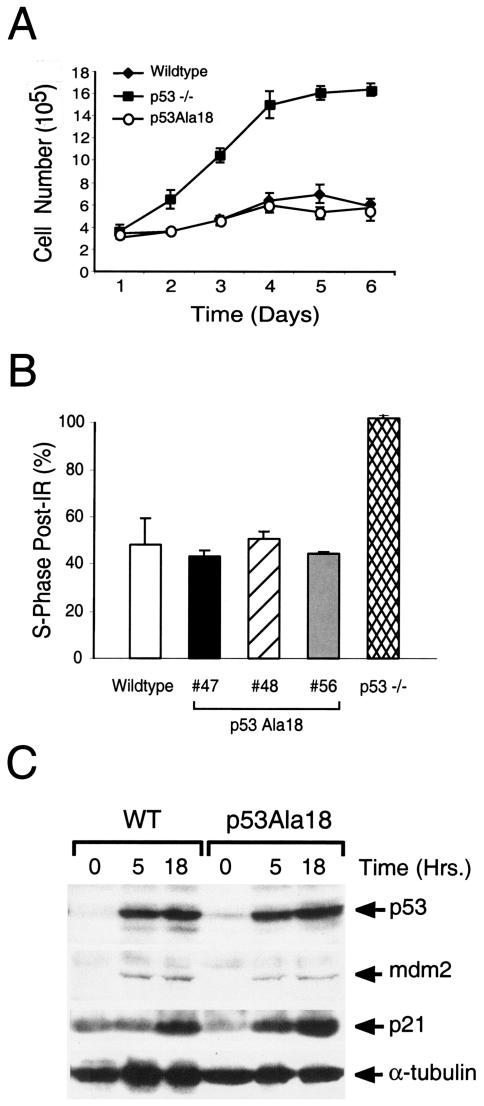
The rate of embryonic fibroblast proliferation (22, 29) and cell cycle arrest in response to ionizing radiation damage (31) is p53 dependent. To explore a role for p53 serine 18 phosphorylation in p53-mediated control of cell growth, we generated murine embryonic fibroblasts (MEFs) from wild-type, p53Ala18/Ala18, and p53-deficient embryos (day 13.5). Studies on bromodeoxyuridine uptake by these cells revealed no difference between wild-type and p53Ala18/Ala18 MEFs (data not shown). The rate of cell proliferation was indistinguishable between these two cell populations (Fig. 4A).
FIG. 4.
(A) Proliferation of wild-type, p53Ala18, and p53-null MEFs. (B) Histograph of percent of MEFs in S phase (treated or untreated). MEFs were treated with ionizing radiation and pulsed with bromodeoxyuridine 15 to 18 h postirradiation. Three independent lines of p53Ala18 MEFs were analyzed. (C) Western analysis of extracts from irradiated (8 Gy) and nonirradiated wild-type MEFs and p53Ala18 MEFs harvested at 5 and 18 h post-ionizing radiation treatment. α-Tubulin was used as a control for the Western blot.
To determine if p53Ser18 phosphorylation alters p53-mediated cell cycling, we analyzed the p53-dependent G1 arrest post-ionizing radiation. We treated cells with 8 Gy of ionizing radiation and analyzed cell cycle profiles at 18 h postirradiation by fluorescence-activated cell sorting analysis. Three lines of p53Ala18/Ala18 MEFs generated from different mice were analyzed. Similar to wild-type MEFs, p53Ala18/Ala18 MEFs displayed a large loss of S phase following ionizing radiation treatment, with values indistinguishable from wild-type cells (Fig. 4B).
Analysis of protein levels revealed no difference between the basal and ionizing radiation-induced amounts of p53 in wild-type and p53Ala18/Ala18 MEFs, and while Mdm2 and p21 levels were both increased by treatment of the cells with ionizing radiation, the levels of Mdm2 and p21 were similar in wild-type and p53Ala18/Ala18 MEFs (Fig. 4C). These data indicate that phosphorylation does not alter the level of p53 protein in MEFs and is not required for induction of p21.
Immortalization of p53Ala18/Ala18 MEFs.
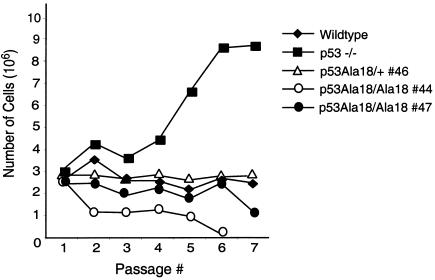
Spontaneous immortalization of embryonic fibroblasts is also dependent upon p53 function, and p53-null MEFs display rapid immortalization in a 3T3 assay, whereas wild-type MEFs do not immortalize or show delayed immortalization in these assays (22). To assess the contribution of p53 serine 18 phosphorylation to immortalization, a modified 3T3 assay was performed with multiple plates of two individual lines of p53Ala18/Ala18 MEFs and one line of p53Ala18/+ MEFs (Fig. 5A). Whereas p53-null MEFs immortalized after passage 4 in the assay and continued to grow rapidly, p53Ala18/Ala18 MEFs stopped dividing by passage 8 and failed to spontaneously immortalize.
FIG. 5.
Immortalization is not induced in p53Ala18 MEFs. 3T3 analysis of two independent p53Ala18/Ala18 MEFs compared to p53Ala18/+, MEFs, and p53-null MEFs.
p53Ala18 allele fails to rescue Mdm2-null mice.
As p53 levels are unchanged by the presence of the p53Ala18 mutation, it is unlikely that phosphorylation alters Mdm2-p53 complex formation. We previously demonstrated the importance of Mdm2-mediated downregulation of p53 activity in early development: Mdm2-null mice die between day 5 and 6 of development but are rescued by deletion of p53 (28, 45). It is presently unclear which p53 functions are inhibited by Mdm2 during embryogenesis. As loss of p21 fails to rescue Mdm2-nullizygous mice, it has been proposed that unregulated p53 apoptosis is responsible for the early demise of the Mdm2-null mice (44). However, the paucity of tissue in the Mdm2 null embryos makes it difficult to determine the precise p53 functions regulated by Mdm2 in this early stage of development. In order to explore whether reduced apoptotic functions of p53Ala18 would rescue Mdm2-null mice, we crossed Mdm2+/− mice with p53Ala18 mice as well as intercrossed Mdm2+/− p53Ala18/Ala18 mice. Genotyping of the Mdm2 and p53Ala18 status of the resulting pups determined that, unlike p53−/−, p53Ala18/Ala18 does not rescue the embryonic lethality of Mdm2-null mice (Table 1).
TABLE 1.
p53Ala18 does not rescue the embryonic lethality of Mdm2-null mice
| Cross | Genotype | No. of offspring with genotype:
|
|
|---|---|---|---|
| Expecteda | Obtained | ||
| Mdm2+/− p53Ala18/Ala18 × Mdm2+/− p53Ala18/Ala18 (n = 15) | Mdm2+/+ p53Ala18/Ala18 | 4 | 3 |
| Mdm2+/− p53Ala18/Ala18 | 8 | 12 | |
| Mdm2−/− p53Ala18/Ala18 | 4 | 0 | |
| Mdm2+/− p53Ala18/+ × Mdm2+/− p53Ala18/+ (n = 96) | Mdm2+/+ p53+/+ | 7 | 18 |
| Mdm2+/+ p53Ala18/+ | 15 | 15 | |
| Mdm2+/+ p53Ala18/Ala18 | 7 | 4 | |
| Mdm2+/− p53+/+ | 15 | 25 | |
| Mdm2+/− p53Ala18/+ | 30 | 24 | |
| Mdm2+/− p53Ala18/Ala18 | 15 | 8 | |
| Mdm2−/− p53+/+ | 0 (lethal) | 0 | |
| Mdm2−/− p53Ala18/+ | 0 (lethal) | 0 | |
| Mdm2−/− p53Ala18/Ala18 | 7 | 0 | |
Calculated assuming p53Ala18/Ala18 rescues Mdm2 deficiency, rounded to nearest whole number.
Absence of spontaneous tumorigenesis in p53Ala18 mice.
To determine if phosphorylation of p53 serine 18 is required for tumor suppression, cohorts of p53Ala18/Ala18 mice were collected and assayed for spontaneous tumor formation. At present, we have 30 p53Ala18/Ala18 mice that are between 40 and 60 weeks of age. Although 100% of p53-null mice and 25% of p53-heterozygous mice (21) succumbed to cancer by 40 weeks of age, we have had no incidence of tumorigenesis in any of our p53Ala18/Ala18 mice, indicating that the reduced apoptotic activity of the mutant p53Ala18 does not alter p53 tumor suppression.
DISCUSSION
It is becoming increasingly apparent from numerous in vitro experiments that p53 is highly posttranslationally modified by phosphorylation. Several studies have demonstrated that p53 serine 18 is phosphorylated in vitro by the ATM and ATR kinases (3, 8, 33, 46, 51, 58). Analysis of p53 protein levels and p53 phosphorylation in ATM-deficient cells has suggested that phosphorylation of this residue by ATM helps regulate the stability of p53 following DNA damage (3, 8, 32, 56). In addition, it has been suggested that modification of this serine regulates p53 stability by altering Mdm2-p53 interactions (55). However, the role of serine phosphorylation in p53 stability has been placed in question by the results of other transient transfection studies with a mutant p53 that lacks all amino-terminal phosphorylation sites (2).
In order to better understand the role of p53 serine 18 phosphorylation in regulating p53 stability and functions, we generated mice bearing a missense mutation (p53Ser18 to Ala18) and analyzed p53 functions in p53Ala18 mice and in primary cells derived from these mice. Our in vivo analysis of p53Ala18 thymocyte and splenocyte apoptosis revealed a clear reduction in p53 apoptotic activity when p53Ser18 was incapable of being phosphorylated. This result is in agreement with a previous report of diminished p53 apoptosis in p53Ala18 ES cells (10) and suggests that phosphorylation of this residue by ATM and ATR contributes to the p53 apoptotic response.
While this article was being prepared for submission, an independent group reported a defect in cell cycle arrest and an apoptotic defect in a p53Ala18 mouse model (9). In contrast, the ability of p53 to regulate cell proliferation and to invoke cell cycle arrest following DNA damage was unaltered in the p53Ala18 MEFs, indicating that modification of this residue does not affect the ability of p53 to regulate cell growth. Thus, apoptosis and cell growth arrest are p53 functions that are distinctly regulated by p53Ser18 phosphorylation.
Interestingly, the amount of p53 protein in wild-type and p53Ala18 thymocytes and MEFs was similar under basal and DNA damaging conditions, indicating that Mdm2 is properly regulating the stability of p53Ala18 in these cells. In addition, p53Ala18 homozygosity failed to rescue Mdm2 deficiency during embryogenesis. Since the absence of Mdm2 is lethal to both wild-type p53 and p53Ala18/Ala18 embryos, Mdm2 must still govern p53Ala18 activity in development. Thus, phosphorylation of serine 18 does not alter Mdm2 regulation of p53 stability in these cells.
The cyclin-dependent kinase inhibitor p21 is a major regulator of the G1 to S transition of the cell cycle and has been shown previously to be upregulated by p53 (6, 13, 16, 20). Cell growth and ionizing radiation-induced G1 arrest was not compromised in p53Ala18 cells in vivo or in MEFs. In agreement with these findings, the levels of p21 were not reduced below wild-type levels in the p53Ala18 MEFs or thymocytes. Apoptosis was compromised in p53Ala18 cells, and the proapoptotic PUMA protein was found to be downregulated in p53Ala18 thymocytes. As p53 protein levels do not differ in p53Ser18 and p53Ala18 cells, these results indicate that phosphorylation of p53Ala18 likely regulates the ability of p53 to transactivate apoptotic genes.
The growth rate of mouse embryonic fibroblasts is significantly increased following deletion of a single p53 allele (22), and p53 haploinsufficiency is sufficient to predispose mice to spontaneous tumorigenesis without loss of heterozygosity for functional p53 in tumors (61). These results indicate that even a partial reduction in p53 activity can alter cell growth in some context and interfere with p53-mediated tumor suppression. However, the reduced levels of p53 apoptotic function in the p53Ala18 mice still induced the embryonic demise of Mdm2-null mice and prevented tumorigenesis in p53Ala18/Ala18 mice. These findings indicate either that cell cycle arrest is an important component of p53-mediated tumor suppression and regulation of development or that p53-induced apoptosis is not rate limiting in these situations.
The role of ATM in cell growth and tumor suppression has been well established (4, 17, 57, 64, 68). The ATM and the related ATR kinase have been demonstrated to regulate p53 activity. In vitro studies have provided evidence suggesting that ATM/ATR-induced phosphorylation of p53 amino-terminal serines regulates p53 function. In addition to phosphorylating p53Ser18, ATM has been proposed to influence Chk2-mediated phosphorylation of p53Ser20 (p53Ser23 in the mouse) following DNA damage (24, 54, 59). However, p53 stability and p53 transactivation of gene expression were found recently to be unaltered in thymocytes and MEFs derived from chimeric mice generated by using p53Ala23 ES cells (67). These findings suggest that p53Ser18 must be the key regulatory target of ATM/ATR phosphorylation. Although p53 apoptosis was partially compromised in our p53Ala18 model, it is clear from our data that ATM and ATR must rely upon targets other than serine 18 to fully regulate p53 activity in response to DNA damage and to suppress tumor formation.
Acknowledgments
We thank Marilyn Keeler and Wynne Morgan for technical help and the UMASS Medical School fluorescence-activated cell sorting facility for their assistance. We are grateful to Steve O'Gorman and Geoffrey Wahl for providing the PC3 ES cells, Sundaresan Venkatachalam for providing plasmids containing p53 genomic DNA fragments, and Ron Wisdom for providing pBABE-HRas. We also thank the members of the Kowalik laboratory and the Kelliher laboratory for useful discussion and Roger Davis for comments on the manuscript and help with timed experiments.
Core facilities used to perform some of these experiments were supported by Program Project grant 5P30DK32520 from the National Institute of Diabetes and Digestive and Kidney Diseases. This research was supported by an NRSA Postdoctoral Fellowship Award to H.K.S. and Public Health Service grant CA-077735 from the National Cancer Institute to S.N.J.
REFERENCES
- 1.Aladjem, M. I., B. T. Spike, L. W. Rodewald, T. J. Hope, M. Klemm, R. Jaenisch, and G. M. Wahl. 1998. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr Biol. 8:145-155. [DOI] [PubMed] [Google Scholar]
- 2.Ashcroft, M., M. H. Kubbutat, and K. H. Vousden. 1999. Regulation of p53 function and stability by phosphorylation. Mol. Cell. Biol. 19:1751-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banin, S., L. Moyal, S. Shieh, Y. Taya, C. W. Anderson, L. Chessa, N. I. Smorodinsky, C. Prives, Y. Reiss, Y. Shiloh, and Y. Ziv. 1998. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science 281:1674-1677. [DOI] [PubMed] [Google Scholar]
- 4.Barlow, C., K. D. Brown, C. X. Deng, D. A. Tagle, and A. Wynshaw-Boris. 1997. Atm selectively regulates distinct p53-dependent cell-cycle checkpoint and apoptotic pathways. Nat. Genet. 17:453-456. [DOI] [PubMed] [Google Scholar]
- 5.Brown, E. J., and D. Baltimore. 2003. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 17:615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brugarolas, J., C. Chandrasekaran, J. I. Gordon, D. Beach, T. Jacks, and G. J. Hannon. 1995. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 377:552-557. [DOI] [PubMed] [Google Scholar]
- 7.Burns, T. F., E. J. Bernhard, and W. El-Deiry. 2001. Tissue specific expression of p53 target genes suggests a key role for KILLER/DR5 in p53-dependent apoptosis in vivo. Oncogene 34:4601-4612. [DOI] [PubMed] [Google Scholar]
- 8.Canman, C. E., D. S. Lim, K. A. Cimprich, Y. Taya, K. Tamai, K. Sakaguchi, E. Appella, M. B. Kastan, and J. D. Siliciano. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281:1677-1679. [DOI] [PubMed] [Google Scholar]
- 9.Chao, C., M. Hergenhahn, M. D. Kaeser, Z. Wu, S. Saito, R. Iggo, M. Hollstein, E. Apella, and Y. Xu. 2003. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J. Biol. Chem. 42:41028-41033. [DOI] [PubMed] [Google Scholar]
- 10.Chao, C., S. Saito, C. W. Anderson, E. Appella, and Y. Xu. 2000. Phosphorylation of murine p53 at ser-18 regulates the p53 responses to DNA damage. Proc. Natl. Acad. Sci. USA 97:11936-11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, J., V. Marechal, and A. J. Levine. 1993. Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol. 13:4107-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clarke, A. R., C. A. Purdie, D. J. Harrison, R. G. Morris, C. C. Bird, M. L. Hooper, and A. H. Wyllie. 1993. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature 362:849-852. [DOI] [PubMed] [Google Scholar]
- 13.Deng, C., P. Zhang, J. W. Harper, S. J. Elledge, and P. Leder. 1995. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82:675-684. [DOI] [PubMed] [Google Scholar]
- 14.Donehower, L. A., M. Harvey, B. L. Slagle, M. J. McArthur, C. A. Montgomery, Jr., J. S. Butel, and A. Bradley. 1992. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature 356:215-221. [DOI] [PubMed] [Google Scholar]
- 15.Dumaz, N., and D. W. Meek. 1999. Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 18:7002-7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El-Deiry, W. S., T. Tokino, V. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, E. Mercer, K. Kinxler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 17.Elson, A., Y. Wang, C. J. Daugherty, C. C. Morton, F. Zhou, J. Campos-Torres, and P. Leder. 1996. Pleiotropic defects in ataxia-telangiectasia protein-deficient mice. Proc. Natl. Acad. Sci. USA 93:13084-13089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiscella, M., S. J. Ullrich, N. Zambrano, M. T. Shields, D. Lin, S. P. Lees-Miller, C. W. Anderson, W. E. Mercer, and E. Appella. 1993. Mutation of the serine 15 phosphorylation site of human p53 reduces the ability of p53 to inhibit cell cycle progression. Oncogene 8:1519-1528. [PubMed] [Google Scholar]
- 19.Fu, L., M. D. Minden, and S. Benchimol. 1996. Translational regulation of human p53 gene expression. EMBO J. 15:4392-4401. [PMC free article] [PubMed] [Google Scholar]
- 20.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 21.Harvey, M., M. J. McArthur, C. A. Montgomery, J. Butel, A. Bradley, and L. Donehower. 1993. Spontanous and carcinogen-induced tumorigenesis in p53-deficient mice. Nat. Genet. 5:225-229. [DOI] [PubMed] [Google Scholar]
- 22.Harvey, M., A. T. Sands, R. S. Weiss, M. E. Hegi, R. W. Wiseman, P. Pantazis, B. C. Giovanella, M. A. Tainsky, A. Bradley, and L. A. Donehower. 1993. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene 8:2457-2467. [PubMed] [Google Scholar]
- 23.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 24.Hirao, A., Y. Y. Kong, S. Matsuoka, A. Wakeham, J. Ruland, H. Yoshida, D. Liu, S. J. Elledge, and T. W. Mak. 2000. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 287:1824-1827. [DOI] [PubMed] [Google Scholar]
- 25.Jeffers, J. R., E. Parganas, Y. Lee, C. Yang, J. Wang, J. Brennan, K. H. MacLean, J. Han, T. Chittenden, J. N. Ihle, P. J. McKinnon, J. L. Cleveland, and G. P. Zambetti. 2003. Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 4:321-328. [DOI] [PubMed] [Google Scholar]
- 26.Jimenez, G. S., M. Nister, J. M. Stommel, M. Beeche, E. A. Barcarse, X. Q. Zhang, S. O'Gorman, and G. M. Wahl. 2000. A transactivation-deficient mouse model provides insights into Trp53 regulation and function. Nat. Genet. 26:37-43. [DOI] [PubMed] [Google Scholar]
- 27.Jones, S. N., A. R. Hancock, H. Vogel, L. A. Donehower, and A. Bradley. 1998. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc. Natl. Acad. Sci. USA 95:15608-15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones, S. N., A. E. Roe, L. A. Donehower, and A. Bradley. 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378:206-208. [DOI] [PubMed] [Google Scholar]
- 29.Jones, S. N., A. T. Sands, A. R. Hancock, H. Vogel, L. A. Donehower, S. P. Linke, G. M. Wahl, and A. Bradley. 1996. The tumorigenic potential and cell growth characteristics of p53-deficient cells are equivalent in the presence or absence of Mdm2. Proc. Natl. Acad. Sci. USA 93:14106-14111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamijo, T., F. Zindy, M. F. Roussel, D. E. Quelle, J. R. Downing, R. A. Ashmun, G. Grosveld, and C. J. Sherr. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91:649-659. [DOI] [PubMed] [Google Scholar]
- 31.Kastan, M. B., Q. Zhan, W. S. el-Deiry, F. Carrier, T. Jacks, W. V. Walsh, B. S. Plunkett, B. Vogelstein, and A. J. Fornace, Jr. 1992. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 71:587-597. [DOI] [PubMed] [Google Scholar]
- 32.Khanna, K. K., H. Beamish, J. Yan, K. Hobson, R. Williams, I. Dunn, and M. F. Lavin. 1995. Nature of G1/S cell cycle checkpoint defect in ataxia-telangiectasia. Oncogene 11:609-618. [PubMed] [Google Scholar]
- 33.Khanna, K. K., K. E. Keating, S. Kozlov, S. Scott, M. Gatei, K. Hobson, Y. Taya, B. Gabrielli, D. Chan, S. P. Lees-Miller, and M. F. Lavin. 1998. ATM associates with and phosphorylates p53: mapping the region of interaction. Nat. Genet. 20:398-400. [DOI] [PubMed] [Google Scholar]
- 34.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 35.Lai, Z., K. V. Ferry, M. A. Diamond, K. E. Wee, Y. B. Kim, J. Ma, T. Yang, P. A. Benfield, R. A. Copeland, and K. R. Auger. 2001. Human mdm2 mediates multiple mono-ubiquitination of p53 by a mechanism requiring enzyme isomerization. J. Biol. Chem. 276:31357-31367. [DOI] [PubMed] [Google Scholar]
- 36.Lambert, P. F., F. Kashanchi, M. F. Radonovich, R. Shiekhattar, and J. N. Brady. 1998. Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 273:33048-33053. [DOI] [PubMed] [Google Scholar]
- 37.Lin, A. W., and S. W. Lowe. 2001. Oncogenic ras activates the ARF-p53 pathway to suppress epithelial cell transformation. Proc. Natl. Acad. Sci. USA 98:5025-5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Linke, S. P., K. C. Clarkin, A. Di Leonardo, A. Tsou, and G. Wahl. 1996. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 10:934-947. [DOI] [PubMed] [Google Scholar]
- 39.Lohrum, M. A., D. B. Woods, R. L. Ludwig, E. Balint, and K. H. Vousden. 2001. C-terminal ubiquitination of p53 contributes to nuclear export. Mol. Cell. Biol. 21:8521-8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowe, S. W., E. M. Schmitt, S. W. Smith, B. A. Osborne, and T. Jacks. 1993. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362:847-849. [DOI] [PubMed] [Google Scholar]
- 41.Marchenko, N. D., A. Zaika, and U. M. Moll. 2000. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J. Biol. Chem. 275:16202-16212. [DOI] [PubMed] [Google Scholar]
- 42.Martin-Caballero, J., J. M. Flores, P. Garcia-Palencia, and M. Serrano. 2001. Tumor susceptibility of p21(Waf1/Cip1)-deficient mice. Cancer Res. 61:6234-6238. [PubMed] [Google Scholar]
- 43.Mayr, G. A., M. Reed, P. Wang, Y. Wang, J. F. Schweds, and P. Tegtmeyer. 1995. Serine phosphorylation in the NH2 terminus of p53 facilitates transactivation. Cancer Res. 55:2410-2417. [PubMed] [Google Scholar]
- 44.Montes de Oca Luna, R., L. L. Amelse, A. Chavez-Reyes, S. C. Evans, J. Brugarolas, T. Jacks, and G. Lozano. 1997. Deletion of p21 cannot substitute for p53 loss in rescue of mdm2 null lethality. Nat. Genet. 16:336-337. [DOI] [PubMed] [Google Scholar]
- 45.Montes de Oca Luna, R., D. S. Wagner, and G. Lozano. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378:203-206. [DOI] [PubMed] [Google Scholar]
- 46.Nakagawa, K., Y. Taya, K. Tamai, and M. Yamaizumi. 1999. Requirement of ATM in phosphorylation of the human p53 protein at serine 15 following DNA double-strand breaks. Mol. Cell. Biol. 19:2828-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nghiem, P., P. K. Park, Y. S. Kim Ys, B. N. Desai, and S. L. Schreiber. 2002. ATR is not required for p53 activation but synergizes with p53 in the replication checkpoint. J. Biol. Chem. 277:4428-4434. [DOI] [PubMed] [Google Scholar]
- 48.Oliner, J. D., J. A. Pietenpol, S. Thiagalingam, J. Gyuris, K. W. Kinzler, and B. Vogelstein. 1993. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 362:857-860. [DOI] [PubMed] [Google Scholar]
- 49.Raman, V., S. A. Martensen, D. Reisman, E. Evron, W. F. Odenwald, E. Jaffee, J. Marks, and S. Sukumar. 2000. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature 405:974-978. [DOI] [PubMed] [Google Scholar]
- 50.Ries, S., C. Biederer, D. Woods, O. Shifman, S. Shirasawa, T. Sasazuki, M. McMahon, M. Oren, and F. McCormick. 2000. Opposing effects of Ras on p53: transcriptional activation of mdm2 and induction of p19ARF. Cell 103:321-330. [DOI] [PubMed] [Google Scholar]
- 51.Saito, S., A. A. Goodarzi, Y. Hagashimoto, Y. Noda, S. P. Lees-Miller, E. Appella, and C. W. Anderson. 2002. ATM mediates phosphorylation at multiple p53 sites, including Ser46, in response to ionizing radiation. J. Biol. Chem. 277:12491-12494. [DOI] [PubMed] [Google Scholar]
- 52.Schuler, M., E. Bossy-Wetzel, J. C. Goldstein, P. Fitzgerald, and D. R. Green. 2000. p53 induces apoptosis by caspase activation through mitochondrial cytochrome c release. J. Biol. Chem. 275:7337-7342. [DOI] [PubMed] [Google Scholar]
- 53.Sherr, C. J., and F. McCormick. 2002. The Rb and p53 pathways in cancer. Cancer Cell 2:103-112. [DOI] [PubMed] [Google Scholar]
- 54.Shieh, S. Y., J. Ahn, K. Tamai, Y. Taya, and C. Prives. 2000. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 14:289-300. [PMC free article] [PubMed] [Google Scholar]
- 55.Shieh, S. Y., M. Ikeda, Y. Taya, and C. Prives. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325-334. [DOI] [PubMed] [Google Scholar]
- 56.Siliciano, J. D., C. E. Canman, Y. Taya, K. Sakaguchi, E. Appella, and M. B. Kastan. 1997. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 11:3471-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spring, K., F. Ahangari, S. P. Scott, P. Waring, D. M. Purdie, P. C. Chen, K. Hourigan, J. Ramsay, P. J. McKinnon, M. Swift, and M. F. Lavin. 2002. Mice heterozygous for mutation in Atm, the gene involved in ataxia-telangiectasia, have heightened susceptibility to cancer. Nat. Genet. 32:185-190. [DOI] [PubMed] [Google Scholar]
- 58.Tibbetts, R. S., K. M. Brumbaugh, J. M. Williams, J. N. Sarkaria, W. A. Cliby, S. Y. Shieh, Y. Taya, C. Prives, and R. T. Abraham. 1999. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 13:152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tominaga, K., H. Morisaki, Y. Kaneko, A. Fujimoto, T. Tanaka, M. Ohtsubo, M. Hirai, H. Okayama, K. Ikeda, and M. Nakanishi. 1999. Role of human Cds1 (Chk2) kinase in DNA damage checkpoint and its regulation by p53. J. Biol. Chem. 274:31463-31467. [DOI] [PubMed] [Google Scholar]
- 60.Unger, T., R. V. Sionov, E. Moallem, C. L. Yee, P. M. Howley, M. Oren, and Y. Haupt. 1999. Mutations in serines 15 and 20 of human p53 impair its apoptotic activity. Oncogene 18:3205-3212. [DOI] [PubMed] [Google Scholar]
- 61.Venkatachalam, S., Y. P. Shi, S. N. Jones, H. Vogel, A. Bradley, D. Pinkel, and L. A. Donehower. 1998. Retention of wild-type p53 in tumors from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. EMBO J. 17:4657-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vousden, K. H. 2002. Activation of the p53 tumor suppressor protein. Biochim. Biophys. Acta 1602:47-59. [DOI] [PubMed] [Google Scholar]
- 63.Webster, G. A., and N. D. Perkins. 1999. Transcriptional cross talk between NF-κB and p53. Mol. Cell. Biol. 19:3485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Westphal, C. H., C. Schmaltz, S. Rowan, A. Elson, D. E. Fisher, and P. Leder. 1997. Genetic interactions between atm and p53 influence cellular proliferation and irradiation-induced cell cycle checkpoints. Cancer Res. 57:1664-1667. [PubMed] [Google Scholar]
- 65.Wright, J. A., K. S. Keegan, D. R. Herendeen, N. J. Bentley, A. M. Carr, M. F. Hoekstra, and P. Concannon. 1998. Protein kinase mutants of human ATR increase sensitivity to UV and ionizing radiation and abrogate cell cycle checkpoint control. Proc. Natl. Acad. Sci. USA 95:7445-7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu, G. S., T. F. Burns, E. R. McDonald, 3rd, W. Jiang, R. Meng, I. D. Krantz, G. Kao, D. D. Gan, J. Y. Zhou, R. Muschel, S. R. Hamilton, N. B. Spinner, S. Markowitz, G. Wu, and W. S. el-Deiry. 1997. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat. Genet 17:141-143. [DOI] [PubMed] [Google Scholar]
- 67.Wu, Z., J. Earle, S. Saito, C. W. Anderson, E. Appella, and Y. Xu. 2002. Mutation of mouse p53 Ser23 and the response to DNA damage. Mol. Cell. Biol. 22:2441-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xie, G., R. C. Habbersett, Y. Jia, S. R. Peterson, B. E. Lehnert, E. M. Bradbury, and J. A. D'Anna. 1998. Requirements for p53 and the ATM gene product in the regulation of G1/S and S phase checkpoints. Oncogene 16:721-736. [DOI] [PubMed] [Google Scholar]
- 69.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]