Abstract
The Arf tumor suppressor inhibits cell cycle progression through both p53-dependent and p53-independent mechanisms, including interference with rRNA processing. Using tandem-affinity-tagged p19Arf, we purified Arf-associated proteins from mouse NIH 3T3 fibroblasts undergoing cell cycle arrest. Tagged p19Arf associated with nucleolar and ribosomal proteins, including nucleophosmin/B23 (NPM), a protein thought to foster the maturation of preribosomal particles. NPM is an abundant protein, only a minor fraction of which binds to p19Arf; however, a significant proportion of p19Arf associates with NPM. The interaction between p19Arf and NPM requires amino acid sequences at the Arf amino terminus, which are also required for Mdm2 binding, as well as the central acidic domain of NPM and an adjacent segment that regulates NPM oligomerization. The interaction between p19Arf and NPM occurs in primary mouse embryonic fibroblasts, including those lacking both Mdm2 and p53. In an NIH 3T3 derivative cell line (MT-Arf) engineered to conditionally express an Arf transgene, induced p19Arf associates with NPM and colocalizes with it in high-molecular-weight complexes (2 to 5 MDa). An NPM mutant lacking its carboxyl-terminal nucleic acid-binding domain oligomerizes with endogenous NPM, inhibits p19Arf from entering into 2- to 5-MDa particles, and overrides the ability of p19Arf to retard rRNA processing.
The Arf tumor suppressor protein binds to the p53 negative regulator Mdm2 to induce a p53 transcriptional response that can lead to either cell cycle arrest or apoptosis (37). Arf can also arrest the proliferation of Mdm2- and p53-null fibroblasts, albeit relatively inefficiently, indicating that p19Arf can interact with other targets (48). Arf is an unusually basic protein (pI > 12) that is highly rich in arginine residues. Apart from Mdm2, a series of proteins have been reported to physically interact with Arf, including the E2F-1 and HIF-1α transcription factors (7, 9, 22), the phosphatase-binding protein spinophilin (46), the peroxisomal protein Pex19P (40), topoisomerase I (18), and cyclin G1 (52), although the biological significance of these interactions, if any, remains unclear. Enforced Arf expression antagonizes the transcriptional activity of NF-κB in a p53- and Mdm2-independent manner (35). In the latter case, direct interactions between p19Arf and NF-κB were not documented. Indeed, Arf can induce many antiproliferative genes and repress others, even in cells that lack functional p53 (19).
The vast majority of Arf protein expressed in cells localizes within nucleoli (30, 33, 50), an organelle responsible for ribosome biogenesis. Ribosomal proteins synthesized in the cytoplasm are imported into nucleoli, where they assemble with rRNA precursors to form preribosomal particles that undergo a series of maturation steps before they are exported to the nucleus and then the cytoplasm. Precursor rRNA transcribed by polymerase I within the nucleolus (47S in mammalian cells) assembles into 90S preribosomal particles, after which several rapid rRNA cleavage steps separate the precursors into large and small ribonucleoprotein subunits, whose maturation continues within the nucleolus (6, 8, 24, 47). rRNA transcription and processing, ribosome assembly, maturation, and transport require hundreds of accessory proteins and small nucleolar RNAs not found within mature ribosomes in the cytoplasm (1, 8, 14, 24, 39, 47), and the functions of most of these regulatory molecules remain obscure.
Expression of p19Arf in nucleoli suggested that it might play a role in ribosome biogenesis, and consistent with this idea, enforced Arf expression was revealed to retard two steps of rRNA processing (41). Specifically, p19Arf slows the processing of early 47S/45S rRNA precursors and also inhibits cleavage of a 32S rRNA intermediate that contains the segmented sequence modules encoding both 28S and 5.8S rRNAs. These effects do not depend on Mdm2 or p53 or on Arf's ability to induce cell cycle arrest per se.
Most of the p19Arf protein expressed in mouse fibroblasts is contained in complexes of very high molecular mass (2 to 5 MDa), consistent with the idea that Arf might enter early preribosomal particles within the nucleolus. We show that p19Arf interacts with several nucleolar proteins implicated in ribosome biogenesis. Among these is an acidic protein, nucleophosmin/B23 (NPM), which oligomerizes with itself and with p19Arf to form complexes that can affect rRNA processing.
MATERIALS AND METHODS
Cells and culture conditions.
NIH 3T3 fibroblasts and MT-Arf cells expressing a zinc-inducible Arf gene (19) were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mM glutamine, and 100 units each of penicillin and streptomycin. Where indicated, MT-Arf cells were treated with 80 μM ZnSO4 for 24 h to induce p19Arf. Mouse embryo fibroblasts (MEFs) from wild-type C57BL/6 mice (17) and from a C57BL/6 × 129 strain lacking the Arf, p53, and Mdm2 genes (48) were cultured in complete medium supplemented with 0.1 mM nonessential amino acids, 55 μM 2-mercaptoethanol, and 10 μg of gentamicin per ml instead of penicillin and streptomycin.
Retrovirus production and infection.
Human kidney 293T cells were transfected as described with helper retrovirus plasmid together with pSRα vectors encoding wild-type or mutant p19Arf proteins (49) or with murine stem cell virus (MSCV)-internal ribosome entry site (IRES)-green fluorescent protein (GFP) vectors encoding tandem affinity purification (TAP)-tagged Arf or Flag-tagged wild-type or mutant human NPM proteins (53) (also see below). Viruses were harvested 24 to 60 h posttransfection, pooled, and stored on ice. Exponentially growing cells (2 × 105 cells per 10-cm-diameter culture dish) were infected twice at 3-h intervals with 2 ml of fresh virus-containing supernatant in complete medium containing 8 μg of Polybrene (Sigma, St. Louis, Mo.) per ml. For large-scale purifications with TAP-tagged Arf, 106 cells in 15-cm-diameter culture dishes were infected once with 7 ml of virus-containing culture supernatant. Where applicable, infection efficiencies were confirmed by flow cytometric analysis of GFP-stained cells.
Tandem affinity purification of tagged Arf protein.
A TAP tag cDNA cassette consisting of two staphylococcal protein A-immunoglobulin G (IgG) binding domains fused to a tobacco etch virus protease site and a calmodulin-binding domain allows sequential affinity binding of TAP-tagged proteins to an IgG column, their recovery by tobacco etch virus protease digestion, and their further purification via Ca2+-dependent calmodulin binding (34). The TAP tag cassette (a kind gift from Kathy Gould, Vanderbilt University School of Medicine, Nashville, Tenn.) was amplified by PCR and inserted as an EcoRI/XhoI fragment into the cloning sites of the MSCV-IRES-GFP retroviral vector. Full-length, N-terminally hemagglutinin (HA)-tagged Arf cDNA (33) was subcloned into the EcoRI site of the derivative plasmid, fusing the TAP tag sequence in frame with the Arf 3′ end. Deletion mutants lacking sequences encoding N-terminal amino acids 2 to 14 (Δ2-14) or 2 to 62 (Δ2-62) (50), each including the HA tag at the N terminus, were subcloned in place of full-length Arf in the same retroviral expression vector.
Cells infected with expression vectors encoding TAP-tagged p19Arf proteins were harvested by trypsinization, neutralized with complete medium, and washed twice in phosphate-buffered saline (PBS). Harvested cells were divided into aliquots containing 2 × 107 cells, pelleted at 1,000 × g, snap frozen in liquid nitrogen, and stored at −80°C until used. Cell pellets were resuspended on ice, each in 1 ml of Tween 20 lysis buffer [50 mM HEPES (pH 7.5), 150 mM NaCl, 1 mM EDTA, 2.5 mM EGTA, 0.1% Tween 20, 1 mM phenylmethylsulfonyl fluoride, 0.2 U of aprotinin per ml, 10 mM β-glycerophosphate, 1 mM NaF, and 0.1 mM NaVO4], sonicated in 2-ml batches twice for 7 s each (at 14% power with a Virsonic 475 sonicator) with a 30-s pause between bursts, and then left on ice for 30 min. Lysates were centrifuged at 14,000 × g at 4°C for 15 min, and the resulting supernatants were filtered through 0.45-μm cellulose-acetate filters to remove insoluble material.
In some experiments, lysates were incubated at 30°C for 10 min with or without 100 μg of RNase A per ml, and insoluble material was again removed. Following protein quantitation with the BCA assay (Pierce, Rockford, Ill.), equal quantities of different protein preparations were diluted to 8 ml in Tween 20 lysis buffer. Tandem affinity purification was performed essentially as described by Puig et al. (31) except that Tween 20 lysis buffer was used to equilibrate and wash the IgG column. Following elution from the second calmodulin column, proteins were concentrated by trichloroacetic acid and deoxycholate precipitation (21) and stored as pellets at −80°C until analyzed.
Identification of TAP-tagged Arf-containing complexes.
Arf-containing complexes were separated on 4 to 12% gradient Bis-Tris-polyacrylamide gels containing sodium dodecyl sulfate. Silver staining was performed by the European Molecular Biology Laboratory method as described (23) except that 0.04% formaldehyde was used in place of 0.04% formalin during development. For Coomassie blue staining, gels were washed twice for 5 min each in water, fixed in 10% methanol-10% glacial acetic acid for 30 min, washed in 30% methanol-10% glacial acetic acid for 20 min, and then incubated in Coomassie blue stain solution (Bio-Rad, Hercules, Calif.) for at least 1 h. Gels stained with Coomassie blue were destained in 30% methanol-10% glacial acetic acid. In some cases, proteins electroblotted onto polyvinylidene fluoride membranes (Millipore, Billerica, Mass.) were stained as per the manufacturer's instructions with Sypro Ruby (Molecular Probes) and visualized with a blue filter on a Storm 860 imager (Amersham Biosciences, Piscataway, N.J.). Proteins stained with Coomassie blue were recovered from gels and analyzed by mass spectrometry exactly as described (25).
Gel filtration chromatography.
Cell lysates were prepared as for TAP tag purification except that neither EDTA nor EGTA was used in the lysis buffer. Lysates were diluted to 2 mg/ml, and 0.5 ml was injected onto a Superose-6 HR 10/30 gel filtration column (Amersham Biosciences). The column was resolved with 50 mM sodium phosphate (pH 7.0) containing 150 mM NaCl by fast protein liquid chromatography with a Bio-Rad BioLogic HR system. Fractions of 1 ml were collected, concentrated by trichloroacetic acid-deoxycholate precipitation, and stored at −80°C until analyzed. Standards of known molecular mass were used to calibrate the column (Bio-Rad).
Generation of NPM mutants.
Full-length human NPM cDNA was provided by Steve Morris (St. Jude Children's Research Hospital). Deletion mutants were generated by PCR with the following PCR primers: sense primers for C1, C2, and C3 (5′-AAAGAATTCGAAGATTCGATGGACATGGAC); antisense primer for C1 (5′-AAACTCGAGTTATTTTTCTATACTTGCTTG); antisense primer for C2 (5′-AAACTCGAGTTAAGATTTCTTCACTGGCGC); antisense primer for C3 (5′-AAACTCGAGTTACACAGCTACTAAGTGCTG); sense primer for N2 (5′-AAAGAATTCGAGGAAGATGCAGAGTCAGAA); sense primer for N3 (5′-AAAGAATTCCCAGTGAAGAAATCTATACGA); and antisense primer for N2 and N3 (5′-AAACTCGAGTTAAAGAGACTTCCTCCACTG). PCR products were digested with EcoRI and XhoI, purified, and cloned into pcDNA3 or MSCV-IRES-GFP vectors containing N-terminal Flag tag coding sequences.
Immunofluorescence.
Cells seeded on coverslips were fixed with methanol-acetone (1:1) for 20 min at −20°C and stained with affinity-purified antibodies to the p19Arf C terminus (33) for 1 h at room temperature. Washed coverslips were then incubated with biotinylated antibodies to rabbit IgG and streptavidin-conjugated Texas Red (both from Amersham Biosciences) (49, 50). To detect the Flag-tagged NPM, cells were fixed with 10% formalin-PBS and stained with M2 anti-Flag antibodies (Sigma Chemicals). For measurement of DNA synthesis, cells were labeled for 1 h with 10 μM 5-bromodeoxyuridine (Sigma) in complete medium, fixed with methanol-acetone, and stained with anti-bromodeoxyuridine plus nuclease (Amersham Biosciences), followed by fluorescein isothiocyanate-conjugated antibodies to mouse IgG as described (49).
Immunoprecipitation, immunoblotting, and RNA end labeling.
Cells were suspended in NET2 buffer (50 mM Tris HCl, pH 7.4, containing 150 mM NaCl and 0.05% Nonidet P-40) and sonicated three times for 30 s. Lysates cleared by sedimentation in a microcentrifuge at 14,000 rpm for 10 min were precipitated for 3 h at 4°C with anti-NPM (Zymed, South San Francisco, Calif.), M2 anti-Flag (Sigma Chemicals), or antibodies to p19Arf. Immune complexes were recovered with protein G- (Zymed) or protein A-Sepharose (Amersham Biosciences) and washed four times with NET2 buffer. Immunoprecipitated complexes and total cell lysates were electrophoretically separated on denaturing polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Millipore). Proteins were detected with antibodies to p19Arf (33), p53 (Ab-7, Oncogene Research Products, San Diego, Calif.), p21Cip1 (C-19, Santa Cruz Biotechnology, Santa Cruz, Calif.), NPM (C-19 Santa Cruz), Flag tag (M2, Sigma Chemicals), or ribosomal protein L7 (Novus Biologicals, NB200-308).
For analysis of coprecipitating RNAs, immune complexes were suspended and incubated for 30 min at 37°C in 7 mM Tris HCl, pH 7.5, containing 0.7 mM EDTA, 20 mM NaCl, 0.7% sodium dodecyl sulfate, and 30 μg of proteinase K per ml (Sigma Chemicals). RNAs were recovered by phenol-chloroform extraction followed by ethanol precipitation in 0.3 M sodium acetate containing 20 μg of glycogen (Roche, Basel, Switzerland), and end-labeled with 40 μCi of [32P]cytidine 3′, 5′-bis(phosphate) ([32]pCp) (3,000 Ci/mmol; ICN, Costa Mesa, Calif.) with T4 RNA ligase (Amersham Biosciences) as described (45).
Analysis of newly synthesized rRNA.
Cells were metabolically labeled with 2.5 μCi of [3H]uridine per ml (30 to 60 Ci/mmol, Amersham Biosciences) in complete medium for 30 min, washed twice with PBS, and incubated in complete medium without the radioactive precursor for 2 h. Total RNAs were isolated with Trizol (Invitrogen, Carlsbad, Calif.), and quantitated by absorbance measurement, and their specific activities were determined by liquid scintillation. RNAs (2,000 cpm per lane) loaded into 1% agarose gels containing formaldehyde were electrophoretically separated and transferred to Hybond N+ membranes (Amersham). Dried membranes were treated with En3Hance (Perkin Elmer Life Sciences, Boston, Mass.) and subjected to autoradiography at −70°C with intensifying screens.
RESULTS
Arf associates with proteins involved in ribosome biogenesis.
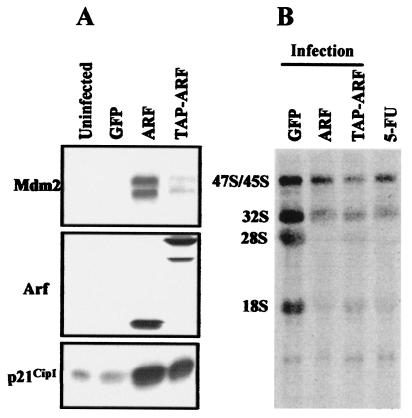
In an effort to identify proteins that associate with p19Arf in growth-arrested cells, we infected NIH 3T3 fibroblasts with a retrovirus expressing p19Arf fused to a tandem affinity purification (TAP) epitope tag (34). Expression of TAP-tagged p19Arf induced p53-responsive genes (Mdm2 and p21Cip1) (Fig. 1A), caused cell cycle arrest (reduction of S-phase fraction from 23% to 4%), and inhibited rRNA processing (Fig. 1B), indicating that the fusion protein remained biologically active. Two days after infection, arrested cells were lysed, and TAP-tagged Arf complexes were purified from 33 mg of protein recovered from ≈2 × 108 cells by use of a modified version of the tandem affinity purification protocol (31, 34). As a control for nonspecific binding, the same amount of protein recovered from uninfected NIH 3T3 cells was carried through the same purification procedure.
FIG. 1.
TAP-tagged-Arf is functional. NIH 3T3 cells were infected with MSCV-IRES-GFP retroviruses expressing Arf, TAP-tagged-Arf, or GFP alone as a negative control. Two days later, cell lysates were immunoblotted with antibodies to Arf, Mdm2, and p21Cip1 (A). Like p19Arf, although to a lesser extent, TAP-tagged Arf induced expression of Mdm2 and p21Cip1. Due to the C-terminal tag, the electrophoretic migration of TAP-tagged Arf on the denaturing gel was retarded. (B) Cells were pulse labeled with [3H]uridine for 30 min and then chased for 2.5 h. Total RNA extracted from cells was separated on a gel, transferred to a membrane, and subjected to fluororadiography. RNA in the 5-FU lane was extracted from uninfected NIH 3T3 cells which were exposed to 5-fluorouridine for 20 min prior to metabolic labeling. Expression of Arf and TAP-tagged Arf and inclusion of 5-fluorouridine in the medium all inhibited rRNA processing.
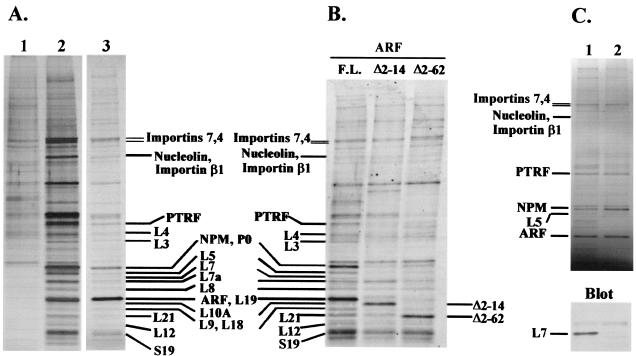
Recovered proteins concentrated by trichloroacetic acid-deoxycholate precipitation were electrophoretically resolved in denaturing 4 to 12% gradient polyacrylamide gels (Fig. 2A). Small portions of purified, gel-separated proteins were stained with silver (sensitivity, ≈1 ng of protein per band) to pinpoint bands that were specifically recovered with TAP-tagged p19Arf. The bulk of the protein (80%) was resolved on gels separately and stained with Coomassie blue (sensitivity, ≈100 ng of protein per band), and specific bands were excised, digested with trypsin, and sequenced by mass spectrometry. Peptides from TAP-tagged p19Arf (GPHYLLPPGAR and KNFIAVSAANR) and 28 other proteins were identified. Of these, 13 were large ribosomal subunit proteins, one was a small ribosomal subunit protein, three were nonribosomal proteins involved in ribosome biogenesis, including nucleolin, nucleophosmin (NPM), and polymerase I and transcript release factor (PTRF), and three were importins (Fig. 2A and Table 1). The fact that many of these proteins localize to the nucleolus gave us some confidence that TAP-tagged p19Arf did not bind indiscriminately to proteins normally resident in the nucleoplasm or cytoplasm.
FIG. 2.
Arf associates with proteins involved in ribosome biogenesis. (A) NIH 3T3 cells were infected with a retrovirus expressing TAP-tagged Arf and lysed 48 h later. Purifications were performed with lysates from cells expressing TAP-tagged Arf (lanes 2 and 3) or from uninfected cells (lane 1). TAP-tagged Arf complexes were electrophoretically resolved on denaturing gels and visualized with silver (lanes 1 and 2, 10% of purified protein loaded) or Coomassie blue (lane 3, 80% of purified protein loaded). Coomassie blue-stained bands were sequenced by mass spectrometry. (B) NIH 3T3 cells were infected with retroviruses expressing TAP-tagged full-length Arf, Arf Δ2-14, or Arf Δ2-62 and lysed 48 h later. Purifications were performed with the same amount of protein from each of the three cell lysates. Purified complexes were resolved on denaturing gels, blotted onto a polyvinylidene difluoride membrane, stained with Sypro Ruby, and visualized with a Storm imager with the blue filter. Bands corresponding to those in panel A are indicated. (C) Lysates were either left untreated (lane 1) or treated with RNase A (lane 2) prior to affinity purification from equal quantities of total protein. Recovered proteins were separated electrophoretically and stained with Coomassie blue. The lower panel shows the results of immunoblotting performed with antibodies to ribosomal protein L7.
TABLE 1.
Arf-associated proteins
| Type | Protein | Accession no. | Peptide(s) | Predicted size (kDa) | pI |
|---|---|---|---|---|---|
| Ribosomal subunits | RP P0 | gi13277927 | GHLENNPALEK | 34 | 5.9 |
| RP L3 | gi16307136 | NNASTDYDLSDK | 46 | 10.2 | |
| RP L4 | gi22001911 | KLDELYGTWR | 47 | 11.0 | |
| RP L5 | gi22002065 | HIMGQNVADYMR, YLMEEDEDAYKKRP | 34 | 9.8 | |
| RP L7 | gi31981515 | KAGNFYVPAEPK | 31 | 10.9 | |
| RP L7a | gi30410942 | TNYNDRYDEIR | 31 | 10.6 | |
| RP L8 | gi6755358 | ASGNYATVISHNPETK | 28 | 11.0 | |
| RP L9 | gi14149647 | KFLDGIYVSEK | 22 | 10.0 | |
| RP L10a | gi6755350 | KYDAFLASESLIK, FPSLLTHNENMVAK | 25 | 10.0 | |
| RP L12 | gi17390751 | HSGNITFDEIVNIAR | 18 | 9.5 | |
| RP L18 | gi25050299 | TNSTFNQVVLK | 22 | 11.8 | |
| RP L19 | gi6677773 | VNLDPNETNEIANANSR | 24 | 11.5 | |
| RP L21 | gi31560385 | VYNVTQHAVGIIVNK | 19 | 10.5 | |
| RP S19 | gi12963511 | RVLQALEGLK | 16 | 10.4 | |
| Ribosome biogenesis associated nonribosomal | Nucleolin | gi128843 | GLSEDTTEETLK | 77 | 4.7 |
| Nucleophosmin | gi6679108 | VDNDENEHQLSLR | 33 | 4.6 | |
| PTRF | gi6679567 | SFTPDHVVYAR | 44 | 5.4 | |
| Importins | Importin 7 | gi31581595 | KQLQDIATLADQR, ENIVEAIIJSPELIR, SDQNLQTALELTR, DGALHMIGSLAEILLK | 119 | 4.7 |
| Importin 4 | gi19745156 | YVRPDDVSLAR, SSSDPSSSPVLQTSLAR | 119 | 4.9 | |
| Importin β1 | gi31543051 | GDQENVHPDVMLVQPR | 97 | 4.7 |
PTRF binds to the 3′ end of pre-rRNA and is required for the termination of transcription by RNA polymerase I, allowing release of both the pre-rRNA transcript and the polymerase I complex from the rDNA template (16). Nucleolin binds to specific RNA stem-loops at several positions in pre-rRNA, and its interaction with one such structure within the 5′ external transcribed spacer sequence is required for the first step of ribosomal processing in vitro (10-12). NPM has been proposed to function in ribosome biogenesis, but its exact role remains unclear (28, 36, 51). Such data suggest that the ability of p19Arf to inhibit rRNA processing (41) may reflect its ability to associate with early preribosomes in the nucleolus.
Mutant forms of Arf lacking residues 2 to 62 (Δ2-62) or 2 to 14 (Δ2-14) are impaired in causing cell cycle arrest (32, 48, 49), and Arf Δ2-14, although localizing to nucleoli, is unable to efficiently inhibit rRNA processing (41). Further purifications were therefore conducted with TAP-tagged Arf Δ2-14 and Arf Δ2-62 in tandem with full-length TAP-tagged Arf in order to determine whether the mutant proteins would associate with nucleolin, PTRF, and NPM. A caveat is that Arf Δ2-14 is a hypomorphic mutant not completely devoid of activity (49). Purified proteins were again resolved on denaturing gels, but were blotted onto a polyvinylidene difluoride membrane and stained with Sypro Ruby to visualize proteins (Fig. 2B). Sypro Ruby is more sensitive than Coomassie blue, and unlike silver, it stains different proteins to the same degree. A band in the position of nucleolin was recovered with all three TAP-tagged Arf “baits,” but mass spectroscopy also identified another protein in this region (importin β1) that occluded our ability to detect quantitative differences in nucleolin itself. By contrast, several p19Arf-associated proteins, including NPM, PTRF, and ribosomal proteins, were seemingly absent or were recovered in significantly lower abundance after purification with mutant TAP-tagged Arf proteins.
To determine whether the association of TAP-tagged Arf with these interacting proteins depended upon the presence of RNA, NIH 3T3 cells were infected with the vector encoding TAP-tagged Arf, and detergent lysates were treated with RNase A before we performed tandem affinity purification. Following RNase treatment, some of the protein in the extracts, including about half of the TAP-tagged Arf, became insoluble and precipitated out of solution. Nonetheless, tandem affinity purification of the remaining protein again revealed recovery of the bands containing nucleolin, PTRF, and NPM at similar molar ratios to the TAP-tagged Arf protein (Fig. 2C, upper panel). By contrast, the intensities of bands corresponding to several ribosomal proteins (e.g., L7) were diminished, whereas others (e.g., L5) were not. By immunoblotting these samples with antibodies to ribosomal protein L7, we were able to confirm its markedly reduced recovery after RNase treatment (Fig. 2C, lower panel). Therefore, the interaction of TAP-tagged Arf with nucleolin, PTRF, NPM, and some ribosomal proteins does not appear to depend upon RNA. The experiments that followed focused on the NPM-Arf interaction.
Nucleophosmin associates both with p19Arf and 5.8S rRNA.
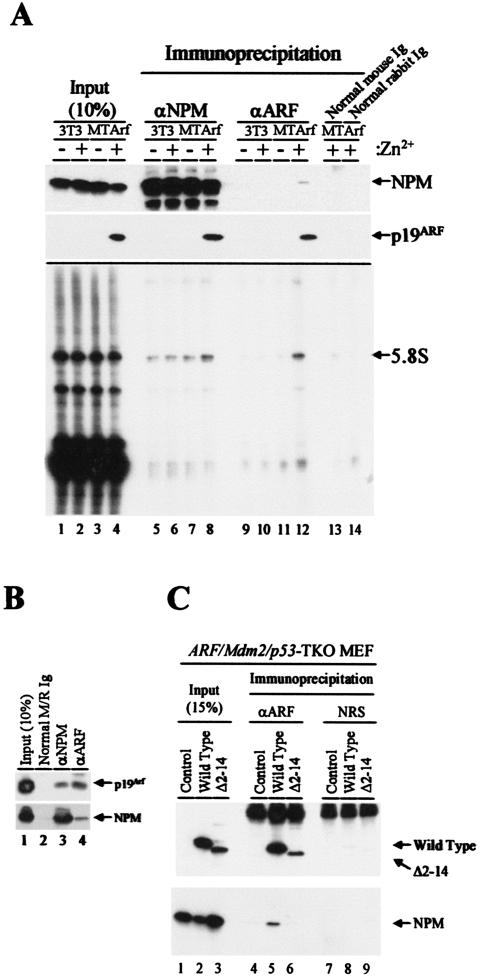
NIH 3T3 fibroblasts which have sustained a deletion of the endogenous Ink4a-Arf locus were previously engineered to express an Arf transgene under control of the zinc-inducible mouse metallothionein promoter (19). Antibody precipitates of NPM from lysates of these zinc-induced and growth-arrested MT-Arf cells contained p19Arf (Fig. 3A, top two panels, lane 8), whereas antibodies to p19Arf reciprocally coprecipitated NPM (lane 12). NPM is a relatively abundant nucleolar protein, so that under the conditions used, several sequential rounds of precipitation are required to recover it all. We estimated that only ≈30% of NPM was recovered in the initial antibody precipitation shown (compare input lanes 1 to 4 with immune precipitates in lanes 5 to 8). Still, >10% of the total p19Arf in the cells coprecipitated with NPM (lane 4 versus 8). Note that p19Arf was not efficiently precipitated with the cognate antibody from mammalian cells either. Only ≈10% of the total p19Arf pool was recovered by direct antibody precipitation in this experiment (Fig. 3A, lane 12 versus 4), indicating that the amount of NPM in complexes with Arf is also likely to be underestimated by this approach. Nonetheless, we could conclude that only a relatively small proportion of the total NPM pool is associated with p19Arf, whereas a significant proportion of p19Arf is bound to NPM.
FIG. 3.
Arf forms complexes with NPM. (A, upper two panels) Lysates prepared from NIH 3T3 and MT-Arf cells treated with zinc sulfate for 24 h (+) or not (−) were precipitated with antibodies to NPM (lanes 5 to 8), p19Arf (lanes 9 to 12), or control IgG (lanes 13 and 14). Denatured immune complexes electrophoretically separated on gels were transferred to a membrane and blotted with the same antibodies. Cell lysates (10%) separated on denaturing gels were directly blotted with antibodies to NPM and p19Arf in order to estimate levels of the expressed proteins (lanes 1 to 4; see text). (A, lower panel) RNAs eluted from immune complexes were labeled with [32P]pCp and T4 RNA ligase, separated on a denaturing gel, and detected by autoradiography. (B) Primary MEFs established from day 15 embryos were propagated on a 3T3 protocol for seven passages. Cell lysates expressing both p19Arf and NPM (lane 1) were precipitated with control antibodies (lane 2) or with antibodies to NPM or Arf (lanes 3 and 4). Proteins separated on denaturing gels were immunoblotted with the same antibodies, indicated at the right of the panel. (C) Arf/Mdm2/p53 triple knockout (TKO) MEFs infected with retroviruses encoding p19Arf or the p19Arf Δ2-14 mutant were lysed and precipitated with antibodies to p19Arf (lanes 4 to 6) or nonimmune rabbit serum (NRS) (lanes 7 to 9) and subjected to immunoblotting with antibodies to Arf (top panel) or NPM (bottom panel). A portion of the cell lysate was used to demonstrate levels of protein expression (lanes 1 to 3) as in panel A.
Because p19Arf coprecipitates with 5.8S rRNA (41), we next tested whether NPM can also associate with this RNA species. RNAs eluted from immunoprecipitated complexes were end labeled with T4 RNA ligase and separated on denaturing gels. As expected, 5.8S rRNA was recovered from p19Arf immunoprecipitates obtained from zinc-induced cells (Fig. 3A, bottom panel, lane 12), but not from control antibody precipitates (lanes 13 and 14). Under the same conditions, NPM coprecipitated with 5.8S rRNA whether or not Arf was induced (Fig. 3A, bottom panel, lanes 5 to 8), although somewhat more 5.8S rRNA was recovered when p19Arf was present (lane 8). Hence, both proteins can interact with rRNA, potentially within the same complex. Whether p19Arf or NPM can also associate with unprocessed 5.8S sequences found within rRNA precursors remains unclear (41).
To ensure that the Arf-NPM interaction observed in MT-Arf cells did not depend upon p19Arf overexpression, we next determined whether endogenous p19Arf and NPM could be coprecipitated from lysates of primary mouse embryo fibroblasts (MEFs). MEFs established from embryonic day 15 embryos were propagated in culture on a 3T3 protocol, a process during which p19Arf progressively accumulates as the proliferative capacity of the cells diminishes (17). Cells at passage 7 were lysed and precipitated with antibodies to NPM or p19Arf, and recovered immune complexes were denatured, separated on gels, and blotted with the same or reciprocal antibodies. Both proteins were again observed to coprecipitate (Fig. 3B, lanes 3 and 4), whereas control antibodies recovered neither protein (lane 2).
We next examined if the NPM-Arf interaction occurs independently of Mdm2 and p53. MEFs prepared from Arf/Mdm2/p53 triple knockout mice were infected with retroviral vectors encoding full-length Arf or Arf (Δ2-14) proteins. Four days after infection, cells expressing wild-type p19Arf ceased proliferating, but those infected with an empty control vector or with the one encoding mutant Arf Δ2-14 continued to divide (48) (data not shown). Arf-NPM complexes were then analyzed by sequential immunoprecipitation and immunoblotting (Fig. 3C). Endogenous NPM coprecipitated with the wild-type p19Arf protein (lane 5) but not with Arf Δ2-14 (lane 6). Since this Arf mutant does not bind to 5.8S rRNA either (41), physical interactions between Arf, NPM, and 5.8S rRNA correlate with p19Arf's ability to inhibit cell proliferation in a p53-independent manner.
NPM acidic and oligomerization domains are required for Arf binding in vivo.
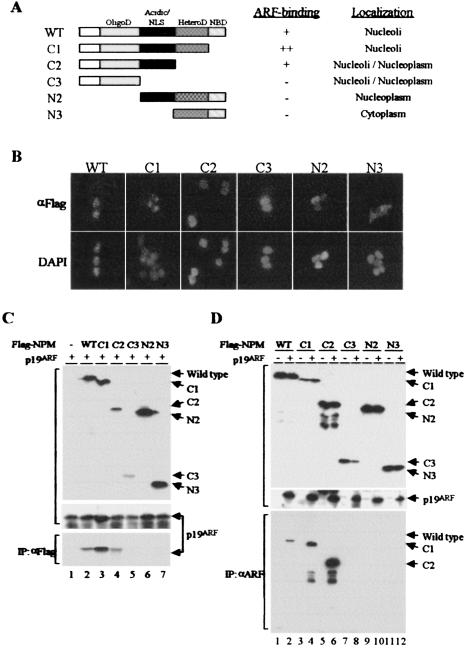
Several functional domains have been mapped within NPM (Fig. 4A) (15). The N-terminal portion of NPM includes an oligomerization domain. The central domain, while highly acidic, is punctuated by two nuclear localization signals. A heterodimerization domain that mediates interactions with other nucleolar proteins lies in the C-terminal moiety and is followed by a nucleic acid-binding domain at the extreme C terminus (15).
FIG. 4.
Mapping of the Arf-interacting domain in NPM. (A) Schematic representation of NPM mutants. The oligomerization domain (OligoD) (shaded), acidic domain including nuclear localization signals (NLS) (solid), heterodimerization domain (HeteroD) (stippled), and nucleic acid-binding domain (NBD) (hatched) are indicated. The results from the experiments shown in B and C are summarized on the right. (B) Localization of NPM mutants in 293T cells. Cells were transfected with pcDNA3 expression vectors containing wild-type (WT) or mutant Flag-tagged NPM variants. Exogenous proteins were detected by immunofluorescence with antibodies to the Flag epitope, and nuclei were visualized with 4′,6′-diamidino-2-phenylindole (DAPI). (C) Lysates prepared from 293T cells transiently transfected with NPM and Arf expression plasmids were separated on denaturing gels and blotted with antibodies to the Flag tag (top panel) or Arf (middle panel) to demonstrate expression of the respective proteins. Immune complexes recovered from lysates precipitated with antibodies to the Flag epitope were denatured, electrophoretically separated, and blotted with antibodies to p19Arf (bottom). (D) Transfected cells as in panel C were similarly immunoblotted to demonstrate expression of NPM variants (top panel) and p19Arf (middle panel). Immune complexes recovered from lysates precipitated with antibodies to p19Arf were blotted with antibodies to the Flag tag (bottom).
We generated a series of deletion mutants of Flag-tagged NPM and transiently expressed them in 293T cells. Analysis of the overexpressed proteins by immunofluorescence revealed that wild-type NPM and the C1 mutant lacking the nucleic acid-binding domain localized to nucleoli (Fig. 4B) just like endogenous NPM, which is frequently used as a nucleolar marker. Further C-terminal deletions (C2 and C3) dispersed the protein throughout the nucleus; mutants with N-terminal deletions were also mislocalized to the nucleoplasm (N2) or cytoplasm (N3).
When cotransfected with an Arf expression plasmid into 293T cells, all NPM variants were detected with antibodies to the Flag tag (Fig. 4C, top panel); p19Arf was coexpressed with all forms of NPM (Fig. 4C, middle panel). Antibodies to Flag-tagged NPM coprecipitated p19Arf, as expected (Fig. 4C, bottom panel, lane 2). Arf also bound to the C1 and C2 mutants (Fig. 4C, bottom panel, lanes 3 and 4) but not to the others (Fig. 4C, bottom panel, lanes 5 to 7). Note that the C1 mutant seemed to associate more avidly than wild-type NPM with p19Arf (Fig. 4C, bottom panel, compare lanes 3 and 2). Reciprocally, antibodies to p19Arf selectively coprecipitated wild-type NPM, C1, and C2 but not the other NPM variants (Fig. 4D). More C1 and C2 than wild-type NPM coprecipitated with p19Arf (Fig. 4D, bottom panel, lanes 4 and 6 versus 2), despite the fact that the expressed levels of full-length NPM exceeded those of the C1 mutant (Fig. 4D, top panel, lanes 3 and 4 versus 1 and 2). These results are summarized in Fig. 4A.
The difference between C2 and C3 is the presence of the central acidic domain (Fig. 4A), and because p19Arf is a highly basic protein (pI > 12), it likely requires this region for binding. Although p19Arf did not detectably interact in vivo with the N2 mutant retaining the NPM acidic domain but lacking the oligomerization domain, this protein was excluded from nucleoli (Fig. 4B). Nonetheless, the NPM oligomerization domain may also contribute to p19Arf binding, given evidence that Arf interacts with NPM oligomers (see below).
NPM isoform lacking its nucleic acid-binding domain can override Arf-induced inhibition of rRNA processing.
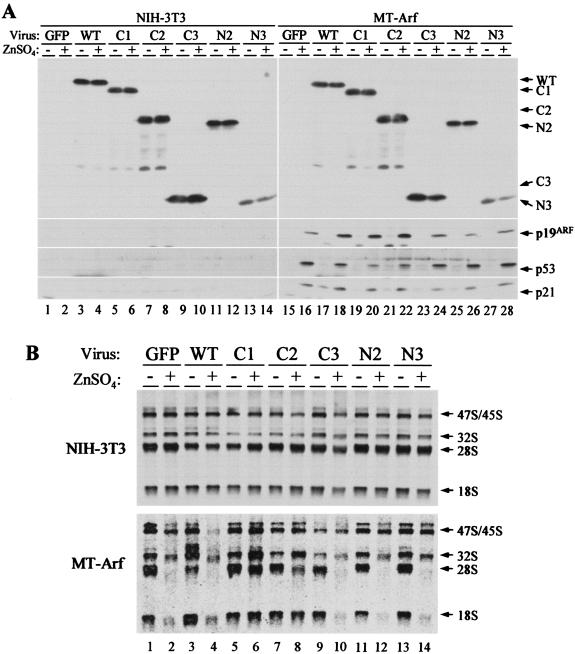
To assess a role for NPM in modulating Arf function, NIH 3T3 and MT-Arf cells were infected with retroviruses encoding wild-type or mutant forms of NPM. Immunoblotting confirmed that similar quantities of Flag-tagged NPM proteins were expressed in both cell lines (Fig. 5A). The expression levels of exogenous proteins except N3 were similar to that of endogenous NPM (data not shown, but see Fig. 7 below). Treatment of MT-Arf cells with zinc sulfate induced p19Arf, p53, and the p53-responsive gene p21Cip1 (Fig. 5A, right, bottom panels). Interestingly, the levels of p19Arf were increased in cells expressing wild-type NPM, C1, or C2 (Fig. 5A, compare lanes 18, 20, and 22 to others). These differences appear to reflect NPM-induced retardation of p19Arf turnover (Mei-Ling Kuo and C. J. Sherr, unpublished observations).
FIG. 5.
NPM modulates Arf functions. (A) NIH 3T3 and MT-Arf cells infected with the indicated retroviruses encoding NPM variants were treated (+) with zinc sulfate for 24 h or left untreated (−). Expression of NPM and p19Arf (in induced MT-Arf cells only) was confirmed by immunoblotting. The same lysates were blotted with antibodies to p53 and the p53-inducible gene product p21Cip1. Lower levels of p53 were detected in cells expressing NPM mutants C1 and C2 (lanes 20 and 22 versus 16), and this may explain the observed modest attenuation of Arf-induced proliferative arrest (Fig. 6). (B) NIH 3T3 cells (top panel) and MT-Arf cells (bottom panel) infected with the indicated retroviruses encoding NPM variants as in A were treated with zinc sulfate (+) or not (−). Newly synthesized rRNAs were analyzed by pulse-chase labeling with [3H]uridine. The indicated rRNA species separated on a gel and detected by fluoroautoradiography are indicated at the right. Note that enforced expression of C1 (lanes 5 and 6) and C2 (lanes 7 and 8) overrides the effects of Arf expression (lanes 6 and 8), whereas wild-type NPM (lane 4) and the other NPM mutants do not.
FIG. 7.
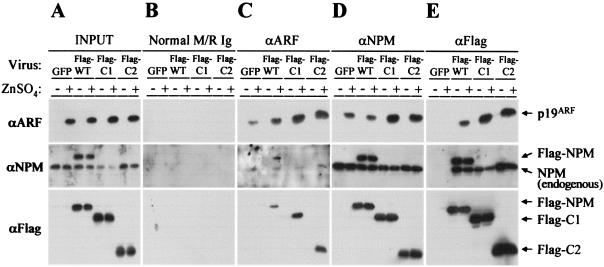
Association of endogenous and exogenous nucleophosmin and Arf. MT-Arf cells infected with retroviruses encoding GFP, full-length NPM (wild type, WT), or the C1 or C2 NPM mutant were treated with zinc sulfate for 24 h (+) or left untreated (−). Following Arf induction, cell lysates were prepared, and 10% of the total protein was separated directly on denaturing gels (A). The remainder was precipitated with the indicated antibodies (panels C to E, top) prior to separation of denatured immune complexes on gels. Control precipitations were performed with a mixture of normal mouse and rabbit IgGs (B). Separated proteins were transferred to membranes and blotted with antibodies to p19Arf (top), NPM (middle), or the Flag epitope (bottom). The monoclonal antibody to NPM used in this experiment reacts with an epitope at the C terminus of the protein and therefore recognizes both endogenously and exogenously expressed full-length NPM, but not C1 or C2. Note that Flag-tagged NPM variants form coprecipitating complexes with endogenously expressed NPM (E).
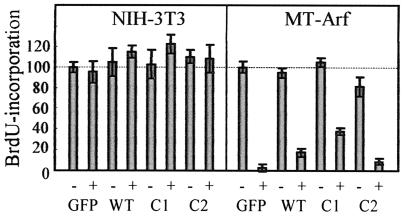
Expression of exogenous NPM, C1, or C2 did not affect DNA synthesis in control NIH 3T3 or in uninduced MT-Arf cells (Fig. 6). As expected, Arf induction by zinc led to proliferative arrest of MT-Arf cells within 24 h. The reduction in DNA synthesis in Arf-induced cells was somewhat attenuated in the presence of excess wild-type NPM, whereas the C1 mutant, which bound to p19Arf more efficiently (Fig. 4C and 4D), had a greater effect in overriding p19Arf-induced arrest (Fig. 6). In contrast, C2 was inactive in this respect. Immunofluorescence confirmed that wild-type NPM and C1 colocalized with Arf to the nucleolus, whereas C2 was again delocalized across the nucleus (Fig. 4B and data not shown). In MT-Arf cells, Arf's ability to rapidly inhibit cell proliferation depends upon p53, and indeed, the p53 level achieved in cells coexpressing C1 was somewhat lower than those seen in cells expressing either a control GFP protein, wild-type NPM (which showed a slight p53 reduction), or the other NPM mutants (Fig. 5A, compare lanes 16, 18, and 20). Since Arf's ability to interact with NPM does not depend upon Mdm2 or p53 (Fig. 3C), the simplest interpretation is that NPM can sequester p19Arf from Mdm2, thereby accelerating p53 turnover, with the C1 mutant being more efficient than full-length NPM in doing so. Despite the modulating effects of overexpressed NPM on Arf-induced arrest, p19Arf remained able to elicit a p53 response.
FIG. 6.
Enforced expression of NPM mutant C1 attenuates Arf-induced cell cycle arrest. NIH 3T3 and MT-Arf cells transfected with NPM and mutants C1 and C2 were pulsed for 1 h with bromodeoxyuridine (BrdU), and incorporation of bromodeoxyuridine into S-phase cells was determined by immunofluorescence. To induce p19Arf in MT-Arf cells, cultures were treated for 24 h with zinc (+) or were left untreated (−) prior to bromodeoxyuridine labeling. Incorporation into control virus-infected cells not treated with zinc was normalized to 100% in each group.
We next tested whether the NPM mutants might affect Arf's ability to inhibit ribosome biogenesis. NIH 3T3 and MT-Arf cells were infected with MSCV-IRES-GFP retroviruses encoding NPM variants, and 3 days later, infected cells were >95% GFP positive. Cells taken postinfection and incubated for 24 h in medium containing or lacking zinc were metabolically labeled with [3H]uridine for 30 min and incubated for another 2 h in the absence of the labeled precursor. Radiolabeled RNA was isolated, separated on a formaldehyde-agarose gel, and analyzed by fluoroautoradiography. As reported previously (41), most of the newly synthesized rRNA (47S/45S) was processed into mature 28S and 18S rRNA within 2 h, and expression of exogenous NPM proteins did not affect rRNA synthesis in NIH 3T3 or uninduced MT-Arf cells (Fig. 5B). Treatment with zinc did not significantly affect rRNA production in NIH 3T3 cells (Fig. 5B, upper panel) but strongly inhibited rRNA maturation in MT-Arf cells, with concomitant expression of p19Arf protein (Fig. 5B, lower panel, compare lanes 2 and 1). Wild-type NPM (Fig. 5B, lower panel, lanes 3 and 4) and mutants that do not bind to p19Arf (C3, N2, and N3, Fig. 5B, lower panel, lanes 9 to 14) did not prevent p19Arf from inhibiting rRNA maturation. However, expression of the C1 and C2 mutants significantly blocked Arf's ability to inhibit rRNA processing (Fig. 5B, lower panel, lanes 5 to 8), with C1 (localizing in nucleoli) exhibiting a more profound effect than C2 (pannuclear localization).
NPM mutants form oligomers with full-length NPM.
As indicated above, the levels of expression of exogenous NPM proteins achieved were marginally greater than that of endogenous NPM in MT-Arf cells (Fig. 7A). Immunoblotting with antibodies to the Flag tag selectively detected the exogenous NPM variants, which exhibited characteristic electrophoretic mobilities on denaturing gels (Fig. 7A, bottom panel). Antibodies to NPM itself recognized both the endogenous and exogenous forms of NPM (Fig. 7A, middle panel), and p19Arf was detected only in induced cells (Fig. 7A, top). Control antibodies precipitated none of these proteins (Fig. 7B), whereas antibodies to p19Arf coprecipitated Flag-tagged (C1 and C2) and untagged forms of NPM (Fig. 7C). In turn, antibodies to NPM coprecipitated p19Arf (Fig. 7D). Moreover, antibodies to the Flag tag also precipitated the endogenously expressed NPM protein whether Arf was induced or not (Fig. 7E), indicating that exogenous NPM as well as the C1 and C2 mutants formed oligomers with endogenous NPM. The p19Arf protein joined these complexes upon induction (Fig. 7E), again with a greater propensity to bind to C1- and C2-containing complexes than those containing only wild-type NPM. Therefore, the NPM C1 and C2 mutants do not sequester p19Arf protein from endogenous NPM but instead exist together in a complex with endogenous NPM and p19Arf.
NPM mutant C1 displaces p19Arf from a high-molecular-weight complex.
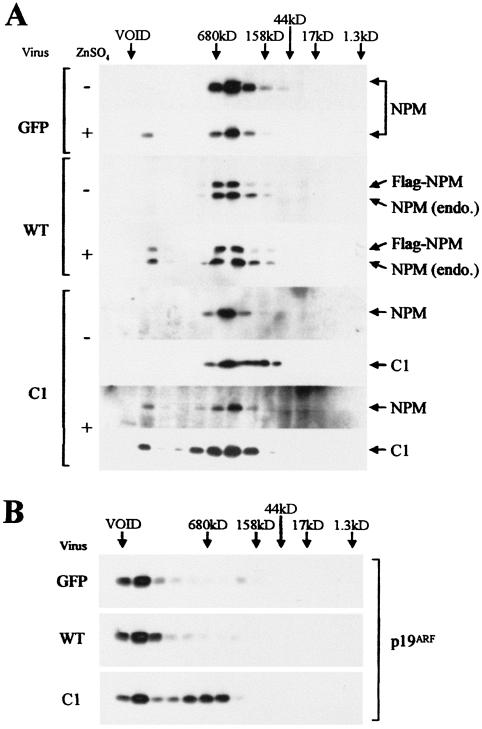
From the results above, we reasoned that the C1 mutant might interfere with Arf's inhibition of rRNA processing by causing some qualitative change in the Arf-NPM complex. Cell lysates prepared from induced or uninduced MT-Arf cells expressing either exogenous wild-type NPM or the C1 mutant were subjected to gel filtration through Superose-6. Fractions were analyzed for the presence of the various proteins by immunoblotting. Endogenous NPM was seen to reside in a complex of ≈500 kDa in uninduced cells transduced with a control GFP vector (Fig. 8A, top panel). Because the parental NIH 3T3 cells have sustained deletions of the Arf locus, the 500-kDa NPM complex must lack p19Arf. Following Arf induction, however, a small proportion of NPM protein moved into a protein complex of much greater mass that was delivered near the column void volume (≈5 MDa). Exogenous wild-type NPM behaved like the endogenous protein (Fig. 8A, middle panel); the C1 mutant exhibited a similar distribution, although the smaller complex was more heterodisperse (Fig. 8A, bottom panel).
FIG. 8.
Gel filtration of complexes containing NPM and Arf. Cell lysates were prepared from MT-Arf cells infected with retroviruses encoding control protein (GFP), wild-type NPM, or the NPM C1 mutant and then treated with zinc sulfate (+) or left untreated (−). (A) Extracted cellular proteins applied and eluted from a Superose-6 column were analyzed by immunoblotting with antibodies to NPM that detect both the endogenous protein and all Flag-tagged variants. Antibodies to the Flag epitope were used to detect the C1 mutant. (B) The distribution of p19Arf recovered from induced MT-Arf cells was determined in parallel.
Figure 8B shows the distribution of p19Arf in the same column fractions. Most of the induced Arf protein cochromatographed with the minor fraction of NPM in the high-molecular-mass complex (≈5 MDa). This is entirely consistent with our previous experiments, which revealed that only a small proportion of total NPM associated with p19Arf (Fig. 3A). However, in the presence of the C1 mutant, more than 60% of the Arf protein was present in the smaller C1-containing complex. These results suggest that an inability of p19Arf to associate with the larger NPM-containing complex prevents its ability to interfere with rRNA processing.
DISCUSSION
By counteracting the activity of Mdm2, p19Arf can trigger p53-dependent cell cycle arrest or apoptosis, depending on cell type and collateral signals. Although it was initially thought that Arf's ability to arrest cell proliferation depended exclusively on p53, subsequent studies showed that p19Arf could also retard the growth of cells lacking p53 (2) or both p53 and Mdm2 (48), albeit much less efficiently than in cells that retain p53 function. The bulk of p19Arf normally resides within the nucleolus (30, 33, 50), Arf being as robust a nucleolar marker as fibrillarin (50) or NPM (20), implying that p19Arf might play a distinct role in this compartment. Indeed, recent experiments indicated that p19Arf could inhibit rRNA processing by retarding the initial cleavages of the 47S/45S rRNA precursors and also by inhibiting a later processing step involving the 32S rRNA precursor that eventually leads to the formation of both 28S and 5.8S rRNAs (41). As we demonstrated previously, Arf can associate with 5.8S rRNA, but we do not know whether it might also interact with larger rRNA precursors. Moreover, the mechanism by which Arf affects rRNA processing remains unclear.
To identify other proteins with which p19Arf may interact, we introduced a TAP-tagged Arf protein into cultured mouse fibroblasts and copurified complexes containing a number of p19Arf-associated proteins. These included NPM, nucleolin, and PTRF (as well as many ribosomal proteins), all of which localize to the nucleolus of mammalian cells. Parallel purifications performed with Arf mutants which are unable to induce either p53-dependent or -independent cell cycle arrest recovered less NPM or PTRF. Since tandem affinity purification was performed with proteins obtained from unfractionated cell lysates, these results provide some assurance that p19Arf physically interacts either directly or indirectly with these nucleolar proteins in situ. Treatment of cell lysates with RNase A prior to tandem affinity purification did not disrupt the interaction of TAP-tagged Arf with NPM, PTRF, or nucleolin, although it did limit the recovery of some ribosomal proteins. Thus, despite the fact that p19Arf, NPM, PTRF, and nucleolin can all interact with RNA, the association of Arf with these other proteins appears not to be RNA dependent. PTRF regulates rRNA transcriptional termination (16), whereas nucleolin controls the earliest processing steps within the 5′ external transcribed spacer sequence of 47S pre-rRNA (10-12). The interaction of p19Arf with complexes containing these proteins therefore implicates Arf as an inhibitor of early preribosome maturation, consistent with its ability to retard 47S/45S rRNA processing.
NPM is also thought to function in preribosome maturation (5, 29, 55). Full-length NPM/B23.1 binds to nucleic acids (4), a function which depends upon the presence of 35 amino acid residues at its C terminus (15). A second NPM isoform, B23.2, formed by alternative splicing of 3′ exons, has a substitution of two amino acids for the C-terminal 37 residues of B23.1; hence, B23.2 does not bind directly to RNA (15). NPM has been reported to act via an associated RNase activity to direct endonucleolytic cleavage of rRNA precursors at a site within the second internal transcribed spacer sequence (ITS-2) located 3′ to the 5.8S rRNA domain in 32S pre-rRNA (36). Like Arf, NPM coprecipitated in immune complexes with 5.8S rRNA from MT-Arf cells whether p19Arf was induced or not, although more 5.8S rRNA was recovered when Arf was expressed. Since, apart from its effects on 47S/45S rRNA, p19Arf also interferes with processing of the 32S intermediate containing 28S and 5.8S rRNAs (41), the Arf-NPM interaction might facilitate the ability of p19Arf to block this later processing step as well. The interaction of p19Arf and NPM requires the integrity of both the acidic and oligomerization domains of NPM as well as amino acid residues 2 to 14 at the extreme Arf N terminus. Intriguingly, deletion of the central domain of NPM enhances its RNase activity (15), suggesting a mechanism by which the binding of Arf to this region might interfere with NPM's RNA-processing function.
The NPM isoforms B23.1 and B23.2 can form oligomers (15, 27, 55), but the RNA binding activity of B23.1 is impaired by its oligomerization with B23.2 (27). The C1 deletion mutant, which lacks the 37 C-terminal amino acids of B23.1, is virtually identical to the B23.2 isoform and therefore cannot bind RNA. Mouse NIH 3T3 cells express abundant endogenous B23.1 but very little B23.2, so that only full-length NPM/B23.1 was routinely detected by immunoblotting. Much of the NPM protein recovered from uninduced MT-Arf cells under our conditions of lysis and extraction was found in high-molecular-mass complexes of ≈500 kDa. Therefore, the bulk of NPM, although localized within nucleoli, was recovered in complexes that are too small to represent preribosomes. Because parental NIH 3T3 cells have sustained deletion of the Arf locus, the 500-kDa complex formed in MT-Arf cells cannot contain endogenous p19Arf. Although these complexes may well contain other as yet unidentified proteins or RNAs, we confirmed that NPM can homo-oligomerize with itself and hetero-oligomerize with truncated, mutant forms of NPM (C1 and C2) that retain the N-terminal sequences required for both oligomer formation and p19Arf binding.
Under the same conditions of extraction, induced p19Arf was recovered near the void volume of Superose-6 columns, and this mobilized some NPM into these higher-molecular-weight complexes (2 to 5 MDa). These results are consistent with observations that only a small proportion of NPM associated with p19Arf, although a significant pool of p19Arf coprecipitated with NPM. However, complexes containing C1, which, like C1 itself, should be impaired in RNA binding (27), redistributed the vast majority of p19Arf from the high-molecular-weight (2- to 5-MDa) to lower-molecular-weight (≈500-kDa) complexes. Importantly, enforced expression of C1 at levels that modestly exceeded the level of endogenous NPM also overrode the ability of p19Arf to retard rRNA processing. Clearly, the simplest interpretation is that Arf joins some NPM in preribosomal particles to negatively regulate rRNA processing and that C1, like B23.2, generates lower-molecular-weight p19Arf-containing complexes that can no longer interact with rRNA.
These results beg the question of whether NPM's nucleic acid binding domain is required for it to enter the larger particles. We attempted to address this question by treating extracts with RNase prior to molecular sieving. Under these conditions, however, much of the Arf protein and some NPM became insoluble and precipitated out of solution. Thus, although NPM itself can prevent thermally induced aggregation of other proteins in vitro (15, 42), this in itself is insufficient to maintain p19Arf solubility. We also used low doses of actinomycin D to selectively inhibit rRNA synthesis, conditions under which nucleolar structures are disrupted and NPM is dispersed (13, 38). Actinomycin D treatment redistributed NPM and p19Arf from 2- to 5-MDa complexes into those of lower molecular mass (≈500 kDa) (data not shown), again consistent with the idea that the larger complex localizes to nucleoli, where its formation is RNA dependent.
Functions other than preribosomal maturation have been attributed to NPM. For example, NPM is phosphorylated by cyclin-dependent kinases (Cdks), specifically during mitosis by cyclin B-Cdk1 (27) and during interphase by cyclin E-Cdk2 (26, 44). Although most NPM localizes to nucleoli, some associates with unduplicated centrosomes during G1 phase, where its cyclin E-Cdk2-mediated phosphorylation takes place. Phosphorylation dissociates NPM from centrosomes and leads to centrosome duplication, a process that can be inhibited by expression of a nonphosphorylatable NPM mutant (26, 44). Although it is conceivable that the binding of p19Arf to NPM might somehow affect this process, primary MEFs lacking Arf tend to remain diploid, whereas cells lacking p53, which are defective in expressing the p53-responsive Cdk2 inhibitor p21Cip1 in response to DNA damage, rapidly become tetraploid (17, 54). Thus, unlike the case for p53, we have no evidence that p19Arf plays a role in regulating centrosome duplication.
Within the first 24 h following zinc induction of p19Arf in MT-Arf cells up to the time that cells underwent proliferative arrest, we observed that a major proportion of induced p19Arf associated with NPM, whereas a considerably smaller fraction stably bound to Mdm2, a much less abundant protein. In this setting, Arf's ability to arrest cell proliferation rapidly is p53 dependent. We noted that p53 accumulation in response to Arf induction was modestly attenuated by enforced NPM expression and even more so by the NPM C1 mutant. Hence, by associating with p19Arf, NPM might limit Arf's interaction with Mdm2 to some degree. In contrast, others reported that NPM could bind directly to p53 to increase its stability and activity following stress (3). The C-terminal heterodimerization and nucleic acid binding domains of NPM were reported to be necessary and sufficient for p53 binding, whereas, as shown here, the nonoverlapping central oligomerization and acidic domains are required for NPM's interaction with p19Arf. Such data raise the formal possibility that NPM might simultaneously form complexes with p19Arf and p53 and, in principle, with Mdm2 as well. However, by molecular sieving analyses, we found that Arf-NPM and Arf-Mdm2 complexes chromatographed in different peak fractions; moreover, antibodies to NPM did not precipitate Mdm2. Although induction or introduction of p19Arf into naive cells can localize Mdm2 to nucleoli, activated p53 remains nucleoplasmic (43, 50). Furthermore, the interaction of NPM and p19Arf occurs equally well in primary MEFs lacking both Mdm2 and p53.
Whether Arf can form complexes with preribosomal particles in the nucleolus and interfere with certain stages of their maturation warrants further study. The ability of p19Arf to interact with NPM and other nucleolar proteins and the finding that certain NPM mutants can override the effects of Arf on rRNA processing reinforce the idea that Arf can play a role in regulating ribosome biosynthesis and, by inference, cell growth as well as cell proliferation.
Addendum in Proof
After acceptance of our paper, Itahana and coworkers also reported that ARF physically interacts with B23/NPM/nucleophosmin (Mol. Cell 12:1151-1164, 2003).
Acknowledgments
We thank Jun Qin and Doug W. Chan for generously performing mass spectroscopic identification of proteins and acknowledge the Hartwell Center at St. Jude Children's Research Hospital for independently confirming some of the key assignments. We thank Kathy Gould for the gift of the TAP tag cDNA cassette; Martine Roussel, Pim den Besten, Mei-Ling Kuo, and other members of the Sherr-Roussel laboratory for many helpful suggestions during the course of this work; and Rose Mathew and Cam Hornsby for technical assistance with some of the experiments.
This work was supported in part by Cancer Center CORE grant CA-21765 and by ALSAC of St. Jude Children's Research Hospital. C.J.S. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Bassler, J., P. Grandi, O. Gadal, T. Leβmann, E. Petfalski, D. Tollervey, J. Lechner, and E. Hurt. 2001. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell 8:517-529. [DOI] [PubMed] [Google Scholar]
- 2.Carnero, A., J. D. Hudson, C. M. Price, and D. H. Beach. 2000. p16INK4A and p19ARF act in overlapping pathways in cellular immortalization. Nat. Cell Biol. 2:148-155. [DOI] [PubMed] [Google Scholar]
- 3.Colombo, E., J.-C. Marine, D. Danovi, B. Falini, and P. G. Pelicci. 2002. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat. Cell Biol. 4:529-533. [DOI] [PubMed] [Google Scholar]
- 4.Dumbar, T. S., G. A. Gentry, and M. O. Olson. 1989. Interaction of nucleolar phosphoprotein B23 with nucleic acids. Biochemistry 28:9495-9501. [DOI] [PubMed] [Google Scholar]
- 5.Dundr, M., and M. O. Olson. 1998. Partially processed pre-rRNA is preserved in association with processing components in nucleolus-derived foci during mitosis. Mol. Biol. Cell 9:2407-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eichler, D. C., and N. Craig. 1994. Processing of eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 49:197-239. [DOI] [PubMed] [Google Scholar]
- 7.Eymin, B., L. Kanaryan, P. Seite, C. Brambilla, E. Brambilla, C. J. Larsen, and S. Gazzeri. 2001. Hum. ARF binds E2F1 and inhibits its transcriptional activity. Oncogene 20:1033-1041. [DOI] [PubMed] [Google Scholar]
- 8.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 9.Fatyol, K., and A. A. Szalay. 2001. The p14ARF tumor suppressor protein facilitates nucleolar sequestration of hypoxia-inducible factor-1α (HIF-1α) and inhibits HIF-1-mediated transcription. J. Biol. Chem. 276:28421-28429. [DOI] [PubMed] [Google Scholar]
- 10.Ghisolfi-Nieto, L., G. Joseph, F. Puvion-Dutilleul, F. Amalric, and P. Bouvet. 1996. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on preribosomal RNA. J. Mol. Biol. 260:34-53. [DOI] [PubMed] [Google Scholar]
- 11.Ginisty, H., F. Amalric, and P. Bouvet. 1998. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 17:1476-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ginisty, H., H. Sicard, B. Roger, and P. Bouvet. 1999. Structure and functions of nucleolin. J. Cell Sci. 112:761-772. [DOI] [PubMed] [Google Scholar]
- 13.Gonda, K., J. Fowler, N. Katoku-Kikyo, J. Haroldson, J. Wudel, and N. Kikyo. 2003. Reversible disassembly of somatic nucleoli by the germ cell proteins FRGY2a and FRGY2b. Nat. Cell Biol. 5:205-210. [DOI] [PubMed] [Google Scholar]
- 14.Harnpicharnchai, P., J. Jakovljevic, E. Horsey, T. Miles, J. Roman, M. Rout, D. Meagher, B. Imai, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and J. L. Woolford, Jr. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8:505-515. [DOI] [PubMed] [Google Scholar]
- 15.Hingorani, K., A. Szebeni, and M. O. Olson. 2000. Mapping the functional domains of nucleolar protein B23. J. Biol. Chem. 275:24451-24457. [DOI] [PubMed] [Google Scholar]
- 16.Jansa, P., S. W. Mason, U. Hoffman-Rohrer, and I. Grummt. 1998. Cloning and functional characterization of PTRF, a novel protein which induces dissociation of paused ternary transcription complexes. EMBO J. 17:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamijo, T., F. Zindy, M. F. Roussel, D. E. Quelle, J. R. Downing, R. A. Ashmun, G. Grosveld, and C. J. Sherr. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91:649-659. [DOI] [PubMed] [Google Scholar]
- 18.Karayan, L., J. F. Riou, P. Seite, J. Migeon, A. Cantereau, and C. J. Larsen. 2001. Human ARF protein interacts with Topoisomerase I and stimulates its activity. Oncogene 20:836-848. [DOI] [PubMed] [Google Scholar]
- 19.Kuo, M.-L., E. J. Duncavage, R. Mathew, W. den Besten, D. Pie, D. Naeve, T. Yamamoto, C. Cheng, C. J. Sherr, and M. F. Roussel. 2002. Arf induces p53-dependent and independent anti-proliferative genes. Cancer Res. 63:1046-1053. [PubMed] [Google Scholar]
- 20.Lohrum, M. A. E., M. Ashcroft, M. H. G. Kubbutat, and K. H. Vousden. 2000. Identification of a cryptic nucleolar-localization signal in MDM2. Nat. Cell Biol. 2:179-181. [DOI] [PubMed] [Google Scholar]
- 21.Marshak, D. R., J. T. Kadonaga, R. R. Burgess, M. W. Knuth, W. A. Brennan, Jr., and S.-H. Lin. 1996. Strategies for protein purification and characterization. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Martelli, F., T. Hamilton, D. P. Silver, N. E. Sharpless, N. Bardeesy, M. Rokas, R. A. DePinho, D. M. Livingston, and S. R. Grossman. 2001. p19ARF targets certain E2F species for degradation. Proc. Natl. Acad. Sci. USA 98:4455-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mortz, E., T. N. Krogh, H. Vorum, and A. Gorg. 2001. Improved silver staining protocols for high sensitivity protein identification using matrix-assisted laser desorption/ionization-time of flight analysis. Proteomics 1:1359-1363. [DOI] [PubMed] [Google Scholar]
- 24.Nissan, T. A., J. Baβler, E. Petfalski, D. Tollervey, and E. Hurt. 2002. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 21:5539-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogryzko, V. V., T. Kotani, X. Zhang, R. L. Schiltz, T. Howard, X.-J. Yang, B. H. Howard, J. Qin, and Y. Nakatani. 1998. Histone-like TAFs within the PCAF histone acetylase complex. Cell 94:35-44. [DOI] [PubMed] [Google Scholar]
- 26.Okuda, M., H. F. Horn, P. Tarapore, Y. Tokuyama, A. G. Smulian, P.-K. Chan, E. S. Knudsen, I. A. Hofmann, J. D. Snyder, K. E. Bove, and K. Fukasawa. 2000. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 103:127-140. [DOI] [PubMed] [Google Scholar]
- 27.Okuwaki, M., M. Tsujimoto, and K. Nagata. 2002. The RNA binding activity of ribosome biogenesis factor, nucleophosmin/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol. Biol. Cell 13:2016-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olson, M. O., M. O. Wallace, A. H. Herrera, L. Marshall-Carlson, and R. C. Hunt. 1986. Preribosomal ribonucleoprotein particles are a major component of a nucleolar matrix fraction. Biochemistry 25:484-491. [DOI] [PubMed] [Google Scholar]
- 29.Pinol-Roma, S. 1999. Association of nonribosomal nucleolar proteins in ribonucleoprotein complexes during interphase and mitosis. Mol. Biol. Cell 10:77-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pomerantz, J., N. Schreiber-Agus, N. J. Liégeois, A. Silverman, L. Alland, L. Chin, J. Potes, K. Chen, I. Orlow, H.-W. Lee, C. Cordon-Cardo, and R. DePinho. 1998. The Ink4a tumor suppressor gene product, p19Arf interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92:713-723. [DOI] [PubMed] [Google Scholar]
- 31.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 32.Quelle, D. E., M. Cheng, R. A. Ashmun, and C. J. Sherr. 1997. Cancer-associated mutations at the INK4a locus cancel cell cycle arrest by p16INK4a but not by the alternative reading frame protein p19ARF. Proc. Natl. Acad. Sci. USA 94:669-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quelle, D. E., F. Zindy, R. A. Ashmun, and C. J. Sherr. 1995. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell 83:993-1000. [DOI] [PubMed] [Google Scholar]
- 34.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 35.Rocha, S., K. J. Campbell, and N. D. Perkins. 2003. p53- and Mdm2-independent repression of NF-ΚB transactivation by the ARF tumor suppressor. Mol. Cell 12:15-25. [DOI] [PubMed] [Google Scholar]
- 36.Savkur, R. S., and M. O. J. Olson. 1998. Preferential cleavage in pre-ribosomal RNA by protein B23 endoribonuclease. Nucleic Acids Res. 26:4508-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherr, C. J. 2001. The INK4a/ARF network in tumour suppression. Nat. Rev. Mol. Cell. Biol. 2:731-737. [DOI] [PubMed] [Google Scholar]
- 38.Simard, R. 1970. The nucleus: action of chemical and physical agents. Int. Rev. Cytol. 28:169-211. [DOI] [PubMed] [Google Scholar]
- 39.Smith, C. M., and J. A. Steitz. 1997. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell 89:669-672. [DOI] [PubMed] [Google Scholar]
- 40.Sugihara, T., S. C. Kaul, J. Kato, R. R. Reddel, H. Nomura, and R. Wadhwa. 2001. Pex19p dampens the p19ARF-p53-p21WAF1 tumor suppressor pathway. J. Biol. Chem. 276:18649-18652. [DOI] [PubMed] [Google Scholar]
- 41.Sugimoto, M., M.-L. Kuo, M. F. Roussel, and C. J. Sherr. 2003. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol. Cell 11:415-424. [DOI] [PubMed] [Google Scholar]
- 42.Szebani, A., and M. O. Olson. 1999. Nucleolar protein B23 has molecular chaperone activities. Protein Sci. 8:905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tao, W., and A. J. Levine. 1999. P19ARF stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc. Natl. Acad. Sci. USA 96:6937-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tokuyama, Y., H. F. Horn, K. Kawamura, P. Tarapore, and K. Fukasawa. 2001. Specific phosphorylation of nucleophosmin on Thr199 by cyclin-dependent kinase-2-cyclin E and its role in centrosome duplication. J. Biol. Chem. 276:21529-21537. [DOI] [PubMed] [Google Scholar]
- 45.Tyc, K., and J. A. Steitz. 1989. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 8:3113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vivo, M., R. A. Calogero, F. Sansone, V. Calabro, T. Parisi, L. Borrelli, S. Saviozzi, and G. La Mantia. 2001. The human tumor suppressor ARF interacts with spinophilin/neurabin II, a type 1 protein-phosphatase-binding protein. J. Biol. Chem. 276:14161-14169. [DOI] [PubMed] [Google Scholar]
- 47.Warner, J. R. 2001. Nascent ribosomes. Cell 107:133-136. [DOI] [PubMed] [Google Scholar]
- 48.Weber, J. D., J. R. Jeffers, J. E. Rehg, D. H. Randle, G. Lozano, M. F. Roussel, C. J. Sherr, and G. P. Zambetti. 2000. p53-independent functions of the p19ARF tumor suppressor. Genes Dev. 14:2358-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weber, J. D., M.-L. Kuo, B. Bothner, E. L. DiGiammarino, R. W. Kriwacki, M. F. Roussel, and C. J. Sherr. 2000. Cooperative signals governing ARF-Mdm2 interaction and nucleolar localization of the complex. Mol. Cell. Biol. 20:2517-2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weber, J. D., L. J. Taylor, M. F. Roussel, C. J. Sherr, and D. Bar-Sagi. 1999. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1:20-26. [DOI] [PubMed] [Google Scholar]
- 51.Yung, B. Y.-M., H. Busch, and P.-K. Chan. 1985. Translocation of nucleolar protein B23 (37 kDa/pI 5.1) induced by selective inhibitors of ribosome synthesis. Biochim. Biophys. Acta 826:167-173. [DOI] [PubMed] [Google Scholar]
- 52.Zhao, L., T. Samuels, S. Winckler, C. Korgaonkar, V. Tompkins, M. C. Horne, and D. E. Quelle. 2003. Cyclin G1 has growth inhibitory activity linked to the ARF-Mdm2-p53 and pRb tumor suppressor pathways. Mol. Cancer Res. 1:195-206. [PubMed] [Google Scholar]
- 53.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 12:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zindy, F., D. E. Quelle, M. F. Roussel, and C. J. Sherr. 1997. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene 15:203-211. [DOI] [PubMed] [Google Scholar]
- 55.Zirwes, R. F., M. S. Schmidt-Zachmann, and W. W. Franke. 1997. Identification of a small, very acidic constitutive nucleolar protein (NO29) as a member of the nuclophosmin family. Proc. Natl. Acad. Sci. USA 94:11387-11392. [DOI] [PMC free article] [PubMed] [Google Scholar]