Abstract
Xeroderma pigmentosum (XP) is a human genetic disease which is caused by defects in nucleotide excision repair. Since this repair pathway is responsible for removing UV irradiation-induced damage to DNA, XP patients are hypersensitive to sunlight and are prone to develop skin cancer. Based on the underlying genetic defect, the disease can be divided into the seven complementation groups XPA through XPG. XPF, in association with ERCC1, constitutes a structure-specific endonuclease that makes an incision 5′ to the photodamage. XPF-ERCC1 has also been implicated in both removal of interstrand DNA cross-links and homology-mediated recombination and in immunoglobulin class switch recombination (CSR). To study the function of XPF in vivo, we inactivated the XPF gene in mice. XPF-deficient mice showed a severe postnatal growth defect and died approximately 3 weeks after birth. Histological examination revealed that the liver of mutant animals contained abnormal cells with enlarged nuclei. Furthermore, embryonic fibroblasts defective in XPF are hypersensitive to UV irradiation and mitomycin C treatment. No defect in CSR was detected, suggesting that the nuclease is dispensable for this recombination process. These phenotypes are identical to those exhibited by the ERCC1-deficient mice, consistent with the functional association of the two proteins. The complex phenotype suggests that XPF-ERCC1 is involved in multiple DNA repair processes.
Xeroderma pigmentosum (XP) is a human genetic disorder which is characterized by sensitivity to sunlight and an increased incidence of skin cancer (3, 4). Based on the underlying genetic defect, the disease can be divided into the seven complementation groups XPA through XPG. The genes defective in each complementation group have been cloned and were found to be components of a nucleotide excision repair pathway which removes UV irradiation-induced DNA damage (primarily [6-4] photoproducts and, only marginally, cyclobutane pyrimidine dimers) as well as helix-distorting DNA modifications induced by carcinogenic chemicals (24). This repair process has been reconstituted in vitro and subject to extensive analyses (1, 15, 16). Based on biochemical studies, several proteins have been proposed to recognize the DNA damage, including XPC-HR23 (26, 33, 39), replication protein A (RPA) (25), or the XPA-RPA complex (36). Studies in vivo support the view that XPC-HR23B binds to the DNA damage and recruits TFIIH, followed by XPA and RPA to form the initiating repair complex (35). The helicase activity of XPB and XPD in THIIH unwinds the DNA duplex that surrounds the DNA adduct (9, 17). The resulting bubble structure is a substrate for two structure-specific endonucleases, XPF-ERCC1 and XPG, which incise on the 5′ and 3′ sides of the DNA adduct, respectively, to remove the lesion as a 24- to 32-base oligonucleotide (13, 22, 31).
Some XP gene products perform additional functions besides nucleotide excision repair. Such is the case for XPF, which has been implicated in removal of interstrand DNA cross-links (10) and in a homology-based recombination process (2, 20). Furthermore, based on in vitro experiments and the phenotype of mice deficient in mismatch repair proteins, this nuclease could potentially be involved in immunoglobulin class switch recombination (CSR) (7, 28, 34). Consistent with the diverse functions of XPF-ERCC1, mice deficient for ERCC1 exhibit complex phenotypes (14, 37). The mutant mice are runted and die approximately 20 days after birth. These phenotypes cannot be attributed to a deficiency in nucleotide excision repair since mice deficient in XPA or XPC develop normally (6, 19, 27). The reason for the growth defect of ERCC1 knockout mice is unclear.
Mice deficient in XPF have not been reported. Since XPF and ERCC1 function as a complex, inactivation of XPF might have the same consequence as an ERCC1 knockout. However, human XPF patients show relatively mild symptoms of photosensitivity with occasional neurological abnormalities (12, 31, 32). One possible explanation for this apparent discrepancy is that the mutations in these patients do not completely eliminate XPF function. In support of this hypothesis, fibroblasts from these patients retain low levels of nucleotide excision repair activity. Given the severe developmental defect of ERCC1 knockout mice, complete inactivation of the XPF gene in humans may be incompatible with postnatal survival.
The severe developmental defect of the ERCC1 knockout mice makes it difficult to use the mouse as a model for UV carcinogenesis studies and to evaluate the potential role of XPF-ERCC1 in other processes such as CSR in adult animals. To circumvent these problems, we introduced a point mutation into the XPF gene, which is compatible with normal development in humans.
MATERIALS AND METHODS
Generation of XPF-deficient mice.
To create the targeting construct, a 4-kb EcoRI-XhoI fragment was used as the 5′ homology, while a 4-kb XhoI-XhoI fragment was used as the 3′ homology. Exon 8 is contained in the XhoI-XhoI fragment. The sequence around the codon for glycine 445 (G-445) (GGG GAC GGC) was mutated to TAG GAT CCC. The mutation changes G-445 into a stop codon and creates a BamHI site. The homology arms were cloned into the pLN-TK targeting vector to create the targeting construct.
The targeting construct was transfected into TC1 embryonic stem (ES) cells by electroporation, and stable integrants were obtained by drug selection with G418 and ganciclovir. Genomic DNA was isolated from the stable clones and screened by diagnostic Southern analysis. Correctly targeted clones were used to generate chimeric mice, which were further bred for germ line transmission. Genotyping was achieved by a PCR assay using primers that flank exon 8 (5′-GCC GGG TGC TGA TCT GTG CA-3′ and 5′-CAG AGG TTT CCC AGG CCT GC-3′).
Pathological studies on the XPF-deficient mice.
To characterize the postnatal development of homozygous animals, the weights of two homozygous animals (around 15 days old) were compared with wild-type and heterozygous littermate controls: in the first set, +/+ mice weighed 11.6 g, +/m mice weighed 11.6 g, and m/m mice weighed 3.1 g; in the second set, +/+ mice weighed 7.7 g, +/m mice weighed 8.9 g, and m/m mice weighed 2.2 g. For histology analysis, the following tissues were dissected from mice 16 to 18 days old: thymus, aorta, heart, brain, bone marrow, esophagus, adrenaline gland, lung, pancreas, stomach, spleen, bladder, kidney, liver, and intestine. The tissues were fixed in Boins solution (Sigma). Sectioned tissues were stained with hematoxylin and eosin. Two homozygous animals were analyzed with their littermate wild-type controls.
RT PCR analysis of XPF mRNA expression.
Total RNA was isolated from mouse embryonic fibroblasts (MEF), and cDNA was synthesized by reverse transcription. XPF cDNA was amplified with primers at the two ends of the open reading frame (5′-CGC GGA TCC GGA AGG GCG CCC ATG GAG CCA GG-3′ and 5′-CGC GGA TCC TCA CTT TCT CAC TCT GCC TTT GGA CA-3′).
Class-switching assay.
Splenocytes were cultured in RPMI medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 U of penicillin-streptomycin/ml, 100 μM β-mercaptoethanol, and 20 μg of lipopolysaccharide (LPS) per ml plus 20 μg of dextran sulfate per ml or 25 ng of recombinant murine interleukin-4 per ml. LPS plus dextran sulfate induces class switching to immunoglobulin G2b (IgG2b) and IgG3, while LPS plus interleukin-4 promotes class switching to IgG1. The culture supernatant was collected 6 days after stimulation, and the concentration of antibodies was determined by enzyme-linked immunosorbent assay (ELISA).
DNA repair assays.
MEF were isolated from day 13.5 embryos and were grown in Dulbecco's modified Eagle's medium supplemented with 15% fetal bovine serum, 2 mM glutamine, 100 U of penicillin-streptomycin/ml, and 100 μM β-mercaptoethanol. For UV irradiation experiments, 5 × 105 cells were plated per well in a 6-well plate. The cells were grown for 2 days to confluence. The cells were irradiated with short-wavelength UV (UVC) under 3 ml of media. The use of media was intended to attenuate the high UV emission level of the light source (2 W/m2 at 254 nm), which kills even wild-type cells after 2 s of irradiation. Based on the UV absorbency of the media, we estimated that the cells were exposed to UVC at a dose rate of 0.25 W/m2. After irradiation, the cells were trypsinized and 5 × 105 cells were plated into a new well of a 6-well plate. The cells were grown for 2 days and were trypsinized. The number of viable cells was counted in the presence of trypan blue.
To assay mitomycin C (MMC) sensitivity, 1 × 105 cells were plated per well in a 6-well plate. After 1 day, the cells were incubated with media containing MMC for 2 h. After the MMC treatment, the media containing MMC were removed, and the cells were washed with phosphate-buffered saline. The cells were grown in fresh media for 2 days and were trypsinized. The number of viable cells was counted in the presence of trypan blue.
RESULTS
Inactivation of the XPF gene results in defective postnatal growth and a short life span.
Based on mutation analyses, the C-terminal half of XPF interacts with ERCC1 and constitutes the nuclease domain (5, 8). The N-terminal half of XPF shows sequence homology to the SF2 superfamily of archaeal RNA helicases. No biochemical activity has been demonstrated for the putative helicase domain. In most XPF patients, the mutations were mapped to the C-terminal half of the protein. In one patient (XP23OS), the entire C-terminal half of the protein (residues 445 to 905) was eliminated due to the insertion of an A between nucleotides 1330 and 1331, which leads to frameshift mutations after K444 and the appearance of a stop codon 38 residues later (12). Since no other XPF message was detected, the patient may be homozygous for this mutation or the other allele may be silenced. In spite of the extensive deletion, the only reported symptom of the patient is photosensitivity of the skin. Thus, this mutation could serve to maximally eliminate XPF activity without affecting normal development. For this reason, we chose to introduce this mutation into mice.
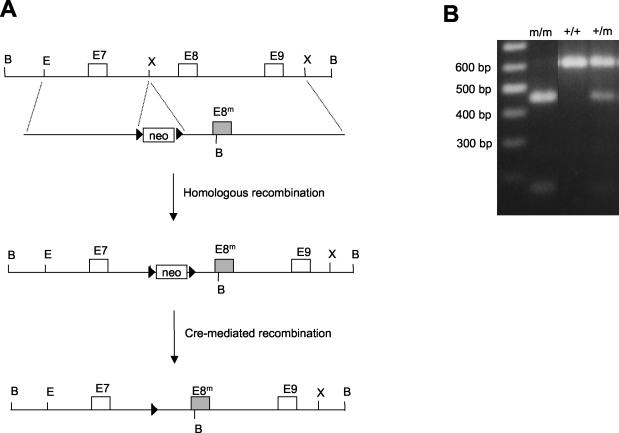
We introduced the mutation into the mouse XPF gene in ES cells by using a homologous recombination-based gene-targeting technique (Fig. 1A). To mimic the frameshift mutation in the human patient, which truncates the XPF protein starting from codon 445, we changed G-445 in exon 8 of the mouse XPF gene into a stop codon in the targeting construct. The mutation also created a novel BamHI site for easy identification (Fig. 1B). We replaced the original exon 8 of the XPF gene with the mutated version through homologous recombination. In addition, a neomycin resistance marker (Neor) was inserted in the intron between exons 7 and 8.
FIG. 1.
(A) This figure shows the strategy to introduce mutation into the XPF gene. The genomic locus around exon 8 is diagramed with relevant restriction enzyme sites shown (B, BamHI; E, EcoRI; X, XhoI). The targeting construct is shown below the genomic locus. The black triangle flanking the Neor marker represents the loxP site. (B) This figure shows the genotyping of mice tail DNA by PCR with primers that flank exon 8. The PCR product was digested with BamHI to distinguish between the wild-type (+, 600 bp) and mutant (m, 440 bp) alleles.
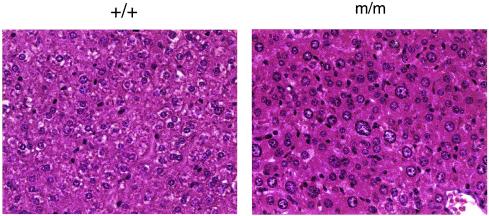
The mutated ES cell was used to generate chimeric mice, which transmitted the mutation through the germ line. The heterozygous mice (+/m) develop normally. Interbreeding between the heterozygous mice produced progeny with genotypes approaching the expected Mendelian ratios (111 mice: +/+, 26%; +/m, 53%; and m/m, 21%). Among the newly born litters, no abnormal pups were observed, suggesting that the homozygous mutation does not affect development up to the neonatal stage. Later on, certain pups were noticed to lag behind their littermates in growth, and these small pups were invariably found by genotyping to be homozygous mutants. At around 15 days after birth, the weight of homozygous mice was 27% of that of wild-type or heterozygous littermates. All the homozygous mice died approximately 3 weeks after birth. We examined the various tissues of the homozygous mice and found that the liver of the mutant animal contains cells with enlarged nuclei (Fig. 2). The other organs appear morphologically normal but significantly smaller.
FIG. 2.
This figure shows hematoxylin- and eosin-stained liver sections. +/+, wild type; m/m, homozygous.
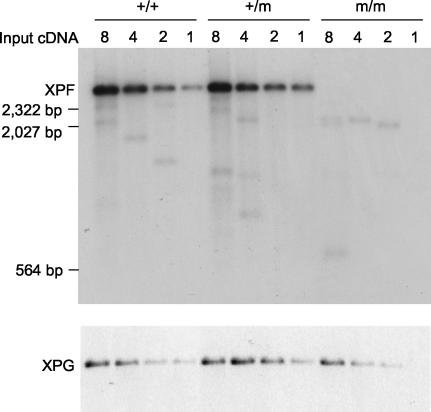
All of these phenotypes are identical to those of ERCC1 knockout mice, suggesting that the nonsense mutation may have completely inactivated the XPF gene (14, 37). We examined the expression of the mutant XPF allele in MEF by reverse transcriptase-mediated PCR (RT PCR) (Fig. 3). XPF message is indeed undetectable in homozygous cells. We also examined whether the mutant XPF allele is expressed in the heterozygous cells. Since the nonsense mutation creates a BamHI site, cDNAs derived from the wild-type and the mutant allele can be distinguished by BamHI digestion. Using this method, we found that all the XPF cDNAs amplified from the heterozygous cells were derived from the wild-type allele (Fig. 3). Thus, the nonsense mutation has severely inhibited the expression of the XPF gene, potentially through nonsense-mediated RNA decay. Based on the titration of RT PCRs, an expression level 10% of wild type should be detectable in this assay. Since no signal for the mutant XPF message was detected, the expression of the mutant XPF allele is decreased by at least 10-fold, and the actual reduction could be even larger. However, similarly mutated XPF mRNA accumulated to normal levels in the human patient. We do not understand the reason for this discrepancy. It is possible that the different nucleotide sequence of the mouse XPF mRNA could account for the distinct consequences of the nonsense mutation. The alternative possibility is that the Neor marker inserted in the intron between exons 7 and 8 interfered with the expression of the XPF gene. To address this possibility, we deleted the integrated Neor marker through the flanking loxP sites via Cre-mediated recombination by breeding XPFm/+ mice with Cre-expressing transgenic mice. We found that deleting the Neor marker had no effect on the mutant phenotype, ruling out the possibility that the inserted Neor marker is responsible for the severe developmental defect. Another possible explanation for the mild symptoms of the human patient might be that the second XPF allele of the patient expresses some (albeit strongly reduced) amount of the normal XPF protein or a XPF protein with a less debilitating mutation.
FIG. 3.
This figure shows the RT PCR analysis of XPF expression. The top panel presents PCR amplification of XPF cDNA from wild-type (+/+), heterozygous (+/m), and homozygous (m/m) MEF RNA. The input cDNA was serially diluted twofold for each sample. The PCR product was digested with BamHI to distinguish between wild-type and mutant XPF message. The PCR product was hybridized with a BamHI-EcoRI fragment from the 5′ end of XPF cDNA. The expected sizes of the wild-type and mutant XPF PCR product are 2,770 bp and 1,380 bp, respectively. The lower panel presents control PCR amplification of XPG cDNA from the same cDNAs.
Class switch recombination in an XPF-deficient mouse is normal.
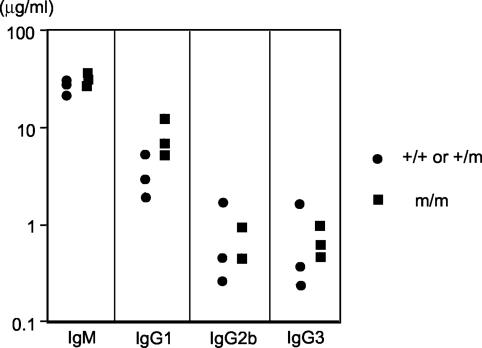
Knockout mice for mismatch repair proteins show defects in CSR (7, 28). Based on genetic studies in yeast, mismatch repair protein MSH2 cooperates with Rad1-Rad10, the yeast homologue of XPF-ERCC1, in cleaving terminal heterology in the single-stranded annealing pathway of double-strand break repair (23). Since a fraction of switch recombination joints appears to be mediated by microhomology, mismatch repair proteins and XPF-ERCC1 may be involved in this process. Furthermore, XPF-ERCC1 is capable of cleaving R-loop structures in vitro, and R loops have been implicated as an intermediate in CSR (30, 34, 40). To evaluate the effect of XPF inactivation on CSR in vivo, we isolated splenocytes from 15-day-old mice and stimulated the splenocytes in vitro with cytokines to induce CSR to various antibody isotypes. Six days later, the culture supernatant was collected and the levels of various antibody isotypes were quantified by ELISA. We found that the level of the different antibody isotypes (IgM, IgG1, IgG2b, and IgG3) secreted by XPF-deficient splenocytes was comparable to that of wild-type or heterozygous cells (Fig. 4). Thus, XPF-ERCC1 is dispensable for CSR. ERCC1 knockout mice were also found to undergo normal CSR (38).
FIG. 4.
This figure shows the ELISA analysis of antibody concentration in the culture supernatant of stimulated splenocytes. Each data point represents analysis of one mouse.
XPF-deficient embryonic fibroblasts are hypersensitive to UV and MMC.
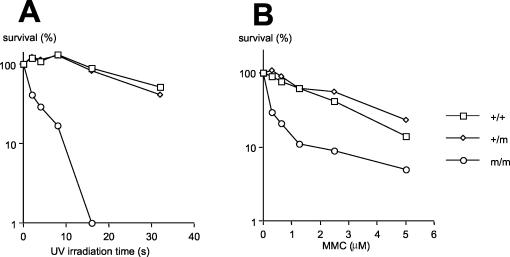
According to in vitro studies, XPF-ERCC1 makes the incision 5′ to the photodamage during nucleotide excision repair. To determine the effect of XPF inactivation on the repair of UV damage in cells, we isolated embryonic fibroblasts from embryos that were 13.5 days old. The size of XPF-deficient embryos is indistinguishable from that of wild-type and heterozygous embryos. Similarly, XPF-deficient embryonic fibroblasts showed no obvious growth defects. However, after irradiation with UVC, XPF-deficient cells show markedly decreased survival compared with wild-type or heterozygous cells (Fig. 5A). The hypersensitivity of XPF-deficient cells to UV irradiation is consistent with the role of XPF in nucleotide excision repair.
FIG. 5.
(A) This figure shows the survival curve of UV irradiation. The data are the average of two experiments. (B) This figure shows the survival curve of MMC treatment. The data are the average of two experiments.
Besides nucleotide excision repair, XPF-ERCC1 has also been implicated in eliminating interstrand DNA cross-links. XPF-ERCC1 has been shown to have cross-link-specific nuclease activity (18). In addition, XPF-ERCC1 is capable of making incisions near a cross-link in a branched DNA substrate (11). Since such branched DNA structure mimics a replication fork stalled by an interstrand cross-link, XPF-ERCC1 could potentially facilitate the removal of the lesion during DNA replication. To assay for this repair function in XPF-deficient cells, we incubated the embryonic fibroblasts with MMC, which generates interstrand DNA cross-links as well as mono-DNA adducts. The XPF-deficient cells are more sensitive to MMC compared with wild-type or heterozygous cells (Fig. 5B). Although the phenotype is consistent with a role of XPF-ERCC1 in repairing interstrand DNA cross-links, it could also be caused by a defect in removing MMC-induced mono-DNA adducts, which are potential substrates for nucleotide excision repair. Comparison with MEF from mice deficient in other nucleotide excision repair proteins such as XPA is needed to resolve this issue.
DISCUSSION
In this study, we present an analysis of mice deficient in XPF. Although designed to generate a partial inactivation of XPF, the mutation results in the complete inhibition of XPF expression. The XPF-deficient mice showed identical phenotypes to ERCC1 knockout mice, suggesting that XPF and ERCC1 function only as a complex. Cells deficient in either XPF or ERCC1 are hypersensitive to UV irradiation and MMC treatment. These phenotypes are readily explained by the well-characterized biochemical activities of the nuclease. On the other hand, the cause of the severe developmental defect of XPF- and ERCC1-deficient mice is unclear. Although the most obvious abnormalities are found in the liver, the defect is unlikely to be cell-type specific. This point was clearly demonstrated in a liver-specific complementation experiment with the ERCC1 knockout mice (29). A liver-specific ERCC1 transgene was shown to correct the liver abnormalities of ERCC1 knockout mice. However, the complemented mutant animal was still below normal size and died prematurely. In addition, cells with enlarged nuclei were found in the kidney of the complemented animal.
The cause of the enlarged nuclei is unknown. Cell cycle analysis revealed that these unusual cells have 4N and 8N DNA content and may be arrested in G2 phase or have undergone endoreduplication (21). Such phenotypes are indicative of problems in DNA replication, which could prevent the cells from entering mitosis by G2 checkpoints. Since XPF-ERCC1 is required for removing interstrand DNA cross-links, it is possible that interstrand DNA cross-links formed by metabolites accumulate in cells deficient in XPF and ERCC1 and impede DNA replication. Another possibility is that XPF-ERCC1 is required to process collapsed replication forks and recombination intermediates, which contain features similar to XPF-ERCC1 substrates.
Acknowledgments
We thank Laurie Davidson, Dan Foy, and Michele Smith for mouse work.
This work was supported by National Institutes of Health grant A13154 (to F.W.A.) and NIH training grant A107512 (to M.T.). F.W.A. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Aboussekhra, A., M. Biggerstaff, M. K. Shivji, J. A. Vilpo, V. Moncollin, V. N. Podust, M. Protic, U. Hubscher, J. M. Egly, and R. D. Wood. 1995. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 80:859-868. [DOI] [PubMed] [Google Scholar]
- 2.Adair, G. M., R. L. Rolig, D. Moore-Faver, M. Zabelshansky, J. H. Wilson, and R. S. Nairn. 2000. Role of ERCC1 in removal of long non-homologous tails during targeted homologous recombination. EMBO J. 19:5552-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berneburg, M., and A. R. Lehmann. 2001. Xeroderma pigmentosum and related disorders: defects in DNA repair and transcription. Adv. Genet. 43:71-102. [DOI] [PubMed] [Google Scholar]
- 4.de Boer, J., and J. H. Hoeijmakers. 2000. Nucleotide excision repair and human syndromes. Carcinogenesis 21:453-460. [DOI] [PubMed] [Google Scholar]
- 5.de Laat, W. L., A. M. Sijbers, H. Odijk, N. G. Jaspers, and J. H. Hoeijmakers. 1998. Mapping of interaction domains between human repair proteins ERCC1 and XPF. Nucleic Acids Res. 26:4146-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Vries, A., C. T. van Oostrom, F. M. Hofhuis, P. M. Dortant, R. J. Berg, F. R. de Gruijl, P. W. Wester, C. F. van Kreijl, P. J. Capel, H. van Steeg, and S. Verbeek. 1995. Increased susceptibility to ultraviolet-B and carcinogens of mice lacking the DNA excision repair gene XPA. Nature 377:169-173. [DOI] [PubMed] [Google Scholar]
- 7.Ehrenstein, M. R., and M. S. Neuberger. 1999. Deficiency in Msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombination: parallels with somatic hypermutation. EMBO J. 18:3484-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enzlin, J. H., and O. D. Scharer. 2002. The active site of the DNA repair endonuclease XPF-ERCC1 forms a highly conserved nuclease motif. EMBO J. 21:2045-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, E., J. G. Moggs, J. R. Hwang, J. M. Egly, and R. D. Wood. 1997. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 16:6559-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoy, C. A., L. H. Thompson, C. L. Mooney, and E. P. Salazar. 1985. Defective DNA cross-link removal in Chinese hamster cell mutants hypersensitive to bifunctional alkylating agents. Cancer Res. 45:1737-1743 [PubMed] [Google Scholar]
- 11.Kuraoka, I., W. R. Kobertz, R. R. Ariza, M. Biggerstaff, J. M. Essigmann, and R. D. Wood. 2000. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. J. Biol. Chem. 275:26632-26636. [DOI] [PubMed] [Google Scholar]
- 12.Matsumura, Y., C. Nishigori, T. Yagi, S. Imamura, and H. Takebe. 1998. Characterization of molecular defects in xeroderma pigmentosum group F in relation to its clinically mild symptoms. Hum. Mol. Genet. 7:969-974. [DOI] [PubMed] [Google Scholar]
- 13.Matsunaga, T., D. Mu, C. H. Park, J. T. Reardon, and A. Sancar. 1995. Human DNA repair excision nuclease. Analysis of the roles of the subunits involved in dual incisions by using anti-XPG and anti-ERCC1 antibodies. J. Biol. Chem. 270:20862-20869. [DOI] [PubMed] [Google Scholar]
- 14.McWhir, J., J. Selfridge, D. J. Harrison, S. Squires, and D. W. Melton. 1993. Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat. Genet. 5:217-224. [DOI] [PubMed] [Google Scholar]
- 15.Mu, D., C. H. Park, T. Matsunaga, D. S. Hsu, J. T. Reardon, and A. Sancar. 1995. Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem. 270:2415-2418. [DOI] [PubMed] [Google Scholar]
- 16.Mu, D., D. S. Hsu, and A. Sancar. 1996. Reaction mechanism of human DNA repair excision nuclease. J. Biol. Chem. 271:8285-8294. [DOI] [PubMed] [Google Scholar]
- 17.Mu, D., M. Wakasugi, D. S. Hsu, and A. Sancar. 1997. Characterization of reaction intermediates of human excision repair nuclease. J. Biol. Chem. 272:28971-28979. [DOI] [PubMed] [Google Scholar]
- 18.Mu, D., T. Bessho, L. V. Nechev, D. J. Chen, T. M. Harris, J. E. Hearst, and A. Sancar. 2000. DNA interstrand cross-links induce futile repair synthesis in mammalian cell extracts. Mol. Cell. Biol. 20:2446-2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakane, H., S. Takeuchi, S. Yuba, M. Saijo, Y. Nakatsu, H. Murai, Y. Nakatsuru, T. Ishikawa, S. Hirota, Y. Kitamura, Y. Kato, Y. Tsunoda, H. Miyauchi, T. Horio, T. Tokunaga, T. Matsunaga, O. Nikaido, Y. Nishimune, Y. Okada, and K. Tanaka. 1995. High incidence of ultraviolet-B or chemical-carcinogen-induced skin tumours in mice lacking the xeroderma pigmentosum group A gene. Nature 377:165-168. [DOI] [PubMed] [Google Scholar]
- 20.Niedernhofer, L. J., J. Essers, G. Weeda, B. Beverloo, J. de Wit, M. Muijtjens, H. Odijk, J. H. Hoeijmakers, and R. Kanaar. 2001. The structure-specific endonuclease Ercc1-Xpf is required for targeted gene replacement in embryonic stem cells. EMBO J. 20:6540-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nunez, F., M. D. Chipchase, A. R. Clarke, and D. W. Melton. 2000. Nucleotide excision repair gene (ERCC1) deficiency causes G(2) arrest in hepatocytes and a reduction in liver binucleation: the role of p53 and p21. FASEB J. 14:1073-1082. [DOI] [PubMed] [Google Scholar]
- 22.O'Donovan, A., A. A. Davies, J. G. Moggs, S. C. West, and R. D. Wood. 1994. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature 371:432-435. [DOI] [PubMed] [Google Scholar]
- 23.Paques, F., and J. E. Haber. 1997. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6765-6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petit, C., and A. Sancar. 1999. Nucleotide excision repair: from E. coli to man. Biochimie 81:15-25. [DOI] [PubMed] [Google Scholar]
- 25.Reardon, J. T., and A. Sancar. 2002. Molecular anatomy of the human excision nuclease assembled at sites of DNA damage. Mol. Cell. Biol. 22:5938-5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riedl, T., F. Hanaoka, and J. M. Egly. 2003. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 22:5293-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sands, A. T., A. Abuin, A. Sanchez, C. J. Conti, and A. Bradley. 1995. High susceptibility to ultraviolet-induced carcinogenesis in mice lacking XPC. Nature 377:162-165. [DOI] [PubMed] [Google Scholar]
- 28.Schrader, C. E., W. Edelmann, R. Kucherlapati, and J. Stavnezer. 1999. Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J. Exp. Med. 190:323-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selfridge, J., K. T. Hsia, N. J. Redhead, and D. W. Melton. 2001. Correction of liver dysfunction in DNA repair-deficient mice with an ERCC1 transgene. Nucleic Acids Res. 29:4541-4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shinkura, R., M. Tian, M. Smith, K. Chua, Y. Fujiwara, and F. W. Alt. 2003. The influence of transcriptional orientation on endogenous switch region function. Nat. Immunol. 4:435-441. [DOI] [PubMed] [Google Scholar]
- 31.Sijbers, A. M., W. L. de Laat, R. R. Ariza, M. Biggerstaff, Y. F. Wei, J. G. Moggs, K. C. Carter, B. K. Shell, E. Evans, M. C. de Jong, S. Rademakers, J. de Rooij, N. G. Jaspers, J. H. Hoeijmakers, and R. D. Wood. 1996. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell 86:811-822. [DOI] [PubMed] [Google Scholar]
- 32.Sijbers, A. M., P. C. van Voorst Vader, J. W. Snoek, A. Raams, N. G. Jaspers, and W. J. Kleijer. 1998. Homozygous R788W point mutation in the XPF gene of a patient with xeroderma pigmentosum and late-onset neurologic disease. J. Investig. Dermatol. 110:832-836. [DOI] [PubMed] [Google Scholar]
- 33.Sugasawa, K., J. M. Ng, C. Masutani, S. Iwai, P. J. van der Spek, A. P. Eker, F. Hanaoka, D. Bootsma, and J. H. Hoeijmakers. 1998. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell 2:223-232. [DOI] [PubMed] [Google Scholar]
- 34.Tian, M., and F. W. Alt. 2000. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J. Biol. Chem. 275:24163-24172. [DOI] [PubMed] [Google Scholar]
- 35.Volker, M., M. J. Mone, P. Karmakar, A. van Hoffen, W. Schul, W. Vermeulen, J. H. Hoeijmakers, R. van Driel, A. A. van Zeeland, and L. H. Mullenders. 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8:213-224. [DOI] [PubMed] [Google Scholar]
- 36.Wakasugi, M., and A. Sancar. 1999. Order of assembly of human DNA repair excision nuclease. J. Biol. Chem. 274:18759-18768. [DOI] [PubMed] [Google Scholar]
- 37.Weeda, G., I. Donker, J. de Wit, H. Morreau, R. Janssens, C. J. Vissers, A. Nigg, H. van Steeg, D. Bootsma, and J. H. Hoeijmakers. 1997. Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr. Biol. 7:427-439. [DOI] [PubMed] [Google Scholar]
- 38.Winter, A. G., K. Samuel, K. T. Hsia, and D. W. Melton. 2003. The repair and recombination enzyme ERCC1 is not required for immunoglobulin class switching. DNA Repair (Amst). 2:561-569. [DOI] [PubMed] [Google Scholar]
- 39.You, J. S., M. Wang, and S. H. Lee. 2003. Biochemical analysis of the damage recognition process in nucleotide excision repair. J. Biol. Chem. 278:7476-7485. [DOI] [PubMed] [Google Scholar]
- 40.Yu, K., F. Chedin, C. L. Hsieh, T. E. Wilson, and M. R. Lieber. 2003. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 4:442-451. [DOI] [PubMed] [Google Scholar]