Abstract
The budding yeast centromere-kinetochore complex ensures high-fidelity chromosome segregation in mitosis and meiosis by mediating the attachment and movement of chromosomes along spindle microtubules. To identify new genes and pathways whose function impinges on chromosome transmission, we developed a genomic haploinsufficiency modifier screen and used ctf13-30, encoding a mutant core kinetochore protein, as the reference point. We demonstrate through a series of secondary screens that the genomic modifier screen is a successful method for identifying genes that encode nonessential proteins required for the fidelity of chromosome segregation. One gene isolated in our screen was RSC2, a nonessential subunit of the RSC chromatin remodeling complex. rsc2 mutants have defects in both chromosome segregation and cohesion, but the localization of kinetochore proteins to centromeres is not affected. We determined that, in the absence of RSC2, cohesin could still associate with chromosomes but fails to achieve proper cohesion between sister chromatids, indicating that RSC has a role in the establishment of cohesion. In addition, numerous subunits of RSC were affinity purified and a new component of RSC, Rtt102, was identified. Our work indicates that only a subset of the nonessential RSC subunits function in maintaining chromosome transmission fidelity.
Maintaining the integrity of the genome through successive generations is the fundamental goal of the chromosome cycle. High-fidelity chromosome transmission requires the execution and coordination of many events, including DNA replication, sister chromatid cohesion, and kinetochore attachment and movement along microtubules. The regulation of the chromosome cycle relies on a multitude of gene products, many of which are highly conserved in eukaryotes.
In Saccharomyces cerevisiae, sister chromatids are held together by the multisubunit cohesin complex (reviewed in references 33 and 54). Cohesin is composed of at least four proteins, Scc1/Mcd1, Smc1, Smc3, and Scc3; it has been proposed that these proteins form a large ring structure around the sister chromatids (16; reviewed in reference 54). In yeast, cohesin is associated with chromosomes from late G1 until the metaphase-to-anaphase transition (32, 51). Scc2 and Scc4 form a complex that is required for loading cohesin onto chromatin in S phase (10, 32, 51). Localization of cohesin to chromatids is not sufficient for sister chromatid cohesion; rather, cohesion is established during DNA replication. Many proteins have been implicated specifically in either the establishment of sister chromatid cohesion in S phase, including Eco1/Ctf7 (46, 51) and proteins of the alternative replication factor C complex, containing Ctf18, Ctf8, and Dcc1 (18, 27), or the maintenance of cohesion during the subsequent G2 and M phases, including Pds5 (19, 36). Cohesin is deposited at discrete points along the chromosome arms at cohesion attachment regions (CARs) and is highly concentrated at yeast centromere DNA (CEN) (reviewed in reference 54). The high concentration of cohesin flanking the yeast centromere is thought to be required for the proper biorientation of the kinetochore toward opposite poles (49).
The yeast CEN is 125 bp and consists of palindromic sequences CDEI and CDEIII flanking an A-T-rich CDEII sequence (reviewed in references 9 and 43). Evidence suggests that the centromeric chromatin includes a histone octamer that contains the histone H3-like protein Cse4 (reviewed in references in 4 and 9). The inner kinetochore complex, CBF3, composed of the four essential proteins Ctf13, Ndc10/Ctf14, Cep3, and Skp1, binds CDEIII (reviewed in references 4 and 9). Point mutations in either Cse4 or components of the CBF3 complex result in chromosome missegregation, indicating that recruitment of CBF3 and Cse4 to CEN DNA is essential for faithful chromosome maintenance. The CEN-binding proteins of the inner kinetochore, Cse4 and the CBF3 complex, create a scaffold onto which the large central and outer kinetochore complexes bind (reviewed in references 4 and 9). Recent advances in the identification of the proteins linking CEN DNA to microtubules have identified more than 30 proteins in the kinetochore. Several additional distinct kinetochore complexes have been identified, including two central kinetochore complexes, the 12-subunit Ctf19 complex and the 4-subunit Ndc80 complex, and the outer kinetochore complex, the 9-subunit Dam1 complex (reviewed in references 4 and 9).
It is clear that, in addition to the proteins of the kinetochore, chromatin proteins are integral to centromere structure and function (reviewed in reference 43). The core CEN DNA is contained within a nuclease-resistant region, and flanking the core CEN are highly phased nucleosome arrays that can extend for more than 2 kb (5). Further, point mutations in CDEIII and Cse4 cause a drastic disruption of nuclease resistance in the core and flanking regions (31, 40). The RSC (remodel the structure of chromatin) nucleosome remodeling complex has also been implicated in contributing to the integrity of CEN DNA (20, 53). RSC is a member of the SWI/SNF family of ATP-dependent chromatin remodelers, and most of the core subunits of RSC are essential for mitotic growth (reviewed in reference 56). Biochemical analysis has determined that RSC contains at least 15 proteins and that RSC is present in distinct complexes (1, 6, 7). For example, highly related proteins Rsc1 and Rsc2 form two distinct complexes with RSC (7). Neither RSC1 nor RSC2 is essential, but loss of both causes lethality (7). Interestingly, RSC1 and RSC2 deletion mutants display many distinct phenotypes (3, 7, 58, 61), suggesting that the RSC1 and RSC2 forms of RSC have different cellular functions.
Mutations in several, but not all, RSC subunits cause a G2/M arrest characterized by large budded cells containing 2N and 4N chromosomes (1, 7, 8, 12, 14, 38, 52, 53), suggesting that RSC has more then one essential function in yeast. Genome-wide localization studies with various components of RSC indicate that RSC localizes to more than 700 intergenic regions (12, 34). Interestingly, there is no significant overlap between RSC targets and genes required for G2/M progression, suggesting that the G2/M phenotype of rsc mutants may be due to general chromatin-modifying activities and not to specific defects in transcription. The disruption of ordered nucleosomes around CENs in rsc mutants suggests that RSC may directly model the nucleosomes of the core kinetochore. However, while it has been shown that the human homolog of RSC1 and RSC2, BAF180, localizes to the kinetochores specifically in prometaphase (59), there are conflicting observations as to whether RSC components do or do not significantly localize to CEN DNA (20, 34). Interestingly, it was shown recently that the human chromatin remodeling protein hSNF2 interacts with the human hRAD21 cohesin complex and that the chromatin remodeling activity of hSNF2 is required to mediate association of cohesin with chromatin (17). Interaction between yeast chromatin remodeling complexes and cohesin has not yet been demonstrated.
To further advance our understanding of the proteins that contribute to genome stability in yeast, we developed a genomic haploinsufficiency modifier screen to isolate genetic modifiers of a hypomorphic mutation of CTF13, a gene that encodes an essential protein of the CBF3 inner kinetochore complex. We demonstrate through a series of secondary screens that the genomic modifier screen is a successful method for isolating genes that have defects in chromosome segregation. Two of the genes isolated were RSC1 and RSC2. We determined that the Rsc2-containing subcomplex of RSC is the primary form of RSC responsible for chromosome stability and proper sister chromatid cohesion. Further, we demonstrate that RSC interacts genetically with cohesin and that RSC contributes the establishment of sister chromatid cohesion. In addition we identify a new subunit of RSC, Rtt102, which, like Rsc2, contributes to chromosome segregation fidelity.
MATERIALS AND METHODS
Yeast strains.
The MATα deletion mutant array (DMA) (57) was purchased from ResGen/Invitrogen. Table 1 lists the genotypes of all additional yeast strains used in this study. Deletion strains and 13-Myc C-terminal tag strains made for this study were designed with PCR-amplified cassettes as previously described (26). Tandem affinity purification (TAP)-tagged strains made for this study were designed with PCR-amplified cassettes as previously described (24).
TABLE 1.
List of yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| YPH972 | MATα ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ1 ura3-52 CFIII(CEN3.L) URA3 SUP11 ctf13-30 | 13 |
| YKB10 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1 ura3-52 CFIII(CEN3.L) URA3 SUP11ctf13-30 | This study |
| YKB11 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1 ura3-52 CFIII(CEN3.L) URA3 SUP11 | This study |
| YPH907 | MATa/α ade2-101/ade2-101 his3Δ200/his3Δ200 leu2Δ1/LEU2 lys2-801/lys2-801 trp1Δ1/trp1Δ1 ura3-52/ura3-52 CFIII(CEN3.L) URA3 SUP11 | P. Hieter |
| YPH500 | MATα ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 | 45 |
| YKB2 | MATa/α ade2-101/ade2-101 his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 lys2-801/lys2-801 trp1Δ63/trp1Δ1 ura3-52/ura3-52 CFIII(CEN3.L) URA3 SUP11 ctf13-30/CTF13 | This study |
| YKB407 | MATα ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1 ura3-52 ctf13-30 | This study |
| YKB51 | MATα ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ1 ura3-52 CFIII(CEN3.L) URA3 SUP11 rsc2ΔkanMX6 | This study |
| YKB120 | MATα ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ1 ura3-52 CFIII(CEN3.L) URA3 SUP11 rsc1ΔkanMX6 | This study |
| YKB46 | MATα ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ1 ura3-52 CFIII(CEN3.L) URA3 SUP11 fyv8ΔkanMX6 | This study |
| YKB41 | MATα ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ1 ura3-52 CFIII(CEN3.L) URA3 SUP11 nis1ΔkanMX6 | This study |
| YKB43 | MATα ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ1 ura3-52 CFIII(CEN3.L) URA3 SUP11 vid21ΔkanMX6 | This study |
| YKB36 | MATα ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ1 ura3-52 CFIII(CEN3.L) URA3 SUP11 nst1ΔkanMX6 | This study |
| YKB412 | MATα ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ1 ura3-52 CFIII(CEN3.L) URA3 SUP11 spo74ΔkanMX6 | This study |
| YPH982 | MATa/α ade2-101/ade2-101 his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 lys2-801/lys2-801 trp1Δ63/trp1Δ63 ura3-52/ura3-52 CFIII(CEN3.L) URA3 SUP11 | P. Hieter |
| YKB408 | MATa/α ade2-101/ade2-101 his3Δ200/his3Δ200 leu2Δ1/LEU2 lys2-801/lys2-801 trp1Δ63/trp1Δ63 ura3-52/ura3-52 CFIII(CEN3.L) URA3 SUP11 rsc2ΔkanMX6/rsc2ΔkanMX6 | This study |
| YKB409 | MATa/α ade2-101/ade2-101 his3Δ200/his3Δ200 leu2Δ1/LEU2 lys2-801/lys2-801 trp1Δ63/trp1Δ63 ura3-52/ura3-52 CFIII(CEN3.L) URA3 SUP11 rsc1ΔkanMX6/rsc1ΔkanMX6 | This study |
| YKB410 | MATa/α ade2-101/ade2-101 his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 lys2-801/lys2-801 trp1Δ63/trp1Δ63 ura3-52/ura3-52 CFIII(CEN3.L) URA3 SUP11 npl6ΔkanMX6/npl6ΔkanMX6 | This study |
| YKB411 | MATa/α ade2-101/ade2-101 his3Δ200/his3Δ200 leu2Δ1/leu2Δ1 lys2-801/lys2-801 trp1Δ63/trp1Δ63 ura3-52/ura3-52 CFIII(CEN3.L) URA3 SUP11 rtt102ΔkanMX6/rtt102ΔkanMX6 | This study |
| YPH1052 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ-63 ura3-52 ndc10-1 | P. Hieter |
| YKB93 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ1 ura3-52 CFIII(CEN3.L) URA3 SUP11 ctf14-42 | 28 |
| YPH1314 | MATα ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ1 ura3-52 ctf19ΔHIS3 | 21 |
| YPH1313 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ1 ura3-5 ctf19ΔLEU2 CFVII(RAD2.d)URA3 SUP11 | 21 |
| YJL158 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 cyh2+ CFVI(CEN6) URA3 SUP11 CYH2s okp1::okp1-5::TRP1 | 35 |
| YVM111 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 ctf3ΔHIS3 | 28 |
| YVM957 | MATα leu2-3 ade2-101 trp1-Δ901 his3-11,15 cse4-1::RSCL1-1H(HIS3) | 28 |
| YVM280 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 mcm22ΔURA3 | 28 |
| YKB147 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1 ura3-52 rsc2ΔkanMX6 cse4-1::HIS3 | This study |
| YKB57 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1 ura3-5 rsc2ΔkanMX6 ctf13-30 YMR090w::HIS3 | This study |
| YKB208 | MATα ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1 ura3-5 rsc2ΔkanMX6 okp1::okp1-5::TRP1 | This study |
| YKB140 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1 ura3-5 rsc2ΔkanMX6 ctf3ΔHIS3 | This study |
| YKB183 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ1 ura3-5 rsc2ΔkanMX6 ctf19ΔLEU2 | This study |
| YKB136 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1 ura3-5 rsc2ΔkanMX6 mcm22ΔURA3 | This study |
| YKB169 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1 ura3-5 rsc1ΔkanMX6 ndc10-1 | This study |
| YKB167 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1 ura3-5 rsc1ΔkanMX6 ctf14-42 | This study |
| YKB219 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1 ura3-5 rsc1ΔkanMX6 cse4-1::HIS3 | This study |
| YKB166 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1 ura3-5 rsc1ΔkanMX6 okp1::okp1-5::TRP | This study |
| YKB217 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1 ura3-5 rsc1ΔkanMX6 ctf3ΔHIS3 | This study |
| YKB185 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1 ura3-5rsc1ΔkanMX6 ctf19ΔLEU2 | This study |
| YKB216 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1 ura3-5 rsc1ΔkanMX6 mcm22ΔURA3 | This study |
| YPH499 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trpΔ63 ura3-5 | P. Hieter |
| YKB49 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trpΔ63 ura3-5 rsc2ΔkanMX6 | This study |
| YVM499 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trpΔ63 ura3-5 NDC10-13MYC::kanMX6 | 28 |
| YKB121 | MATα ade2-101 his3Δ200 leu2Δ1 lys2-801 trpΔ63 ura3-5 NDC10-13MYC:kanMX6 rsc2ΔkanMX6 | This study |
| YVM218 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trpΔ63 ura3-5 CTF3-13MYC:TRP1 | 28 |
| YKB127 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trpΔ63 ura3-5 CTF3-13MYC:TRP1 rsc2ΔkanMX6 | This study |
| YVM1141 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trpΔ63 ura3-5 CSE4-3HA:URA3 | 28 |
| YKB124 | MATα ade2-101 his3Δ200 leu2Δ1 lys2-801 trpΔ63 ura3-5 CSE4-3HA:URA3 rsc2ΔkanMX6 | This study |
| K5832 | MATaade2-1can1-100 his3-11,15 leu2-3,112 ura3-5 scc1-73 | 32 |
| K5828 | MATaade2-1can1-100 his3-11,15 leu2-3,112 ura3-5 scc2-4 | 32 |
| K5824 | MATaade2-1can1-100 his3-11,15 leu2-3,112 ura3-5 smc3-42 | 32 |
| YBS514 | MATα ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 ctf7Δ1::HIS3 ctf7-203::LEU2 | 46 |
| YMM1093 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 ctf8ΔHIS3 | M. Mayer |
| YPH1492 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 ctf18ΔHIS3 | 27 |
| YKB405 | MATaade2 his3 leu2 ura3 scc1-73 rsc2ΔkanMX6 | This study |
| YKB172 | MATaade2 his3 leu2 ura3 scc1-73 rsc1ΔkanMX6 | This study |
| YKB151 | MATaade2 his3 leu2 ura3 scc2-4 rsc2ΔkanMX6 | This study |
| YKB173 | MATaade2 his3 leu2 ura3 scc2-4 rsc1ΔkanMX6 | This study |
| YKB404 | MATaade2 his3 leu2 ura3 smc3-42 rsc2ΔkanMX6 | This study |
| YKB142 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 ctf8ΔHIS3 rsc2ΔkanMX6 | This study |
| YKB160 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 ctf8ΔHIS3 rsc1ΔkanMX6 | This study |
| YKB210 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 ctf18ΔHIS3 rsc2ΔkanMX6 | This study/PICK> |
| YKB214 | MATα ade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 ctf18ΔHIS3 rsc1ΔkanMX6 | This study |
| YKB413 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 ctf7ΔHIS3 ctf7-203::LEU2 rsc2ΔkanMX6 | This study |
| YPH1477 | MATaade2-1 trp1-1 can1-100 his3-11,15 leu2::LEU2tetR-GFP ura3::3XURA3tet0112 PDS1-13MYC::TRP1 | 27 |
| YKB235 | MATaade2-1 trp1-1 can1-100 his3-11,15 leu2::LEU2tetR-GFP ura3::3XURA3tet0112 PDS1-13MYC::TRP1 rsc2ΔkanMX6 | This study |
| YKB177 | MATaade2-1 trp1-1 can1-100 his3-11,15 leu2::LEU2tetR-GFP ura3::3XURA3tet0112 PDS1-13MYC::TRP1 rsc1ΔkanMX6 | This study |
| YCK245 | MATaura3-1 leu2-3,112 his3-11,15 trp1-1 ade2-1 can1-100 | 24 |
| NJK182 | As YCK245 but RTT102-TAP::TRP1 | This study |
| NJK192 | As YCK245 but RSC58-TAP::TRP1 | This study |
| NJK203 | As YCK245 but RSC3-TAP::TRP1 | This study |
| NJK208 | As YCK245 but RSC2-TAP::TRP1 | This study |
| NJK210 | As YCK245 but RSC4-TAP::TRP1 | This study |
| NJK211 | As YCK245 but RSC8-TAP::TRP1 | This study |
| NJK212 | As YCK245 but NPL6-TAP::TRP1 | This study |
| NJK213 | As YCK245 but RSC6-TAP::TRP1 | This study |
| K7562 | MATα ade2-1 can1-100 leu2-3,112 his3-11,14 ura3 SMC-3HA6::HIS3 | 51 |
| YKB353 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 SMC3-HA6::HIS3 | This study |
| YKB355 | MATaade2-101 his3Δ200 leu2Δ1 lys2-801 trp1Δ63 ura3-52 SMC3-HA6::HIS3 rsc2ΔkanMX6 | This study |
| K8502 | MATaura trp ade pep4ΔLEU2 SCC1-HA6::HIS3 | 51 |
| YKB341 | MATaura trp ade leu his SCC1-HA6::HIS3 rsc2ΔkanMX6 | This study |
| YKB342 | MATaura trp ade leu his SCC1-HA6::HIS3 | This study |
| YKB426 | MATaade2-1 trp1-1 can1-100 his3-11,15 leu2::LEU2tetR-GFP ura3::3XURA3tet0112 PDS1-13MYC::TRP1 scc1-73 | This study |
ctf13-30/CTF13 genomic modifier screen.
The DMA was transferred by hand with a 96-floating-pin replicator (VP408FH; V & P Scientific Inc.) and a colony copier (VP380) from frozen stocks and propagated on rich medium containing G418 (200 mg/liter; Gibco BRL). All growth was conducted at 25°C. Replicator sterilization procedures were performed as previously described (50). Mid-log-phase cultures of YKB10 and YKB11 grown in synthetic complete medium lacking uracil 3 to an optical density at 600 nm of 10 (SC-Ura) were plated on omnitrays containing yeast extract-peptone-dextrose (YPD) to create mating lawns. For the mating reaction the DMAs were pinned on top of the lawns of YKB10 or YKB11 and placed at 25°C overnight to mate. The resulting MATa/α diploids were pinned onto SC−Ura−Trp to select for diploids. After 1 day of growth, the diploids were selected for a second time by repinning onto a second SC−Ura−Trp plate. The following day the diploids were inoculated into 96-well plates containing 100 μl of liquid SC−Ura−Trp. The strains were grown overnight to saturation to compensate for any differences in growth rates. The next day, a multichannel electronic pipette (Ranin; EPDS) was used to dilute the strains to 200 cells/μl in 96-well plates, and 5 μl of each strain was deposited onto six different plates: two YPD plates, two YPD plates containing benomyl (5 and 8 μl/ml), and two YPD plate containing nocodazole (2.5 and 4 μl/ml). One YPD plate was grown at 37°C, while the remaining five plates were grown at 25°C. The growth behavior of the ctf13-30/CTF13 XΔ/X strain was compared directly to that of the XΔ/X strain over a period of 4 days.
SDL assay.
The inducible plasmids used for the synthetic dosage lethality (SDL) screen were pGAL1-SKP1 (bPH562) (11), pGAL1-CTF13 (pKF88) (25), pGAL1-NDC10/CTF14 (pKH2) (29), and pRS415GEU1 (vector control pBH546) (11). SDL analysis was performed essentially as previously described (29) with the following exception: the plasmids were transformed into the 36 MATα deletion strains (ResGen/Invitrogen) listed in Table 2 and into YPH499 and YKB407.
TABLE 2.
Summary of ctf13-30/CTF13 modifier screen
| Open reading frame | Gene name | Biological processa |
|---|---|---|
| Enhancers of Ts and benomyl and nocodazole sensitivity | ||
| YDR359C | VID21 | Unknown |
| YKL054C | DEF1 | Response to DNA damage |
| YML094W | GIM5 | Tubulin folding |
| YNL091W | NST1 | Salinity response |
| Enhancer of Ts and nocodazole sensitivity | ||
| YLR357W | RSC2 | Chromatin modeling |
| Enhancers of benomyl and nocodazole sensitivity | ||
| YER095W | RAD51 | Recombinase involved in double-strand break repair and meiosis |
| YGR056W | RSC1 | Chromatin modeling |
| YGR092W | DBF2 | Kinase involved in nuclear division |
| YIL165C | YIL165C | Unknown |
| YKR082W | NUP133 | Nuclear pore protein |
| YMR214W | SCJ1 | Protein folding |
| YNL078W | NIS1 | Regulation of mitosis |
| YNL298W | CLA4 | Kinase involved in cytokinesis and polarity establishment |
| YNL307C | MCK1 | Kinase involved in mitosis, meiosis, and stress response |
| YOL012C | HTZ1 | Histone-related protein involved in silencing |
| YPL193W | RSA1 | Ribosomal large-subunit assembly and maintenance |
| Enhancers of benomyl sensitivity | ||
| YBR188C | NTC20 | mRNA splicing |
| YDR309C | GIC2 | Establishment of polarity |
| YHR191C | CTF8 | Sister chromatid cohesion |
| YKL204W | EAP1 | Negative regulation of translation |
| YOL081W | IRA2 | RAS protein signal transduction |
| YOL104C | NDJ1 | Synapsis |
| Enhancers of nocodazole sensitivity | ||
| YBR189W | RPS9B | Protein synthesis |
| YCR008W | SAT4 | Kinase involved in G1/S transition of cell cycle |
| YGL121C | GPG1 | Signal transduction |
| YGR196C | FYV8 | Unknown |
| YNL294C | RIM21 | Invasive growth and sporulation |
| YOR296W | YOR296W | Unknown |
| Suppressors of benomyl and nocodazole sensitivity | ||
| YDL047W | SIT4 | Phosphatase involved in G1/S transition of cell cycle |
| YGR056W | PAC10 | Tubulin folding |
| Suppressors of nocodazole sensitivity | ||
| YBL067C | UBP13 | Hypothetical deubiquitinating enzyme |
| YGL170C | SPO74 | Sporulation |
| YJL188C | BUD19 | Bud site selection |
| YMR252C | YMR252C | Unknown |
| YPL018W | CTF19 | Chromosome segregation |
| Suppressors of benomyl sensitivity | ||
| YDR488C | PAC11 | Microtubule-based process |
| YLR370C | ARC18 | Actin filament organization |
Biological process adapted from the Saccharomyces Genome Database report.
Chromosome missegregation assay.
Colony color sector analysis was performed as previously described (23, 47). For qualitative analysis the tested strains along with wild-type and ctf13-30 control strains were plated to single colonies onto limiting adenine plates and colonies were grown at 25, 30, 33, and 35°C for 3 days before the plates were placed at 4°C for optimal red pigment development. Quantitative half-sector analysis was performed as previously described (21, 23). In brief, homozygous diploid strains (YPH907, wild type; YKB408, rsc2Δ; YKB409, rsc1Δ; YKB410, npl6Δ; YKB411, rtt102Δ) containing a single SUP11-marked chromosome fragment were plated to single colonies on solid media containing limiting adenine and grown at 30°C for 3 days before the plates were placed at 4°C for optimal red pigment development.
ChIP assays.
Chromatin immunoprecipitation (ChIP) experiments were performed as previously described (2) with the following alterations. For all ChIP assays, the cells were cross-linked with 1% formaldehyde for 15 min at room temperature. Cell extracts were sonicated until chromatin was sheared to an average size of 500 bp. Immunoprecipitations (IPs) were performed with 20 μl of anti-Myc (9E10) or anti-hemagglutinin (HA) (HA.11) affinity matrix (Covance). Two milligrams of lysate was used per IP. The templates used for PCRs ranged from 1/2,000 to 1/10,000 of total chromatin and 1/100 to 1/50 of total immunoprecipitate, depending on the linear range. Primers used for PCR analysis were previously described (30).
Cell cycle synchronization.
For α-factor block-and-release experiments cultures were grown at 25°C to an optical density at 600 nm of 0.2 and α-factor was added directly to the medium to a final concentration of 5 μM. Cultures were incubated with α-factor for 2 h until at least 95% of the cells were arrested in G1 phase, as determined by microscopy. Cells were released from α-factor arrest into YPD containing 15 μg of nocodazole/ml. After 120 min at least 90% of the cells had undergone S phase and were arrested in G2/M. Cells were pelleted and resuspended in YPD media containing 15 μg of nocodazole/ml prewarmed to 37°C. Samples were taken at various times, and position in the cell cycle was assessed by flow cytometry analysis of DNA content and bud morphology.
Sister chromatid cohesion assays.
Sister chromatid cohesion assays were performed as described previously (27). For sister chromatid cohesion assays performed on G2/M- or G1-arrested cells, cells were arrested in G2/M with 15 μg of nocodazole (Sigma)/ml or in G1 with 5 μg of α-factor (Diagnostic Chemicals Limited)/ml for 3 h at 37°C. Sister chromatid cohesion assays performed on α-factor block and release are described in the previous section. Cells were fixed with an equal volume of 4% paraformaldehyde, washed once with SK (1 M sorbitol, 0.05 M K2PO4), and resuspended in SK for cohesion assessment. For each sample a minimum of 100 cells were assessed for sister chromatid cohesion.
Chromatin spreads.
Chromatin spreads were performed as previously described (32). Mouse anti-HA (12CA5; Boehringer Mannheim) antibodies were used at 1:2,500, and Cy3-conjugated goat anti-mouse (Jackson ImmunoResearch Laboratories, Inc.) antibodies were used at 1:3,000.
TAP-tagged-protein purification and identification.
Purification of TAP-tagged proteins and identification of associated proteins by matrix-assisted laser desorption ionization-time of flight in (MALDI-ToF) mass spectrometry (MS) and liquid chromatography-tandem MS (LC/MS/MS) were performed essentially as described previously (24).
RESULTS
ctf13-30/CTF13 genomic modifier screen.
To identify new genes and pathways whose function impinges on chromosome transmission, we developed a genomic haploinsufficiency modifier screen using the yeast gene deletion set. Genetic modifier screens have been utilized extensively for Drosophila melanogaster and other multicellular organisms where the mutation of interest is dosage sensitive; however, few screens of this nature have been conducted with yeast (48, 55). Two challenges facing traditional, random-mutagenesis-based screens are establishing comprehensive mutagenesis of the genome and the subsequent cloning and confirmation of the second-site alleles. To circumvent these issues, We developed a yeast modifier screen that is accomplished by mating a reference mutant strain to the yeast deletion set. A mutant allele of the CBF3 complex, ctf13-30, that is sensitive to both temperature and microtubule destabilizing drugs was chosen to test the procedure (13, 47).
The genomic modifier screen procedure is outlined in Fig. 1. Briefly, the yeast MATα DMA was mated to both wild-type (CTF13) and mutant (ctf13-30) MATa haploids, and diploid cells were selected. The resultant 4,700 singly heterozygous (xΔ/X CTF13/CTF13) strains, the 4,700 doubly heterozygous diploid (xΔ/X ctf13-30/CTF13) strains, and the single ctf13-30 heterozygous control (ctf13-30/CTF13 X/X) strain were screened for sensitivity to either temperature or microtubule-destabilizing agents benomyl and nocodazole. Deletions that, when heterozygous, modified the behavior of heterozygous ctf13-30/CTF13 strains were sought. By comparing in parallel the behavior of the singly heterozygous diploid to that of the doubly heterozygous diploid, heterozygous deletions that genetically interact with heterozygous ctf13-30/CTF13 are readily identified.
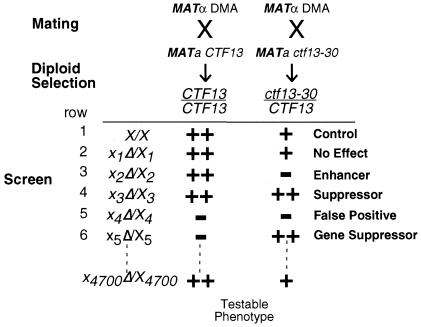
FIG. 1.
Schematic outline of the yeast genomic modifier screen. In our screen we use growth as a testable phenotype. ++, wild-type growth; +, mild slow growth, −, no growth or slow growth. X1, deletion of gene 1, X2, deletion of gene 2, etc. Heterozygous ctf13-30/CTF13 causes a mild slow-growth phenotype compared to that of wild-type (X/X) strains. Heterozygous deletions that modified the phenotype produced by heterozygous ctf13-30/CTF13 were sought. Most unlinked heterozygous deletions do not affect the mild growth defect of heterozygous ctf13-30/CTF13 mutants (row 2). Enhancers are genes whose heterozygous deletion in combination with ctf13-30/CTF13 causes a more severe slow-growth phenotype of ctf13-30/CTF13 mutants (row 3). Suppressors are genes whose heterozygous deletion in combination with ctf13-30/CTF13 causes a rescue of the slow-growth phenotype of ctf13-30/CTF13 mutants (row 4). In the ctf13-30/CTF13 example described in the text the mild slow-growth phenotype of ctf13-30/CTF13 mutants compared to that of wild-type cells did not allow efficient detection of traditional suppressors. Numerous single heterozygous deletions produce haploinsufficiency (rows 5 and 6). False positives are detected in cases where the single heterozygous deletion itself causes a severe growth defect (row 5). In some cases the haploinsufficiency phenotype displayed by a single heterozygous diploid was suppressed by the addition of heterozygous ctf13-30/CTF13 (row 6). These represent genetic interactions in the opposite direction, and we have called these gene suppressors.
We performed the ctf13-30/CTF13 modifier screen twice, and Table 2 lists the 36 deletion strains that were isolated in both screens. Only four genes, VID21, DEF1, GIM5, and NST1, were enhancers of all three phenotypes that were tested. The ctf13-30/CTF13 modifier screen identified many genes whose functions are known to be important for chromosome transmission, including CTF8 (18, 27), CTF19 (21), MCK1 (37, 44), and RAD51 (22), demonstrating the value of performing this type of genomic modifier screen.
Secondary SDL and chromosome loss screens.
An SDL interaction occurs when overexpression of a cloned wild-type “reference” gene is lethal or causes a slow-growth defect in a “target” mutant strain but is viable in a wild-type strain (reviewed in reference 29). If the target mutant exhibits a conditional phenotype, a dosage suppression screen can be conducted in parallel at the nonpermissive condition. SDL screens in which known kinetochore genes have been overexpressed in a collection of chromosome transmission fidelity mutants have been particularly successful in isolating new kinetochore components (21, 25, 28). Therefore, to identify genes more likely to be defective in kinetochore function, we performed an SDL screen on the corresponding 36 haploid deletion strains identified as haploinsufficiency modifiers of ctf13-30/CTF13. We chose to inducibly overexpress three genes of the CBF3 complex, CTF13, SKP1, and NDC10. As previously reported, overexpression of CTF13 and SKP1 had no effect on the growth of a wild-type strain but suppressed the temperature sensitivity (Ts) phenotype of a ctf13-30 strain (Table 3) (11, 29). In addition, the SDL screen demonstrated that 11 out of the 36 deletion strains isolated in the ctf13-30/CTF13 modifier screen displayed sensitivity to overexpression of at least one of the kinetochore proteins (Table 3).
TABLE 3.
SDL and chromosome transmission fidelity analysis of ctf13-30/CTF13 modifiers
| Straina | Result at indicated temp (°C) of:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SDL screenb
|
CTF screenc | ||||||||||||
| Vector
|
SKP1
|
CTF13
|
NDC10
|
||||||||||
| 25 | 30 | 35 | 25 | 30 | 35 | 25 | 30 | 35 | 25 | 30 | 35 | ||
| Wt | + | + | + | + | + | + | + | + | + | + | + | + | No |
| ctf13-30 | + | + | − | + | + | DS | + | + | DS | + | + | − | Yes |
| ctf8Δ | + | + | + | + | + | + | + | + | SG | SG | SDL | SDL | Yes* |
| fyv8Δ | + | + | + | + | + | SG | + | + | SDL | + | + | SDL | Mild |
| gim5Δ | + | + | SG | + | + | SG | + | + | + | SDL | SDL | SDL | ND |
| nis1Δ | + | + | + | + | + | + | SG | SG | SDL | + | SG | SDL | No |
| nst1Δ | + | + | + | + | + | + | SDL | SG | SDL | + | + | + | Mild |
| pac10Δ | + | + | − | + | + | DS | + | + | − | + | + | − | ND |
| rad51Δ | + | + | + | + | + | + | SDL | + | SDL | + | + | SDL | Yes* |
| rsc1Δ | + | + | + | + | + | + | SG | SG | SG | + | + | + | No |
| rsc2Δ | + | + | − | + | + | DS | + | + | DS | + | + | DS | Yes |
| spo74Δ | + | + | + | + | + | + | SG | SG | SG | + | + | + | Mild |
| vid21Δ | + | + | SG | + | + | SG | + | + | SDL | SDL | SDL | SDL | Yes |
Strains used for the SDL assay were the wild type (Wt; YPH499), the ctf13-30 strain (YKB407), and deletion strains from the MATα deletion set.
+, normal growth; −, no growth; DS, dosage suppression; SG, slow growth.
Strains used for the chromosome transmission fidelity (CTF) screen contained a nonessential chromosome fragment. Strains included the wild type (YKB11) and the ctf13-30 strain (YKB10). Deletions of the tested strains were made in a YKB11 background. Yes, high degree of sector formation; mild, weak sector formation; ND, not done; no, no sector formation; *, chromosome loss previously reported.
Our ability to isolate mutants in the ctf13-30/CTF13 modifier screen and the subsequent sensitivity of 11 of the strains to overexpression of core kinetochore proteins suggest that these mutants may have defects in chromosome segregation. Indeed, both ctf8Δ and rad51Δ have been previously shown to have chromosome transmission defects (22, 47). To determine whether any of the remaining genes also have chromosome segregation defects, we deleted seven of the genes in a strain containing a nonessential chromosome fragment (23). The rate of red sector formation, or marker chromosome loss, was assessed by comparison to sector formation in a wild-type strain and a ctf13-30 strain at various temperatures. As shown in Table 3, two of the strains, the nis1Δ and rsc1Δ strains, did not have detectable marker chromosome loss rates, suggesting that the genes do not significantly affect the loss of chromosomes. fyv8Δ, nst1Δ, and spo74Δ strains exhibited mild chromosome loss defects at elevated temperatures. In contrast, rsc2Δ and vid21Δ cells had easily detectable chromosome missegregation defects at 25°C, and the degree of chromosome loss increased at higher temperatures.
RSC2 has a greater role in chromosome stability then RSC1.
Our ctf13-30/CTF13 modifier and SDL secondary screens identified two nonessential components of the chromatin remodeling complex RSC, Rsc1 and Rsc2. Consistent with the model that Rsc1 and Rsc2 have related, but not identical, functions (3, 7, 58, 61), our qualitative sectoring assay suggested that only rsc2Δ mutants have detectable chromosome loss defects. To further characterize the roles of Rsc1 and Rsc2 in chromosome stability, we quantified chromosome missegregation in rsc1Δ and rsc2Δ diploid strains using colony half-sector analysis (23). Consistent with the qualitative assay performed previously, rsc1Δ homozygous diploids did not display chromosome loss or nondisjunction rates higher than those displayed by the wild-type strain (Table 4). However rsc2Δ homozygous diploids had a 10-fold increase in chromosome loss events and a 19.5-fold increase in chromosome nondisjunction compared to wild-type diploids.
TABLE 4.
Rates of chromosome missegregation events in RSC componentsa
| Genotype | Rate of:
|
Total colonies | |
|---|---|---|---|
| Chromosome loss (1:0 events) | Nondisjunction (2:0 events) | ||
| +/+ | 8.7 × 10−5 (1.0) | 8.7 × 10−5 (1.0) | 22,800 |
| rsc1Δ rsc1Δ | 5.4 × 10−5 (0.6) | 1.1 × 10−4 (1.3) | 18,450 |
| rsc2Δ rsc2Δ | 8.8 × 10−4 (10.1) | 1.7 × 10−3 (19.5) | 19,320 |
| npl6Δ npl6Δ | 2.1 × 10−4 (2.4) | 1.6 × 10−4 (1.8) | 18,072 |
| rtt102Δ rtt102Δ | 4.4 × 10−4 (5.1) | 1.8 × 10−4 (2.1) | 27,072 |
Mutants were plated to single colonies, and visual sectoring phenotypes were scored. Colonies scored as half-sectored were ≥50% red (23). Strains used were YPH982, YKB408, YKB409, YKB410, and YKB411. Chromosome loss or 1:0 events were scored as colonies that were half red and half pink; nondisjunction or 2:0 events were scored as colonies that were half red and half white. Numbers in parentheses are factors of increase in rates of missegregation events above wild-type rates.
To further define the differences between Rsc1 and Rsc2 in chromosome segregation, we performed genetic analysis to look for synthetic phenotypes between rsc1Δ and rsc2Δ mutants and known kinetochore mutants. We found that deletion of RSC2 is synthetically lethal with mutations of NCD10 (ndc10-1 and ctf14-42) and is conditionally synthetic lethal (CSL), causing significant lowering of the permissive temperature, with cse4-1, okp1-5, and ctf13-30 (Table 5). These synthetic interactions of rsc2Δ with core kinetochore mutant genes are consistent with recently reported synthetic interactions between alleles with mutations in two essential RSC subunits, specifically sth1-3 and sfh1-1, and core kinetochore mutant genes (20). Deletion of RSC2 in combination with nonessential components of the central kinetochore (CTF3, CTF19, and MCM22) did not produce any synthetic interactions. rsc1Δ only displayed CSL with ndc10-1 and ctf14-42. The wild-type rates of chromosome missegregation events in rsc1Δ strains and weak genetic interactions of rsc1Δ with kinetochore mutant genes suggest that the Rsc2 isoform of the RSC complex is predominantly required for faithful chromosome maintenance in yeast.
TABLE 5.
Genetic interactions
| Mutant genotype | Phenotypea |
|---|---|
| rsc2Δ ndc10-1 | SL |
| rsc1Δ ndc10-1 | CSL |
| rsc2Δ ctf14-42 | SL |
| rsc1Δ ctf14-42 | CSL |
| rsc2Δ cse4-1 | CSL |
| rsc1Δ cse4-1 | Viable |
| rsc2Δ ctf13-30 | CSL |
| rsc1Δ ctf13-30 | Viable |
| rsc2Δ okp1-5 | CSL |
| rsc1Δ okp1-5 | Viable |
| rsc2Δ ctf3Δ | Viable |
| rsc1Δ ctf3Δ | Viable |
| rsc2Δ ctf19Δ | Viable |
| rsc1Δ ctf19Δ | Viable |
| rsc2Δ mcm22Δ | Viable |
| rsc1Δ mcm22Δ | Viable |
| rsc2Δ ctf8Δ | CSL |
| rsc1Δ ctf8Δ | Viable |
| rsc2Δ ctf18Δ | CSL |
| rsc1Δ ctf18Δ | Viable |
| rsc2Δ scc1-73 | CSL |
| rsc1Δ scc1-73 | Viable |
| rsc2Δ scc2-4 | CSL |
| rsc1Δ scc2-4 | Viable |
| rsc2Δ smc3-4 | CSL |
| rsc2Δ ctf7-203 | CSL |
SL, synthetic lethality. CSL, spores are viable at 25°C but die at a lower restrictive temperature than the restrictive temperature of either single mutant.
Rsc2 is not required for localization of Ndc10, Ctf3, or Cse4 to centromeres.
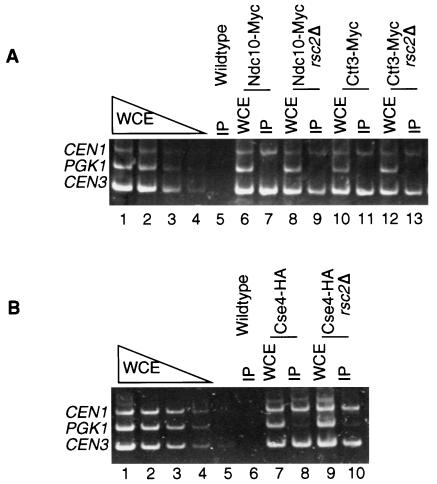
Our genetic interactions and chromosome missegregation assays suggested that Rsc2 has a role in establishing or maintaining chromosome structures. To determine whether Rsc2 is required for proper maintenance or assembly of the kinetochore complex, we performed ChIP assays to assess the ability of inner kinetochore proteins Ndc10 and Cse4 and central kinetochore protein Ctf3 to localize to centromeres in the absence of Rsc2. Myc-tagged-Ndc10 and -Ctf3 strains and HA-tagged-Cse4 strains in either wild-type or rsc2Δ backgrounds were grown at 25°C to mid-log phase and subsequently shifted to 37°C, a temperature which causes severe growth inhibition in rsc2Δ cells (7). After 3 h at 37°C, chromatin was prepared, followed by IP of the tagged kinetochore proteins and PCR analysis to determine if CEN DNA was present in the immunoprecipitates (Fig. 2). The samples for both total chromatin and immunoprecipitates were titrated to determine the linear range for PCR. As expected, CEN3 and CEN1 were amplified specifically in Ndc10-Myc, Ctf3-Myc, and Cse4-HA immunoprecipitates from wild-type cell extracts (Fig. 2A, lanes 7 and 11, and B, lane 8), but not from an untagged-strain immunoprecipitate (Fig. 2A, lane 5, and B, lane 6). Disruption of RSC2 did not alter the localization of Ndc10-Myc, Ctf3-Myc, or Cse4-HA (Fig. 2A, lanes 9 and 13, and B, lane 10). These results indicated that Rsc2 is not required for association of the kinetochore proteins Ndc10, Cse4, and Ctf3 with CEN DNA.
FIG. 2.
Ndc10, Ctf3, and Cse4 coimmunoprecipitation with CEN DNA is not dependent on Rsc2. Anti-Myc or anti-HA ChIP assays were performed with chromatin extracts of wild-type (RSC2) or rsc2Δ mutant cells expressing either Ndc10-Myc (YVM499 and YKB121), Ctf3-Myc (YVM 218 and YKB127), or Cse4-HA (YVM1141 and YKB124), as well as with cells expressing no tag (YPH499). Cells were grown to mid-log phase in YPD medium at 25°C, and then the culture was shifted to 37°C for 3 h before chromatin was isolated for IP. Multiplex PCR was performed to amplify the centromeric DNA of CEN1, CEN3, and a non-CEN locus, PGK1, for both whole-cell extracts (WCE) and immunoprecipitates.
Deletion of RSC2 has synthetic interactions with mutations in genes that function in sister chromatid cohesion.
Since Rsc2 does not appear to be required for localization of kinetochore proteins to CENs, we decided to explore the possibility that Rsc2 may contribute to sister chromatid cohesion. Therefore, we tested for genetic interactions of rsc2Δ with mutations in proteins required for various aspects of sister chromatid cohesion. We found that deletion of RSC2 is CSL with two different mutations in genes encoding cohesin subunits, scc1-73 and smc3-4 (Table 5). Further, we determined that deletion of RSC2 in combination with ctf8Δ, ctf18Δ, and scc2-4 drastically reduced the permissive growth temperature to 30°C. Interestingly, we recovered few rsc2Δ ctf7-203 mutants out of more than 40 tetrad dissections, and the mutants that we did recover had a permissive growth temperature of only 25°C. Therefore, RSC2 exhibits specific genetic interactions with genes in the cohesin complex and genes involved in the loading and establishment of sister chromatid cohesion, suggesting that Rsc2 may have a role in sister chromatid cohesion.
RSC2 is required for sister chromatid cohesion.
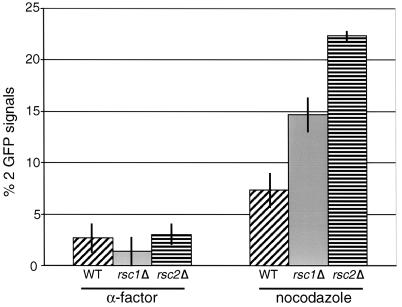
To determine whether RSC2 has a role in sister chromatid cohesion, RSC2 was deleted in a strain expressing a Tet repressor-green fluorescent protein (GFP) fusion protein and containing Tet operator repeats integrated 35 kb from the centromere of chromosome V (27, 32). Since rsc2Δ strains have a slow-growth phenotype at 37°C, we grew the cells at 25°C to mid-log phase and then arrested the cells at 37°C for 3 h in G1 with α-factor or in G2/M with nocodazole. As expected, only 7% of wild-type cells arrested in G2/M had two GFP signals, indicating that 93% of the sister chromatids remained paired prior to anaphase (Fig. 3). In contrast, 22% of the rsc2Δ cells arrested in G2/M had two GFP signals, indicating a defect in sister chromatid cohesion. The presence of Pds1-13Myc, monitored by indirect immunofluorescence, confirmed that the nocodazole-arrested cells had not progressed to anaphase (data not shown). Nearly identical results were obtained in testing for a chromatid cohesion defect on the right arm of chromosome IV (data not shown). Consistent with the milder defects rsc1Δ strains have in chromosome missegregation and genetic interactions with kinetochore mutants, 14% of rsc1Δ cells arrested in G2/M had two GFP signals. We conclude that the RSC complex is required for proper sister chromatid cohesion.
FIG. 3.
rsc2Δ mutant cells exhibit defects in sister chromatid cohesion. The numbers of GFP signals were scored in wild-type cells (YPH1477), three independent isolates of rsc2Δ cells (YKB235a to -c), and three independent isolates of rsc1Δ cells (YKB177) arrested for 3 h at 37°C in G1 with α-factor or in G2 with nocodazole. The data shown represent the averages of three independent experiments. One hundred cells were counted for each sample.
Deletion of RSC2 causes a defect in establishing stable cohesion.
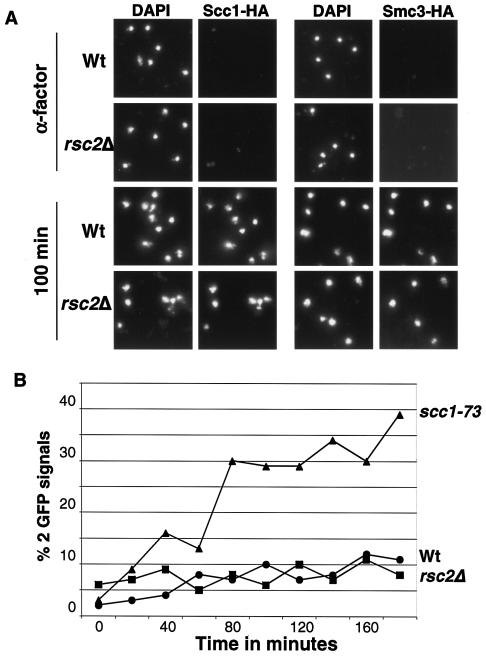
Our genetic and functional data suggested that RSC may be involved in either the loading of cohesin on chromosomes in G1/S, the establishment of sister chromatid cohesion in S phase, or the maintenance of sister cohesion in G2/M. Association of cohesin subunits with chromatin in chromosome spreads can be used to identify defects in cohesin loading onto chromosomes (51). The amount of Scc1-HA6 bound to chromosomes in spreads is strongly reduced in scc2-1 mutant strains. In contrast, eco1-1 and pds5 mutants do not display reduced association of cohesin bound to chromosomes in spreads (19, 51). To determine whether rsc2Δ mutations affect the association of cohesin subunits with chromatin, we constructed rsc2Δ and wild-type strains expressing HA-tagged versions of either Scc1 or Smc3. Cells were synchronized in G1 by α-factor treatment at 25°C and released at 37°C, and samples were taken every 20 min to analyze the association of Scc1-HA or Smc3-HA with chromatin by chromosome spreading. Figure 4A shows representative chromosome spreads from the α-factor time point (early G1) and 100-min time point after release of the α-factor block, when most cells have progressed through S phase but have not yet progressed to anaphase (data not shown). As expected, Smc3-HA and Scc1-HA were not detected on chromatin from wild-type cells in early G1 but were tightly associated with chromatin from late G1 until metaphase (32, 51). Deletion of RSC2 did not detectably affect the association of Smc3-HA or Scc1-HA with chromatin (Fig. 4A). ChIP studies also indicated that Smc3-HA localization to cohesin sites was not affected by deletion of RSC2 (data not shown). Therefore, our data suggest that, like eco1 and pds5 mutants, rsc2Δ mutants are defective in sister chromatid cohesion despite the presence of cohesin on chromosomes.
FIG. 4.
rsc2Δ mutant cells exhibit defects in establishment of sister chromatid cohesion. (A) Wild-type cells (Wt; RSC2) and rsc2Δ mutants expressing either Smc3-HA (YKB353 or YKB355) or Scc1-HA (YKB342 or YKB341) were blocked in G1 by α-factor treatment and released at 37°C. Chromosome spreads shown are of cells were taken prior to release or at 100 min postrelease. DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole). Chromatin-associated Scc1-HA and Smc3-HA were detected by indirect immunofluorescence. (B) Wild-type (YPH1477; circle), scc1-73 (YKB426; triangle), and rsc2Δ (YKB235; square) cells were arrested with α-factor and released into nocodazole media at 25°C. Cultures were shifted to 37°C after >90% of cells had budded (time zero), samples were taken every 20 min for 180 min, and the percentages of sister chromatid separation were scored. One hundred cells were scored per sample; two separate experiments were performed with nearly identical results.
The previous experiment did not address whether Rsc2 is needed to establish cohesion during S phase or whether Rsc2 was required to maintain sister chromatid cohesion during G2 or M phase. To address this, we released wild-type, scc1-73, and rsc2Δ cells with GFP-marked chromosome V arms from an α-factor-induced G1 arrest into nocodazole at 25°C. Two hours after release from the pheromone, when >90% of the cells had completed replication and were budded, all three cultures were shifted to 37°C and incubated for 3 h. Sister chromatid cohesion was monitored by counting the number of cells that had one or two GFP dots. As previously reported, the number of wild-type cells with two GFP dots was low at the start of the temperature shift and remained low during the course of the experiment, while the number of scc1-73 mutant cells with separated arms increased rapidly during the shift (Fig. 4B) (36, 51). rsc2Δ cells displayed a sister chromatid separation pattern similar to that of wild-type cells, with almost no increases in separation over time (Fig. 4B). The pattern displayed by rsc2Δ cells is similar to results previously reported for eco1-1 cells (51). Our data imply that, like Eco1, Rsc2 is required to establish sister cohesion prior to M phase but is not required to maintain sister cohesion during M phase.
Rtt102, a novel RSC-associated protein.
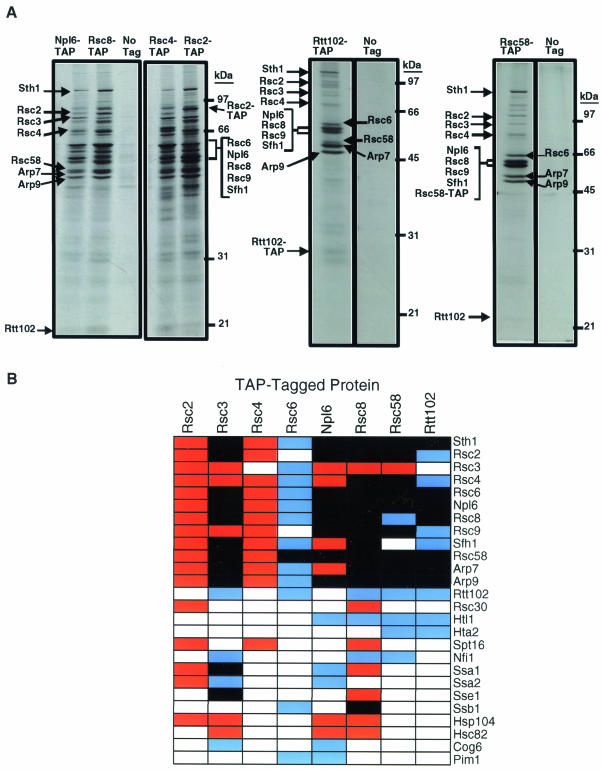
In an attempt to identify additional RSC subunits and/or RSC-associated proteins that may also contribute to the fidelity of chromosome transmission, we performed a series of affinity purification experiments to identify proteins that physically associate with RSC. We placed TAP tags containing a calmodulin-binding peptide and Staphylococcus aureus protein A, separated by a tobacco etch virus protease cleavage site, at the C termini of five known components of RSC: Rsc2, Rsc3, Rsc4, Rsc6, and Rsc8. We then performed affinity purifications of the various RSC components. An aliquot of the purified material was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and silver stained, and the specifically copurifying proteins were excised from the gel and identified by trypsin digestion and MALDI-ToF MS. Since the RSC complex has been implicated in a variety of cellular functions, we suspected that interacting protein complexes involved in chromosome maintenance might be recovered in substoichiometric amounts. Therefore, the second aliquot of the preparation was subjected directly to trypsin digestion and LC/MS/MS in an attempt to identify proteins recovered in low yield that interact with RSC. Each tagged RSC subunit copurified with a core set of RSC proteins (Fig. 5), indicating that our purification procedure was successful in isolating an intact RSC complex. Although either MS procedure alone did not identify all the RSC subunits for a given preparation, the combination of the two procedures missed few known RSC subunits in any single preparation. We were not, however, able to reproducibly identify Rsc1 when other TAP-tagged RSC subunits were purified (Fig. 5A).
FIG. 5.
Purification of RSC protein complexes and associated proteins. TAPs of the RSC complex were carried out on strains containing either no tagged proteins or TAP-tagged versions of Rsc2 (NJK208), Rsc3 (NJK203), Rsc4 (NJK210), Rsc6 (NJK213), Np16 (NJK212), Rsc8 (NJK211), Rsc58 (NJK192), and Rtt102 (NJK182). (A) Silver-stained sodium dodecyl sulfate gels of some of the RSC affinity purifications. Proteins copurifying with the TAP-tagged proteins were identified by either MALDI-ToFMS or LC/MS/MS. (B) Clustered summary of the proteins identified copurifying with the TAP-tagged versions of RSC proteins. Red boxes, proteins identified by MALDI-ToF MS; blue boxes, proteins identified by LC/MS/MS; black boxes, proteins identified by both methods. Proteins were listed only if they were identified as copurifying with at least two TAP-tagged baits.
Recently, Np16, Rsc58 (39), and Htl1 (38) have been implicated as new components of RSC. Our purifications confirmed that they indeed copurify with previously known tagged RSC components (Fig. 5). Further, both TAP-tagged Np16 and Rsc58 copurified with components of the RSC complex, confirming that Np16 and Rsc58 are components of RSC. Rtt102, a protein implicated in regulation of Ty transposition (41), copurified with four TAP-tagged RSC components, suggesting that Rtt102 might also be a component of RSC. To determine whether Rtt102 is a true component of RSC, we performed the reciprocal purification experiment with TAP-tagged Rtt102 as the bait. Twelve subunits of RSC copurified with Rtt102, confirming that Rtt102 is indeed a subunit of the RSC complex (Fig. 5).
To determine whether deletions of the new nonessential components of RSC. RTT102 or NPL6, are required for faithful chromosome segregation, we quantified chromosome missegregation events in npl6Δ and rtt102Δ diploid strains using colony half-sector analysis (Table 4). While strains with deletions in NPL6 did not display significant elevations in chromosome missegregation rates, strains with deletions in RTT102 showed a modest fivefold increase in chromosome loss events. In addition, we tested for genetic interactions of rtt102Δ with mutations in proteins required for various aspects of sister chromatid cohesion and segregation. Our data suggest that only a subset of the nonessential RSC subunits, including Rsc2 and Rtt102, have a role in chromosome maintenance.
DISCUSSION
Genomic haploinsufficiency modifier screens.
To isolate genes whose function impinges on chromosome transmission fidelity in yeast, we designed a genomic haploinsufficiency modifier screen and performed the screen using a Ts allele of CTF13, ctf13-30. The ctf13-30/CTF13 modifier screen identified 36 genes whose haploinsufficiency modified the behavior of a ctf13-13/CTF13 heterozygote. Secondary SDL and chromosome segregation assays determined that many of the deletion strains isolated in the modifier screen have direct effects on chromosome stability. We determined that deletions of RSC2 and RSC1 are not synthetically lethal with ctf13-30 (Table 5). Furthermore, synthetic genetic array analysis (SGA) (50) has been conducted on ctf13-30, and none of the genes identified in our modifier screen were observed to be synthetically lethal or synthetically sick with ctf13-30 (K. Baetz and V. Measday, unpublished data). This indicates that SGA and genomic haploinsufficiency modifier screens are complementary approaches in identifying genetic interactions. Further, since genomic haploinsufficiency modifier screens require only the selection of diploids and do not require the time-consuming steps of sporulation and subsequent haploid selection, they can be conducted in a shorter period of time than SGA screens. The success of the haploinsufficiency modifier screen in identifying genes whose function impinges on chromosome transmission fidelity and the complementary results with SGA analysis suggest that systematic screening for genetic modifiers in heterozygous diploids will be advantageous in isolating additional alleles of interest.
Rtt102 is a new component of the RSC complex.
Our systematic high-throughput method of TAP tag purification was successful in purifying 14 out of 15 known proteins of the RSC complex and confirmed the presence of the recently identified Rsc58, Np16, and Htl1 proteins in the RSC complex. In addition, our reciprocal TAP tag purification experiments identified Rtt102 as a new component of RSC. As was the case for Htl1 (38), Rtt102 may not have been identified in previous characterizations of RSC components because it may exist in substoichiometric amounts within the RSC complex. More likely, the small size of Rtt102 and its relatively poor staining with silver may have limited detection of Rtt102 in the RSC complex by other investigators. RTT102 was identified in a screen for regulators of Ty1 transposition (RTT); however no other components of RSC were identified in the RTT screen (41). This suggests that Rtt102 may have additional roles distinct from its roles in the RSC complex.
A role for distinct RSC subcomplexes in chromosome maintenance.
The RSC complex has previously been implicated in chromosome maintenance (20, 53). Our work, however, suggests that only a subset of RSC subunits are responsible for RSC's role in chromosome maintenance and further highlights the functional differences between Rsc1 and Rsc2. Although both RSC1 and RSC2 were isolated in the ctf13-30/CTF13 modifier screen, rsc2Δ mutants have a higher rate of chromosome missegregation, numerous genetic interactions with chromosome stability mutants, and a significant defect in sister chromatid cohesion. It has also been shown that RSC2, not RSC1, is essential for 2μm plasmid maintenance (58). While RSC2 may play the predominant role in faithful chromosome segregation, the identification of RSC1 in our screens and the mild sister chromatid cohesion defects of rsc1 mutants indicate that the RSC1 form of RSC may have functions that impinge on chromosome transmission. As is the case for RSC1 and RSC2, the nonessential RSC subunits RTT102 and NPL6 make different contributions to chromosome stability. Deletions of RTT102 cause a reproducible fivefold increase in chromosome missegregation events at 30°C, while deletions of NPL6 produce nearly wild-type rates of chromosome missegregation. The moderate increase of chromosome missegregation in rtt102Δ deletion strains is similar to that in many other ctf mutants (47), CAF-1 and Hir protein mutants (42), and essential RSC component mutants (20, 53).
It will be interesting to determine the minimal RSC complex necessary for chromosome segregation by systematic assessment of the contribution of each RSC subunit to chromosome maintenance. One intriguing hypothesis is that the nonessential or substoichiometric components of RSC may provide the specificity of RSC's multiple functions in vivo, possibly by localizing RSC to discrete loci. Genome-wide localizations of Rsc1 and Rsc2 to chromosome regions in vivo are nearly identical (34), but these experiments were performed using log-phase cells, and it is doubtful that subtle changes in Rsc1 and Rsc2 localization throughout the cell cycle would have been identified. Alternatively, substoichiometric subunits may specifically localize RSC to distinct regions of the chromosome, requiring chromatin modification for chromosome maintenance. The broad distribution of RSC on chromatin (12, 34) may indicate that it is not the localization of RSC per se that changes but rather the activity of RSC that is modified at distinct positions along the chromosome. Perhaps spatial restriction of RSC activation may be mediated through a nonessential component of RSC, such as Rsc2 or Rtt102.
The RSC chromatin-remodeling complex is required for establishment of cohesion.
The isolation of RSC2 and RSC1 in a kinetochore mutant ctf13-30/CTF13 modifier screen and the genetic interaction of rsc2Δ and rsc1Δ with kinetochore mutant genes suggest that RSC could function at the kinetochore. Indeed, the centromeric nucleosome structure in both sth1 and sfh1 mutants is disrupted (20, 53), suggesting that RSC is required for maintaining the centromere and centromere-flanking nucleosome structure. It has recently been hypothesized that RSC could function at kinetochores in either the assembly or the proper positioning of Cse4-containing nucleosomes on centromeric DNA (20). There is still little evidence, however, directly linking RSC with Cse4. Indeed, Cse4 is still localized to CEN DNA in rsc mutants (Fig. 2) (20). Functional Cse4 is required for Ctf3 localization to CEN DNA (28). However, we show that Ctf3 localization to CEN DNA is not affected in rsc2Δ strains, suggesting that RSC is not required for Cse4-dependent loading of central kinetochore proteins onto CEN DNA. Furthermore, while overexpression of CSE4 can partially suppress phenotypes of the sfh1-1 RSC mutant (20), it is unlikely that this suppression is specific to CSE4, for we show that overexpression of core kinetochore components SKP1, CTF13, and NDC10 also suppresses the Ts growth defects of rsc2Δ strains. In addition, while it was elegantly demonstrated that histones H2B, H3, and H4 interact directly with the Sth1 subunit of RSC (20), no such direct interactions have been demonstrated for Cse4 and the RSC complex. Although we cannot rule out the hypothesis that the role of RSC in chromosome maintenance is to regulate Cse4 function at CEN DNA, we believe that RSC may have other functions that contribute to the fidelity of chromosome transmission.
Our data suggest that RSC contributes to the fidelity of chromosome transmission through sister chromatid cohesion. We show that rsc2Δ has synthetic interactions with mutations in cohesin and mutations affecting both cohesin loading and the establishment of cohesion (Table 5) and that rsc2Δ cells have defects in sister chromatid cohesion (Fig. 3). Defects in sister chromatid cohesion have also been reported for sth1 and sfh1 mutants (20). Together, the defects of rsc mutants in premature separation of chromosome arms prior to anaphase indicate that RSC is required for proper sister chromatid cohesion. Our chromosome spread assays determined that, despite sister chromatid cohesion defects, rsc2Δ mutants have high levels of cohesin associated with chromatin (Fig. 4A). Both eco1 and pds5 mutants also display sister chromatid cohesion defects in the presence of high levels of cohesin. This suggests that, like Eco1 and Pds5, RSC is not involved in the loading of cohesin but rather in either the establishment of cohesion in S phase or the maintenance of cohesion in G2/M. We determined that, although rsc2Δ cells blocked in nocodazole at 37°C display sister chromatid cohesion defects (Fig. 3), rsc2Δ cells blocked in nocodazole at 25°C prior to temperature shifting to 37°C maintain cohesion (Fig. 4B). Our data imply that, like Eco1, Rsc2 is needed to establish cohesion but that it is not necessary to maintain cohesion during G2 and M phases.
How RSC contributes to cohesion establishment is unknown, as are the exact mechanisms by which Eco1 and the alternative RFC (Ctf18) contribute to cohesion establishment (reviewed in reference 54). The defects in sister chromatid cohesion could be due to RSC-dependent transcriptional defects on either genes encoding cohesin or regulators of cohesin loading, establishment, or maintenance. However, microarray analysis of rsc mutants does not indicate that this is the case (1, 34). Since many rsc mutants arrest at the G2/M boundary with 2N DNA content (1, 8, 12, 14, 38, 53), it has been proposed that the activity of RSC may be required for the establishment or maintenance of chromosome structure after replication (15). The strong genetic interactions between RSC2 and the essential cohesion establishment factor ECO1/CTF7 and the alternative replication factor C (Ctf18) complex factors CTF18 and CTF8, along with defects in rsc2Δ mutants in establishing cohesion, suggest a model in which RSC functions to pair sister chromatids as they emerge from the DNA replication fork. Although ordered nucleosome arrays at arm CAR sites have not been reported, proper positioning of nucleosomes at arm CAR sites might be required for cohesion. One possibility is that RSC may be necessary for mediating a nucleosomal higher-ordered structure required for cohesion establishment as DNA emerges from the replication machinery. The mechanisms of cohesin binding to arms and centromeres appear distinct, as cohesin localization to centromeres is dependent on various centromere DNA elements and kinetochore proteins (reviewed in references 54 and 60). However, the proposed role of RSC in mediating ordered nucleosome arrays at CEN DNA may reflect RSC's requirement for establishment of cohesion at centromeres and not localization of Cse4. Intriguingly, the human chromatin remodeling protein hSNF2 has been shown to directly interact with the human hRad21 cohesion complex and the chromatin remodeling activity of hSNF2 is required to mediate association of cohesin with chromatin (17). Though we have shown that Rsc2 is not required for the loading of cohesin onto chromatin, the human results suggest that yeast chromatin remodeling complexes might also directly interact with cohesin. Further studies will elucidate how the chromatin remodeling complex RSC, and possibly other chromatin remodeling complexes in yeast, contribute to the regulation of cohesion of sister chromatids.
Acknowledgments
We thank Liz Conibear, Douglas McCallum, and Vivien Measday for critical reading of this manuscript.
K.K.B. was supported by a Canadian Institute of Health Research (CIHR) postdoctoral fellowship and a Michael Smith Foundation for Health Reaserch postdoctoral fellowship, and N.J.K. was supported by a CIHR doctoral fellowship. This work was supported by grants to J.G. from CIHR and to P.H. from NIH (grant CA16591).
REFERENCES
- 1.Angus-Hill, M. L., A. Schlichter, D. Roberts, H. Erdjument-Bromage, P. Tempst, and B. R. Cairns. 2001. A Rsc3/Rsc30 zinc cluster dimer reveals novel roles for the chromatin remodeler RSC in gene expression and cell cycle control. Mol. Cell 7:741-751. [DOI] [PubMed] [Google Scholar]
- 2.Baetz, K., J. Moffat, J. Haynes, M. Chang, and B. Andrews. 2001. Transcriptional coregulation by the cell integrity mitogen-activated protein kinase Slt2 and the cell cycle regulator Swi4. Mol. Cell. Biol. 21:6515-6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett, C. B., L. K. Lewis, G. Karthikeyan, K. S. Lobachev, Y. H. Jin, J. F. Sterling, J. R. Snipe, and M. A. Resnick. 2001. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 29:426-434. [DOI] [PubMed] [Google Scholar]
- 4.Biggins, S., and C. E. Walczak. 2003. Captivating capture: how microtubules attach to kinetochores. Curr. Biol. 13:R449-R460. [DOI] [PubMed] [Google Scholar]
- 5.Bloom, K. S., and J. Carbon. 1982. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell 29:305-317. [DOI] [PubMed] [Google Scholar]
- 6.Cairns, B. R., Y. Lorch, Y. Li, M. Zhang, L. Lacomis, H. Erdjument-Bromage, P. Tempst, J. Du, B. Laurent, and R. D. Kornberg. 1996. RSC, an essential, abundant chromatin-remodeling complex. Cell 87:1249-1260. [DOI] [PubMed] [Google Scholar]
- 7.Cairns, B. R., A. Schlichter, H. Erdjument-Bromage, P. Tempst, R. D. Kornberg, and F. Winston. 1999. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell 4:715-723. [DOI] [PubMed] [Google Scholar]
- 8.Cao, Y., B. R. Cairns, R. D. Kornberg, and B. C. Laurent. 1997. Sfh1p, a component of a novel chromatin-remodeling complex, is required for cell cycle progression. Mol. Cell. Biol. 17:3323-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheeseman, I. M., D. G. Drubin, and G. Barnes. 2002. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157:199-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciosk, R., M. Shirayama, A. Shevchenko, T. Tanaka, A. Toth, and K. Nasmyth. 2000. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol. Cell 5:243-254. [DOI] [PubMed] [Google Scholar]
- 11.Connelly, C., and P. Hieter. 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86:275-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damelin, M., I. Simon, T. I. Moy, B. Wilson, S. Komili, P. Tempst, F. P. Roth, R. A. Young, B. R. Cairns, and P. A. Silver. 2002. The genome-wide localization of Rsc9, a component of the RSC chromatin-remodeling complex, changes in response to stress. Mol. Cell 9:563-573. [DOI] [PubMed] [Google Scholar]
- 13.Doheny, K. F., P. K. Sorger, A. A. Hyman, S. Tugendreich, F. Spencer, and P. Hieter. 1993. Identification of essential components of the S. cerevisiae kinetochore. Cell 73:761-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du, J., I. Nasir, B. K. Benton, M. P. Kladde, and B. C. Laurent. 1998. Sth1p, a Saccharomyces cerevisiae Snf2p/Swi2p homolog, is an essential ATPase in RSC and differs from Snf/Swi in its interactions with histones and chromatin-associated proteins. Genetics 150:987-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fyodorov, D. V., and J. T. Kadonaga. 2001. The many faces of chromatin remodeling: SWItching beyond transcription. Cell 106:523-525. [DOI] [PubMed] [Google Scholar]
- 16.Gruber, S., C. H. Haering, and K. Nasmyth. 2003. Chromosomal cohesin forms a ring. Cell 112:765-777. [DOI] [PubMed] [Google Scholar]
- 17.Hakimi, M. A., D. A. Bochar, J. A. Schmiesing, Y. Dong, O. G. Barak, D. W. Speicher, K. Yokomori, and R. Shiekhattar. 2002. A chromatin remodelling complex that loads cohesin onto human chromosomes. Nature 418:994-998. [DOI] [PubMed] [Google Scholar]
- 18.Hanna, J. S., E. S. Kroll, V. Lundblad, and F. A. Spencer. 2001. Saccharomyces cerevisiae CTF18 and CTF4 are required for sister chromatid cohesion. Mol. Cell. Biol. 21:3144-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartman, T., K. Stead, D. Koshland, and V. Guacci. 2000. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J. Cell Biol. 151:613-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu, J. M., J. Huang, P. B. Meluh, and B. C. Laurent. 2003. The yeast RSC chromatin-remodeling complex is required for kinetochore function in chromosome segregation. Mol. Cell. Biol. 23:3202-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hyland, K. M., J. Kingsbury, D. Koshland, and P. Hieter. 1999. Ctf19p: a novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J. Cell Biol. 145:15-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein, H. L. 2001. Spontaneous chromosome loss in Saccharomyces cerevisiae is suppressed by DNA damage checkpoint functions. Genetics 159:1501-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koshland, D., and P. Hieter. 1987. Visual assay for chromosome ploidy. Methods Enzymol. 155:351-372. [DOI] [PubMed] [Google Scholar]
- 24.Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor, G. Cagney, A. Emili, A. Shilatifard, S. Buratowski, and J. F. Greenblatt. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22:6979-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kroll, E. S., K. M. Hyland, P. Hieter, and J. J. Li 1996. Establishing genetic interactions by a synthetic dosage lethality phenotype. Genetics 143:95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 27.Mayer, M. L., S. P. Gygi, R. Aebersold, and P. Hieter. 2001. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): an alternative RFC complex required for sister chromatid cohesion in S. cerevisiae Mol. Cell 7:959-970. [DOI] [PubMed] [Google Scholar]
- 28.Measday, V., D. W. Hailey, I. Pot, S. A. Givan, K. M. Hyland, G. Cagney, S. Fields, T. N. Davis, and P. Hieter. 2002. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 16:101-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Measday, V., and P. Hieter. 2002. Synthetic dosage lethality. Methods Enzymol. 350:316-326. [DOI] [PubMed] [Google Scholar]
- 30.Meluh, P. B., and D. Koshland. 1997. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11:3401-3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meluh, P. B., P. Yang, L. Glowczewski, D. Koshland, and M. M. Smith. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell 94:607-613. [DOI] [PubMed] [Google Scholar]
- 32.Michaelis, C., R. Ciosk, and K. Nasmyth. 1997. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell 91:35-45. [DOI] [PubMed] [Google Scholar]
- 33.Nasmyth, K. 2001. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu. Rev. Genet. 35:673-745. [DOI] [PubMed] [Google Scholar]
- 34.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2002. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 16:806-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortiz, J., O. Stemmann, S. Rank, and J. Lechner. 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13:1140-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Panizza, S., T. Tanaka, A. Hochwagen, F. Eisenhaber, and K. Nasmyth. 2000. Pds5 cooperates with cohesin in maintaining sister chromatid cohesion. Curr. Biol. 10:1557-1564. [DOI] [PubMed] [Google Scholar]
- 37.Puziss, J. W., T. A. Hardy, R. B. Johnson, P. J. Roach, and P. Hieter. 1994. MDS1, a dosage suppressor of an mck1 mutant, encodes a putative yeast homolog of glycogen synthase kinase 3. Mol. Cell. Biol. 14:831-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Romeo, M. J., M. L. Angus-Hill, A. K. Sobering, Y. Kamada, B. R. Cairns, and D. E. Levin. 2002. HTL1 encodes a novel factor that interacts with the RSC chromatin remodeling complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:8165-8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanders, S. L., J. Jennings, A. Canutescu, A. J. Link, and P. A. Weil. 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22:4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saunders, M., M. Fitzgerald-Hayes, and K. Bloom. 1988. Chromatin structure of altered yeast centromeres. Proc. Natl. Acad. Sci. USA 85:175-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scholes, D. T., M. Banerjee, B. Bowen, and M. J. Curcio. 2001. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159:1449-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharp, J. A., A. A. Franco, M. A. Osley, and P. D. Kaufman. 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16:85-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharp, J. A., and P. D. Kaufman. 2003. Chromatin proteins are determinants of centromere function. Curr. Top. Microbiol. Immunol. 274:23-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shero, J. H., and P. Hieter. 1991. A suppressor of a centromere DNA mutation encodes a putative protein kinase (MCK1). Genes Dev. 5:549-560. [DOI] [PubMed] [Google Scholar]
- 45.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skibbens, R. V., L. B. Corson, D. Koshland, and P. Hieter. 1999. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 13:307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spencer, F., S. L. Gerring, C. Connelly, and P. Hieter. 1990. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics 124:237-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stearns, T., and D. Botstein. 1988. Unlinked noncomplementation: isolation of new conditional-lethal mutations in each of the tubulin genes of Saccharomyces cerevisiae. Genetics 119:249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka, T., J. Fuchs, J. Loidl, and K. Nasmyth. 2000. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2:492-499. [DOI] [PubMed] [Google Scholar]
- 50.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 51.Toth, A., R. Ciosk, F. Uhlmann, M. Galova, A. Schleiffer, and K. Nasmyth. 1999. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication Genes Dev. 13:320-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Treich, I., L. Ho, and M. Carlson. 1998. Direct interaction between Rsc6 and Rsc8/Swh3, two proteins that are conserved in SWI/SNF-related complexes. Nucleic Acids Res. 26:3739-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsuchiya, E., T. Hosotani, and T. Miyakawa. 1998. A mutation in NPS1/STH1, an essential gene encoding a component of a novel chromatin-remodeling complex RSC, alters the chromatin structure of Saccharomyces cerevisiae centromeres. Nucleic Acids Res. 26:3286-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uhlmann, F. 2003. Chromosome cohesion and separation: from men and molecules. Curr. Biol. 13:R104-R114. [DOI] [PubMed] [Google Scholar]
- 55.Vinh, D. B., M. D. Welch, A. K. Corsi, K. F. Wertman, and D. G. Drubin. 1993. Genetic evidence for functional interactions between actin noncomplementing (Anc) gene products and actin cytoskeletal proteins in Saccharomyces cerevisiae. Genetics 135:275-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, W. 2003. The SWI/SNF family of ATP-dependent chromatin remodelers: similar mechanisms for diverse functions. Curr. Top. Microbiol. Immunol. 274:143-169. [DOI] [PubMed] [Google Scholar]
- 57.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 58.Wong, M. C. V. L., S. R. S. Scott-Drew, M. J. Hayes, P. J. Howard, and J. A. H. Murray. 2002. RSC2, encoding a component of the RSC nucleosome remodeling complex, is essential for 2μm plasmid maintenance in Saccharomyces cerevisiae Mol. Cell. Biol. 22:4218-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xue, Y., J. C. Canman, C. S. Lee, Z. Nie, D. Yang, G. T. Moreno, M. K. Young, E. D. Salmon, and W. Wang. 2000. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. USA 97:13015-13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yokomori, K. 2003. SMC protein complexes and the maintenance of chromosome integrity. Curr. Top. Microbiol. Immunol. 274:79-112. [DOI] [PubMed] [Google Scholar]
- 61.Yukawa, M., H. Koyama, K. Miyahara, and E. Tsuchiya. 2002. Functional differences between RSC1 and RSC2, components of a for growth essential chromatin-remodeling complex of Saccharomyces cerevisiae, during the sporulation process. FEMS Yeast Res. 2:87-91. [DOI] [PubMed] [Google Scholar]